Abstract
The budding yeast Pichia pastoris contains ordered Golgi stacks next to discrete transitional endoplasmic reticulum (tER) sites, making this organism ideal for structure–function studies of the secretory pathway. Here, we have used P. pastoris to test various models for Golgi trafficking. The experimental approach was to analyze P. pastoris tER-Golgi units by using cryofixed and freeze-substituted cells for electron microscope tomography, immunoelectron microscopy, and serial thin section analysis of entire cells. We find that tER sites and the adjacent Golgi stacks are enclosed in a ribosome-excluding “matrix.” Each stack contains three to four cisternae, which can be classified as cis, medial, trans, or trans-Golgi network (TGN). No membrane continuities between compartments were detected. This work provides three major new insights. First, two types of transport vesicles accumulate at the tER-Golgi interface. Morphological analysis indicates that the center of the tER-Golgi interface contains COPII vesicles, whereas the periphery contains COPI vesicles. Second, fenestrae are absent from cis cisternae, but are present in medial through TGN cisternae. The number and distribution of the fenestrae suggest that they form at the edges of the medial cisternae and then migrate inward. Third, intact TGN cisternae apparently peel off from the Golgi stacks and persist for some time in the cytosol, and these “free-floating” TGN cisternae produce clathrin-coated vesicles. These observations are most readily explained by assuming that Golgi cisternae form at the cis face of the stack, progressively mature, and ultimately dissociate from the trans face of the stack.
INTRODUCTION
The Golgi apparatus consists of flattened membrane cisternae that are usually organized into stacks (Berger and Roth, 1997). Newly synthesized biosynthetic cargo molecules exit the endoplasmic reticulum (ER) in COPII-coated vesicles and enter the cis cisterna of the Golgi (Farquhar and Hauri, 1997; Barlowe, 2002). These cargo molecules then occur in medial and trans-cisternae. In the trans-Golgi network (TGN), the cargo molecules are sorted into different types of carriers for delivery to the plasma membrane and other destinations (Mellman and Simons, 1992). Although this basic scheme is well established, the pathway of intra-Golgi transport is still being investigated. Anterograde Golgi transport has been proposed to occur via vesicular intermediates, membrane continuities, cisternal maturation, or a combination of these mechanisms (Beznoussenko and Mironov, 2002). The extent to which a given mechanism operates may vary with the cell type and the stage of the cell cycle (Pelham and Rothman, 2000; Marsh and Howell, 2002). Considerable evidence now favors cisternal maturation as a major route for intra-Golgi transport (Pelham, 2001; Storrie and Nilsson, 2002), but the generality and several key predictions of this model remain to be verified.
We are using electron microscopy to test predictions of the different Golgi-trafficking models. Because some of the relevant questions involve structural details such as the connectivity between membrane compartments, we use cryofixation and freeze substitution to maximize the fidelity of cellular preservation (Gilkey and Staehelin, 1986). Computerized tomography provides enhanced resolution for three-dimensional (3D) analysis (McEwen and Marko, 1999). Our previous work along these lines yielded valuable qualitative and quantitative data about Golgi organization in mammalian cells (Ladinsky et al., 1999; Marsh et al., 2001).
Although the mammalian Golgi is well characterized, it has several limitations for morphological analysis. Mammalian Golgi stacks are large and are laterally interconnected into a ribbon (Rambourg and Clermont, 1997), so the tomographic reconstruction of an entire Golgi unit would be difficult. The mammalian Golgi ribbon is physically distant from transitional endoplasmic reticulum (tER) sites (Palade, 1975; Bannykh et al., 1996; Hammond and Glick, 2000), and as a result, the early intermediates in ER-to-Golgi transport are absent from tomograms of mammalian Golgi stacks. For these reasons, we decided to examine Golgi organization in a simple unicellular eukaryote.
A suitable organism for this purpose is the budding yeast Pichia pastoris. Unlike Saccharomyces cerevisiae, P. pastoris contains stacked Golgi organelles (Gould et al., 1992; Glick, 1996). A typical P. pastoris cell has three to five separate Golgi stacks, each of which is adjacent to a discrete tER site (Rossanese et al., 1999). Thus, an entire tER-Golgi unit can be reconstructed in a single tomogram. tER sites and Golgi stacks in P. pastoris have been characterized by fluorescence microscopy (Rossanese et al., 1999), and the dynamic behavior of these compartments was recently documented by in vivo imaging (Bevis et al., 2002). Finally, P. pastoris is amenable to experimental manipulations, which are facilitated by the close relatedness of this yeast to S. cerevisiae (Gould et al., 1992; Rossanese et al., 1999).
