Abstract
The histone H3 variant centromere protein A (CENP-A) is central to centromere formation throughout eukaryotes. A long-standing question in centromere biology has been the organization of CENP-A at the centromere and its implications for the structure of centromeric chromatin. In this study, we describe the three-dimensional localization of CENP-A at the inner kinetochore plate through serial-section transmission electron microscopy of human mitotic chromosomes. At the kinetochores of normal centromeres and at a neocentromere, CENP-A occupies a compact domain at the inner kinetochore plate, stretching across two thirds of the length of the constriction but encompassing only one third of the constriction width and height. Within this domain, evidence of substructure is apparent. Combined with previous chromatin immunoprecipitation results (Saffery, R., H. Sumer, S. Hassan, L.H. Wong, J.M. Craig, K. Todokoro, M. Anderson, A. Stafford, and K.H.A. Choo. 2003. Mol. Cell. 12:509–516; Chueh, A.C., L.H. Wong, N. Wong, and K.H.A. Choo. 2005. Hum. Mol. Genet. 14:85–93), our data suggest that centromeric chromatin is arranged in a coiled 30-nm fiber that is itself coiled or folded to form a higher order structure.
Introduction
Throughout eukaryotes, the centromere is the fundamental structure that governs the segregation of sister chromatids during cell division. The centromere comprises the underlying DNA of the primary constriction, often consisting of repetitive “satellite” DNA, and the kinetochore, a trilaminar structure composed of centromere proteins (CENPs) and chromatin.
Central to the formation of a functional kinetochore is CENP-A, a histone H3 paralogue that replaces H3 in a subset of the nucleosomes within the centromere. The protein is vital for the recruitment of all other CENPs (Howman et al., 2000; Oegema et al., 2001; Liu et al., 2006), including a large CENP-A–interacting complex (Foltz et al., 2006; Okada et al., 2006).
A key question in understanding the structure of the centromere has been the organization of CENP-A–containing chromatin. Until now, the major attempts to determine the physical binding domain of CENP-A at the centromere have come from light microscopy (LM; Warburton et al., 1997; Blower et al., 2002; Sullivan and Karpen, 2004). Using deconvolution LM, the protein has been suggested to be present throughout the outer centromeric chromatin in a cylindrical domain spanning approximately half the width and the entire height and length of the centromere (Blower et al., 2002; Sullivan and Karpen, 2004); in other words, half of the constriction DNA. The “repeat-subunit” model of centromeric chromatin organization based on these data has suggested a linear coil of chromatin running between the chromatid arms (Blower et al., 2002).
As well as being studied through LM, the distribution of CENP-A at the centromere has also been studied through chromatin immunoprecipitation (ChIP) at several neocentromeres (Lo et al., 2001a,2001b; Alonso et al., 2003; Chueh et al., 2005; Cardone et al., 2006; Alonso et al., 2007; Capozzi et al., 2008). Neocentromeres are ectopic centromeres that can spontaneously form at nonrepetitive euchromatic regions of the genome, and the lack of repetitive α-satellite (or alphoid) DNA at these centromeres has made them prime candidates for centromere research (for review see Marshall et al., 2008). Interestingly, a quantitative study of CENP-A levels on alphoid centromeres and neocentromeres has shown that neocentromeres bind significantly less CENP-A (Irvine et al., 2004). Such a result has raised the possibility that the CENP-A binding domain of neocentromeres is physically smaller than that found at alphoid centromeres.
In this study, we present detailed EM experiments of the three-dimensional binding domain of CENP-A investigated in intact metaphase cells, cytospun chromosome spreads, and on FACS-sorted populations of individual chromosomes. We describe the localization of CENP-A at both normal human centromeres and at the mardel(10) neocentromere, marking the first time immuno-EM has been used on a neocentromere. Relating this structural data back to ChIP data gathered at this same neocentromere, we suggest a complex higher order structure of chromatin folding at the primary constriction.
Results and discussion
CENP-A at human centromeres occupies an unusually compact domain at the inner kinetochore plate
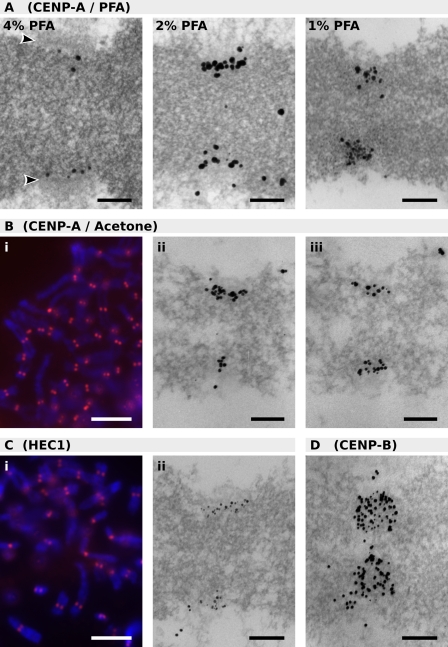
To obtain a higher resolution picture of centromeric organization, we investigated the localization of CENP-A through ultrathin serial-section EM. Several different precipitative and cross-linking fixation techniques were initially investigated to find the best compromise between labeling efficiency and morphological preservation. Although the best morphological preservation of chromosomes is typically obtained using glutaraldehyde, even very low (0.02% wt/vol in buffer) concentrations of this fixative prevented any epitope recognition by the CENP-A antibody (unpublished data). High concentrations of PFA (4%) also resulted in excellent morphological preservation but low antigen recognition, whereas concentrations ≤1% resulted in poor morphological preservation. A good compromise was achieved with 2% PFA (Fig. 1 A). Excellent antibody labeling was also obtained using the precipitative fixative acetone (Figs. 1 B and 2 A), which proved more amenable than PFA to labeling cytospun chromosome spreads and unblocked mitotic cells (Fig. 2 G). Although this fixative appeared to cause a slight loosening of the chromosome structure, a high degree of antigenicity was preserved. The outer kinetochore plate and fibrous corona were still detectable using acetone fixation, which was confirmed through the localization of the known outer plate protein HEC1 (Fig. 1 C; DeLuca et al., 2005), although the outer plate appeared to have collapsed against the chromatin. The ability of the antibody to penetrate throughout the centromeric chromatin using this preservation method was also demonstrated through the localization of the known inner CENP-B (Fig. 1 D; Cooke et al., 1990).
