Abstract
This new century's biology promises more of everything—more genes, more organisms, more species and, in short, more data. The flood of data challenges us to find better and quicker ways to summarize and analyse. Here, we present preliminary results and proofs of concept from three of our research projects that are motivated by our search for solutions to the perils of plenty. First, we discuss how models of evolution can accommodate change to better reflect the dynamics of sequence diversity, particularly when it is becoming a lot easier to obtain sequences at different times and across intervals where the probability of new mutations contributing to this diversity is high. Second, we describe our work on the use of a single locus for species delimitation; this research targets the new DNA-barcoding approach that aims to catalogue the entirety of life. We have developed a single-locus test based on the coalescent that tests the null hypothesis of panmixis. Finally, we discuss new sequencing technologies, the types of data available and the efficacy of alignment-free methods to estimate pairwise distances for phylogenetic analyses.
Keywords: ancient DNA, cryptic species, DNA barcoding, next generation sequencing
1. Introduction
New automated sequencing technologies are capable of sequencing a bacterial genome in 24 hours and a human genome in two months (Wheeler et al. 2008). These times—and the associated costs of sequencing—are expected to decrease substantially. Additionally, the ease with which genetic information can now be obtained means that areas of research once thought to be beyond our reach are now available. We plan on cataloguing the entirety of life using a DNA barcode. Ancient DNA delivers the genetic material of our ancestors, while environmental metagenomics provides us with a snapshot of whole communities of organisms we never knew existed. We are discovering new genes, new proteins and new organisms. What do we do with all these genes?
The sensitivity of our methods to amplify and sequence DNA means that we now have the ability to obtain sequences from sub-fossil remains. There have been several high-profile studies with mitochondrial DNA sequences obtained from the ancient remains of a whole range of organisms including penguins (Lambert et al. 2002), bison (Shapiro et al. 2004), and chickens (Storey et al. 2007), and one study where portions of the genome of the mammoth were obtained using short-read sequencing (Poinar et al. 2006). We, and other researchers, have been involved in the development of evolutionary and phylogenetic methods to model the evolution of sequences sampled serially over time (e.g. Drummond et al. 2003; Liu & Fu 2007). The significant fact with serially sampled sequences is that mutations can emerge over the sampling intervals, and this means that the dynamics of mutation play an important role in understanding sequence diversity. How should one accommodate the changes in evolutionary dynamics and rates that act across all lineages over time? In fact, there have recently been a few studies that have proposed that substitution rates appear to change as a curvilinear function of time (Ho et al. 2005, 2007). Here, we show how such mathematically well-defined changes in substitution rates may be suitably modelled.
The plan to use a single genetic locus as a unique DNA identifier for a species is an attractive idea, and is one which the Consortium for the Barcode of Life (CBOL; www.barcoding.si.edu) uses as its raison d'etre. For CBOL, a 684nt region at the 5′ end of the cytochrome oxidase 1 gene of the mitochondrial genome is the region of choice. In large part, this region has been very successful at uniquely identifying species across a range of taxonomic groups (Hebert et al. 2003). In some instances, morphologically identical species from which several CO1 sequences were obtained have indicated the presence of several apparently genetically distinct clades (e.g. Hebert et al. 2004). Are these genetically distinct but morphologically identical groups, in fact, ‘cryptic’ species? Here, we propose a test of cryptic species identification based on the coalescent (Kingman 1982a,b) that may be applied to genetic data from a single locus. The coalescent is a continuous-time approximation of the times to common ancestry of individuals and lineages sampled from a single population. Genealogies generated under the coalescent can frequently look highly structured and, thus, may mislead researchers to think that cryptic species exist when, in fact, they do not.
Finally, we look at the new sequencing technologies themselves. ‘Pyrosequencing’ is a ‘sequencing by synthesis’ method (Ronaghi et al. 1998; Margulies et al. 2005) that is capable of providing up to several million bases of shotgun-fragmented DNA sequences of lengths from 100 to 300 nucleotides. Other short-read sequencing technologies, using different chemistries, have since become available, and they provide up to 109 sequences of less than 50 nucleotides per fragment in each run (Sundquist et al. 2007).
Preliminary results from the Global Oceanographic Sampling (GOS; Rusch et al. 2007) of marine bacteria reveal a wealth of new genes, open reading frames and putative proteins (Yooseph et al. 2007). Eisen (2007) has suggested that to rapidly identify environmental shotgun sequencing (ESS) data, alignment-free methods may be useful. These types of methods fall into two broad classes: word-frequency spectral comparisons and the use of compression algorithms. More will be said about these methods, but they have been applied to gene and genome sequences with varying levels of success (e.g. Blaisdell 1986, 1989; Höhl et al. 2006; Ferragina et al. 2007). They have not yet been applied to ESS. How do these methods fare?
The ideas presented here represent preliminary forays in our search for solutions and are not fully formed, tried and tested. Some of these ideas, on further exploration, may prove fruitful, others may not.