In this study, we have used P. pastoris to place morphological constraints on models of Golgi function, and more specifically, to test three predictions of the cisternal maturation model. First, the precursor of a new cis cisterna is predicted to be a group of COPII and COPI vesicles. Second, cisternal morphology and composition are predicted to change progressively across the Golgi stack, in a manner that is consistent with a precursor–product relationship. Third, TGN cisternae are predicted to dissociate from the stack as they mature into secretory carriers. Our results confirm all three predictions. In addition, a comparison of P. pastoris with mammalian cells has highlighted both conserved and cell type-specific aspects of Golgi organization.
MATERIALS AND METHODS
Strains and Cultures
The P. pastoris strain PPY12 (Gould et al., 1992) and its derivatives were used for all experiments. Strain PPY12-S7G-OM, which has the chromosomal SEC7 gene replaced with SEC7-GFPx3 and the chromosomal OCH1 gene (accession no. E12456) replaced with OCH1-mycx8 (Soderholm et al., 2001), was used for immunoelectron microscopy and fluorescence microscopy. Cells were cultured in YPD (1% yeast extract, 2% peptone, 2% glucose). For electron microscopy, yeast cultures were subjected to high-pressure freezing at an OD600 of 0.3–0.5. Immunofluorescence microscopy of fixed cells was performed as described in Rossanese et al. (1999).
High-Pressure Freezing and Freeze Substitution
Cells were concentrated using a 0.22-μm filter (Millipore, Bedford, MA) and the wet cell paste was transferred to sample holders. The samples were frozen in a Baltec HPM10 high-pressure freezer (Technotrade, Manchester, NH) and transferred to liquid N2 for storage. For ultrastructural work, samples were subjected to freeze substitution in 0.5% glutaraldehyde and 0.2% tannic acid in acetone at -80°C for 3 d, rinsed for 1 d with -80°C acetone, fixed with 2% OsO4 for 12 h, and then slowly warmed up to room temperature. After several acetone rinses, the samples were infiltrated with Epon resin (Ted Pella, Redding, CA) and polymerized at 60°C as described in Otegui et al. (2001). For immunoelectron microscopy, samples were freeze substituted with 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone at -80°C for 5 d and then slowly warmed up to -20°C. The samples were rinsed for 5 h with -20°C acetone and then infiltrated with Lowicryl HM20 resin (Electron Microscopy Sciences, Ft. Washington, PA) and polymerized with UV light at -50°C as described in Otegui et al. (2001).
Immunolabeling
Ultrathin sections were collected on formvar-coated nickel grids. The sections were blocked for 30 min with 2.5% nonfat milk in phosphate-buffered saline with 0.1% Tween 20 (PBST), incubated with primary antibody for 3 h, rinsed with PBST, incubated for 1 h with 10- or 15-nm gold-conjugated goat anti-rabbit antibody at 1:20 dilution in PBST, rinsed with PBST and water, and finally stained with 2% uranyl acetate in 70% methanol and Reynold's lead citrate. The anti-green fluorescent protein (GFP) (Seedorf et al., 1999) and anti-myc (9E10; Covance, Denver, PA) antibodies were both used at 1:200 dilution in PBST and 2.5% nonfat milk.
High-Voltage Electron Microscopy and Dual-Axis Tilt Series Imaging
Tomograms and modeling of 3D structures was carried out as described in Otegui et al. (2001) and Marsh et al. (2001). In brief, ribbons of serial 250- or 300-nm thick sections were cut and post-stained with 2% uranyl acetate in 70% methanol and Reynold's lead citrate. Colloidal gold markers (10 nm) were deposited on both sides of the sections for use as fiducial points during subsequent image alignment. Selected cells were imaged at 12,000× using a JEM-1000 (JEOL, Tokyo, Japan) high-voltage microscope operating at 750 kV. The specimens were tilted at 1.5° intervals over a range of 120° about two orthogonal axes, and images were captured with a digital camera (Gatan, Pleasanton, CA). Areas larger than the 1.42 × 1.42 μm covered by the camera were captured by combining adjacent images into larger montaged images. The reconstructed 3D data covered volumes ranging between 1.42 × 1.42 × 0.25 μm and 1.42 × 2.84 × 0.75 μm.