Figure 1.
LM and EM of proteins at the centromere. (A) EM of chromosomes fixed with PFA and labeled for CENP-A. Arrowheads denote the outer kinetochore plate. (B) Acetone-fixed chromosomes labeled for CENP-A and illustrated via LM (i) and EM (ii and iii). Adjacent serial sections are shown in ii and iii. (C) Acetone-fixed chromosomes labeled for HEC1 and illustrated via LM (i) and EM (ii). (D) EM of acetone-fixed chromosome labeled with CENP-B. Electron micrographs are cropped to illustrate the centromere. Section thickness: A, 70 nm; B–D, 60 nm. Bars: (A, B [ii and iii], C [ii], and D) 200 nm; (B [i] and C [i]) 5 μm.
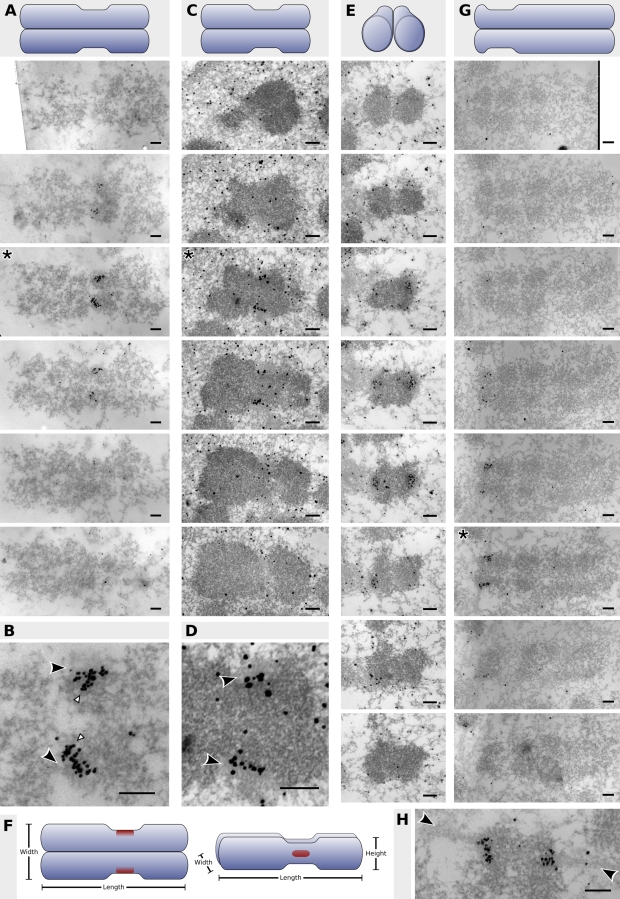
Figure 2.
Localization of CENP-A to the inner kinetochore of human chromosomes. Each panel represents a complete set of serial sections through the centromere, and a schematic of the orientation of each chromosome precedes each set. (A) 60-nm serial sections through an acetone-fixed, colcemid-blocked mitotic chromosome. (C and E) 70-nm serial sections through PFA-fixed, colcemid-blocked mitotic chromosomes in different orientations. (G) 50-nm serial sections through an acetone-fixed unblocked mitotic chromosome. Details of the centromere regions (asterisks) from A, C, and G are shown in B, D, and H, respectively. Black arrowheads in B and D point to the kinetochore outer plate; white arrowheads demarcate putative chromatin fibers illustrated in Fig. 4. Arrowheads in H point to microtubules attached to the kinetochore. (F) A stylized representation of the CENP-A binding domain (red) on human chromosomes drawn to the proportions observed in this study (Table I). Bars, 200 nm.
The localization of CENP-A was initially studied on acetone-fixed cytospun spreads of colcemid-blocked mitotic cells, which possess the advantage of having the majority of chromosomes clearly separated and parallel with the plane of the section (Fig. 1 B), and PFA-fixed, colcemid-blocked whole mitotic cells. Serial sections through representative chromosomes positioned in the plane of the section are illustrated in Fig. 2 (A and C), with details of the sections containing the strongest labeling illustrated in Fig. 2 (B and D), respectively. On each chromosome, the CENP-A domain was measured in terms of length (the x axis, running along the length of the chromatids), width (the y axis, running between the sister kinetochores), and height (the z axis, running through the serial sections; Fig. 2 F). As well as empirical measurements, the proportion of the primary constriction occupied by CENP-A was calculated in each dimension.
