Abstract
Oceanic islands have played a central role in biogeography and evolutionary biology. Here, we review molecular studies of the endemic terrestrial fauna of the Hawaiian archipelago. For some groups, monophyly and presumed single origin of the Hawaiian radiations have been confirmed (achatinelline tree snails, drepanidine honeycreepers, drosophilid flies, Havaika spiders, Hylaeus bees, Laupala crickets). Other radiations are derived from multiple colonizations (Tetragnatha and Theridion spiders, succineid snails, possibly Dicranomyia crane flies, Porzana rails). The geographic origins of many invertebrate groups remain obscure, largely because of inadequate sampling of possible source regions. Those of vertebrates are better known, probably because few lineages have radiated, diversity is far lower and morphological taxonomy permits identification of probable source regions. Most birds, and the bat, have New World origins. Within the archipelago, most radiations follow, to some degree, a progression rule pattern, speciating as they colonize newer from older islands sequentially, although speciation often also occurs within islands. Most invertebrates are single-island endemics. However, among multi-island species studied, complex patterns of diversification are exhibited, reflecting heightened dispersal potential (succineids, Dicranomyia). Instances of Hawaiian taxa colonizing other regions are being discovered (Scaptomyza flies, succineids). Taxonomy has also been elucidated by molecular studies (Achatinella snails, drosophilids). While molecular studies on Hawaiian fauna have burgeoned since the mid-1990s, much remains unknown. Yet the Hawaiian fauna is in peril: more than 70 per cent of the birds and possibly 90 per cent of the snails are extinct. Conservation is imperative if this unique fauna is to continue shedding light on profound evolutionary and biogeographic questions.
Keywords: arthropods, birds, dispersal, Hawaii, snails, speciation
1. Introduction
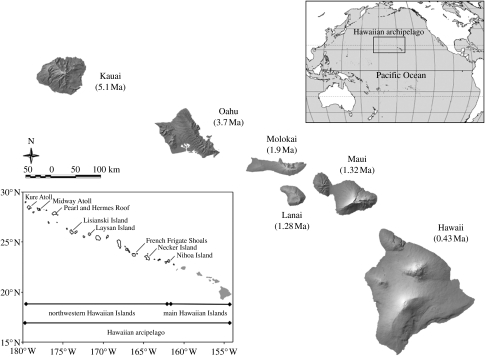
The Hawaiian archipelago (figure 1) consists of a sequence of oceanic islands formed as the Pacific plate moves north-westwards over a stationary plume or ‘hot spot’ in the Earth's mantle, which periodically sends magma up through the plate, creating a chain of volcanoes, each sequentially younger than the one that preceded it, and that will have moved north-westwards away from the hot spot. Eventually, each island subsides and erodes, becoming a low atoll, then a submerged seamount, and is finally subducted as the Pacific plate slides under the adjacent tectonic plate (Price & Clague 2002). The present islands are divided into the younger ‘high’ islands, and the older northwestern islands, which have become low atolls or small eroded islets and pinnacles. The oldest northwestern-most island is Kure Atoll (29 Ma) and the oldest high island is Kauai (5.1 Ma), with the youngest island, Hawaii itself, being less than 0.5 Ma and still being formed (figure 1). This review focuses on the high islands, which harbour spectacular arrays of endemic species of plants and animals (Ziegler 2002).
Figure 1.
Map of the Hawaiian Islands. The upper right figure shows the position of the Hawaiian archipelago in the Pacific basin. The lower left figure shows the entire Hawaiian archipelago, including the northwestern Hawaiian Islands, with Kure Atoll (29 Ma) in the far north-west corner. The scale bar beneath Kauai refers to the central map of the main Hawaiian Islands.
The biodiversity and evolutionary radiations of the Hawaiian Islands are arguably more spectacular than those of the Galápagos, and one can only surmise what would have been Darwin's delight and awe had he visited Hawaii. Inspired by The Origin, the Reverend John Thomas Gulick, born in Hawaii in 1832, became fascinated by this marvellous biodiversity, especially the brightly coloured and highly variable tree snails of the island of Oahu. During the 1870s, on a trip to England, he corresponded with Darwin and met him at Down House, discussing the spectacular diversity exhibited by the snails. Based partly on his studies of Hawaiian snails, Gulick's book, Evolution, racial and habitudinal (Gulick 1905), and some of his other publications include arguably the first exposition of the founder effect, genetic drift and the shifting balance concept (Carson 1987a). Since his time, the biological radiations of the Hawaiian Islands have come to be considered some of the paramount examples of natural evolutionary experiments (Simon 1987), in which replicate clades on sequentially produced and precisely dated islands have lent major insights into many aspects of evolutionary biology including speciation (Carson 1987b), biogeography (Wagner & Funk 1995; Cowie & Holland 2006), sexual selection (Kaneshiro 1988, 2006; Mendelson & Shaw 2005) and ecosystem processes (Vitousek 2002, 2004).
With the advent and increasingly widespread application of molecular phylogeographic methods, details of historical biogeography and systematics such as geographic origin, monophyly and clade bifurcation timing and order have become tractable issues. Factors with potential to erase or confound molecular phylogenetic signal, however, include extinction, repeated long-distance dispersal, hybridization and restricted distributions. Studies focusing on elucidation of historical biogeographic patterns in the Hawaiian Islands are beginning to reveal some common patterns. Perhaps most notable in this regard is the watershed compilation of Wagner & Funk (1995), which dealt with the biogeography of a suite of Hawaiian plant and animal taxa. The approach adopted was explicitly cladistic and some general patterns emerged, including adherence of numerous Hawaiian lineages to an island version of the so-called ‘progression rule’ (Hennig 1966), which identifies centres of origin with the youngest members of a monophyletic lineage on the geographic periphery, in this case the geologically younger islands. Thus, following initial colonization of the oldest island in a sequentially produced chain of islands, dispersal, accompanied by lineage bifurcation, occurs from older to younger islands sequentially down the chain. At the time of Wagner & Funk's (1995) compilation, molecular techniques were only beginning to become routine for addressing biogeographic questions and most of the studies in that volume were primarily or entirely morphological. This short review aims to revisit and update the biogeography of the Hawaiian terrestrial fauna in light of the numerous molecular studies that have been undertaken since Wagner & Funk's (1995) landmark contribution. In attempting to be as comprehensive as possible in the space available, few studies can be reviewed in the depth they deserve. However, what follows summarizes much of the work done since Wagner & Funk (1995), provides access to the large and scattered literature, and offers some thoughts on future directions and difficulties.
Most early molecular studies used a single, usually mitochondrial DNA (mtDNA) marker. Increasingly, however, use of multiple markers, including nuclear DNA (nDNA) sequences, has become the norm, permitting more robust interpretation of phylogenetic trees at both deep and shallow levels, and easier identification of hybrids. Use of multiple markers together with non-molecular systematic characters is widely agreed to be the preferred approach to phylogenetic reconstruction (Rubinoff & Holland 2005) and, in recent years, technical advances in DNA amplification, sequencing and analysis as well as cost reductions has allowed collection of larger molecular datasets than was previously feasible.
Most studies of terrestrial Hawaiian taxa have involved invertebrates, no doubt because of the relative ease of collection of large numbers of samples, generally fewer legal restrictions and the far larger number of invertebrate radiations. Nonetheless, there have been a number of interesting studies of the origins and radiations of birds, including fossil taxa, and of the single native terrestrial mammal species, a bat (there are no native Hawaiian terrestrial reptiles or amphibians).
2. Origins of the fauna
Earlier literature that addressed the phylogenetic and biogeographic origins of the Hawaiian fauna often tacitly assumed that each major group of Hawaiian taxa, e.g. all Hawaiian members of a particular family, was monophyletic and derived from a single colonizing species. While this seems reasonable for those major groups that are endemic to the islands, e.g. the land snail family Amastridae or the Hawaiian honeycreepers, the Drepanidinae, there is less justification for such an a priori assumption in a group that is not endemic to the islands, e.g. the land snail family Succineidae or the fruit fly genus Drosophila.
For some groups, monophyly and therefore presumed single origin of the Hawaiian taxa of a more widespread group has been confirmed. For instance, there are approximately 60 Hawaiian species of bees in the worldwide genus Hylaeus, all belonging to the subgenus Nesoprosopis, which is otherwise known primarily from Japan, with one species extending westwards into Europe and a number of undescribed species in China (Magnacca & Danforth 2006). The entire Hawaiian radiation has been shown, using the mtDNA markers COI, COII and tRNA-leucine, combined with morphological characters, to be monophyletic—the result of a single colonization. Assuming the subgeneric assignment to be correct, the Hawaiian radiation probably has an eastern Asian origin.
Similarly, a combined mtDNA and nDNA study of the Hawaiian species of the spider genus Havaika (nine nominal species), which also occurs in the Marquesas Islands of the south Pacific (Gillespie et al. 2008), demonstrated the monophyly of the Hawaiian radiation but did not explicitly infer its geographic origin (Arnedo & Gillespie 2006).
Increasingly, however, there is evidence of multiple colonizations for certain endemic Hawaiian species that belong to larger groups that are more widely distributed. Gillespie et al. (1994), using a small fragment of the 12S ribosomal RNA gene, demonstrated multiple origins of Hawaiian tetragnathid spiders, with two separate colonizations leading to independent species radiations and two additional colonizations that lead to no significant radiations. Although based on somewhat limited global sampling, and using only mtDNA (COI and 16S) markers, Gillespie (2002) suggested that Hawaiian Tetragnatha were derived from an American source.