3D Reconstruction from Dual-Axis Tilt Series
Images were aligned as described in Ladinsky et al. (1999). Tomograms were computed for each tilt series using an R-weighted back-projection algorithm (Gilbert, 1972). Next, tomograms from orthogonal tilt series were combined to yield dual-axis tomograms by using a warping procedure (Mastronarde, 1997). Dual-axis tomograms from serial sections could be combined, producing serial tomograms of volumes thicker than individual sections.
3D Modeling
Tomograms were displayed and analyzed using the IMOD software package (Kremer et al., 1996) on a Silicon Graphics computer (Silicon Graphics, Mountain View, CA). Membranes and ribosomes were modeled as described in Ladinsky et al. (1999), and a surface mesh was rendered to fit the modeled contours. Quantitative data about the 3D models were extracted using the program imodinfo, which is part of the IMOD software package.
RESULTS
Cryofixation of P. pastoris Cells Yields Excellent Preservation of Secretory Compartments
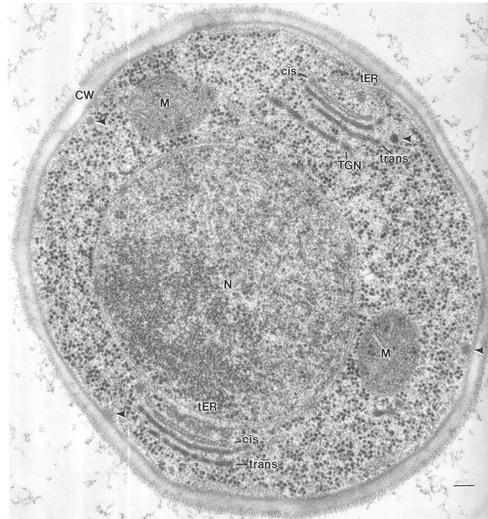
Our initial goal was to study the basic organization of tER-Golgi units in P. pastoris. Cells were high-pressure frozen and freeze substituted to obtain the best possible structural preservation of tER sites, Golgi stacks, and other cellular structures (Figure 1). The tER sites were identified by the presence of clustered budding profiles and free vesicles adjacent to the nuclear envelope or the cortical ER. The Golgi stacks were located next to tER sites and exhibited a distinct polarity due to a differential staining of the lumenal contents of the cisternae by OsO4. Most likely, these differences in staining reflect a progressively heavier glycosylation of glycoproteins during passage through the Golgi (Zhang and Staehelin, 1992; Rambourg et al., 1993; Munro, 2001). Individual vesicles were observed throughout the cytoplasm, and secretory granules were clustered under emerging buds at the plasma membrane.
Figure 1.
Electron micrograph of a high-pressure frozen and freeze substituted P. pastoris cell. Golgi stacks and tER sites are clearly defined with discernible membrane bilayers. The Golgi stacks have a clear polarity in this OsO4-stained material, with the cis cisternae staining faintly and the trans cisternae staining intensely. The trans and TGN cisternae seem to have fenestrae. CW, cell wall; M, mitochondrion; N, nucleus; arrows, vesicles. Bar, 100 nm.
