Abstract
We present a heuristic approach to the DNA assignment problem based on phylogenetic inferences using constrained neighbour joining and non-parametric bootstrapping. We show that this method performs as well as the more computationally intensive full Bayesian approach in an analysis of 500 insect DNA sequences obtained from GenBank. We also analyse a previously published dataset of environmental DNA sequences from soil from New Zealand and Siberia, and use these data to illustrate the fact that statistical approaches to the DNA assignment problem allow for more appropriate criteria for determining the taxonomic level at which a particular DNA sequence can be assigned.
Keywords: assignment, barcoding, phylogenetics, neighbour joining
1. Introduction
DNA barcoding is the use of DNA sequences for identifying unknown biological specimens. A DNA sequence is obtained for a particular marker, typically cytochrome oxidase I in animals, and this sequence is compared to a DNA database to determine to which species or other taxonomic unit the specimen belongs. DNA barcoding is, in one form or another, widely used in conservation genetics and molecular ecology (e.g. Duminil et al. 2006; Rubinoff 2006; Ward et al. 2008), but is also used in a number of other areas including forensic applications (e.g. Dawnay et al. 2007) and ancient DNA studies (e.g. Willerslev et al. 2007). It has often been associated with methods for delineating and defining species based on DNA evidence (e.g. Floyd et al. 2002; Hebert et al. 2003; Remigio & Hebert 2003; Moritz & Cicero 2004). However, in this paper, we will solely consider the statistical question of how to assign DNA sequences to a priori defined taxonomical units. This fundamental statistical problem has been addressed in a number of studies (e.g. Matz & Nielsen 2005; Meyer & Paulay 2005; Steinke et al. 2005; Nielsen & Matz 2006; Abdo & Golding 2007; Munch et al. 2008). We recently proposed a Bayesian approach based on a combination of automated database searches, alignment and Bayesian phylogenetic inference (Munch et al. 2008). The objective of this approach is to approximate the posterior probability that the unknown specimen belongs to a specific species or taxonomic group. This is done by first obtaining a number of sequences with high homology to the unknown specimen using database searches, aligning these sequences to each other and the unknown specimen, and then determining the posterior probability of membership of a particular group using a Markov chain Monte Carlo (MCMC) approach similar to the one commonly used in phylogenetic inference (e.g. Yang & Rannala 1997; Huelsenbeck & Ronquist 2001). Under the assumption that the sequences in the alignment include all relevant species, the MCMC output can be directly processed to give the desired probabilities of taxon membership. This method was implemented in a computer program ‘Statistical Assignment Package’ (SAP; Munch et al. 2008), and was used in several applications, including the analysis of hundreds of ancient DNA sequences from ice cores from the Greenlandic ice (Johnson et al. 2007; Willerslev et al. 2007).
While the method in SAP was found to have good statistical performance on real and simulated datasets (Munch et al. 2008), it may not be easily applicable to large-scale datasets, such as the datasets produced in metagenomics applications. In such applications, thousands or hundred of thousands of sequences are being analysed, rendering MCMC-based approaches computationally intractable. In this paper, therefore, we explore the possibility of using the neighbour-joining algorithm (Saitou & Nei 1987) in combination with bootstrapping (Felsenstein 1985) as a heuristic approach to approximate the posterior probabilities. An alternative approach is to interpret bootstrap proportions in a frequentist framework to make assignments based on hypothesis testing (e.g. Nielsen & Matz 2006). The Bayesian interpretation of bootstrap proportions used here has the advantage that it allows for the possibility of using decision theory to devise criteria for assignment (Abdo & Golding 2007). We will show that while there often are large differences between posterior probabilities and bootstrap proportions, the neighbour joining with bootstrap approach nonetheless performs quite well as a method for DNA barcoding inference. As with all other approaches, the inferences are only as good as the database used. The method does not model species not represented in the database, and can lead to wrong inferences if the database is not representative.