2. Serially sampled sequences and varying rates of evolution
Over the last decade, more sensitive methods have meant that relatively large samples of ancient DNA sequences from sub-fossils may be obtained. Over the intervals between sub-fossil samples, there is a significant probability that mutations will emerge, but the best models to explain the dynamics of mutation may not be those that rely on the constancy of rate parameters. Phylogenies of serially sequence samples permit us to decouple time from substitution rate and, therefore, measure branch lengths in units of chronological time (Rambaut 2000). In fact, because these phylogenies are anchored in time, it has also been possible to measure the changes in substitution parameters over time (Drummond & Rodrigo 2000; Drummond et al. 2001) although, to date, changes in substitution rates have been modelled as stepwise functions and, as such, constant over a given interval. Recent work, however, by Ho et al. (2005, 2007) has suggested that there is an apparent curvilinearity to rates estimated over time. Why this is the case (and whether this is a real phenomenon) is still a question under discussion. However, in broader terms, we do expect to find changes in evolutionary processes over time (e.g. Galtier & Guoy 1998; Lemey et al. 2007).
Assume we have sequences sampled serially from a single population for which there is exact information on sampling times and a known phylogeny. The distance of each terminal node of the tree to the root is no longer required to be equal. The parameters of the tree are the substitution rate μ, the vector of timestamps τ and the (n−1 for a bifurcating tree) internal node heights h measured in units of substitutions from the root of the tree.
For a given phylogeny, , for which only the topology is known, we may estimate the joint likelihood of μ and H, the vector of internal node heights on , as the conditional probability of obtaining the sequence data, S, given μ, , H and τ, the vector of times, as well as the model of substitution implied by the instantaneous substitution rate matrix, Q,
| (2.1) |
This likelihood is calculated in the standard manner (Felsenstein 1981; Goldman 1990; Rodriguez et al. 1990) for phylogenetic trees, and is the product of sitewise likelihoods (which follows from the assumption that sites are independent and identically distributed). The addition of μ and τ enters the calculations as constraints on the distances of the branch tips from the root: for serial samples, the distance of each sequence to the root is proportional to its timestamp.
To calculate Li(μ, H, Q), the likelihood at site i of the alignment, we need to be able to calculate, for each branch of with length T, the probability, P(T), of moving from nucleotide m to n for all .
(a) Varying substitution rate, μ, as a function of time
It is biologically plausible for population-wide substitution rates to change over time, perhaps as a consequence of selection and fixation, longer generation times or an improvement in nucleotide error correction and repair. It is natural to start by letting the substitution rate depend on time, i.e. . The only constraint we set on μ(t) is that it is an integrable function.
To obtain P(T), break the interval T into N sub-intervals ti (i=1, …, N) of size Δt, and approximate μ over each small sub-interval with . Over each interval, ti, we therefore assume a constant substitution rate that changes in a stepwise manner to a new substitution rate in the next small interval, ti+1. Consequently, over the interval T, we obtain
| (2.2) |
As N→∞ and Δt→0, , and we obtain
| (2.3) |
(b) Varying the instantaneous rate matrix, Q, as a function of time: the commutable case
A time-dependent μ is in fact a special case of the general case of commutable models of substitution. Let each element of Q change independently as a function of time, while holding the substitution rate, μ, constant, i.e. , where . If for any i and j, (i.e. the matrices are commutable), then a similar result to that described in the previous section can be obtained as follows. Once again, we partition T into N sub-intervals ti (i=1, …, N) of size Δt, and set the instantaneous rate of change over the interval ti to Q(ti).
| (2.4) |
The commutability of Q over T makes the last step possible since is true only when A and B commute.
Again, as Δt→0,
| (2.5) |
where the integral over Q is to be understood as an element-by-element integration. As before, this assumes that the functions that apply to the elements of Q are integrable. Not all models of evolution generate instantaneous rate matrices that are commutable, but many standard models do. These include the Jukes–Cantor, Kimura two-parameter (K2P), Hasegawa–Kishino–Yano and Felsenstein 81 and 84 models.
(c) Varying the instantaneous rate matrix, Q, as a function of time: the non-commutable case
If Q is not commutable, then . To obtain P(T), we use the expansion of the matrix exponential, as follows:
| (2.6) |
As Δt→0, we discard all terms of O(Δt2). While this is the simplest approximation available, it has the advantage of running quickly and it is easy to code. It is worth noting that this approach also works if Q(t) is a complex function for which the integral is difficult to compute analytically.
(d) The challenge
Preliminary simulations indicate that the methods outlined above work as well as any that are presently available to estimate substitution rates for serially sampled sequences (data not shown), but in a sense, this is irrelevant: the mathematics of the method stands on its own, and performance will be a consequence of sampling. And therein lies the first challenge: we do not yet have a good idea on how we should sample the data in the first place. To date, only a few studies have examined the experimental design and power (Seo et al. 2002; Drummond et al. 2003; Liu & Fu 2007). Although it is true that the sampling of ancient DNA is frequently a catch-as-catch-can strategy, there are many instances when targeted sampling is possible (e.g. Lambert et al. 2002; Shapiro et al. 2004). In this case, it is not obvious how one should apportion sampling effort. Should we sample more sequences per time point and fewer time points, or more time points with fewer sequences per time point?