In another group of Hawaiian spiders in the cosmopolitan genus Theridion, Arnedo et al. (2007) analysed sequence data from two mtDNA (COI and 16S) and three nDNA (18S, 28S and histone H3) markers. Two independent colonizations were also inferred, with at least one originating in Central or South America, while the other, arguably incorrectly placed in Theridion, appeared to be related to the holarctic genus Rugathodes.
Similarly, Rundell et al. (2004), using COI, revealed two distinct reciprocally monophyletic lineages and demonstrated at least two origins of the Succineidae, a globally distributed land snail family, in the Hawaiian Islands: an older colonization, initially of the island of Kauai, which is the oldest of the main Hawaiian Islands (figure 1), and a more recent colonization of the island of Hawaii, the youngest island. Rundell et al. (2004) speculated that the latter radiation had an eastern Asian origin. The origins of the older clade remained obscure. Holland & Cowie (in press) verified these results in a multilocus study with expanded Hawaiian and global sampling and suggested a South Pacific origin for the older Hawaiian clade.
The origins of the well-known achatinellid land snails, an essentially Pacific island endemic family, which includes the spectacular Hawaiian tree snails (subfamily Achatinellinae), remain less clear. The family is monophyletic, based on the geographically limited sampling undertaken so far (Holland & Hadfield 2004; Wade et al. 2006), and the single species that has been sampled outside Hawaii is sister to a monophyletic Hawaiian radiation (Holland & Hadfield 2004), suggesting a single origin of the Hawaiian radiation. However, the lack of molecular analysis of other non-Hawaiian achatinellid taxa that are traditionally placed in genera represented in Hawaii (Cooke & Kondo 1961) suggests that there may have been multiple colonizations of Hawaii from elsewhere in the Pacific. The Achatinellidae belong to a well-supported clade of ‘orthurethran’ taxa that include the Hawaiian endemic family Amastridae, the New Caledonian endemic family Draparnaudiidae, the Pacific island (excluding Hawaii) endemic family Partulidae and other more globally distributed families (Holland & Hadfield 2004; Wade et al. 2006). However, relationships within the Orthurethra are not well understood (Wade et al. 2006) and the ultimate geographic origin of the Achatinellidae (and the other Pacific island taxa) remains obscure, although fossils of questionable achatinellid affinity have been found in Europe and North America (Solem 1976; Solem & Yochelson 1979).
In other Hawaiian invertebrate groups that have been analysed genetically, it has sometimes been possible to infer the number of independent colonizations, although as in the Achatinellidae, the absence of broader geographic sampling of groups that are not endemic to Hawaii precludes drawing firm conclusions. For example, while Hawaiian crane flies (Diptera, Limoniidae, Dicranomyia) may be monophyletic, suggesting a single colonization, although with a possible second colonization by one species (Nitta & O'Grady 2008), lack of sampling of Dicranomyia from elsewhere in its wide Pacific distribution does not permit definitive confirmation.
In an effort to elucidate diversification patterns in the Hawaiian damselflies as well as to place them within a larger geographic context, Jordan et al. (2003) used nDNA and mtDNA from multiple individuals of 20 of the 23 described species of the endemic genus Megalagrion. They included eight out-group species (in two genera), and although Megalagrion was clearly monophyletic, no one species or genus emerged as the sister to the Hawaiian genus. As in many Hawaiian groups, the out-groups were distant; the out-group choice influenced the in-group topology but the problem could be resolved by fitting a model of evolution independently to the in-group and constraining in-group topology during out-group rooting. More out-group sampling may reveal an appropriate sister group.
Similarly, while there are two well-supported monophyletic clades of Hawaiian tettigoniid crickets, or katydids (Banza spp.), placement of a single species (Banza nihoa) is equivocal and combined with lack of sampling of related taxa from the possible original source areas (Banza seems closely related to both New and Old World taxa) precludes determination of the number of Hawaiian colonizations (Shapiro et al. 2006).
Based on an amplified fragment length polymorphism (AFLP) analysis of 25 species of endemic Hawaiian crickets (Laupala), Mendelson & Shaw (2005) suggested that the genus is monophyletic, although they did not sample widely beyond Hawaii and therefore did not address its geographic origin.
Despite the relatively huge amount of research on the Hawaiian drosophilids, and while they have increasingly been accepted as having arisen from a single colonization (O'Grady 2002; Remsen & O'Grady 2002; O'Grady & DeSalle 2008), their geographic origin has remained elusive although with the possibility of east Asia having been mentioned from time to time (Remsen & DeSalle 1998; Davis 2000). Again, adequate sampling of potential source regions has not been undertaken.
The Hawaiian vertebrate fauna, although far less diverse than the insects or land snails, has nevertheless prompted a number of molecular studies addressing their origins, although much remains unknown. Fleischer & MacIntosh (2001) reviewed molecular studies of birds, mostly using mtDNA markers, largely from the perspective of understanding their phylogenetic relationships and origins. The only other native terrestrial vertebrate is a bat. The following summary of information on the birds is derived primarily from the review of Fleisher & MacIntosh (2001).
The most spectacular radiation of birds is the Hawaiian honeycreepers (Drepanidinae) with over 50 species. They appear to be monophyletic and derived from a New World cardueline finch. Other groups have radiated far less spectacularly and since over 70 per cent of the endemic avifauna is extinct (Boyer 2008), molecular analyses are challenging. Nevertheless, a combination of analyses of extant taxa and considerable success in obtaining sequences from bones of extinct taxa has given significant insight into the phylogenetic and geographic origins of the various groups and non-radiating species. The extinct Hawaiian flightless rails, Porzana spp. (perhaps more than 12 species), may have resulted from two independent colonizations from Asia or the western Pacific by volant species (see also Slikas et al. 2002). The Hawaiian thrush, Myadestes obscurus, the only extant representative of a radiation of five species, is of New World (Caribbean or western North American) origin. The Hawaiian crow, Corvus hawaiiensis, the only extant of four corvid species, seems more closely related to North American ravens than to other Pacific island corvids. The extinct flightless ducks, or ‘moa-nalos’, were previously thought to be forms of geese. However, they are related to New World dabbling ducks, perhaps South American Anas or Anas relatives, although they split from them a long time ago, arriving and becoming flightless and gigantic before the island of Hawaii, from which they are absent, came into being (see also Sorenson et al. 1999). However, the extant ducks, the Hawaiian duck, Anas wyvilliana, and the Laysan duck, Anas laysanensis, appear to represent independent colonizations, the former being most closely related to North American mallards or mottled ducks and the latter to the South Pacific black duck clade. The two extinct flightless ibis species, Apteribis spp., are most closely related to the New World white ibis, Eudocimus alba. The Hawaiian goose (Branta sandvicensis), along with a fossil radiation of Hawaiian Branta spp., is nested within the widespread Nearctic Canada goose (Branta canadensis) clade, rendering the latter paraphyletic, but confirming the North American origin of Hawaiian Branta (see also Paxinos et al. 2002). The Hawaiian hawk, Buteo solitarius, groups with New World not Old World Buteo species. The ‘elepaio’, Chasiempis sandwichensis, is related to other Polynesian flycatchers (Monarcha spp.).
In contrast to most of these taxa that have New World or in some cases south Pacific origins, the Hawaiian eagle, a relatively recent but prehuman arrival in the islands, is Palaearctic in origin and has differentiated little from the white-tailed eagle, Haliaeetus albicilla (see also Fleischer et al. 2000). Molecular approaches have not been used to address the origins of other Hawaiian bird taxa, the knowledge of which remains based on the morphological taxonomy.
The single native terrestrial mammal in Hawaii, the Hawaiian hoary bat, Lasiurus cinereus semotus, has been shown using mtDNA restriction site mapping to be more closely related to the North American than the South American subspecies of the hoary bat (Morales & Bickham 1995), suggesting a relatively recent colonization of the Hawaiian Islands from North America.
3. Phylogenetics and biogeography
While rather few molecular studies have explicitly and satisfactorily addressed the origins of the Hawaiian fauna, perhaps with the exception of the birds, numerous studies have focused on the evolutionary radiations and the patterns of diversification within the island chain following initial colonization. Many of these patterns correspond to a greater or lesser degree with the progression rule, although there may be back colonizations from younger to older islands, major radiations within rather than among islands, and considerable stochastic dispersal, especially among more vagile or actively dispersing taxa.
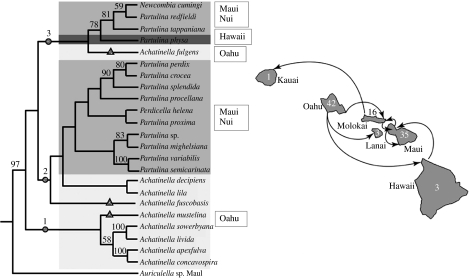
Thacker & Hadfield (2000), using 16S, and Holland & Hadfield (2004), using COI, addressed the diversification of the achatinelline tree snails, with essentially consistent results between the two studies. The phylogenetic reconstructions suggested Oahu as the first island to be colonized, with subsequent colonization and diversification that in general followed the progression rule but with a number of instances of back colonization (figure 2). However, the enigmatic finding (Gage 1996) of a single fossil species of the achatinelline genus Newcombia on Kauai (no other achatinellines are known from Kauai and Newcombia is otherwise known only from Maui and Molokai) probably represents a back colonization from Maui/Molokai, skipping Oahu (figure 2), rather than a Kauai origin of the achatinelline radiation, because Newcombia is a more derived rather than basal taxon within the Achatinellinae (Holland & Hadfield 2004).
Figure 2.