2. Material and methods
SAP implements automatic assignment of sample sequences to taxa based on the position of the sample sequence in the phylogeny of life. In the first presentation of SAP (Munch et al. 2008) a Bayesian approach was taken, using MCMC to estimate the posterior probabilities that the sample sequence forms a monophyletic group together with a particular monophyletic clade.
Ideally, all available homologues available in the database should be included in such analyses. However, due to the computational complexity of running the MCMC analysis, a heuristic is instead applied to compile a representative set of sequence homologues. We use BLAST searches against GenBank to identify homologues and retrieve sequences and taxonomic annotation for each one, disregarding homologues with insufficient annotation. By including only homologues with a BLAST score of at least half that of the best matching homologue, we exclude the bulk of sequence homologues representing taxa whose probability of grouping with the sample sequence is not appreciably large.
In many cases, however, even this cut-off does not limit the number of homologues to a set that can be handled practically by the MCMC approach. In these cases, we use a heuristic to compile a limited set with the best possible taxonomic coverage: we include only the best matching sequence homologue for each species. A maximum of 30 different species homologues are included in this manner. If allowed by the BLAST score cut-off, up to 20 homologues providing further taxonomic diversity are added progressively, including up to 10 genera, 6 families, 5 orders, 3 classes and 2 phyla in the set. If the BLAST score cut-off is reached before 50 homologues have been included in the set, additional sequences are added for the species already represented in the set by including homologues previously rejected as suboptimal representatives for the species.
Based on the alignment of the compiled set of homologues, phylogenetic trees are then sampled from a Markov chain with stationary density of trees given by the posterior probability of trees (e.g. Yang & Rannala 1997; Huelsenbeck & Ronquist 2001). A backbone topological constraint is imposed to increase the MCMC convergence and to provide the method with information regarding known phylogenetic relationships. The constraints are generated from the retrieved taxonomic annotation.
The taxonomic annotation is mapped onto each sampled tree. In this way, each clade in the tree is associated with the taxon with lowest taxonomic rank, which includes all sequences in the clade. The sister clade to the sample sequence is then identified by assuming the rooting implicit from the taxonomic annotation. In cases where the position of the root relative to the sample sequence cannot be deduced from the taxonomic annotation, the entire tree is considered the sister clade. An estimate of the posterior probability of assignment to a species or taxonomic group is then obtained as the fraction of sampled trees where the sister clade is a member of this species or group.
While this method was found to have desirable statistical properties, it may be prohibitively slow for large metagenomic datasets. We here propose a fast heuristic alternative: sampling of trees using neighbour joining (Saitou & Nei 1987) and non-parametric bootstrapping (Felsenstein 1985). We will use a constrained version of the neighbour-joining algorithm, and in order to discuss properties of this algorithm, we will first review how the standard neighbour-joining algorithm works.
Neighbour joining progressively selects taxon pairs from a set of taxa and constructs a new subtree that joins the pair. The root of the new subtree replaces the two nodes that are joined, reducing the taxon set by one. Pairs are selected by minimizing the following criterion:
| (2.1) |
where L is the number of taxa left to be joined and d(i,j) is the distance between sequence i and j, here calculated using Kimura's (1980) two-parameter model. If i and j are joined creating the new node p, then the distances d(i,p) and d(j,p) are calculated using
| (2.2) |
When p replaces i and j in the distance matrix, the new distances, from p to the remaining taxa, are calculated using
| (2.3) |
We can interpret the neighbour-joining algorithm as a greedy optimization algorithm of the balanced minimum evolution criterion given by
| (2.4) |
where o(i,j) is the sum of the number of outgoing branches from internal nodes on the directed path from i to j (Desper & Gascuel 2004; Semple & Steel 2004). For a binary tree, o(i,j) reduces to 2n where n is the number of internal nodes connecting i and j. Because the neighbour-joining algorithm does not search the whole space of possible trees, it is not guaranteed to return the tree maximizing l.
The algorithm is outlined below.
- Initialization
- Define T as the set of L leaf nodes.
- Calculate the sequence distances between all pairs in T.