The combined results from nine sectioned acetone-fixed chromosomes and five sectioned PFA-fixed chromosomes are presented in Table I. Although the physical height of the CENP-A domain in PFA-fixed chromosomes was found to be slightly greater using this fixation/preparation method (a mean of 151 nm compared with 125 nm for the acetone-fixed sample; Mann-Whitney U test: P = 0.02), no significant difference was observed in the length and width dimensions (Mann-Whitney U test: length, P = 0.30; width, P = 0.70). More importantly, no significant difference was observed in the proportions of the domain relative to the constriction (Mann-Whitney U test: length, P = 0.90; width, P = 0.28; height, P = 0.73) between acetone- and PFA-fixed chromosomes, demonstrating consistency between the fixation methods.
Table I. The dimensions and proportions of the CENP-A binding domain on human chromosomes.
| Sample | Inner kinetochore length |
Proportion of constriction lengtha |
Inner kinetochore heightb |
Proportion of constriction heightc |
Inner kinetochore widthd |
Proportion of constriction widthe |
|---|---|---|---|---|---|---|
| nm | nm | nm | ||||
| Acetone fixed (colcemid; n = 9) |
233 ± 16 | 0.68 ± 0.04 | 125 ± 5 | 0.34 ± 0.02 | 102 ± 6 | 0.33 ± 0.02 |
| Acetone fixed (unblocked; n = 4) |
248 ± 18 | 0.74 ± 0.02 | 147 ± 5 | 0.34 ± 0.02 | 85 ± 9 | 0.29 ± 0.04 |
| PFA fixed (n = 5) |
236 ± 9 | 0.62 ± 0.03 | 151 ± 8 | 0.35 ± 0.02 | 96 ± 9 | 0.29 ± 0.02 |
| Sorted mardel(10) (n = 6) |
245 ± 11 | 0.70 ± 0.03 | 135 ± 7 | 0.33 ± 0.01 | 152 ± 12 | 0.31 ± 0.03 |
| Sorted chromosome 18 (n = 5) |
242 ± 28 | 0.66 ± 0.03 | 131 ± 7 | 0.32 ± 0.01 | 148 ± 6 | 0.32 ± 0.02 |
Values are expressed as the mean ± the standard error of the mean.
Calculated as the length of the CENP-A binding domain divided by the length of the constriction.
Calculated as the mean number of sections exhibiting CENP-A labeling multiplied by the mean section thickness.
Calculated as the number of sections exhibiting CENP-A labeling divided by the total number of sections containing the constriction.
The maximum width of labeling observed within a complete set of sections.
Calculated as the maximum kinetochore width divided by the maximum constriction width.
The data indicate that CENP-A occupies a relatively small proportion of the centromeric chromatin (Table I). Although the size of the protein-binding domain shows some variation between chromosomes, in general, CENP-A stretches over the length of the inner kinetochore in a domain spanning ∼235 nm and occupying two thirds of the length of the constriction. However, protein labeling was visible in only one third of the serial sections through the height of the constriction, representing a span of between 125 and 150 nm based on the section thickness. Although the protein was predominantly found at the inner kinetochore plate (determined relative to the localization of HEC1; Fig. 1 C), labeling was observed to extend a maximum of ∼100 nm into the underlying chromatin in the width dimension, representing one third of the width of the chromatid at the primary constriction. The total extent of centromeric chromatin bound by CENP-A thus represents two thirds of the constriction length, one third of the constriction height, and one third of the constriction width.
The chromosome in Fig. 2 E is orientated longitudinally within the plane of the section. Again, it can be seen that the CENP-A binding domain occupies approximately one third of the height of the constriction. Fig. 2 F shows a schematic summary of the aforementioned results from both acetone- and PFA-fixed preparations.
Finally, the distribution of CENP-A was examined in acetone-fixed unblocked metaphase cells (Fig. 2 G). The position of bound microtubules adjacent to the label indicates that CENP-A localizes predominantly at the inner kinetochore plate (Fig. 2 H). No significant difference was observed between blocked and unblocked chromosomes fixed with acetone except in the empirical height of the domain (Mann-Whitney U test: P = 0.01; again, no significant difference was observed in the proportion of the constriction height), and no significant difference was observed between unblocked metaphase chromosomes and blocked mitotic chromosomes fixed in PFA (Table I).
Our results indicate that CENP-A occupies a much narrower domain at the constriction than previously observed by deconvolution LM (Blower et al., 2002; Sullivan and Karpen, 2004). Although the CENP-A domain spans two thirds of the constriction length, the protein occupies only one third of the constriction height, which is in direct contrast to previous LM data that suggested the protein was present throughout the entire height of the centromere (Blower et al., 2002). Furthermore, the protein occupies only a small proportion (between 6 and 8%) of the total volume of the constriction DNA. Such a discrepancy between the EM data and deconvolution LM is not surprising considering that the dimensions of the CENP-A binding domain in both width and height fall below the resolution limits for optical microscopy.
The CENP-A domain occupies the same extent of chromatin at a human neocentromere
A previous study had demonstrated that significantly less CENP-A is present at the kinetochores of neocentromeres, including the well-studied mardel(10) neocentromere (Irvine et al., 2004). This neocentromere was shown to bind only one third of the amount of CENP-A present at most alphoid centromeres. Therefore, we were interested in determining whether the CENP-A binding domain on this neocentromere was physically smaller or in any way different from that found at alphoid centromeres.