Over long time spans, molecular sequences do not evolve in a homogeneous, time-reversible and stationary manner. As more ancient DNA sequences become available, the second challenge will be to model gene and genome evolution in biologically realistic ways.
3. Testing for cryptic species using a single genetic locus
In 2004, Hebert et al. published a paper that they claimed demonstrated the power of DNA barcoding to clarify the taxonomy of a group of butterflies, Astraptes fulgerator, that are morphologically similar as adults. The single partial mtDNA fragment they used—part of the cytochrome oxidase 1 gene—indicated at least 10 genetically distinct clades on a tree of 466 individuals. Hebert et al. (2004) argued a case for recognizing these clades as morphologically cryptic species.
However, a collection of sequences sampled randomly from a population in which individuals can interbreed freely (i.e. a panmictic population) can frequently appear to have a degree of cladistic structure. Even if the individuals are morphologically identical and there is no a priori reason to believe that the group can be divided further, it is still possible to encounter a cladistic pattern and within- and between-clade genetic distances that appear to reinforce the view that the group is an assemblage of genetically distinct ‘species’. How then can we distinguish between a sound hypothesis of cryptic speciation and a spurious one?
Obviously, one approach is to obtain sequences from other, unlinked, loci: if phylogenies are congruent across loci, and this congruence is unlikely to be due to chance, then the hypothesis that cryptic species are present will be warranted. However, it is useful to know, given a single locus, whether it is worth further time and effort to test the hypothesis of cryptic speciation.
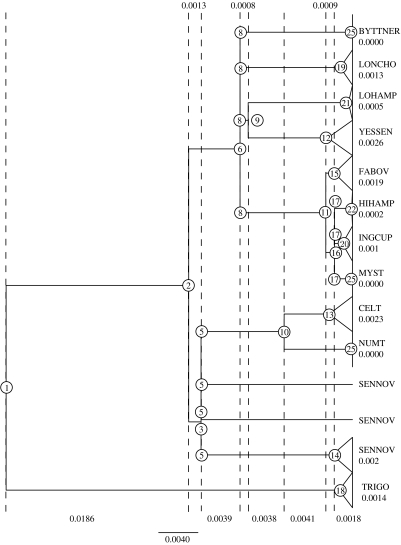
Here, we propose a simple, single-locus test that is based on the coalescent. The coalescent is a mathematical description of the genealogy of a sample of sequences from a Wright–Fisher population. Kingman (1982a,b) showed that the times to common ancestry of pairs of lineages, measured from present to past, can be approximated by exponential random variables with the expected time proportional to 2N/i(i−1), where N is the effective size of the population and i is the number of lineages that have yet to coalesce as we move from the tips to the root of the tree. The method we have developed tests the null hypothesis that the apparent ‘distinctiveness’ of any specified clade is a consequence of the coalescent process acting on a single, panmictic population of constant size. More formally, for any given node on a genealogy that defines a putative (cryptic) species or other taxonomic unit, we calculate the ratio, M, of the sum of the intervals spanning the node to the tips and the sum of intervals between the node and the root (or its most recent ancestor; figure 1), and we compute the probability of obtaining that value of M under a standard coalescent model. This statistic represents an intuitive measure of what people believe is an indication of taxonomic distinctiveness—indeed, those who advocate the use of DNA barcoding set such a threshold to assign sequences to species and higher taxonomic units (Ratnasingham & Hebert 2007).
Figure 1.
Neighbour-joining tree of 466 mtDNA partial cytochrome oxidase 1 sequences with maximum-likelihood branch lengths optimized assuming a molecular clock. Putative ‘species’ groups are labelled with names assigned by Hebert et al. (2004) and distances from the tips to their respective ‘species-defining’ nodes. Dashed lines indicate coalescent intervals, along with interval lengths in substitution units.
(a) The probability density function of M
To compute the distribution of M, we need the probability density function (PDF) of the sum, sk, of k independent exponential random variables, each with a unique mean , and this is given by (Khuong & Kong 2006)
| (3.1) |
M is the ratio of two sums, representing the distances of the ‘species-defining’ node, x, to the tips of the tree (st), and to its ancestor, a (sa). Note that under the coalescent, we assume that time can be measured according to the ticking of the molecular clock; consequently, under the null hypothesis of a single panmictic, constant-sized population, these distances are composed of (n−1−a) and a exponential random variables, respectively, where n is the number of sequences in the sample. Each of these intervals has an expected time (in substitutions) equal to , where (μ is the mutation rate, and the proportionality constant depends on whether the population is haploid or diploid) and i is the number of lineages that have yet to coalesce. The probability of having a value of M=st/sa less than the observed value, m, is
| (3.2) |
where . In fact, p(M≤m) is invariant under Θ because the terms of Θ in the numerator cancel the terms in the denominator. Hence, we can rewrite λr=r(r−1).