- Iteration
- Identify the pair i, j in T for which Q(i,j) is minimal.
- Define a new node p and compute the distance d(p,k) to the k other nodes in T using equation (2.3).
- Add p to T and calculate lengths of edges i, p and j, p using equation (2.2).
- Remove i and j from T.
- Termination
- When T consists of two leaves i and j, add the remaining edge between them with length d(i,j).
Each iteration step requires only the recalculation of one row in the Q matrix leaving the initial calculation of sequence distances and the identification of the minimal entry in Q the only operations with O(L2) complexity.
The constrained version of the algorithm is simply implemented by replacing the first operation in the iteration by
Identify the pair i, j among all pairs in T not violating the backbone constraint, for which Q(i,j) is minimal.
We immediately note that this constrained algorithm preserves some of the desirable properties of neighbour joining. Firstly, the interpretation of the neighbour-joining algorithm as a greedy optimization algorithm of the minimum evolution criterion in equation (2.4) is preserved—but now subject to the backbone constraint. The constraints do not affect the calculations of distances. Secondly, and quite trivially, if the backbone constraint imposed is correct, the usual arguments for statistical consistency of the neighbour-joining method (e.g. Gascuel 1997) are also preserved. However, the use of topological constraints does not necessarily ensure high support for the correct assignment in cases where the constraints are not themselves supported by the sequence data.
With a fully specified constraint the computational complexity of identifying the pair to join is now linear in L.
3. Results
(a) Benchmark analysis
To compare the neighbour-joining approach with the already implemented MCMC approach, we used a benchmark set by selecting at random 500 sequences annotated as Insecta from GenBank disregarding sole representatives of a species. These sequences were assigned using the MCMC and neighbour-joining approach, disregarding the homologue itself when found in GenBank.
To examine the correspondence between the bootstrap proportions and the posterior probabilities, we examined the correlation between the highest estimated posterior probability using MCMC with the one using bootstrapped neighbour joining. We used 1000 bootstrap samples and 1 000 000 iterations of the MCMC approach to estimate bootstrap proportions and posterior probabilities, respectively. For further description of models and parameter settings used in the MCMC approach, see Munch et al. (2008).
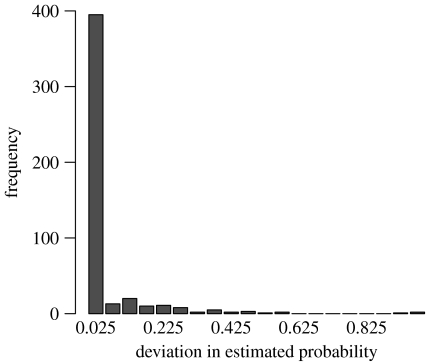
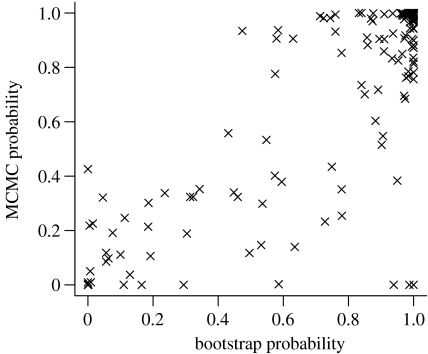
The average difference between the assignment scores estimated by the two approaches is 5 per cent. As shown in figure 1, the estimated values are in good agreement when the estimated probabilities are large. For estimated probabilities between 0.8 and 1.0, the average deviation between the two is only 2.6 per cent. For the lower values, however, the correlation is not good, as illustrated in figure 2.
Figure 1.
Histogram showing the difference in probabilities of assignment to the correct species estimated using the neighbour joining and MCMC.
Figure 2.
Estimated probabilities of assignment to the correct species using neighbour joining are plotted against the estimate obtained using MCMC.