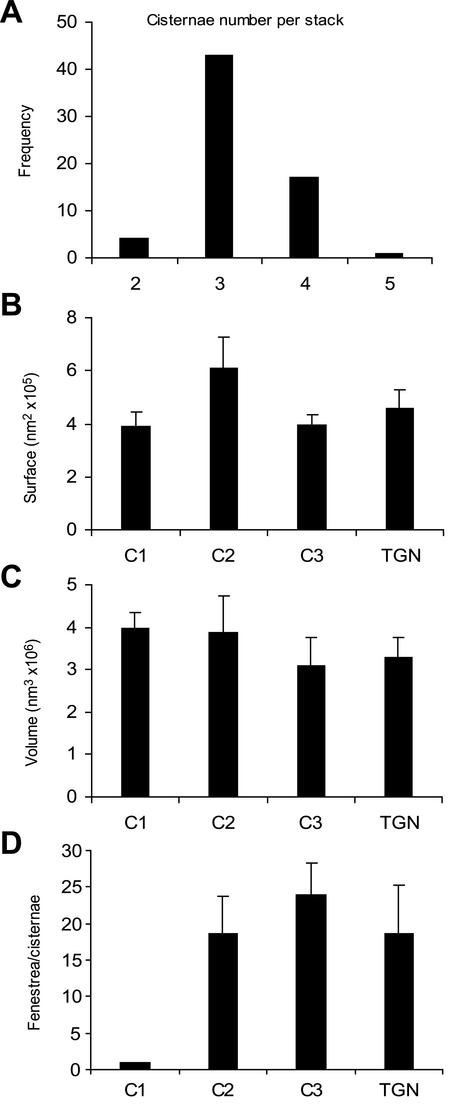
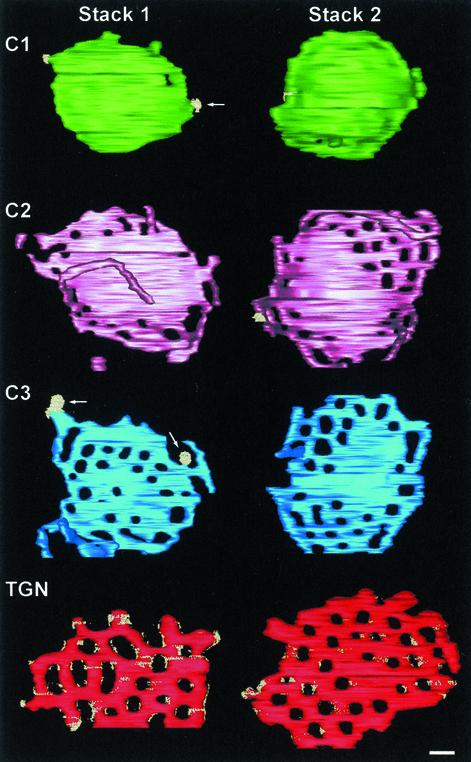
Serial thin-section analysis of 67 Golgi stacks demonstrated that most contained three or four cisternae (Figure 2A). For convenience, we have classified these cisternae as cis, medial, trans, or TGN based on morphological and biochemical criteria (see below). Such a classification may be somewhat imprecise, because the cisternal maturation model implies that Golgi cisternae change continuously, but even so it is useful to classify cisternae with regard to their approximate stage of maturation. According to our definitions, the Golgi stacks with three cisternae contained a cis (C1), a medial (C2), and a trans (C3) cisterna, and the stacks with four cisternae also contained a TGN (C4) cisterna.
Figure 2.
Quantitative data from the serial thin section analysis and the tomographic 3D models. (A) The number of cisternae per Golgi stack was counted in 67 stacks. Electron micrographs of serial ultrathin sections were used to ensure that all cisternae were counted. The majority of the stacks have three or four cisternae. (B–D) These values are averages of data from three reconstructed Golgi stacks, two of which are shown in Figures 8 and 9. (B and C) The surface areas and volumes of individual cisternae were calculated using the IMOD software. Interpolated gaps were not included and the truncated edges added to the uncertainty. Despite these caveats, the data reveal no significant change in either surface area or volume across the stacks. (D) The number of fenestrae increases dramatically from C1 to C2, but stays relatively constant across the rest of the stack. The error bars indicate SD.
Polarity of P. pastoris Golgi Stacks Reflects a Biochemical Compartmentation
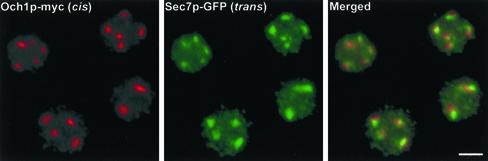
The next step was to determine whether the differential lumenal staining of the cisternae reflects a biochemical compartmentation of the Golgi. To address this question, we examined the distribution of proteins that were predicted to mark either the cis or the trans/TGN side of the stack. A P. pastoris strain was made with the genomic copies of the OCH1 and the SEC7 genes replaced with the gene fusions OCH1-myc and SEC7-GFP, respectively. Och1p is a mannosyltransferase of the cis Golgi (Nakayama et al., 1992; Gaynor et al., 1994), and in a previous study we expressed a tagged version of S. cerevisiae Och1p in P. pastoris (Rossanese et al., 1999). Because endogenous Och1p is likely to be a more reliable cis Golgi marker, we have now replaced the P. pastoris OCH1 gene with a tagged version. Sec7p is a peripheral membrane protein of the late Golgi (Franzusoff et al., 1991), and we replaced the P. pastoris SEC7 gene with SEC7-GFP (Soderholm et al., 2001) in the strain expressing OCH1-myc. By immunofluorescence microscopy, Och1p-myc and Sec7p-GFP were localized to punctate structures typical of the P. pastoris Golgi (Figure 3; Rossanese et al., 1999). In merged images, the two markers were adjacent or partially overlapping, consistent with localization to opposite sides of the Golgi stacks.