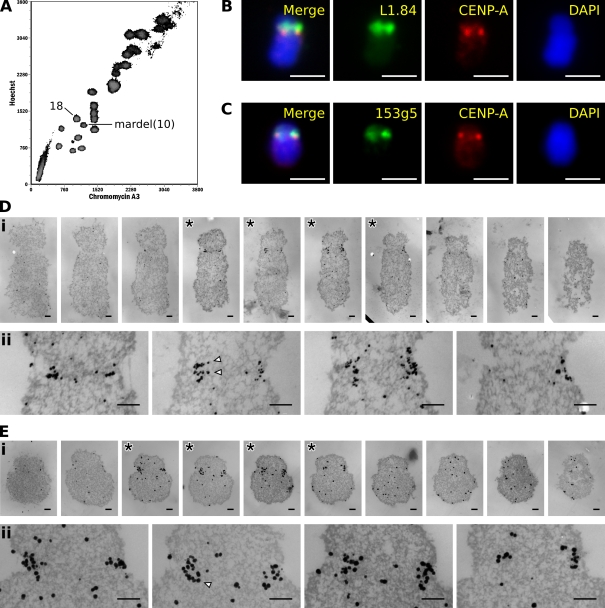
Because the mardel(10) chromosome is morphologically difficult to identify via EM, the chromosome was isolated using flow cytometry (Fig. 3 A). To verify that the sorting process did not produce a distortion of the kinetochore structure, a population of chromosome 18 was isolated as a control. Purity of the sorted mardel(10) and chromosome 18 populations was 92% and 97%, respectively, as assessed by immuno-FISH with anti–CENP-A antibodies and FISH probes targeted to the relevant centromeres (Fig. 3, B and C).
Figure 3.
Localization of CENP-A to the centromeres of flow-sorted chromosomes. (A) Flow-karyotype profile of chromosomes from a human 14ZBHT cell line with the chromosome 18 and mardel(10) populations indicated. (B) Immuno-FISH example of a sorted chromosome 18 using a chromosome 18–specific alphoid probe L1.84. (C) Immuno-FISH example of a sorted mardel(10) chromosome using a bacterial artificial chromosome probe 153g5 specific for the neocentromere. (D) 45-nm serial sections through chromosome 18 (i) and detail of sections exhibiting labeling (asterisks; ii). (E) 45-nm serial sections through a mardel(10) chromosome (i) and detail of sections exhibiting labeling (asterisks; ii). Arrowheads demarcate putative chromatin fibers illustrated in Fig. 4. Bars: (B and C) 2 μm; (D and E) 200 nm.
Ultrathin serial sections through a typical chromosome 18 (Fig. 3 D) and mardel(10) chromosome (Fig. 3 E) are shown. Although the limits of the constriction may be difficult to recognize in any one section, inspection of numerous serial sections enables us to define the limits with some confidence. In concordance with previous observations (Irvine et al., 2004), levels of CENP-A labeling were found to be significantly reduced on the mardel(10) neocentromere compared with the chromosome 18 alphoid centromere (a mean of 35 ± 6 labeled antibody molecules [n = 6] per mardel(10) kinetochore compared with 73 ± 7 labeled antibody molecules [n = 5] per chromosome 18 kinetochore; Student's t test: P = 0.004).
Surprisingly, despite the reduction of CENP-A at the mardel(10) neocentromere, the same distinctive CENP-A domain encompassing two thirds of the constriction length, one third of the constriction width, and one third of the constriction height was observed for both chromosome populations (Table I). No significant difference was found between the labeling domain on the chromosome 18 alphoid centromere or the mardel(10) neocentromere in any dimension, neither between the physical measurements (Student's t test: length, P = 0.93; width, P = 0.80; height, P = 0.64) nor between the relative proportions of the constriction bound by the protein (Student's t test on arcsine-transformed data: length, P = 0.41; width, P = 0.83; height, P = 0.84). (When compared with unsorted chromosome populations, both groups of sorted chromosomes were found to have expanded in the width dimension of the constriction, a result that appears to be related to the combination of polyamine buffer isolation and subsequent acetone fixation. However, as the aforementioned analysis indicates, this expansion was accompanied by that of the CENP-A domain and resulted in no net alteration in the relative proportion of the CENP-A and constriction widths.)
Such a result indicates that the same extent of centromeric chromatin is bound by CENP-A on an alphoid centromere and a neocentromere despite the clear reduction in CENP-A at the neocentromere (Irvine et al., 2004; and this study). The two datasets together suggest that the mardel(10) kinetochore is not physically smaller than an alphoid kinetochore but rather that CENP-A is less frequently incorporated into neocentromeric chromatin. In support of this observation, recent ChIP evidence suggests that CENP-A nucleosomes within a neocentromere are not present in large homologous tracts but are interspersed with H3-containing nucleosomes on a single nucleosome basis (Alonso et al., 2007).
Clustering of CENP-A at the inner kinetochore plate has implications for the structure of centromeric chromatin
A clearly recognizable trend in all acetone-fixed material was a distinct clustering of label. Such clusters gave the impression of labeled chromatin fibers, stretching from the kinetochore plate into the internal chromatin (see arrowheads in Fig. 2 B and Fig. 3, D [ii] and E [ii]). These structures may have resulted from an apparent loosening of the chromatin structure in acetone-fixed samples, allowing the underlying unit of centromeric chromatin to become more definable.