Posterior probabilities and bootstrap proportions are not expected to match closely, because they measure different quantities (e.g. Alfaro et al. 2003; Douady et al. 2003; Huelsenbeck & Rannala 2004). In addition, they use different models of nucleotide substitution in the two approaches, and the high variance in the estimates due to a relatively small number of bootstrap replicates and a relatively small number of MCMC iterations may also contribute to the discrepancy. However, it is clear that, for high posterior probabilities, the neighbour-joining approach can be interpreted as a fast heuristic approximation of the Bayesian approach.
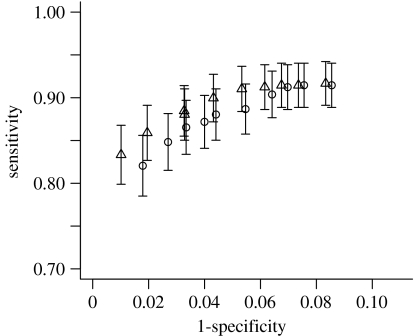
Given a specified measure of confidence, assignments can be accepted or rejected based on a criterion of minimal required assignment probability/bootstrap proportion. The performance of the two approaches can be compared in a receiver operating characteristic (ROC) plot, where sensitivity is plotted against specificity for the range of most to least stringent assignment criterion. As shown in figure 3, the two approaches show only insignificant differences in performance.
Figure 3.
ROC curves summarizing the trade-off between sensitivity and specificity in the range of most to least stringent assignment criteria used. Sensitivity is the fraction of all sequences that are correctly assigned and specificity is the fraction of assignments that are correct. Vertical bars represent confidence intervals of the sensitivity statistic. Triangles, NJ; circles, MCMC.
Posterior probabilities estimated by bootstrap proportions are often found to be conservative compared to probabilities computed using MCMC (e.g. Alfaro et al. 2003; Douady et al. 2003; Huelsenbeck & Rannala 2004). In the cases examined here, the specificity of the neighbour-joining approach for the 0.8 cut-off, above which the estimates correlate well, is 0.98. The sensitivity is 0.86, which is not a drastic reduction from the maximum of 0.91 when accepting all assignments irrespective of assignment probability.
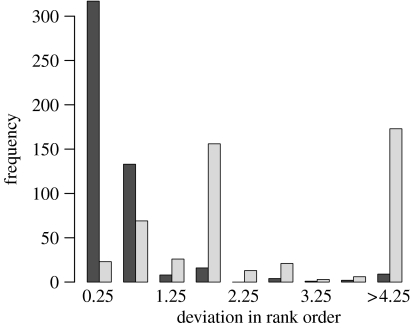
To further analyse the correspondence between results from the two approaches, we examined the rank orders of species in the assignment of each sample sequence. The histogram in figure 4 shows the frequencies of average deviation in rank order from the one obtained from the MCMC approach. For comparison, the histogram also plots this measure for rankings obtained from a BLAST search. The ordering of species obtained using neighbour joining deviates only slightly from that obtained with MCMC. By contrast, the ranking from BLAST shows a much poorer correlation.
Figure 4.
Histogram illustrating the agreement in terms of rank order obtained by sorting the set of homologues by the assignment probability associated obtained neighbour joining with bootstrapping and maximum likelihood and MCMC. The histograms show the average difference in rank order for neighbour joining and BLAST from the one obtained using MCMC.
(b) Re-analysis of ancient DNA environmental samples
To illustrate the use of our method, we re-analysed environmental ancient DNA data from a previously published analysis of permafrost and temperate sediments from Siberia and New Zealand (Willerslev et al. 2007). This study established that genetic records of palaeocommunities may be preserved in sediments formed in the Holocene and Pleistocene. The Siberian permafrost samples were obtained from cores drilled in former western Beringia. The New Zealand samples included dry sediments from a subalpine cave in the Clutha Valley, Otago, and sand from the interior and exterior of a bone of an extinct moa, Euryapteryx curtus, collected in situ from a coastal dune deposit in Northland. DNA was extracted and 130 bp fragments of the chloroplast rbcL gene and 100–280 bp fragments of the vertebrate mitochondrial 16S, 18S, cytochrome b and control region genes were obtained using PCR.