For a specified node, equation (3.2) gives a closed-form solution to the probability, under the coalescent, of observing a ratio of node-to-tip distance to node-to-ancestor distance smaller than m. If this probability is sufficiently high (say, greater than 0.05), one would provisionally accept the null hypothesis that the observed ratio is a consequence of the conditions of the Wright–Fisher population model, these conditions being panmixis, and the absence of selection, changes in population size or population subdivision. In other words, the hypothesis that the specified node identifies a cryptic species would not be statistically significant. There is a technical issue with the computation of equation (3.2) that relates to computational precision. We have found, using several programming languages (e.g. C++, Java and VisualBasic.Net), that the value of p(M≤m) cannot be calculated reliably when a+t>45. With the matrix computing language Matlab, it is possible to calculate p(M≤m) for higher values, using its ‘variable precision arithmetic’ function, but there is a significant increase in computational time. However, since p(M≤m) decreases monotonically as a+t increases, we have taken the approach that if p(M≤m)<α at a+t=40, we say that the M ratio is significantly different from that expected under the coalescent.
A reasonable objection to this test is that it relies on only one of many possible coalescent models. For instance, one can imagine a different model of a single panmictic species with a population size that has decreased from some time in the past. The genealogical consequence of this type of dynamic is a tree with short coalescent intervals towards the tips and long coalescent intervals closer to the root. This pattern is likely to show up as statistically significant with the test we propose here, and one would be fooled into thinking that cryptic species are present when, in fact, they are not. Similarly, since most hypotheses of cryptic species are proposed after the tree is constructed, there needs to be some a posteriori correction of the level of significance. An uncorrected p-value will be too liberal. Our response to both of these criticisms is to admit that the test is liberal. However, it means that if we cannot reject the null hypothesis, then it makes it even harder to believe that cryptic species are present based on the phylogeny alone.
(b) Application to the data of Hebert et al. (2004)
We applied our test to the 466 sequences obtained by Hebert et al. (2004). We began by building a neighbour-joining tree in PAUP* (Swofford 1999) using K2P distances. We next optimized the branch lengths of the tree using a clock-constrained maximum-likelihood procedure, also with PAUP*. A molecular clock allows us to recover the order of the coalescent nodes on the tree. For each putative cryptic species within A. fulgerator (labelled using the nomenclature of Hebert et al. (2004)), we measured M as the ratio of the distance between the putative species-defining node and the tips of the tree to the distance between that node and its most recent common ancestor. For example, for the clade SENNOV, the species-defining node is the fourteenth coalescent node from the base of the tree, and the ancestral node is the fifth coalescent node from the base of the tree (figure 1). The results (table 1) indicate that of the 12 groups proposed by Hebert et al., TRIGO, FABOV and INGCUP have p-values greater than 5 per cent (although the last is right on the cusp of statistical significance and, given the problem with precision noted above, should probably be considered a statistically supported group). MYST, NUMT and BYTTNER have identical sequences and they return a p-value equal to 0, although one would be justifiably cautious in quoting this value. This leaves 6 of the 12 species for which the null hypothesis has been conclusively rejected.
Table 1.
M ratios and associated p-values for the different putative species suggested by Hebert et al. (2004).
| name | distance to putative species node | number of species nodea | distance between species node and ancestral node | number of ancestral nodeb | M | pc |
|---|---|---|---|---|---|---|
| TRIGO | 0.0014 | 18 | 0.0353 | 1 | 0.0413 | <0.58 |
| SENNOVd | 0.0021 | 14 | 0.0139 | 5 | 0.1438 | <0.05 |
| BYTTNER | 0.0000 | 25e | 0.0115 | 8e | 0.0000 | – |
| LONCHO | 0.0013 | 19 | 0.0100 | 8e | 0.1275 | <0.05 |
| LOHAMP | 0.0005 | 21 | 0.0100 | 9 | 0.0500 | <0.05 |
| YESSEN | 0.0026 | 12 | 0.0079 | 9 | 0.3291 | <0.05 |
| NUMT | 0.0000 | 25e | 0.0067 | 10e | 0.0000 | – |
| CELT | 0.0023 | 13 | 0.0023 | 10e | 0.5227 | <0.05 |
| FABOV | 0.0019 | 15 | 0.0007 | 11 | 2.7143 | <0.74 |
| HIHAMP | 0.0002 | 22 | 0.0016 | 17e | 0.1250 | <0.05 |
| INGCUP | 0.0010 | 20 | 0.0008 | 17e | 1.2500 | <0.06 |
| MYST | 0.0000 | 25e | 0.0018 | 0.0018 | 0.0000 | – |
See figure 1 for the numbers of the nodes corresponding to each putative species. These numbers represent coalescent events counting from the root towards the tips, with the root node specified as ‘1’.
This is the first ancestral node of the ‘species’ node.
Calculation of p-values encounters precision problems and can only be reliably obtained for n<45. Consequently, all p-values are obtained with an n=40.