We considered that the width of these putative fibers could be important for elucidating the form of chromatin present at the centromere. Therefore, we measured the widths of 40 fibers taken from 19 different chromosomes where the labeled cluster was spatially isolated, the structure was significantly longer than it was wide, and the appearance of a contiguous unit of label was unambiguous from the serial sections. These putative fibers measure a mean of 32.6 nm in width (standard deviation, 5.6 nm; standard error, 0.9 nm; n = 40); representative examples are shown in Fig. 4. The width of these fibers fits well with observed widths of the 30-nm fiber in vitro (between 25 and 33 nm; Dorigo et al., 2004; Robinson et al., 2006), and it is tempting to suggest that this may be evidence for the existence of the 30-nm chromatin fiber in vivo at centromeres.
Figure 4.
Examples of labeled putative chromatin fibers. The distance between parallel lines represents the measured width of the fiber, which is listed below each example. Five of the fibers depicted are cropped and rotated from centromeres in Figs. 2 and 3. Bar, 200 nm.
Such an observation would appear to be contrary to recent evidence of half-octameric CENP-A nucleosomes in Drosophila melanogaster, which are reportedly present in a 10-nm fiber configuration (Dalal et al., 2007). However, the same study also described an increased proportion of octameric CENP-A nucleosomes in a mitotic-enriched fraction of cells, and current evidence in human cells suggests that CENP-A nucleosomes are present throughout the cell cycle in an octameric form (Shelby et al., 1997; Orthaus et al., 2008). Thus, it appears more likely that a 10-nm fiber, half-octamer configuration of CENP-A may be specific to Drosophila cells in interphase.
Centromeric chromatin is present in a complex higher order structure
The localization of CENP-A at a neocentromere has a wider implication for the higher order structure of centromeric chromatin. Previous models have suggested that the DNA fiber at the primary constriction is arranged in a linear coil (Zinkowski et al., 1991; Blower et al., 2002). This repeat-subunit model has been built on deconvolution LM data for CENP-A and data that suggested an interspersed pattern of CENP-A and H3 nucleosomes on stretched chromatin fibers (Blower et al., 2002). A similar arrangement of nucleosomes has been observed via CENP-A ChIP at the mardel(10) neocentromere (Chueh et al., 2005). With both three-dimensional structural data and ChIP data for CENP-A at this neocentromere, we can now ask how well this model predicts the structure of the kinetochore and primary constriction.
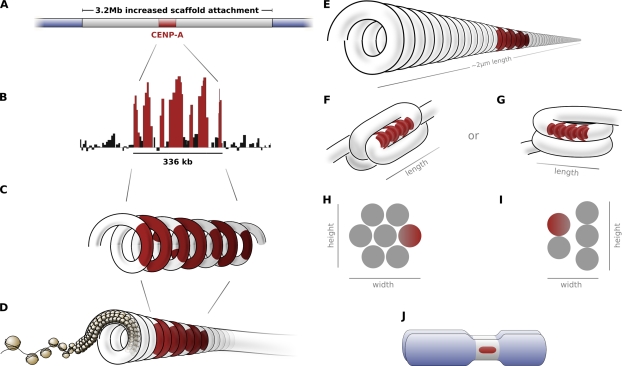
At the mardel(10) neocentromere, CENP-A occupies a 336-kb domain (Lo et al., 2001a; Chueh et al., 2005) within a larger 3.2-Mb domain of increased scaffold and matrix protein attachment (Saffery et al., 2003). Considering that scaffold proteins are also enriched at human centromeric α-satellite DNA (Strissel et al., 1996) and that the higher order structure of the chromosome becomes more condensed at the centromere both in a biochemical sense (Gilbert and Allan, 2001) and visibly, it is logical to infer that the scaffold enrichment region on the mardel(10) chromosome represents the DNA within the primary constriction. At this neocentromere, therefore, CENP-A is present within only slightly more than one tenth of the centromeric chromatin (Fig. 5 A).
Figure 5.
Higher order chromatin structure at the centromere. (A and B) The CENP-A binding domain (red) at the mardel(10) neocentromere occupies less than one tenth of the constriction DNA (silver) as determined by previous ChIP studies (A; Lo et al., 2001a; Saffery et al., 2003) and exhibits a periodic distribution at higher resolutions (B; adapted from Chueh et al., 2005). (C–E) Arranged into the coiled repeat-subunit model (C and D) of Zinkowski et al. (1991) and Blower et al. (2002), a three-dimensional domain is predicted that covers only one tenth of the constriction length (E). (F–J) However, folding (F) or coiling (G) the resulting intermediate chromatin fiber yields higher order structures (shown in H and I) that fit the distribution of CENP-A observed in this study (J).
Based on the models of Blower et al. (2002) and Zinkowski et al. (1991), each peak in the mardel(10) ChIP data (Fig. 5 B) should represent a separate coil of the repeat-subunit model (Fig. 5 C), bringing the isolated regions of CENP-A binding into a contiguous unit (Fig. 5 D) at the kinetochore inner plate. Such a model fits well with the fine structural localization of CENP-A to the inner kinetochore presented in this study. However, if this model is extended for the remainder of the DNA of the primary constriction, CENP-A will only occupy one tenth of the constriction length (Fig. 5 E). This is contrary to our observation that approximately two thirds of the constriction length is bound by CENP-A, both at the mardel(10) neocentromere and at alphoid centromeres (Table I). Furthermore, based on the number of chromatin coils required in the CENP-A domain and the relative length of the scaffold enrichment region, the physical length predicted by the repeat-subunit model would be somewhere between 700 nm (in the case of a coiled 10-nm fiber) and >2 μm (in the case of a coiled 30-nm fiber), dimensions not observed at the mardel(10) centromere or indeed at any human primary constriction.