As a method for statistical assignment was not available at the time of publication these data were analysed using BLAST searches against GenBank, assigning each sample sequence to the taxon represented by the highest scoring hit or the taxonomic rank shared by multiple equally good hits. For putative vertebrate sequences, the assignments were further supported by consensus neighbour-joining phylogenies of the sample sequences and their best BLAST hits in GenBank.
This analysis probably represents the approach to sequence identification, or DNA barcoding assignment, most commonly used up to now. Assignment using BLAST, however, is associated with a number of problems. BLAST searches the database for similar sequences using a local alignment heuristic. This means that the ranking of the identified homologues is based on local and not a global alignment to the sample sequence. Most importantly, however, assignments using BLAST are not associated with any measure of confidence in assignment. Since the E-values only state the probability of retrieving a similarly good hit by chance from the database, the relative size of these offers no information about the reliability of assignment to the species or other taxonomic group represented by the best BLAST hit.
Using bootstrap consensus phylogenies may often yield conservative results. In some cases, the sample sequence may group equally well with multiple individual species from the same genus, while each specific topology has low bootstrap support. Even though the sample sequence in each case groups with a member of the genus, the consensus tree will not show high support for any particular monophyletic group containing only the sample sequence and members of the genus. This problem will be particularly severe if monophyly for the genus is not strongly supported by the data. However, by calculating bootstrap proportions as described in §2, the support for the different topologies may add together to provide strong support for an assignment at the genus level.
The sequences from the original analysis were retrieved from GenBank and statistical assignment was performed disregarding sequences from the dataset when found in GenBank. Assignments were accepted at a significance level of 0.8. Six of the 11 animal species originally identified by Willerslev et al. (2003) were also found in our analyses (Mammuthus primigenius (woolly mammoth), Bison spp. (bison), Ovibos moschatus (muskox), Rangifer tarandus (reindeer), Pachyornis elephantopus (heavy footed moa) and Megalapteryx didinus (upland moa)). One of the 11 species, Cyanoramphus novaezelandiae (New Zealand parakeet) could not be tested in this analysis, as the C. novaezelandiae sequence used in the original phylogeny still remains unpublished. At the genus level, additional three of the original animal taxa were found: Equus (horse), Euryapteryx (incl. E. curtus (costal moa)) and Lepus (hare). The remaining species Lemmus lemmus (Norway lemming) originally assigned to the genus Neotoma (wood rats)—a group no longer present in Beringia but found during Pleistocene times—obtained only a significant assignment probability at the level of order.
Although the findings from the two analyses overlap, the re-analysis shows that identifications based on consensus neighbour-joining phylogenies that include only the GenBank sequences showing the highest scores in a BLAST search may falsely inflate confidence in assignment to taxa represented by these sequences.
The plant rbcL chloroplast sequences that were originally identified to order and family levels through the consensus taxa from GenBank, based on those sequences with the highest BLAST scores, showed less consistent results. Of the 28 families and 23 orders originally identified, we could identify only 11 of the families and 14 of the orders at the 0.8 significance level. Additionally, we identified six new families not found in the original paper (Mniaceae, Oleaceae, Scapaniaceae, Apiaceae, Plantaginaceae and Santalaceae). Of these, the first three are cosmopolitan and the remaining are found in New Zealand among other places. Thus, our results suggest that a simple BLAST search provides only poorly supported sequence identifications.
One of the plant sequences analysed could be identified to species level, Mida salicifolia—a species indigenous to New Zealand. Additionally, two plant genera could be identified: Nothofagus (southern beeches)—a genus of approximately 35 species of trees and shrubs native to the temperate oceanic to tropical Southern Hemisphere, including New Zealand, and Plantago, a genus of approximately 200 species of small inconspicuous plants commonly called plantains of which most are herbaceous. Plantago is found all over the world, especially in wet areas such as seepages or bogs, or in alpine and semi-alpine or coastal areas, including Asia and New Zealand. Thus, our method provides in some cases identifications to lower taxonomic levels such as genus and species, even for short pieces (120 bp) of the fairly conservative trnL chloroplast region.