Inter-island colonization routes for the achatinelline tree snails inferred from the mtDNA phylogeny of Holland & Hadfield (2004). Arrows correspond to nodes and clades in the tree, with the exception of the arrow from Molokai to Kauai, which represents subfossil Newcombia sp. shells on Kauai (the only achatinelline on that island). Numbers on islands are numbers of endemic achatinelline species on each island (Cowie et al. 1995). Although the phylogeny also suggests a number of within-island diversification events, here we focus on the among-island colonizations. The tree was produced using corrected maximum likelihood with PAUP (Swofford 2002) and is based on 675 bp of mtDNA (COI; Holland & Hadfield 2004). Circles, main clades; triangles, seeding lineages.
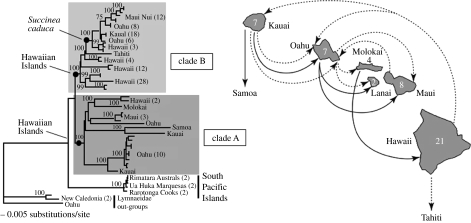
Within the two succineid land snail radiations identified by Rundell et al. (2004) and Holland & Cowie (in press) using nuclear and mitochondrial markers, the older radiation roughly followed the progression rule with the initial colonization site being Kauai. However, the more recent colonization of the island of Hawaii, the youngest island, has so far resulted only in a radiation on that island, with the exception (Holland & Cowie 2007) of the single species Succinea caduca, which occurs on all the main Hawaiian Islands and is derived from this radiation (figure 3, and see below).
Figure 3.
Inter-island colonization patterns for endemic Hawaiian succineid amber snails inferred from molecular phylogenetic data (Holland & Cowie in press). Arrows are based on nodes and clades in the tree, solid lines representing clade A and dotted lines representing clade B (Holland & Cowie in press). Numbers on islands are numbers of endemic succineid species on each island (Cowie et al. 1995). The two arrows, one from each of the two lineages, pointing southwards, one from the Island of Hawaii labelled ‘Tahiti’, the other from Kauai labelled ‘Samoa’, each represent a species from the South Pacific, suggesting historical colonization of these localities from the Hawaiian Islands. The phylogenetic tree was generated using corrected maximum likelihood in PAUP (Swofford 2002), based on three gene fragments, two mitochondrial and one nuclear (COI, 16S and 18S; 1740 bp). Succineid out-groups were selected based on their basal position in a global succineid phylogeny (Holland & Cowie in press). The shaded boxes highlight the two well-supported Hawaiian lineages, each of which maintains its integrity in the global phylogeny, with multiple lineages forming clades between them. The multi-island Hawaiian endemic species Succinea caduca is the only representative of clade B that occurs on islands other than the island of Hawaii (Holland & Cowie 2007).
The radiation of Hylaeus bees (Magnacca & Danforth 2006) similarly seems to have resulted from an initial colonization of the youngest island, Hawaii, in this case, however, followed by dispersal and speciation north-westwards throughout the island chain. For the most part, speciation has been between islands (sister species occurring on different islands) rather than within islands (sister species occurring on the same island), with the exception of the large radiation on the island of Hawaii, the location of the original colonization of the islands.
Jordan et al. (2003) analysed various mtDNA markers and the nuclear EF1α in 20 of the 23 species in the endemic monophyletic Hawaiian damselfly genus Megalagrion. The radiation is not a single uniformly speciating clade but is composed of a number of well-supported subclades exhibiting different patterns and rates of diversification. Nevertheless, most speciation, in four clades, is inter-island and follows the progression rule. One clade may have colonized in the opposite direction, from the youngest to older islands. Each clade roughly constitutes a group of species with similar ecology (breeding habitat). However, one clade diversified within Kauai to occupy each available breeding habitat.
Among the spiders, both clades of Tetragnatha exhibit patterns roughly conforming to the progression rule inasmuch as the most basal taxa occur on Kauai and Oahu with more derived taxa on younger islands (Gillespie et al. 1997; Pons & Gillespie 2004). However, in the ‘spiny leg’ Tetragnatha clade, while there is an overall progression rule tendency, the species on a single island tend to be most closely related to each other. This contrasts with a previous morphological analysis (Gillespie & Croom 1995) that showed that no sister taxa occurred on the same island. The favoured explanation for the disagreement (Gillespie et al. 1997) is that the morphology, including colour, was convergent between spiders on different islands. The monophyletic Hawaiian spider genus Orsonwelles also exhibits a pattern of initial colonization of Kauai followed by a somewhat ambiguous pattern that at least does not contradict the progression rule, although the pattern is predominantly within-island speciation (Hormiga et al. 2003). In Havaika, however, the lineages do not clearly follow the progression rule (Arnedo & Gillespie 2006). The original colonization appears to have occurred after Kauai, Oahu and Maui Nui had formed, and following colonization of one of these islands dispersal to other islands in all directions was rapid. Diversification appears to have been predominantly among islands rather than within islands.
Among the Hawaiian drosophilids, molecular analysis of the Drosophila planitibia group based on two mitochondrial and four nuclear markers (Bonacum et al. 2005) suggested an origin 6.1 Ma on an island older than Kauai, but with major diversification beginning on Kauai and subsequent colonization and diversification occurring as younger islands became available. While the overall pattern follows the progression rule, speciation has been both within and between islands, some back colonizations have occurred, and the patterns of diversification on the Maui Nui group (the combined islands of Molokai, Maui, Lanai and Kahoolawe, which have for most of their history been joined into a single island, e.g. Price & Elliott-Fisk 2004) is especially complex, probably involving both vicariance and dispersal. Notable is a basal clade containing only a single species from each of the islands of Kauai and Hawaii, the suggestion being that the absence of members of this clade from intermediate islands is most probably a result of extinction. By contrast, molecular analysis of the Drosophila haleakalae species group revealed no progression rule pattern (O'Grady & Zilversmit 2004).
The endemic Hawaiian moth genus Hyposmocoma (Cosmopterigidae) rivals the drosophilids in number of species with more than 350 recognized and many more undescribed (Rubinoff 2008). Study of a small subset of this diversity, the aquatic, so-called ‘cone-cased’ taxa, showed them to follow a progression rule pattern with a small radiation of three taxa on Kauai giving rise to a single species on each of Oahu, Molokai and Maui. The hugely diverse Hawaiian Cosmopterigidae are ripe for further investigation.
Shaw (2002) compared phylogenies derived from nDNA and mtDNA data in the almost 40 endemic Hawaiian Laupala species (crickets). Both partitions revealed a pattern of colonization from older to younger islands but conflicted in the detail. The nuclear data suggested a pattern of inter-island speciation and multiple colonizations and largely conformed to a previous morphological analysis (Otte 1994). By contrast, the mtDNA tree suggested in general that the species on each island had arisen from a single colonization. Shaw (2002) downplayed the utility of mtDNA and argued that the conflict could be explained by hybridization. Rubinoff & Holland (2005) re-analysed Shaw's (2002) data and showed that the conflict was over-stated, as many of the nodes in the mtDNA tree were not well supported, suggesting that the mtDNA data merely lacked appropriate phylogenetically informative sites and were therefore not comparable to the more robust nDNA data. A subsequent investigation (Mendelson et al. 2004) using nDNA AFLPs further supported an overall pattern of colonization from older to younger islands but contradicted the previous nDNA-based interpretation (Shaw 2002) of multiple origins of Laupala cerasina on the island of Hawaii. The clear lesson from these conflicting studies is that all approaches have value and that rather than discounting any one partition a better approach is to formally assess the congruence of the partitions, combine them if congruence tests suggest this is appropriate and seek explanations if they do not (Rubinoff & Holland 2005).
Less diverse groups have generally been less well studied. Shapiro et al. (2006) analysed approximately 2 kb of mtDNA and nDNA sequences from the radiation of crickets (katydids) in the genus Banza (one to three species per island) but were unable to draw firm biogeographic conclusions, other than to suggest that perhaps the radiation originated on Oahu, diversifying into two clades, neither of which exhibited clear progression rule patterns. Nitta & O'Grady (2008), using four mtDNA markers, investigated the biogeography of the 13 species of Hawaiian Dicranomyia (crane flies), the majority of which are not single-island endemics, and found no evidence of the progression rule, and indeed were unable to resolve the ancestral island for the majority of nodes in their phylogeny. They explained this lack of pattern as resulting from the relative ease with which crane flies, though large insects, are blown from island to island passively. They did, however, detect some evidence for progression rule diversification within species (see below).
Rivera et al. (2002) attempted to determine the origins of troglobitic (obligately cave dwelling) terrestrial isopods from their epigean progenitors. Two groups of cave isopods, derived from independent colonizations of the Hawaiian Islands, were confirmed in their analysis of COI. In one of these groups (Littorophiloscia), the single cave species is derived from the single epigean species. However, in the other group (Hawaiioscia) there are four cave-adapted species, one on each of Kauai, Oahu, Molokai and Maui. Rivera et al. (2002) argued that these species evolved from a formerly widespread (and now possibly extinct) surface species or group of closely related species, rather than dispersal of cave-adapted species between islands, but because of the absence of the surface ancestor(s) this issue remains unresolved.
The only major extant Hawaiian vertebrate radiation is the honeycreepers (Drepanidinae). A species from Kauai is the most basal and in general within each of the various small clades the progression rule is roughly followed and, as might be expected for such vagile organisms, there has been no within-island speciation (Fleischer et al. 1998, 2001).
The progression rule pattern provides a predictive theoretical framework against which lineage age, bifurcation order and colonization timing can be better understood, especially when applied within a comparative approach. Overall, many groups examined to date follow, at least roughly, a progression rule pattern of diversification, wherein the most basal taxa occur on the oldest island in the distribution of the lineage. However, a number of biogeographic patterns in terrestrial Hawaiian radiations are complex (Funk & Wagner 1995; Holland & Hadfield 2004). While some endemic groups clearly originated on Kauai, others appear to have first colonized either Oahu (achatinelline tree snails), Maui Nui (Laupala cerasina group, crickets) or the island of Hawaii (Hylaeus bees and one lineage of succineid snails: clade B, figure 2). In a few groups (Havaika spiders, Banza crickets, Dicranomyia crane flies), neither the original island colonized nor the subsequent pattern of diversification is clear.