To account for the CENP-A binding domain at the mardel(10) neocentromere, the coiled chromatin fiber of the repeat-subunit model must itself be present in a higher order configuration such that one tenth of the centromeric chromatin spans two thirds of the constriction length. This arrangement implies that the coiled centromeric chromatin is folded (Fig. 5 F) or looped (Fig. 5 G) back upon itself multiple times in the length dimension to form a multilayered structure. By folding the coiled fiber back upon itself (Fig. 5, F and H), the proportions of one third of the height and one third of the constriction width are predicted. Although the looped model (Fig. 5, G and I) is not quite as accurate (predicting a CENP-A domain still occupying one third of the constriction height but more than one third of the constriction width), it remains a possible structural alternative.
Both models, but especially the looped model in Fig. 5 G, fit with previous transmission EM experiments showing a higher order structure in partially decondensed chromosomes or centromeres. The presence of an intermediate 100–130-nm DNA fiber (analogous to the coiled 30-nm fiber in Fig. 5 D) has been previously observed both at the distended centromeres of chromosomes treated with 5-azacytidine (Rattner and Lin, 1987) and during chromosome arm condensation and decondensation (Belmont and Bruce, 1994; Kireeva et al., 2004).
With both models (Fig. 5, F and G), we would suggest that the CENP-A chromatin at the inner kinetochore plate creates an unfoldable unit that is intimately connected with the final structure. Considering the large number of proteins now shown to interact with CENP-A at the inner plate (Foltz et al., 2006; Okada et al., 2006), it may be conjectured that the chromatin loops are held together in a tight protein-dense structure that forms a natural impediment to folding or coiling. The expanse of the CENP-A domain may thus define the physical length of the constriction during chromosome condensation at mitosis.
In summary, defining the precise three-dimensional binding domain of CENP-A at human centromeres has shown that the protein occupies an unexpectedly small and compact region of chromatin at the centromere. We further demonstrate that the CENP-A binding domain at a neocentromere has the same size and proportions as that found at alphoid centromeres. Collectively, our results imply that current models of chromatin compaction at the metaphase centromere are unable to predict the proportions of the CENP-A domain observed in this study and that centromeric chromatin is arranged in a coiled 30-nm fiber that is itself coiled or folded to achieve the final structure of the primary constriction.
Materials and methods
Cell culture
Human HT1080 or 14ZBHT (an HT1080 variant line containing the mardel(10) chromosome) cell lines were cultured in DME with 10% FCS supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin antibiotics. Colcemid (Invitrogen) was added to the culture medium at a concentration of 10 μM before harvesting.
Cytospun spread preparations
Human HT1080 cells were blocked in colcemid for 1 h, and mitotic cells were harvested via shake off. Cells were washed in PBS and resuspended at a concentration of 1.6 × 105 cells/ml in hypotonic solution (75 mM KCl) for 10 min at room temperature. The 200-μl cell suspension was then cytospun (Shandon Cytospin 3; Life Sciences International) onto glass coverslips at 1,000 rpm for 8 min. Coverslips were immediately fixed in liquid chromatography–grade acetone (Merck) for 10 min before being washed three times in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, and 0.1% [vol/vol] Triton X-100) for 5 min per wash. Immunolabeling was performed.
Whole colcemid-blocked cell preparations
HT1080 cells were blocked in colcemid for 1 h, and mitotic cells were harvested via shake off. Cells were washed in PBS and fixed in 2% PFA in PBS for 10 min at room temperature. Cells were again washed in PBS and cytospun onto glass coverslips at 500 rpm, a speed at which no morphological distortion was apparent, for 5 min before permeabilization in KCM buffer.
Unblocked mitotic cell preparations
HT1080 mitotic cells were harvested via shake off. Cells were washed in PBS, resuspended in PBS, and cytospun onto glass coverslips at 1,000 rpm for 5 min. Coverslips were immediately fixed in liquid chromatography–grade acetone for 10 min before being washed three times in KCM buffer for 5 min per wash.
Immunocytochemistry
Preparations were labeled with either a mouse monoclonal CENP-A antibody (MBL International), a mouse monoclonal HEC1 antibody (9G3; Abcam), or a rabbit polyclonal CENP-B antibody (BL1104; Abcam); all primary antibodies were diluted 1:100 in KCMB (KCM buffer containing 1% BSA) and incubated with the sample for 1 h at 37°C or for 1.5 h at room temperature. Samples were washed three times in KCM and incubated with a gold-labeled secondary antibody (Ultrasmall [AURION]; Nanogold [Nanoprobes]) at 1:50 dilution in KCMB for either 20 h at 4°C, 2 h at 37°C, or 1.5 h at room temperature. Samples were washed three times in KCM and processed for EM by fixation in 2.5% glutaraldehyde/1% PFA in 0.2 M phosphate buffer, pH 7.2, at 4°C for 20 h.
As an alternative to the final fixation step, for LM, duplicate preparations were labeled with a donkey anti–goat Alexa Fluor 594 antibody (Invitrogen) in KCMB for 1 h at 37°C or 1.5 h at room temperature. Samples were washed three times in KCM and fixed in 4% PFA in PBS, washed in PBS, mounted in Vectorshield containing DAPI, and observed under a microscope (AxioImager M1; Carl Zeiss, Inc.) with a 100× NA 1.4 objective lens. Images were captured using a camera (AxioCam; Carl Zeiss, Inc.) and AxioVision software (Carl Zeiss, Inc.).