In summary, the re-analysis emphasizes the value of a measure of confidence in assignment whereby insufficiently supported assignments can be rejected. It also shows that a statistical assignment approach allows for a greater sensitivity and resolution in describing the original community than does a conservative approach accounting for some of the uncertainties related to assignment using BLAST.
4. Discussion
In this paper, we present a heuristic approach for DNA assignment based on neighbour joining and bootstrapping. The method may be interpreted as a rough and fast approximation to a full Bayesian approach. In fact, in the empirical study of insect sequences retrieved from GenBank, we found that the method performed as well or better than the full Bayesian approach, although it is possible that analyses of other examples might have led to different results. In our evaluation, it is possible that the Bayesian approach in some individual cases may have been challenged by poor MCMC convergence. A distinct disadvantage of the Bayesian approach is that it has to rely on automated convergence diagnostics, when large amounts of datasets have to be analysed. There is, therefore, always a possibility that the analysis of a few datasets will fail due to improperly assessed convergence. For this and other computational reasons, the neighbour-joining-based methods appear to be an attractive alternative in the analysis of very large datasets.
There are a number of caveats to DNA barcoding. The first, and most important, is errors associated with incomplete databases. A DNA barcoding inference is only as good as the database on which it is based. In theory, one could statistically correct for the possibility of unobserved species; however, this would require modelling of the distribution of unsampled species. Also, the inferences made here are based on purely phylogenetic criteria, and they largely ignore interspecific variation and the possibility of incomplete lineage sorting. In cases where assignment is made to one of several closely related species, it may be more desirable to use methods that explicitly model population genetic variation (e.g. Matz & Nielsen 2005; Nielsen & Matz 2006; Abdo & Golding 2007).
In the approach to tree sampling taken here, the most time-consuming task is the calculation of the distance matrix. For many databases, however, such as the Barcoding of Life Database, all the database sequences are pre-aligned and distances between these may thus be locally precomputed. The sample sequence can then be aligned to the database alignment using a profile-hidden Markov model, leaving only the distances between the sample sequence and the database sequences to be calculated. This potentially allows thousands or even millions of homologues to be included in the analysis.
We used the new method on previously published datasets of ancient DNA sequences. Assignment of such sequences is particularly difficult because ancient DNA sequences often are fragmented and, therefore, very short. The advantage of statistical approaches, such as the one presented here, is that they allow the calculation of measures of statistical confidence in the assignment. This allows us to determine to which taxonomic level a specific sequence can be assigned. In our automated method, users will be provided with such assignment confidence measures based on previously published taxonomies and DNA sequences available in public databases. The program for doing this is available at http://fisher.berkeley.edu/cteg/software/munch.
Acknowledgments
This work was funded by the Lundbeck Foundation.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Statistical and computational challenges in molecular phylogenetics and evolution’.