This putative species group, SENNOV, ignores the two singleton species that appear as outliers to the main clade.
On the phylogenetic tree, these are multifurcating nodes. Under the coalescent, no two coalescent events can occur simultaneously. Consequently, we have labelled these nodes with identical coalescent ranks, equal to the highest coalescent node possible given the number of bifurcations that can be resolved from the reconstructed multifurcation.
It is important to understand what it means when we reject the null hypothesis constructed here. Each test is an independent test of a specified clade. The ratio of intra- and extra-clade distances is compared against a null distribution of ratios that is not conditioned on the existing coalescent intervals of the reconstructed phylogeny. This p-value is simply the probability of obtaining the observed ratio from the space of all possible topologies and all possible coalescent intervals (under a constant-sized Wright–Fisher population model).
One could argue that such a test is inappropriate because significant results for some clades necessarily change the form of the null hypothesis as it is applied to other clades. In our example, for instance, both HIHAMP and YESSEN have rejected the null hypothesis of panmixis. This should mean that, logically, FABOV should also be treated as a separate species, but it does not reject the null hypothesis. Similarly, if all species except one closer to the tips are not significantly supported by our test, then this surely means that the taxonomic validity of the significant clade is at least logically in doubt. These are problems that we have yet to solve.
Hebert et al. (2004) provided other evidence for the presence of several cryptic species within the group, including the colour variation in caterpillars, and the different preferences for food plants. The question that needs to be asked is whether such corroborative evidence is obtained after the groups are ‘identified’, in which case there is cause to wonder how easy it would be to obtain equally corroborative evidence for any random grouping of individuals. It is difficult to know how to design appropriate a posteriori corrections for corroborative evidence.
(c) The challenge
There are several issues that we believe make the identification of cryptic species a particularly difficult problem. First, as with any hypothesis test, failure to reject the null hypothesis does not mean that the alternative is false, only that there is insufficient evidence of its truth. As we accumulate more genetic information, we are likely to see more genetic variation among morphologically identical individuals. Similarly, we need to work out how corroborating evidence (collected after the genetic analyses) should be handled. The challenge is to develop a suite of methods that tests the hypothesis of cryptic speciation rigorously and critically. Rosenberg (2007) has developed a statistical method to test whether the observed monophyly of a specified group could have occurred by chance. The test takes account of the number of sequences in the specified group. Pons et al. (2006) have developed an explicit model that combines a Yule branching process to describe a species tree and the coalescent for intraspecific genealogies to locate the ‘switch points’ on a phylogeny that correspond to the transitions from Yule to coalescent processes. Fontaneto et al. (2007), building on the analysis by Pons et al. (2006), designed a test to compare the likelihoods of a phylogeny fitted assuming a coalescent process against that fitted with an estimated number of independently evolving clusters. Together with the method outlined here, we may have a near-complete toolbox of such tests.
A second challenge is to determine the best approach to deal with the false discovery rate (FDR) that is inherent in any procedure that constructs a hypothesis (in this case, of cryptic species) a posteriori. A good FDR correction recovers the frequency with which patterns that fool us into thinking they are biologically significant emerge by chance alone. How are we to do this with the identification of cryptic species? The test we have developed here is, strictly speaking, an a priori test applied to an a posteriori diagnosis. We apply the test assuming that the particular group we are testing is one we had intended to test all along—it is only with this assumption that we are able to test each node in isolation. However, in reality, we test nodes that appear after the phylogeny has been constructed.
Biologists use both cladistic (i.e. monophyly) and phenetic (i.e. ‘genetic distinctiveness’) criteria to identify the taxa hidden within the trees. To correct for the FDR, it would seem that we need to merge Rosenberg's (2007) test and the test we have developed here. Alternatively, the tests by Pons et al. and Fontaneto et al. have the advantage of treating all clades simultaneously, and so may be less prone to false discovery.
4. Next generation sequencing and phylogenetic analyses
Next generation sequencing (NGS) involves the parallel sequencing of hundreds of thousands of short fragments of DNA varying in lengths from 30 nucleotides to 250 nucleotides depending on the particular technology. The total amount of DNA sequenced per run may be as much as 109 nucleotides, and each run typically takes less than a week including preparation time. NGS works very well for resequencing genomes that have already been sequenced by traditional methods, and less well for the de novo assembly of previously unsequenced genomes. If this continues to be the case, an obvious challenge presents itself: is there a rapid way of determining the evolutionary relatedness of genomes (or other fragments) sequenced using NGS without the need for time-consuming, and potentially inaccurate, assembly methods?
One approach that has been suggested by Eisen (2007) involves the use of compression or word-frequency algorithms. These methods are useful because no alignment is necessary to obtain pairwise distances between sequence datasets. The use of lossless compression algorithms to measure the shared information content between two molecular sequences and establish evolutionary relatedness has received increasing attention in the literature over the last few years. Word-frequency methods that measure the distance between frequency spectra of all possible k-words of two molecular sequences date back to Blaisdell (1989). Recently, Höhl et al. (2006) examined the performance of several methods to accurately reconstruct evolutionary distances of complete molecular sequences and showed that some performed relatively well. Here, we look at how these methods perform with short-read sequences.