FISH
Coverslips containing chromosomes previously labeled with CENP-A were soaked off slides in PBS. The coverslips were washed several times in PBS and refixed in ice-cold methanol–acetic acid (3:1) for 10 min before being allowed to dry completely at room temperature and aging for 1 d. DNA probes were created via nick translation (Roche) with biotin–deoxy-UTP (Boehringer Ingelheim) according to the manufacturer's protocol. Coverslips were codenatured with 5 μl of the DNA probe dissolved in hybridization buffer (50% formamide, 2× SSC, and 10% dextran sulfate) at 71°C for 5 min before incubating overnight at 37°C. Two posthybridization washes were performed in 2× SSC at room temperature for 5 min followed by three washes in 0.1× SSC at 60°C for 7 min. Slides were placed in 2× SSC for 2 min at room temperature and subsequently blocked in 80 μl TNB (10% blocking agent, 150 mM NaCl, and 100 mM Tris, pH 7.5) for 2 h at 37°C. Avidin-FITC (1:500 in 50 μl TNB) was added followed by further incubation at 37°C for 1 h, three washes in 4× SSC/0.5% Tween, and another incubation at 37°C for 1 h with goat anti–avidin-FITC (1:200 in 50 μl TNB). Three final washes in 4× SSC/0.5% Tween were then performed. Slides were mounted in Vectorshield containing DAPI and observed under a microscope (AxioPlan 2; Carl Zeiss, Inc.).
Chromosome soup preparation
Chromosomes were isolated from 14ZBHT cells using the following procedure. Cells were blocked in colcemid for 6 h, and mitotic cells were collected via shake off. The harvested cells were pelleted at 300 g for 5 min before being washed in PBS and pelleted again. Cells were then resuspended in hypotonic solution (0.075 M KCl) for 10–15 min before gently being pelleted at 300 g for 5 min and gently being resuspended in ice-cold polyamine buffer (80 mM KCl, 0.5 mM EGTA, 2 mM EDTA, 15 mM Tris-HCl, pH 7.5, 0.5 mM spermidine, 0.2 mM spermine, 30 mM DTT, and 0.25% Triton X-100) and being incubated on ice for 10 min. The suspension was then vortexed for 10 s followed by a further 10-min incubation on ice until the resulting chromosome soup exhibited a high degree of single chromosomes. In subsequent procedures, the chromosome soup was kept on ice at all times.
Chromosome sorting
Chromosome soup was labeled with 40 μg/ml chromomycin A3 and 5 μg/ml Hoechst dyes in the presence of 10 mM MgSO4 overnight, with 100 mM sodium citrate and 250 mM sodium sulphite being added to the solution 1 h before sorting. Chromosomes were sorted back into 500 μl polyamine buffer using a MoFlo FACS machine (Dako). Sorted chromosome populations were gently cytospun at 400 rpm for 4 min onto coverslips for subsequent labeling and FISH analysis. To assess the purity of each sorted population, a minimum of 200 individual chromosomes were scored by immuno-FISH.
EM
Coverslips containing labeled and fixed chromosomes were prepared and washed three times in distilled H2O (dH2O) before being postfixed in 2% (wt/vol) OsO4 for 15 min at 0°C. Coverslips were again washed three times in dH2O and silver enhanced for 50 min at room temperature using an R-Gent SE-EM kit (AURION) according to the manufacturer's protocol. Samples were subsequently dehydrated, infiltrated, and embedded in Procure 812 resin (Electron Microscopy Services) for 48 h at 60°C. Coverslips were subsequently detached from resin blocks containing the chromosome spreads or whole cells via repeated freeze–thawing in LN2.
Ultrathin serial sections were cut using an ultramicrotome (Ultracut-E; Leica). Because of the positioning of the sample at the blockface, care was taken to align the block before sectioning to obtain serial sections from the first section of the block. Sections were collected on formvar-filmed 2 × 1.5-mm slot grids and contrasted by staining for 5 min in 5% uranyl acetate in butanol-saturated dH2O. Samples were observed under a transmission electron microscope (1200EX; JEOL) at 80 keV, and images were captured on film (Kodak) and subsequently scanned at high resolution (800 dpi).
The microscope magnification was calibrated using latex beads of known sizes (Electron Microscopy Services) and a diffraction grating replica (Electron Microscopy Services). Section thickness was estimated during sectioning via section interference colors and was measured more accurately in the microscope using the “small-fold” technique (Weibel, 1979). Stated section thicknesses represent a mean measurement of the thickness of multiple sections rounded to the nearest 5 nm; calculations of the height of the CENP-A–labeling domain were made with unrounded values.
Measurements of the physical lengths of CENP-A labeling and the constriction were made by measuring pixel distances using the GNU Image Manipulation Program (http://www.gimp.org/); to eliminate the effects of silver-enhanced particle size variation on measurements, all distances involving silver-enhanced particles were measured from the center of the particle.
Statistical analysis
Statistics were performed using the R software package (http://www.r-project.org/). Comparisons between measurements taken from random chromosomes were performed using the nonparametric Mann-Whitney U test; comparisons between sorted populations of chromosomes (where the data were confirmed to be normally distributed using the Shapiro-Wilk test) were performed using a two-tailed unequal variance Student's t test. Ratio datasets were converted to a normal distribution for parametric tests using an arcsine transform. α was considered to be 0.05 for all tests.