The time of the initial colonization and the vagility of the organisms are crucial variables in determining the current patterns of distribution, and especially of adherence to the progression rule pattern of diversification. Thus, if lineage age, or colonization time, is roughly coincident with the age of an older island, and vagility is sufficient for propagules to reach islands that formed subsequently but rare enough to maintain species integrity, there is potential for diversification to adhere to the progression rule. For example, if the initial colonization occurred prior to the subaerial rise of Oahu then a progression rule pattern originating on Kauai or an older island would be possible, given appropriate levels of vagility. However, if the initial colonization occurred after the formation of, for instance, Oahu and Maui Nui, there is no reason other than chance to expect Kauai to be the island first colonized. And if it was even more recent, the island of Hawaii could be the site of initial colonization, in which case a progression rule pattern of diversification from oldest to youngest island would not be expected.
Dispersal ability also appears to play an important role in the degree to which monophyletic terrestrial radiations with multi-island distributions follow the progression rule. If a radiation has a strong active or passive dispersal mechanism (as suggested for Dicranomyia, birds, flying insects), a distinct progression rule pattern is not predicted regardless of the initial island colonized.
Identifying tractable, suitably diverse and broadly distributed lineages for which comprehensive phylogenies can be developed, and determining the timing of the initial colonization (e.g. Price & Clague 2002) is thus of key significance in grounding our understanding of these patterns of diversification.
4. Phylogeography and population structure
The majority of the species of most groups of terrestrial Hawaiian organisms are single-island endemics (Simon 1987), e.g. land snails, approximately 90 per cent (Cowie 1995; Cowie et al. 1995), and drosophilids, more than 90 per cent (Nitta & O'Grady 2008). Higher levels of endemism are expected in less vagile organisms and some Hawaiian taxa have reduced active dispersal capacity compared with their continental relatives. Nevertheless, some species do occur on more than one island, some occur on all main islands and certain groups (e.g. crane flies) exhibit much lower levels of single-island endemism. Recent work on a number of diverse taxa has detected intraspecific phylogeographic structure both within and among islands.
Drosophila grimshawi is unique among Drosophila in both its distribution throughout the main Hawaiian Islands and its apparent diversification pattern, which does not follow the prediction that inter-island colonization leads to speciation. Morphological and molecular data (Piano et al. 1997) recover two congruent clades. One clade originated on Kauai and via dispersal gave rise to two lineages, one on Oahu and one on Hawaii (the derived species D. pullipes). The second clade occurs only on the islands of Maui Nui, where gene flow was enhanced when the islands were connected, but more recent sea-level rise has resulted in their present vicariant differentiation.
Of the 13 Hawaiian Dicranomyia species (crane flies), 10 occur on more than one island and 7 occur on all the main high islands. Nitta & O'Grady (2008) found a complex series of phylogeographic patterns within these species, with some exhibiting population structuring reflecting an origin on Kauai and subsequent diversification involving back colonization and island skipping, while others appear to have originated on younger islands and colonized older islands subsequently. The variability in these patterns was suggested as being related to the relative ease with which crane flies are blown from island to island rather than their own active dispersal.
The damselflies Megalagrion xanthomelas and Megalagrion pacificum are currently each found on five of the Hawaiian Islands including all three of the Maui Nui complex. Jordan et al. (2005) confirmed the reciprocal monophyly of the two species using both nuclear and mitochondrial markers and studied their phylogeography using the mitochondrial COII gene and 157 individuals from 25 populations. Contrary to Funk & Wagner (1995), Roderick & Gillespie (1998) and Craddock (2000), they did not find a strong footprint of vicariance on Maui Nui but neither did they find panmixia. Rather, they proposed an intermediate scenario in which overland dispersal was common but overwater dispersal was rare, a similar interpretation to that of Piano et al. (1997) for D. grimshawi and Holland & Cowie (2007) for S. caduca (see below). This study highlights the importance of Pleistocene land bridges to historical gene flow in Megalagrion and suggests that the repeated bottlenecks that occurred as Maui Nui changed from nearly twice its current size during stands of low sea level to the much smaller area we see today, resulted in the low genetic diversity observed on these islands compared to the island of Hawaii.
The land snail S. caduca is the only one of 42 Hawaiian succineid species that occurs on all the main Hawaiian Islands (Cowie et al. 1995). Holland & Cowie (2007) showed that dispersal and gene flow among islands have been sufficiently frequent to prevent speciation. Nevertheless, older islands and volcanoes such as Kauai and west Oahu tend to harbour more deeply diverged populations, whereas populations from younger islands and volcanoes, such as east Oahu, Molokai, Lanai and Maui, are genetically more homogeneous. This pattern was manifested in broken haplotype networks for west Oahu and Kauai, and a single continuous network for snails from east Oahu and Maui Nui (Molokai, Maui and Lanai), islands that once shared forested land bridge connections (Carson & Clague 1995). This pattern suggests that incipient speciation may be occurring on older islands and demonstrates the importance of Pleistocene land bridges in enhancing recent inter-island gene flow between east Oahu and Maui Nui (figure 3). In a multilocus phylogenetic analysis of the Hawaiian succineids, S. caduca from six islands is nested within the island of Hawaii clade (Holland & Cowie in press). Intraspecific divergence values in the S. caduca lineage suggest that the species is older than the island of Hawaii (0.43 Ma), which harbours the species that comprise the basal clade. However, the overall topology shows that all island of Hawaii taxa are sister to S. caduca. This pattern suggests that a number of succineid species originated on an older island, but that either extinction of the older components of the lineages on other islands has occurred or that older populations were not sampled during this study.
Among those Hawaiian honeycreepers that occur on more than one island, some show among island differentiation of mtDNA and others do not, as might be expected for volant species (Tarr & Fleischer 1995). Notably, the amakihi (Loxops stejnegeri from Kauai, Loxops virens from Oahu, Maui and Hawaii) follows a very clear progression rule pattern of diversification, based on Cytb (Fleischer et al. 1998).
Among certain single-island endemic species, there is evidence of deep phylogeographic structuring. For instance, three species of Tetragnatha spiders on the island of Hawaii exhibit intraspecific patterns of genetic structure reflecting fragmentation of habitat by lava flows resulting from relatively recent and ongoing volcanic activity (Vandergast et al. 2004). A study of the endemic and endangered Oahu tree snail Achatinella mustelina, revealed deep genetic breaks corresponding to the eroded topography of the 3.7 Ma Waianae Mountain range (Holland & Hadfield 2002). Clusters of similar haplotypes follow mountain ridge lines for distances of more than 8 km. Molecular divergence values of over 5 per cent among populations suggest that this species is several million years old, and that genetically divergent populations are reproductively isolated and possibly undergoing incipient speciation. The phylogeographic evidence suggests that this species once had a broad panmictic distribution that has been reduced to fragmented forest patches along mountain ridges.
Crickets (Laupala) exhibit complex and not fully understood patterns of molecular variation (see above). Within L. cerasina on the island of Hawaii, however, there is a clear phylogeographic break separating northern and southern populations. Maui is the origin of these populations, but it remains unclear whether they have resulted from one (Mendelson et al. 2004) or two colonizations (Shaw 2002).
Whether Hawaiian species are single-island endemics or are more widespread fundamentally relates to their vagility. However, many factors may influence vagility: habitat preference, life history, ease of passive transport (e.g. clinging to birds), behavioural traits, etc. Similarly, the degree of phylogeographic structuring both among and within islands is related to vagility. However, in most cases, the level of vagility is assumed based on the distribution and phylogeographic structuring, introducing a certain circularity. To truly understand such structuring, we need to understand the basic ecology of the species in question in a comparative framework that includes other species in the group. Why, for instance, is S. caduca the only succineid land snail species on all the main islands? What is special about its ecology, life history or behaviour? In what way does it differ from single-island endemic succineids? Elucidation of the factors, both abiotic and biological, inherent to lineages that fail to radiate will be an important step forward in understanding the causative mechanisms that ultimately generate biodiversity.
5. The Hawaiian Islands as a source of colonization
Generally, there has tended to be an assumption that islands serve as dispersal sinks and evolutionary dead ends, with unidirectional movement of species from continents to islands over evolutionary time scales (e.g. Wilson 1961). Since they are often considered to be some of the most remote islands in the world, farthest from a continental land mass, the Hawaiian Islands have often been assumed to be the end of the road for biogeographic dispersal. Many Hawaiian species, once they had arrived, colonized and evolved, have indeed been thought of as dead ends—flightless birds and insects, plants that have lost avian and aerial seed dispersal mechanisms (Ziegler 2002)—that gave rise to no further dispersal.
Increasingly, however, instances are being discovered of dispersal out of the Hawaiian Islands resulting in colonization of distant archipelagos and even continents. Rundell et al. (2004) demonstrated that a species of Succinea from Tahiti clusters within the island of Hawaii clade of succineid land snails (see above), indicating its origin in Hawaii. This was confirmed by Holland & Cowie (in press), who also showed that a succineid from Samoa clusters within the other major clade of Hawaiian Island species, specifically those on the island of Kauai, indicating the Samoan species’ origin on Kauai.
Most spectacularly, O'Grady & DeSalle (2008), resolving in part the long-standing controversy about the generic nomenclature of Hawaiian drosophilids (see below), have shown that Hawaiian Scaptomyza originated in the Hawaiian Islands, diversified widely there and subsequently colonized other islands and continents across the globe.