(a) Simulations
We applied word-frequency and compression algorithms to datasets consisting of:
16S complete rDNA sequences of 35 bacteria spanning a wide range of phyla and with a range of GC contents from the Ribosomal Database Project (Maidak et al. 1997),
the same 16S rDNA sequences, cut into random short fragments of length 250 (±50) each with 3× coverage, using the program ReadSim (Schmid & Huson 2007; http://www-ab.informatik.uni-tuebingen.de/software/readsim/welcome.html) with a relatively high error rate of approximately 4 per cent, and
full genomes of the same bacteria as in (i).
We began with a simple word-counting algorithm as presented in Höhl et al. (2006). We created vectors of the frequencies with which all possible words of length k(4≤k≤8) occur in each sequence. We calculated the pairwise distances between vectors using either the squared Euclidean distance or the Manhattan distance.
We used 22 compression algorithms listed by Ferragina et al. (2007) in their paper describing the use of these methods in sequence classification. As a measure to build the distance matrix, we used the Universal Compression Dissimilarity (UCD) distance (Ferragina et al. 2007),
| (4.1) |
where |c(xy)| signifies the size of the compressed file containing sequences (or sequence sets) x and y; and |c(x)| is the size of the compressed file containing only sequence x. The pairwise distances were estimated using the software developed by Ferragina et al. (source: http://www.math.unipa.it/∼raffaele/kolmogorov/).
To visualize the results, we plotted the pairwise distances obtained using either the word-frequency or compression algorithms against distances obtained by PAUP* (Swofford 1999) using models of substitution identified by ModelTest (Posada & Crandall 1998). If a method performs well, then we expect that there will be a monotonic relationship between the estimated distances and the evolutionary distances obtained using standard phylogenetic algorithms. We do not expect to see a linear relationship, of course, because there is no correction for multiple substitutions, but a simple transform will suffice to straighten any simple curvilinear trend, should one be obtained.
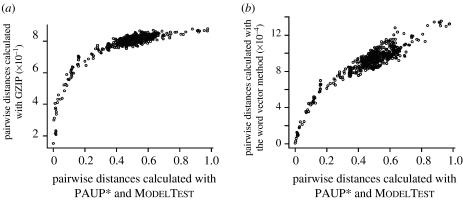
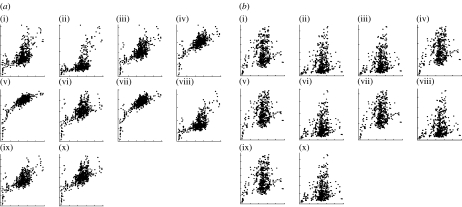
When we applied the algorithms to full-length 16S rDNA sequences, we obtained reasonably good estimates of distances for most of the methods (figure 2). With simulated short-read fragments from 16S rDNA sequences, there was considerably more scatter to the distance plots, but again, a definite monotonicity between compression/word-frequency distances and evolutionary distances (see figure 3a for word-frequency graphs; compression algorithms have similar distributions).
Figure 2.
Plots of pairwise distances of 35 bacterial 16S rDNA sequences obtained using the (a) best compression or (b) word-frequency methods (y-axis) against ML distances obtained using PAUP* and ModelTest. (a) GZIP was the best-performing algorithm, as measured by the Robinson–Foulds distance (Robinson & Foulds 1979) between the reconstructed NJ tree using GZIP distances and the NJ tree using ML distances. (b) Euclidean distances with six-word vectors were the best, as judged by the Robinson–Foulds distance between the reconstructed NJ tree using these distances and the NJ tree using ML distances.
Figure 3.
A comparison between word-frequency distances using (a) simulated short reads of 16S rDNA sequences and (b) whole genomes of the same species. Note the dramatic difference in the estimated distances. Graphs (i)–(x) show distances using either the Manhattan or the Euclidean metric with different word lengths as follows: (i) Manhattan word length 4, (ii) Euclidean word length 4, (iii) Euclidean word length 6, (iv) Manhattan word length 6, (v) Manhattan word length 8, (vi) Euclidean word length 8, (vii) Manhattan word length 7, (viii) Euclidean word length 5, (ix) Manhattan word length 5, and (x) Euclidean word length 7.
By contrast, when we used genomic DNA as our input for compression or word-frequency algorithms, the distances obtained did not fit well with the 16S rDNA evolutionary distances that were used as the benchmark (see figure 3b for word-frequency graphs; compression algorithms have similar distributions). There may be several reasons for this—differences in GC content across the genomes, stretches of nucleotide repeats and lateral gene transfer. We have not identified which of these possible factors challenge the ability of these methods to work well.