Acknowledgments
We thank N. Carter for advice on chromosome sorting, M. Burton for FACS, and L. Wong and E. Earle for critical comments on the manuscript.
This work was supported by the National Health and Medical Research Council of Australia and the National Institutes of Health. K.H. Andy Choo is a Senior Principal Research Fellow (I.D. 334300) of the National Health and Medical Research Council.
Abbreviations used in this paper: CENP, centromere protein; ChIP, chromatin immunoprecipitation; dH2O, distilled H2O; LM, light microscopy.
References
- Alonso, A., R. Mahmood, S. Li, F. Cheung, K. Yoda, and P.E. Warburton. 2003. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12:2711–2721. [DOI] [PubMed] [Google Scholar]
- Alonso, A., B. Fritz, D. Hasson, G. Abrusan, F. Cheung, K. Yoda, B. Radlwimmer, A. Ladurner, and P. Warburton. 2007. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 8:R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, A.S., and K. Bruce. 1994. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D., B.A. Sullivan, and G.H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi, O., S. Purgato, L.V. di Cantogno, E. Grosso, R. Ciccone, O. Zuffardi, G.D. Valle, and M. Rocchi. 2008. Evolutionary and clinical neocentromeres: two faces of the same coin? Chromosoma. 117:339–344. [DOI] [PubMed] [Google Scholar]
- Cardone, M.F., A. Alonso, M. Pazienza, M. Ventura, G. Montemurro, L. Carbone, P.J. de Jong, R. Stanyon, P. D'Addabbo, N. Archidiacono, et al. 2006. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 7:R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh, A.C., L.H. Wong, N. Wong, and K.H.A. Choo. 2005. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14:85–93. [DOI] [PubMed] [Google Scholar]
- Cooke, C.A., R.L. Bernat, and W.C. Earnshaw. 1990. CENP-B: a major human centromere protein located beneath the kinetochore. J. Cell Biol. 110:1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal, Y., H. Wang, S. Lindsay, and S. Henikoff. 2007. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5:e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca, J.G., Y. Dong, P. Hergert, J. Strauss, J.M. Hickey, E.D. Salmon, and B.F. McEwen. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo, B., T. Schalch, A. Kulangara, S. Duda, R.R. Schroeder, and T.J. Richmond. 2004. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 306:1571–1573. [DOI] [PubMed] [Google Scholar]
- Foltz, D.R., L.E.T. Jansen, B.E. Black, A.O. Bailey, J.R. Yates, and D.W. Cleveland. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., and J. Allan. 2001. Distinctive higher-order chromatin structure at mammalian centromeres. Proc. Natl. Acad. Sci. USA. 98:11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman, E.V., K.J. Fowler, A.J. Newson, S. Redward, A.C. MacDonald, P. Kalitsis, and K.H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 97:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, D.V., D.J. Amor, J. Perry, N. Sirvent, F. Pedeutour, K.H.A. Choo, and R. Saffery. 2004. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 12:805–815. [DOI] [PubMed] [Google Scholar]
- Kireeva, N., M. Lakonishok, I. Kireev, T. Hirano, and A.S. Belmont. 2004. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 166:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.-T., J.B. Rattner, S.A. Jablonski, and T.J. Yen. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W., J.M. Craig, R. Saffery, P. Kalitsis, D.V. Irvine, E. Earle, D.J. Magliano, and K.H. Choo. 2001. a. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20:2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W., D.J. Magliano, M.C. Sibson, P. Kalitsis, J.M. Craig, and K.H. Choo. 2001. b. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11:448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, O.J., A.C. Chueh, L.H. Wong, and K.H.A. Choo. 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82:261–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, M., I.M. Cheeseman, T. Hori, K. Okawa, I.X. McLeod, J.R. Yates, A. Desai, and T. Fukagawa. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. [DOI] [PubMed] [Google Scholar]
- Orthaus, S., C. Biskup, B. Hoffmann, C. Hoischen, S. Ohndorf, K. Benndorf, and S. Diekmann. 2008. Assembly of the inner kinetochore proteins CENP-A and CENP-B in living human cells. Chembiochem. 9:77–92. [DOI] [PubMed] [Google Scholar]
- Rattner, J.B., and C.C. Lin. 1987. The higher order structure of the centromere. Genome. 29:588–593. [DOI] [PubMed] [Google Scholar]
- Robinson, P.J.J., L. Fairall, V.A.T. Huynh, and D. Rhodes. 2006. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. USA. 103:6506–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery, R., H. Sumer, S. Hassan, L.H. Wong, J.M. Craig, K. Todokoro, M. Anderson, A. Stafford, and K.H.A. Choo. 2003. Transcription within a functional human centromere. Mol. Cell. 12:509–516. [DOI] [PubMed] [Google Scholar]
- Shelby, R.D., O. Vafa, and K.F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel, P.L., R. Espinosa, J.D. Rowley, and H. Swift. 1996. Scaffold attachment regions in centromere-associated DNA. Chromosoma. 105:122–133. [DOI] [PubMed] [Google Scholar]
- Sullivan, B.A., and G.H. Karpen. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, P.E., C.A. Cooke, S. Bourassa, O. Vafa, B.A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K.F. Sullivan, et al. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901–904. [DOI] [PubMed] [Google Scholar]
- Weibel, E.R. 1979. Stereological Methods. Vol. 1. Academic Press, New York/London. 415 pp.
- Zinkowski, R.P., J. Meyne, and B.R. Brinkley. 1991. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]