Future efforts to sample Pacific wide and more globally will permit us to ascertain how common such ‘out-of-Hawaii’ colonizations have been and whether the Hawaiian Islands, and perhaps by analogy other oceanic archipelagos, are biogeographically not the remote dead ends that has often been assumed.
6. Systematics
Traditionally, taxonomy has been based on morphology, with molecular phylogenetic analysis used to refine the morphological taxonomy and elucidate the evolutionary patterns and processes. Increasingly, however, especially in groups lacking sufficient readily assessable morphological characters or those exhibiting multiple convergences, molecular analysis is being used to define lineages a priori, thereby guiding the selection of morphologically useful characters and permitting subsequent morphological characterization of these difficult groups. Taking this approach to its extreme has seen the advent of DNA ‘barcoding’, the use of a fragment of a single mtDNA marker (usually COI) as a means of identifying and categorizing all biodiversity (Hebert et al. 2003). The Hawaiian Islands, with their marvellous diversity in many groups, are undoubtedly an excellent place to do such molecular work, at least as a preliminary to more comprehensive systematics study. However, the use of mtDNA data in taxonomy and systematics has recently seen considerable debate, with some authors severely criticizing mtDNA-based approaches (Shaw 2002; Ballard & Whitlock 2004). A more measured, middle ground seems to be emerging (Rubinoff & Holland 2005; Rubinoff 2006; Trewick 2008) in which use of multiple mitochondrial and nuclear molecular markers as well as morphological, cytological, behavioural and other data to make taxonomic and phylogenetic inferences and determinations, with a concomitant recognition that there is scientific value in exploring areas where data partitions conflict.
Within the Hawaiian terrestrial fauna, molecular data have permitted important insight into the basic taxonomy and systematics of a number of groups. Holland & Hadfield (2007), for instance, synonymized a large number of nominal subspecies (established on the basis of shell colour and pattern) of the tree snail A. mustelina based on the analysis of COI, which showed that the monophyletic clades in the gene tree contained representatives of multiple subspecies, that haplotypes were shared among more than one subspecies, and that none of the nominal subspecies was monophyletic.
The generic classification of the Hawaiian drosophilids has prompted considerable controversy (O'Grady 2002; van der Linde et al. 2007; O'Grady et al. 2008; Thompson et al. 2008). Early molecular analyses (DeSalle 1995; Russo et al. 1995) that showed them to be monophyletic referred them to three genera, Drosophila, Scaptomyza and Engiscaptomyza. O'Grady et al. (2003) revised the genera and subgenera of Hawaiian drosophilids, based on the morphological and molecular grounds, subsuming all within the two genera Drosophila and Scaptomyza, the latter including Engiscaptomyza as a subgenus. Both Drosophila and Scaptomyza are widespread globally, so if the Hawaiian drosophilids indeed are derived from a single colonization, this renders these genera paraphyletic, unless Hawaii were the source of the non-Hawaiian members of one of these groups, i.e. Scaptomyza (Russo et al. 1995), which still leaves Drosophila paraphyletic. (More generally, Drosophila is paraphyletic because a number of other, non-Hawaiian genera are also embedded within it; Remsen & O'Grady 2002; van der Linde et al. 2007). An alternative that has not been formally addressed is the possibility of separate introductions to Hawaii of Drosophila and Scaptomyza, as proposed by Thomas & Hunt (1991), followed by hybridization rendering the Hawaiian taxa apparently monophyletic in molecular phylogenetic analyses (K. Y. Kaneshiro 2008, personal communication; see also O'Grady & Zilversmit 2004). While hybridization in the drosophilids has not been formally demonstrated based on molecular evidence, it has been proposed to explain phylogeographic and systematic discrepancies in Laupala crickets (Shaw 2002) and Megalagrion damselflies (Jordan et al. 2003). However, recent molecular data confirm that the Hawaiian drosophilids (Drosophila and Scaptomyza) are monophyletic and nested within the globally distributed genus Drosophila (O'Grady 2002; O'Grady & DeSalle 2008). This, therefore, indeed renders Drosophila paraphyletic with regard to Scaptomyza (including non-Hawaiian species), which nests within it. This nomenclatural problem is yet to be resolved.
Especially among the birds, molecular analysis has been used in the support of basic systematics, including the specific or subspecific distinction between the Laysan and Nihoa millerbirds (Fleischer et al. 2007), the phylogenetic position of the very recently extinct po‘o-uli (Fleischer et al. 2001) and the conspecificity of the now extinct Hawaiian eagle with the Palaearctic white-tailed eagle (Fleischer et al. 2000).
In some groups, molecular analysis has contradicted traditional, morphology-based taxonomy and will probably lead to major taxonomic revisions. For instance, the classification of the spider genus Havaika into nine nominal species based on morphology proved incongruent with the molecular data of Arnedo & Gillespie (2006) who demonstrated four monophyletic lineages occurring on multiple islands, with each lineage exhibiting diversification primarily among islands.
In general, however, despite the frequent discovery of cryptic species as a result of molecular analysis of invertebrates (e.g. Holland et al. 2004; Bickford et al. 2007), rather few cases, implied for instance by Holland & Hadfield (2002) and Shaw (2002), seem to have been found in systematics studies of the Hawaiian fauna (but see Schmitz et al. 2008).
Many other taxonomic questions remain unresolved. A major problem in some groups is that many Hawaiian species when described originally were placed in poorly differentiated, global catch-all genera, e.g. spiders in Theridion (Arnedo et al. 2007) and snails in Succinea (Rundell et al. 2004; Holland & Cowie in press), and their correct placement in these or other genera (including new genera) depends on research that has yet to be done. Using molecular phylogenetic analysis to guide such basic systematics as well as to help resolve other difficult taxonomic questions will be increasingly important in analyses of the complex terrestrial Hawaiian fauna.
7. Discussion and conclusions
The advent and increasing ease of use of molecular approaches to phylogenetics, biogeography and phylogeography has led to a burgeoning of studies on the Hawaiian biota that extend our knowledge dramatically beyond the landmark compilation of Wagner & Funk (1995). From biochemical advances to development of new sophisticated instrumentation and software, molecular biology is undergoing revolutionary advances in the nature and quantity of genetic data that can be accessed, compiled and analysed. In addition, recent linkages established between high throughput DNA technologies and emerging computational techniques are resulting in an unprecedented ability to analyse large datasets and recover supertrees for thousands of evolutionary units, accomplishments that were unthinkable at the time of publication of Wagner & Funk (1995). Other statistical advances and mathematical approaches are beginning to focus on improved molecular clock estimation and have begun to allow improved insight into the timing of major colonization and cladogenic events (Price & Clague 2002).
However, our knowledge remains unbalanced. We know more about the geographic and phylogenetic origins of Hawaiian vertebrates than invertebrates, while there has been far more research on the intra-archipelago biogeography and phylogeography of invertebrates. This probably reflects the interplay of the generally larger size of vertebrates, essentially birds, compared with invertebrates, the greater vagility of the birds, and the far far greater species richness of invertebrates, which permits robust analyses of biogeographic patterns that are not possible with just three or four species.
As many of the endemic Hawaiian bird species resulted from single, non-radiating colonizations, the obvious question to ask is where did they come from? A few samples from Asia and North and South America, guided by the usually well-known morphological taxonomy, will probably provide the answer—and that is why we have the answer for most of the avian lineages in Hawaii. By contrast, with a radiation of, say, 1000 species of drosophilids, these questions of origins are not so easily addressed and the immediate ones that spring to mind tend to be more locally focused, for instance on the patterns and processes of diversification within and among the islands. And, with so much invertebrate diversity, contrasting with just one or a few species per vertebrate lineage, such questions may indeed only be relevant in these diverse invertebrate groups.
Thus, the geographic origins of many of the invertebrate groups are unknown (e.g. drosophilids, Megalagrion, Laupala and others for which only vague speculation is possible) and this reflects the lack of basic understanding of the groups globally and therefore necessarily insufficient sampling of the enormous potential source regions (the entire Pacific Rim and Pacific islands). Questions of the geographic origins and pathways of colonization, in many cases probably in a stepping-stone fashion, island hopping across the Pacific, remain a wide open area of investigation. Eventually, we may be able to discern generalized patterns related to ocean currents, bird migration routes and prevailing wind patterns (Cowie & Holland 2006), but these are only just beginning to be elucidated.
As the number and resolution of molecular phylogeographic studies continues to increase, so does our ability to address detailed patterns and to determine shared as well as unique features of the diversification process in various radiations. For example, when considering the relationships among speciation, species diversity and number of colonization events, we may ask the question ‘do lineages that have colonized the Hawaiian Islands multiple times have higher numbers of species than those derived from a single event?’ We are thus beginning to understand the importance of factors such as the number of long-distance versus inter-island colonizations in the determination of biodiversity in endemic Hawaiian lineages. Similar questions can be addressed regarding the forces driving generation of deeper levels of diversity (genera, subfamilies, etc.).
Whether a radiation resulted from a single or multiple colonizations is perhaps not an especially deterministic issue on its own, in terms of generating biodiversity. For instance, the achatinelline land snails (a subfamily; the subfamily Auriculellinae may have arisen from the same single colonization) probably arose from a single origin, but the Hawaiian succineids (a family) from two or more. But these two succineid colonizations resulted in two radiations, perhaps representing two global subfamilies. At the subfamily level, therefore, there has been only one radiation per lineage, as in the achatinellines. But while the achatinelline radiation resulted in more than twice the species diversity of the two succineid radiations combined (Holland & Hadfield 2004; Holland & Cowie in press), the succineids are far more ecologically diverse. Hawaiian birds (many orders and families) resulted from numerous colonizations but drepanidines from only one. The answer to the question thus depends in part on the taxonomic level studied. However, why the achatinelline subfamilial radiation includes far more species (99) than either succineid subfamilial radiation/colonization (total 42) (Cowie et al. 1995) is indeed a question worth addressing, as it relates to the underlying reasons why some groups are more likely to speciate than others. For instance, why are there only two endemic Hawaiian butterflies but well over a thousand moths (Ziegler 2002), 13 crane flies but probably a thousand drosophilids (Kaneshiro 2006; Nitta & O'Grady 2008), and so on?