(b) The challenge
Our analyses suggest that the use of alignment-free compression and word-frequency algorithms to construct pairwise distances between sets of short-read sequences is quite feasible (but see Höhl & Ragan 2007 for caveats). However, we have only shown that this is possible with a single locus. Our results indicate that when we try to apply these methods to whole genomes, they fail. We have suggested that this may be owing to the different evolutionary dynamics across genomes. Clearly, this suggestion needs to be tested.
A further challenge involves the use of short-read sequencing in metagenomic studies and ESS. If only a single gene region is obtained from ESS (e.g. the 16S rDNA region), then it may be possible to compare communities using alignment-free methods.
5. Discussion
In this paper, we first looked at how our models of evolution can incorporate change as a fundamental evolutionary process, as we accumulate more and different types of data. Next, given the impetus to develop a single marker of species identity, we looked at how to avoid the dangers of falsely assuming that observed cladistic structure is indicative of real biological separation. Finally, we looked directly at one of the benefactors of all this genetic largesse. New sequencing technologies deliver large numbers of short fragments that may be difficult to assemble and align. We explore the use of alignment-free methods and show that their use holds some promise.
With large collections of data, the emergence of patterns by chance alone becomes more probable. Sound and rigorous tests of the significance of emergent patterns are going to be required over the next few years. Additionally, the seductive appeal of large amounts of data seems to rest on a relatively contemporary belief that biological understanding emerges only when we have a handle on the componentry and mechanics of the organisms we study. And yet, if the last 200 years has taught us anything, it is that much insight emerges when we view the world at a sufficiently high level. Evolution, Mendelian genetics and the structure of DNA—the foundations on which modern biological science are built—emerged without the luxury of the vast quantities of data we now have. Consequently, we believe that the firmest challenge for twenty-first century biology is to work out what information we need to keep, what we need to ignore and how to summarize effectively and appropriately.
Acknowledgments
We thank Howard Ross, Alexei Drummond, David Bryant and other staff and students at the Bioinformatics Institute and the Allan Wilson Centre for Molecular Ecology and Evolution for contributing to many fruitful discussions that touched on the ideas in this manuscript.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Statistical and computational challenges in molecular phylogenetics and evolution’.
References
- Blaisdell B.E. A measure of the similarity of sets of sequences not requiring sequence alignment. Proc. Natl Acad. Sci. USA. 1986;83:5155–5159. doi: 10.1073/pnas.83.14.5155. doi:10.1073/pnas.83.14.5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell B.E. Effectiveness of measures requiring and not requiring prior sequence alignment for estimating the dissimilarity of natural sequences. J. Mol. Evol. 1989;29:526–537. doi: 10.1007/BF02602924. doi:10.1007/BF02602924 [DOI] [PubMed] [Google Scholar]
- Drummond A, Rodrigo A.G. Reconstructing genealogies of serial samples under the assumption of a molecular clock using serial-sample UPGMA (sUPGMA) Mol. Biol. Evol. 2000;17:1807–1815. doi: 10.1093/oxfordjournals.molbev.a026281. [DOI] [PubMed] [Google Scholar]
- Drummond A.J, Forsberg R, Rodrigo A.G. Estimating stepwise changes in substitution rates using serial samples. Mol. Biol. Evol. 2001;18:1365–1371. doi: 10.1093/oxfordjournals.molbev.a003920. [DOI] [PubMed] [Google Scholar]
- Drummond A.J, Pybus O.G, Rambaut A, Forsberg R, Rodrigo A.G. Measurably evolving populations. Trends Ecol. Evol. 2003;18:481–488. doi:10.1016/S0169-5347(03)00216-7 [Google Scholar]
- Eisen J.A. Environmental shotgun sequencing: the potential and challenges of random and fragmented sampling of the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. doi:10.1371/journal.pbio.0050082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. doi:10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Ferragina P, Giancarlo R, Greco V, Manzini G, Valiente G. Compression-based classification of biological sequences and structures via the Universal Similarity Metric: experimental assessment. BMC Bioinform. 2007;8:252. doi: 10.1186/1471-2105-8-252. doi:10.1186/1471-2105-8-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaneto D, Herniou E, Bonschetti C, Caprioli M, Melone G, Ricci C, Barraclough T.G. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 2007;5:e87. doi: 10.1371/journal.pbio.0050087. doi:10.1371/journal.pbio.0050087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Guoy M. Inferring patterns and process: maximum likelihood implementation of a nonhomogeneous model of DNA sequence evolution. Mol. Biol. Evol. 1998;15:871–879. doi: 10.1093/oxfordjournals.molbev.a025991. [DOI] [PubMed] [Google Scholar]