References
- Abdo Z, Golding G.B. A step toward barcoding life: a model-based, decision-theoretic method to assign genes to preexisting species groups. Syst. Biol. 2007;56:44–56. doi: 10.1080/10635150601167005. doi:10.1080/10635150601167005 [DOI] [PubMed] [Google Scholar]
- Alfaro M.E, Zoller S, Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003;20:255–266. doi: 10.1093/molbev/msg028. doi:10.1093/molbev/msg028 [DOI] [PubMed] [Google Scholar]
- Dawnay N, Ogden R, McEwing R, Carvalho G.R, Thorpe R.S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. 2007;173:1–6. doi: 10.1016/j.forsciint.2006.09.013. doi:10.1016/j.forsciint.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Desper R, Gascuel O. Theoretical foundation of the balanced minimum evolution method of phylogenetic inference and its relationship to weighted least-squares tree fitting. Mol. Biol. Evol. 2004;21:587–598. doi: 10.1093/molbev/msh049. doi:10.1093/molbev/msh049 [DOI] [PubMed] [Google Scholar]
- Douady C.J, Delsuc F, Boucher Y, Ford Doolittle W, Douzery E.J.P. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 2003;20:248–252. doi: 10.1093/molbev/msg042. doi:10.1093/molbev/msg042 [DOI] [PubMed] [Google Scholar]
- Duminil J, Caron H, Scotti I, Cazal S.-O, Petit R.J. Blind population genetics survey of tropical rainforest trees. Mol. Ecol. 2006;15:3505–3513. doi: 10.1111/j.1365-294X.2006.03040.x. doi:10.1111/j.1365-294X.2006.03040.x [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. doi:10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Floyd R, Abebe E, Papert A, Blaxter M. Molecular barcodes for soil nematode identification. Mol. Ecol. 2002;11:839–850. doi: 10.1046/j.1365-294x.2002.01485.x. doi:10.1046/j.1365-294X.2002.01485.x [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Hebert P, Cywinska A, Ball S, Dewaard J. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J, Rannala B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst. Biol. 2004;53:904–913. doi: 10.1080/10635150490522629. doi:10.1080/10635150490522629 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Johnson S.S, et al. Ancient bacteria show evidence of DNA repair. Proc. Natl Acad. Sci. USA. 2007;104:14 401–14 405. doi: 10.1073/pnas.0706787104. doi:10.1073/pnas.0706787104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. doi:10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Matz M, Nielsen R. A likelihood ratio test for species membership based on DNA sequence data. Phil. Trans. R Soc. B. 2005;360:1969–1974. doi: 10.1098/rstb.2005.1728. doi:10.1098/rstb.2005.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. doi:10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Cicero C. DNA barcoding: promise and pitfalls. PLoS Biol. 2004;2:e354. doi: 10.1371/journal.pbio.0020354. doi:10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch, K., Boomsma, W., Huelsenbeck, J., Willerslev, E. & Nielsen, R. 2008 Statistical assignment of DNA sequences using Bayesian phylogenetics. Syst. Biol 57, 750–757. (doi:10.1080/10635150802422316) [DOI] [PubMed]
- Nielsen R, Matz M. Statistical approaches for DNA barcoding. Syst. Biol. 2006;55:162–169. doi: 10.1080/10635150500431239. doi:10.1080/10635150500431239 [DOI] [PubMed] [Google Scholar]
- Remigio E, Hebert P. Testing the utility of partial COI sequences for phylogenetic estimates of gastropod relationships. Mol. Phylogenet. Evol. 2003;29:641–647. doi: 10.1016/s1055-7903(03)00140-4. doi:10.1016/S1055-7903(03)00140-4 [DOI] [PubMed] [Google Scholar]
- Rubinoff D. Utility of mitochondrial DNA barcodes in species conservation. Conserv. Biol. 2006;20:1026–1033. doi: 10.1111/j.1523-1739.2006.00372.x. doi:10.1111/j.1523-1739.2006.00542.x [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Semple C, Steel M. Cyclic permutations and evolutionary trees. Adv. Appl. Math. 2004;32:669–680. doi:10.1016/S0196-8858(03)00098-8 [Google Scholar]
- Steinke D, Vences M, Salzburger W, Meyer A. TaxI: a software tool for DNA barcoding using distance methods. Phil. Trans. R. Soc. B. 2005;360:1975–1980. doi: 10.1098/rstb.2005.1729. doi:10.1098/rstb.2005.1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R, Holmes B, White W, Last P. DNA barcoding Australasian chondrichthyans: results and potential uses in conservation. Marine Freshw. Res. 2008;59:57–71. doi:10.1071/MF07148 [Google Scholar]
- Willerslev E, et al. Diverse plant and animal genetic records from holocene and pleistocene sediments. Science. 2003;300:791–795. doi: 10.1126/science.1084114. doi:10.1126/science.1084114 [DOI] [PubMed] [Google Scholar]
- Willerslev E, et al. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science. 2007;317:111–114. doi: 10.1126/science.1141758. doi:10.1126/science.1141758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo Method. Mol. Biol. Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]