Such questions represent just a few of the exciting new research directions we foresee in island biology, particularly in the areas of comparative phylogeography and coevolutionary studies. Comparative phylogeographic studies have begun to reveal pervasive and unanticipated patterns suggesting that cryptic biological diversification has played a more important role in the development of oceanic island assemblages than traditional taxonomy indicates, and that these processes are not as readily apparent in analyses of single lineages. Comparative phylogeography can contribute significantly to understanding the relationships between geological history and biodiversity, and of biogeographic provenance and fates of island lineages. In a comparative framework, phylogeography can be used to evaluate both biogeographic patterns and evolutionary processes, providing a fundamental basis for new insights into the regional factors generating and maintaining biodiversity.
Coevolutionary studies seek to understand how interspecific interactions are shaped by reciprocal natural selection, undergo concerted evolutionary change and persist through space and time (Thompson 2005). When two lineages interact intimately during diversification, it is probable that speciation events in both groups are contemporaneous (Page 2002). Coevolution on oceanic islands is an essentially unexplored frontier, involving multiple interacting populations co-distributed across complex and diverse island environments, subject to events such as climate change, El Niño southern oscillations, volcanic eruptions and invasion by colonizing species. Although co-speciating lineages in the Hawaiian Islands are yet to be investigated using molecular data, they nevertheless hold potential for exciting new discoveries.
However, a major challenge to a comprehensive understanding of the phylogenetics, systematics and evolution of Hawaiian taxa is the sheer diversity of many invertebrate lineages. There are over 5000 recognized endemic insect species (Eldredge & Evenhuis 2003), but this number could easily double as many groups include numerous undescribed species (e.g. Kaneshiro 2006; Rubinoff 2008).
Further confounding any systematics assessment or phylogenetic analysis of the terrestrial Hawaiian fauna is the recent accelerated anthropogenic extinction rate. The Hawaiian Islands are notorious for the extreme levels of extinction among many groups: over 70 per cent of the birds (Boyer 2008), up to 90 per cent of the land snails (Cowie 2001; Lydeard et al. 2004) and undocumented numbers of arthropods (Wagner & Funk 1995, p. 421), which are generally understudied and for which extinction is particularly difficult to document (Dunn 2005). Also, most of the species that remain are confined to severely contracted ranges in relatively inaccessible, high-elevation habitats rendering collection difficult. Missing taxa in phylogenetic analyses can lead to spurious interpretations of phylogenetic trees, so the level of extinction in Hawaii is of particular concern in phylogenetic reconstruction. This problem has been noted above, for instance, in explaining the distribution of species in the D. planitibia group, the age of the island of Hawaii clade of succineid land snails and the evolution of troglobitic isopods. Great steps forward have, however, been made by the development of the capability to extract DNA from museum specimens, notably of birds, and hopefully in the future of additional Hawaiian taxa.
Molecular studies of the Hawaiian fauna have burgeoned since the mid-1990s. Many questions have been posed, some have been answered and others have arisen unexpectedly as the research unfolded. We know much more about the Hawaiian fauna than we did when Wagner & Funk (1995) published their watershed book. But much is still to be learned. If this unsurpassed evolutionary laboratory is to give up the secrets that Darwin only guessed at and that Gulick began but tentatively to unfold, research must be undertaken with focus and determination before it is too late, and biologists must integrate their research rigorously into conservation, public education and the socio-political process (Cowie 2004).
Acknowledgments
We thank Rob Fleischer and Sheila Conant for help with avian literature, Neal Evenhuis, Ken Kaneshiro and Patrick O'Grady for drosophilid literature and discussion of drosophilids, and Chris Simon and Rob DeSalle for their comments on the manuscript. Hiromi Nagatsuka assisted with the figures. Our work on succineid land snails was supported by NSF grant DEB-0316308. B.S.H. was supported in part by a grant to Dan Rubinoff and NSF grant OIA0554657 during preparation of this manuscript.
Footnotes
One contribution of 15 to a Theme Issue ‘Evolution on Pacific islands: Darwin's legacy’.
References
- Arnedo M.A, Gillespie R.G. Species diversification patterns in the Polynesian jumping spider genus Havaika Prószyński, 2001 (Araneae, Salticidae) Mol. Phylogenet. Evol. 2006;41:472–495. doi: 10.1016/j.ympev.2006.05.012. doi:10.1016/j.ympev.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Arnedo M.A, Agnarsson I, Gillespie R.G. Molecular insights into the phylogenetic structure of the spider genus Theridion (Araneae, Theridiidae) and the origin of the Hawaiian Theridion-like fauna. Zool. Scripta. 2007;36:337–352. doi:10.1111/j.1463-6409.2007.00280.x [Google Scholar]
- Ballard J.W, Whitlock M.C. The incomplete history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. doi:10.1046/j.1365-294X.2003.02063.x [DOI] [PubMed] [Google Scholar]
- Bickford D, Lohman D.J, Sodhi N.S, Ng P.K.L, Meier R, Winker K, Ingram K.K, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. doi:10.1016/j.tree.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Bonacum J, O'Grady P.M, Kambysellis M, DeSalle R. Phylogeny and age of diversification of the planitibia species group of the Hawaiian Drosophila. Mol. Phylogenet. Evol. 2005;37:73–82. doi: 10.1016/j.ympev.2005.03.008. doi:10.1016/j.ympev.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Boyer A.G. Extinction patterns in the avifauna of the Hawaiian Islands. Divers. Distrib. 2008;14:509–517. doi:10.1111/j.1472-4642.2007.00459.x [Google Scholar]
- Carson H.L. The process whereby species originate. BioScience. 1987a;37:715–720. doi:10.2307/1310468 [Google Scholar]
- Carson H.L. Colonization and speciation. In: Gray A.J, Crawley M.J, Edwards P.J, editors. Colonization, succession and stability. Blackwell; Oxford, UK: 1987b. pp. 187–205. [Google Scholar]
- Carson H.L, Clague D.A. Geology and biogeography of the Hawaiian islands. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography. Smithsonian Institution Press; Washington, DC: 1995. pp. 14–29. [Google Scholar]
- Cooke C.M, Jr, Kondo Y. [Printed date 1960] Revision of Tornatellinidae and Achatinellidae (Gastropoda Pulmonata) Bernice P. Bishop Mus. Bull. 1961;221:1–303. [Google Scholar]
- Cowie R.H. Variation in species diversity and shell shape in Hawaiian land snails: in situ speciation and ecological relationships. Evolution. 1995;49:1191–1202. doi: 10.1111/j.1558-5646.1995.tb04446.x. doi:10.2307/2410444 [DOI] [PubMed] [Google Scholar]
- Cowie R.H. Invertebrate invasions on Pacific islands and the replacement of unique native faunas: a synthesis of the land and freshwater snails. Biol. Invasions. 2001;3:119–136. doi:10.1023/A:1014529019000 [Google Scholar]
- Cowie R.H. Disappearing snails and alien invasions: the biodiversity/conservation interface in the Pacific. J. Conchol. Spec. Publ. 2004;3:23–37. [Google Scholar]
- Cowie R.H, Holland B.S. Dispersal is fundamental to biogeography and the evolution of biodiversity on oceanic islands. J. Biogeogr. 2006;33:193–198. doi:10.1111/j.1365-2699.2005.01383.x [Google Scholar]
- Cowie R.H, Evenhuis N.L, Christensen C.C. Backhuys Publishers; Leiden, The Netherlands: 1995. Catalog of the native land and freshwater molluscs of the Hawaiian Islands. [Google Scholar]
- Craddock E.M. Speciation processes in the adaptive radiation of Hawaiian plants and animals. Evol. Biol. 2000;31:1–53. [Google Scholar]
- Davis T. On the relationship between the Scaptomyza and the Hawaiian Drosophila. Hereditas. 2000;132:257–259. doi: 10.1111/j.1601-5223.2000.00257.x. doi:10.1111/j.1601-5223.2000.00257.x [DOI] [PubMed] [Google Scholar]
- DeSalle R. Molecular approaches to biogeographic analysis of Hawaiian Drosophilidae. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography. Smithsonian Institution Press; Washington, DC: 1995. pp. 72–89. [Google Scholar]