- Goldman N. Maximum likelihood inferences of phylogenetic trees, with special reference to the Poisson process model of DNA substitutions and to parsimony analysis. Syst. Zool. 1990;39:345–361. doi:10.2307/2992355 [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the semitropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004;101:14 812–14 817. doi: 10.1073/pnas.0406166101. doi:10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.Y.W, Phillips M.J, Cooper A, Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. doi:10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Ho S.Y.W, Shapiro B, Phillips M.J, Cooper A, Drummond A.J. Evidence for time dependency of molecular rate estimates. Syst. Biol. 2007;56:515–522. doi: 10.1080/10635150701435401. doi:10.1080/10635150701435401 [DOI] [PubMed] [Google Scholar]
- Höhl M, Ragan M.A. Is multiple-sequence alignment required for accurate inference of phylogeny? BMC Syst. Biol. 2007;56:206–221. doi: 10.1080/10635150701294741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhl M, Rigoutsos I, Ragan M.A. Pattern-based phylogenetic distance estimation and tree reconstruction. Evol. Bioinform. 2006;2:357–373. [PMC free article] [PubMed] [Google Scholar]
- Khuong H.V, Kong H.Y. General expression for pdf of a sum of independent exponential random variables. IEEE Commun. Lett. 2006;10:159–161. doi:10.1109/LCOMM.2006.1603370 [Google Scholar]
- Kingman J.F.C. The coalescent. Stoch. Process. Appl. 1982a;13:235–248. doi:10.1016/0304-4149(82)90011-4 [Google Scholar]
- Kingman J.F.C. On the genealogy of large populations. J. Appl. Probab. 1982b;19A:27–43. doi:10.2307/3213548 [Google Scholar]
- Lambert D.M, Ritchie P.A, Millar C.D, Holland B, Drummond A.J, Baroni C. Rates of evolution in ancient DNA from Adélie penguins. Science. 2002;295:2270–2273. doi: 10.1126/science.1068105. doi:10.1126/science.1068105 [DOI] [PubMed] [Google Scholar]
- Lemey P, Kosakovsky Pond S.L, Drummond A.J, Pybus O.G, Shapiro B. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. doi:10.1371/journal.pcbi.0030029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu Y.X. Test of genetical isochronism for longitudinal samples of DNA sequences. Genetics. 2007;176:327–342. doi: 10.1534/genetics.106.065037. doi:10.1534/genetics.106.065037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidak B.L, Olsen G.J, Larsen N, Overbeek R, McCaughey M.J, Woese C.R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. doi:10.1093/nar/25.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:326–327. doi: 10.1038/nature03959. doi:10.1038/437326a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar N.N, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. doi:10.1126/science.1123360 [DOI] [PubMed] [Google Scholar]
- Pons J, Barraclough T.G, Gomez-Zurita J, Cardoso A, Duran D.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. doi:10.1080/10635150600852011 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut A. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. doi: 10.1093/bioinformatics/16.4.395. doi:10.1093/bioinformatics/16.4.395 [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert P.D.N. BOLD: the barcode of life data system. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. doi:10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D. F. & Foulds, L. R. 1979 Comparison of weighted labelled trees. In Proc. 6th Australian Conference Combinatorial Mathematics Lecture Notes Mathematics, vol. 748, pp. 119–126. Berlin, Germany: Springer.
- Rodriguez F, Oliver J.F, Marin A, Medina J.R. The general stochastic model of nucleotide substitution. J. Theor. Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. doi:10.1016/S0022-5193(05)80104-3 [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. doi:10.1126/science.281.5375.363 [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A. Statistical tests for taxonomic distinctiveness from observations of monophyly. Evolution. 2007;61:317–323. doi: 10.1111/j.1558-5646.2007.00023.x. doi:10.1111/j.1558-5646.2007.00023.x [DOI] [PubMed] [Google Scholar]
- Rusch D.B, Halpern A.L, Sutton G, Heidelberg K.B, Williamson S. The Sorcerer II gobal ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, R. & Huson, D. 2007 ReadSim–a simulator for Sanger and 454 Sequencing. Software distributed at http://www-ab.informatik.uni-tuebingen.de/software/readsim/welcome.html
- Seo T.K, Thorne J.L, Hasegawa M, Kishino H. A viral sampling design for testing the molecular clock and for estimating evolutionary rates and divergence times. Bioinformatics. 2002;18:115–123. doi: 10.1093/bioinformatics/18.1.115. doi:10.1093/bioinformatics/18.1.115 [DOI] [PubMed] [Google Scholar]
- Shapiro B, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. doi:10.1126/science.1101074 [DOI] [PubMed] [Google Scholar]
- Storey A.A, et al. Radiocarbon and DNA evidence for a pre-Columbian introduction of Polynesian chickens to Chile. Proc. Natl Acad. Sci. USA. 2007;104:10 335–10 339. doi: 10.1073/pnas.0703993104. doi:10.1073/pnas.0703993104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist A, Ronaghi M, Tang H, Pevzner P, Batzoglou S. Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS ONE. 2007;2:e484. doi: 10.1371/journal.pone.0000484. doi:10.1371/journal.pone.0000484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 1999. PAUP*. Phylogenetic analysis using parsimony (* and other methods) [Google Scholar]
- Wheeler D.A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. doi:10.1038/nature06884 [DOI] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch D.B, Halpern A.L, Williamson S.J. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. doi:10.1371/journal.pbio.0050016 [DOI] [PMC free article] [PubMed] [Google Scholar]