- Dunn R.R. Modern insect extinctions, the neglected majority. Conserv. Biol. 2005;19:1030–1036. doi:10.1111/j.1523-1739.2005.00078.x [Google Scholar]
- Eldredge L.G, Evenhuis N.L. Hawaii's biodiversity: a detailed assessment of the numbers of species in the Hawaiian Islands. Bishop Mus. Occas. Pap. 2003;76:1–28. [Google Scholar]
- Fleischer R.C, McIntosh C.E. Molecular systematics and biogeography of the Hawaiian avifauna. Stud. Avian Biol. 2001;22:51–60. [Google Scholar]
- Fleischer R.C, McIntosh C.E, Tarr C.L. Evolution on a volcanic conveyor belt: using phylogeographic reconstructions and K–Ar based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Mol. Ecol. 1998;7:533–545. doi: 10.1046/j.1365-294x.1998.00364.x. doi:10.1046/j.1365-294x.1998.00364.x [DOI] [PubMed] [Google Scholar]
- Fleischer R.C, Olson S, James H.F, Cooper A.C. The identity of the extinct Hawaiian eagle (Haliaeetus) as determined by mitochondrial DNA sequence. Auk. 2000;117:1051–1056. doi:10.1642/0004-8038(2000)117[1051:IOTEHE]2.0.CO;2 [Google Scholar]
- Fleischer R.C, Tarr C.L, James H.F, Slikas B, McIntosh C.E. Phylogenetic placement of the Po‘o-uli Melamprosops phaeosoma based on mitochondrial DNA sequence and osteological characters. Stud. Avian Biol. 2001;22:98–103. [Google Scholar]
- Fleischer R.C, Slikas B, Beadell J, Atkins C, McIntosh C.E, Conant S. Genetic variability and taxonomic status of the Nihoa and Laysan millerbirds. Condor. 2007;109:954–962. doi:10.1650/0010-5422(2007)109[954:GVATSO]2.0.CO;2 [Google Scholar]
- Funk V.A, Wagner W.L. Biogeographic patterns in the Hawaiian Islands. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography. Smithsonian Institution Press; Washington, DC: 1995. pp. 379–419. [Google Scholar]
- Gage R.P., II First record of the land snail subfamily Achatinellinae on Kauai. Bishop Mus. Occas. Pap. 1996;46:21–25. [Google Scholar]
- Gillespie R.G. Biogeography of spiders on remote oceanic islands of the Pacific: archipelagoes as stepping stones? J. Biogeogr. 2002;29:655–662. doi:10.1046/j.1365-2699.2002.00714.x [Google Scholar]
- Gillespie R.G, Croom H.B. Comparison of speciation mechanisms in web-building and non-web-building groups within a lineage of spiders. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography. Smithsonian Institution Press; Washington, DC: 1995. pp. 121–146. [Google Scholar]
- Gillespie R.G, Palumbi S.R, Croom H.B. Multiple origins of a spider radiation in Hawaii. Proc. Natl Acad. Sci. USA. 1994;91:2290–2294. doi: 10.1073/pnas.91.6.2290. doi:10.1073/pnas.91.6.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie R.G, Croom H.B, Hasty G.L. Phylogenetic relationships and adaptive shifts among major clades of Tetragnatha spiders (Araneae: Tetragnathidae) in Hawai'i. Pac. Sci. 1997;51:380–394. [Google Scholar]
- Gillespie R.G, Claridge E.M, Goodacre S.L. Biogeography of the fauna of French Polynesia: diversification within and between a series of hot spot archipelagos. Phil. Trans. R. Soc. B. 2008;363:3335–3346. doi: 10.1098/rstb.2008.0124. doi:10.1098/rstb.2008.0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick J.T. Evolution, racial and habitudinal. Carnegie Inst. Wash. Publ. 1905;25:i–xii. (See also pp. 1–269, pII. 1–5). [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. University of Illinois Press; Urbana, IL: 1966. Phylogenetic systematics. [Google Scholar]
- Holland B.S, Cowie R.H. A geographic mosaic of passive dispersal: population structure in the endemic Hawaiian amber snail Succinea caduca (Mighels, 1845) Mol. Ecol. 2007;16:2422–2435. doi: 10.1111/j.1365-294X.2007.03246.x. doi:10.1111/j.1365-294X.2007.03246.x [DOI] [PubMed] [Google Scholar]
- Holland, B. S. & Cowie, R. H. In press. Land snail models in island biogeography: a tale of two snails. Am. Malacol. Bull
- Holland B.S, Hadfield M.G. Islands within an island: phylogeography and conservation genetics of the endangered Hawaiian tree snail Achatinella mustelina. Mol. Ecol. 2002;11:365–375. doi: 10.1046/j.1365-294x.2002.01464.x. doi:10.1046/j.1365-294X.2002.01464.x [DOI] [PubMed] [Google Scholar]
- Holland B.S, Hadfield M.G. Origin and diversification of the endemic Hawaiian tree snails (Achatinellinae: Achatinellidae) based on molecular evidence. Mol. Phylogenet. Evol. 2004;32:588–600. doi: 10.1016/j.ympev.2004.01.003. doi:10.1016/j.ympev.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Holland B.S, Hadfield M.G. Molecular systematics of the endangered Oahu tree snail Achatinella mustelina: synonymization of subspecies and estimation of gene flow between chiral morphs. Pac. Sci. 2007;61:53–66. doi:10.1353/psc.2007.0007 [Google Scholar]
- Holland B.S, Dawson M.N, Crow G.L, Hofmann D.K. Global phylogeography of Cassiopea (Scyphozoa: Rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian islands. Mar. Biol. 2004;145:1119–1128. doi:10.1007/s00227-004-1409-4 [Google Scholar]
- Hormiga G, Arnedo M.A, Gillespie R.G. Speciation on a conveyor belt: sequential colonization of the Hawaiian Islands by Orsonwelles spiders (Araneae: Linyphiidae) Syst. Biol. 2003;52:70–88. doi: 10.1080/10635150390132786. doi:10.1080/10635150390132786 [DOI] [PubMed] [Google Scholar]
- Jordan S, Simon C, Polhemus D. Molecular systematics and adaptive radiation of Hawaii's endemic damselfly genus Megalagrion (Odonata: Coenagrionidae) Syst. Biol. 2003;52:89–109. doi: 10.1080/10635150390132803. doi:10.1080/10635150390132803 [DOI] [PubMed] [Google Scholar]
- Jordan S, Simon C, Foote D, Englund R.A. Phylogeographic patterns of Hawaiian Megalagrion damselflies (Odonata: Coenagrionidae) correlate with Pleistocene island boundaries. Mol. Ecol. 2005;14:3457–3470. doi: 10.1111/j.1365-294X.2005.02669.x. doi:10.1111/j.1365-294X.2005.02669.x [DOI] [PubMed] [Google Scholar]
- Kaneshiro K.Y. Speciation in the Hawaiian Drosophila. Sexual selection appears to play an important role. BioScience. 1988;38:258–263. doi:10.2307/1310849 [Google Scholar]
- Kaneshiro K.Y. Dynamics of sexual selection in the Hawaiian Drosophilidae: a paradigm for evolutionary change. Proc. Hawaii. Entomol. Soc. 2006;38:1–19. [Google Scholar]
- Lydeard C, et al. The global decline of nonmarine mollusks. BioScience. 2004;54:321–330. doi:10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2 [Google Scholar]
- Magnacca K.N, Danforth B.N. Evolution and biogeography of native Hawaiian Hylaeus bees (Hymenoptera: Colletidae) Cladistics. 2006;22:393–411. doi:10.1111/j.1096-0031.2006.00119.x [Google Scholar]
- Mendelson T.C, Shaw K.L. Rapid speciation in an arthropod. Nature. 2005;433:375–376. doi: 10.1038/433375a. doi:10.1038/433375a [DOI] [PubMed] [Google Scholar]
- Mendelson T.C, Siegel A.M, Shaw K.L. Testing geographical pathways of speciation in a recent island radiation. Mol. Ecol. 2004;13:3787–3796. doi: 10.1111/j.1365-294X.2004.02375.x. doi:10.1111/j.1365-294X.2004.02375.x [DOI] [PubMed] [Google Scholar]
- Morales J.C, Bickham J.W. Molecular systematics of the genus Lasiurus (Chiroptera: Vespertillionidae) based on restriction-site maps of the mitochondrial ribosomal genes. J. Mammal. 1995;76:730–749. doi:10.2307/1382744 [Google Scholar]
- Nitta J.H, O'Grady P.M. Mitochondrial phylogeny of the endemic Hawaiian craneflies (Diptera, Limoniidae, Dicranomyia): implications for biogeography and species formation. Mol. Phylogenet. Evol. 2008;46:1182–1190. doi: 10.1016/j.ympev.2007.12.021. doi:10.1016/j.ympev.2007.12.021 [DOI] [PubMed] [Google Scholar]
- O'Grady P.M. Notes on the nomenclature of the endemic Hawaiian Drosophilidae. Bishop Mus. Occas. Pap. 2002;69:36–40. [Google Scholar]
- O'Grady, P. & DeSalle, R. 2008 Out of Hawaii: the origin and biogeography of the genus Scaptomyza (Diptera: Drosophilidae). Biol. Lett 4, 195–199. (doi:10.1098/rsbl.2007.0575) [DOI] [PMC free article] [PubMed]
- O'Grady P.M, Zilversmit M. Phylogenetic relationships within the Drosophila haleakalae species group inferred by molecular and morphological characters (Diptera: Drosophilidae) Bishop Mus. Bull. Entomol. 2004;12:117–134. [Google Scholar]
- O'Grady P.M, Bonacum J, Val F.C. The placement of the Engiscaptomyza, Grimshawomyia, and Titanochaeta, three clades of endemic Hawaiian Drosophilidae. Zootaxa. 2003;159:1–16. [Google Scholar]
- O'Grady, P. M. et al 2008 Comment on the proposed conservation of usage of Drosophila Fallén, 1823 (Insecta, Diptera) (Case 3407; see BZN 64(4): 238–242). Bull. Zool. Nomencl 65, 141–144.
- Otte D. The Orthopterists' Society, Academy of Natural Sciences of Philadelphia; Philadelphia, PA: 1994. The crickets of Hawaii: origin, systematics and evolution. [Google Scholar]
- Page R.D.M, editor. Tangled trees: phylogeny, cospeciation and coevolution. University of Chicago Press; Chicago, IL: 2002. [Google Scholar]
- Paxinos E.E, James H.F, Olson S.L, Sorenson M.D, Jackson J, Fleischer R.C. mtDNA from fossils reveals a radiation of Hawaiian geese recently derived from the Canada goose (Branta canadensis) Proc. Natl Acad. Sci. USA. 2002;99:1399–1404. doi: 10.1073/pnas.032166399. doi:10.1073/pnas.032166399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F, Craddock E.M, Kambysellis M.P. Phylogeny of the island populations of the Hawaiian Drosophila grimshawi complex: evidence from combined data. Mol. Phylogenet. Evol. 1997;7:173–184. doi: 10.1006/mpev.1996.0387. doi:10.1006/mpev.1996.0387 [DOI] [PubMed] [Google Scholar]
- Pons J, Gillespie R.G. Evolution of satellite DNAs in a radiation of endemic Hawaiian spiders: does concerted evolution of highly repetitive sequences reflect evolutionary history? J. Mol. Evol. 2004;59:632–641. doi: 10.1007/s00239-004-2655-2. doi:10.1007/s00239-004-2655-2 [DOI] [PubMed] [Google Scholar]
- Price J.P, Clague D.A. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc. R. Soc. B. 2002;269:2429–2435. doi: 10.1098/rspb.2002.2175. doi:10.1098/rspb.2002.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.P, Elliott-Fisk D. Topographic history of the Maui Nui complex, Hawaii, and its implications for biogeography. Pac. Sci. 2004;58:27–45. doi:10.1353/psc.2004.0008 [Google Scholar]
- Remsen J, DeSalle R. Character congruence of multiple data partitions and the origin of the Hawaiian Drosophilidae. Mol. Phylogenet. Evol. 1998;9:225–235. doi: 10.1006/mpev.1997.0484. doi:10.1006/mpev.1997.0484 [DOI] [PubMed] [Google Scholar]
- Remsen J, O'Grady P.M. Phylogeny of Drosophilinae (Diptera: Drosophilidae), with comments on combined analysis and character support. Mol. Phylogenet. Evol. 2002;24:249–264. doi: 10.1016/s1055-7903(02)00226-9. doi:10.1016/S1055-7903(02)00226-9 [DOI] [PubMed] [Google Scholar]
- Rivera M.A.J, Howarth F.G, Taiti S, Roderick G.K. Evolution in Hawaiian cave-adapted isopods (Oniscidea: Philosciidae): vicariant speciation or adaptive shifts? Mol. Phylogenet. Evol. 2002;25:1–9. doi: 10.1016/s1055-7903(02)00353-6. doi:10.1016/S1055-7903(02)00353-6 [DOI] [PubMed] [Google Scholar]
- Roderick G.K, Gillespie R.G. Speciation and phylogeography of Hawaiian terrestrial arthropods. Mol. Ecol. 1998;7:519–531. doi: 10.1046/j.1365-294x.1998.00309.x. doi:10.1046/j.1365-294x.1998.00309.x [DOI] [PubMed] [Google Scholar]
- Rubinoff D. DNA barcoding evolves into the familiar. Conserv. Biol. 2006;20:1548–1549. doi: 10.1111/j.1523-1739.2006.00542.x. doi:10.1111/j.1523-1739.2006.00542.x [DOI] [PubMed] [Google Scholar]
- Rubinoff D. Phylogeography and ecology of an endemic radiation of Hawaiian aquatic case-bearing moths (Hyposmocoma: Cosmopterigidae) Phil. Trans. R. Soc. B. 2008;363:3459–3465. doi: 10.1098/rstb.2008.0115. doi:10.1098/rstb.2008.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinoff D, Holland B.S. Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and systematic inference. Syst. Biol. 2005;54:952–961. doi: 10.1080/10635150500234674. doi:10.1080/10635150500234674 [DOI] [PubMed] [Google Scholar]
- Rundell R.J, Holland B.S, Cowie R.H. Molecular phylogeny and biogeography of the endemic Hawaiian Succineidae (Gastropoda: Pulmonata) Mol. Phylogenet. Evol. 2004;31:246–255. doi: 10.1016/j.ympev.2003.07.014. doi:10.1016/j.ympev.2003.07.014 [DOI] [PubMed] [Google Scholar]
- Russo C.A.M, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Schmitz P, Cibois A, Landry B. Cryptic differentiation in the endemic micromoth Galagete darwini (Lepidoptera, Autostichidae) on Galápagos volcanoes. Phil. Trans. R. Soc. B. 2008;363:3453–3458. doi: 10.1098/rstb.2008.0117. doi:10.1098/rstb.2008.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L.H, Strazanac J.S, Roderick G.K. Molecular phylogeny of Banza (Orthoptera, Tettigoniidae), the endemic katydids of the Hawaiian archipelago. Mol. Phylogenet. Evol. 2006;41:53–63. doi: 10.1016/j.ympev.2006.04.006. doi:10.1016/j.ympev.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Shaw K.L. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc. Natl Acad. Sci. USA. 2002;99:16 122–16 127. doi: 10.1073/pnas.242585899. doi:10.1073/pnas.242585899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C. Hawaiian evolutionary biology: an introduction. Trends Ecol. Evol. 1987;2:175–178. doi: 10.1016/0169-5347(87)90015-2. doi:10.1016/0169-5347(87)90015-2 [DOI] [PubMed] [Google Scholar]
- Slikas B, Olson S.L, Fleischer R.C. Rapid, independent evolution of flightlessness in four species of Pacific island rails (Rallidae): an analysis based on mitochondrial sequence data. J. Avian Biol. 2002;33:5–14. doi:10.1034/j.1600-048X.2002.330103.x [Google Scholar]
- Solem A. Biogeographic significance of land snails, paleozoic to recent. In: Gray J, Boucot A.J, editors. Historical biogeography, plate tectonics, and the changing environment. Oregon State University Press; Corvallis, OR: 1976. pp. 277–287. [Google Scholar]
- Solem A, Yochelson E.L. North American paleozoic land snails, with a summary of other non-marine snails. Prof. Pap. U.S. Geol. Surv. 1979;1072:1–42. [Google Scholar]
- Sorenson M.D, Cooper A, Paxinos E.E, Quinn T.W, James H.F, Olson S.L, Fleischer R.C. Relationships of the extinct moa-nalos, flightless Hawaiian waterfowl, based on ancient DNA. Proc. R. Soc. B. 1999;266:2187–2193. doi: 10.1098/rspb.1999.0907. doi:10.1098/rspb.1999.0907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. Sunderland, MA: Sinauer Associates.
- Tarr C.L, Fleischer R.C. Evolutionary relationships of the Hawaiian honeycreepers (Aves, Drepanidae) In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography. Smithsonian Institution; Washington, DC: 1995. pp. 147–159. [Google Scholar]
- Thacker R.W, Hadfield M.G. Mitochondrial phylogeny of extant Hawaiian tree snails. Mol. Phylogenet. Evol. 2000;16:263–270. doi: 10.1006/mpev.2000.0793. doi:10.1006/mpev.2000.0793 [DOI] [PubMed] [Google Scholar]
- Thomas R.H, Hunt J.A. The molecular evolution of the alcohol dehydrogenase locus and the phylogeny of the Hawaiian Drosophila. Mol. Biol. Evol. 1991;8:687–702. doi: 10.1093/oxfordjournals.molbev.a040678. [DOI] [PubMed] [Google Scholar]
- Thompson F.C, Evenhuis N.L, Pape T, Pont A. Comments on the proposed conservation of usage of Drosophila Fallén, 1823 (Insecta, Diptera) (Case 3407; see BZN 64: 238–242) Bull. Zool. Nomencl. 2008;65:140–141. [Google Scholar]
- Thompson J.N. University of Chicago Press; Chicago, IL: 2005. The geographic mosaic of coevolution. [Google Scholar]
- Trewick S.A. DNA barcoding is not enough: mismatch of taxonomy and genealogy in New Zealand grasshoppers (Orthoptera: Acrididae) Cladistics. 2008;24:240–254. doi:10.1111/j.1096-0031.2007.00174.x [Google Scholar]
- Vandergast A.G, Gillespie R.G, Roderick G.K. Influence of volcanic activity on the population genetic structure of Hawaiian Tetragnatha spiders: fragmentation, rapid population growth and the potential for accelerated evolution. Mol. Ecol. 2004;13:1729–1743. doi: 10.1111/j.1365-294X.2004.02179.x. doi:10.1111/j.1365-294X.2004.02179.x [DOI] [PubMed] [Google Scholar]
- van der Linde K, Bächli G, Toda M.J, Zhang W.-X, Hu Y.-G, Spicer G.S. Case 3407: Drosophila Fallén, 1832 (Insecta, Diptera): proposed conservation of usage. Bull. Zool. Nomencl. 2007;64:238–242. [Google Scholar]
- Vitousek P.M. Oceanic islands as model systems for ecological studies. J. Biogeogr. 2002;29:573–582. doi:10.1046/j.1365-2699.2002.00707.x [Google Scholar]
- Vitousek P.M. Princeton University Press; Princeton, NJ: 2004. Nutrient cycling and limitation; Hawai'i as a model system. [Google Scholar]
- Wade C.M, Mordan P.B, Naggs F. Evolutionary relationships among the pulmonate land snails and slugs (Pulmonata, Stylommatophora) Biol. J. Linn. Soc. 2006;87:593–610. doi:10.1111/j.1095-8312.2006.00596.x [Google Scholar]
- Wagner W.L, Funk V.A. Smithsonian Institution Press; Washington, DC: 1995. Hawaiian biogeography. [Google Scholar]
- Wilson E.O. The nature of the taxon cycle in the Melanesian ant fauna. Am. Nat. 1961;95:169–193. doi:10.1086/282174 [Google Scholar]
- Ziegler A.C. University of Hawaii Press; Honolulu, HI: 2002. Hawaiian natural history, ecology and evolution. [Google Scholar]