Figure 3.
Immunofluorescence localization of Och1p-myc and Sec7p-GFP. Each panel shows the same cells with the indicated markers. The cis marker Och1p-myc and the trans/TGN marker Sec7p-GFP are adjacent, but not completely overlapping. Bar, 2 μm.
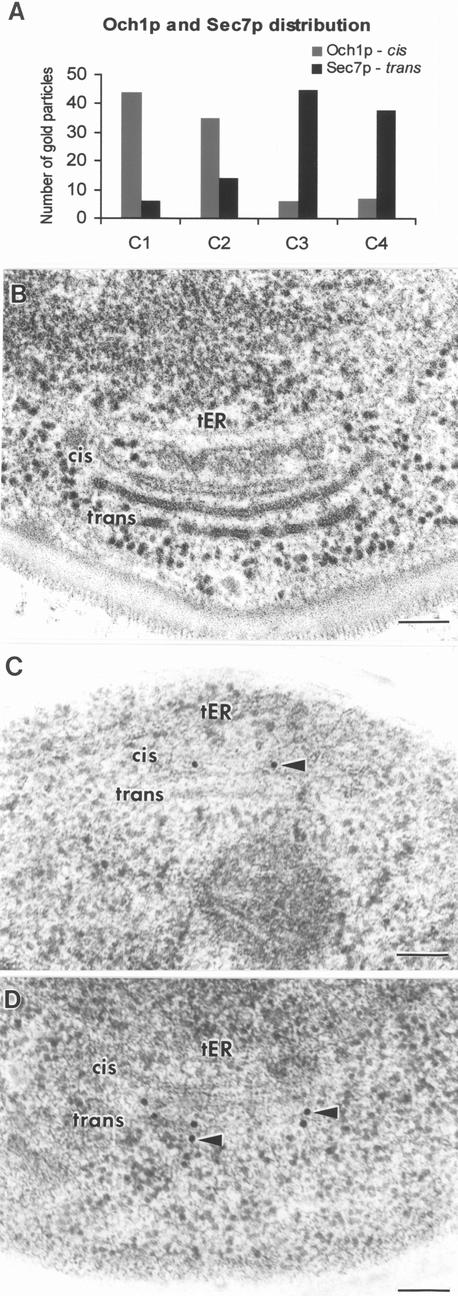
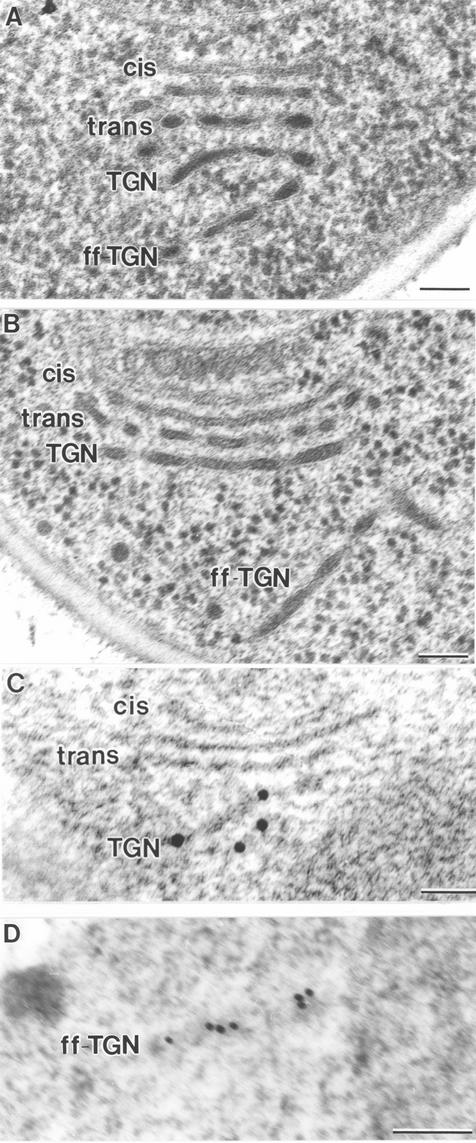
To confirm this interpretation of the fluorescence micrographs, the fusion proteins were immunolocalized in ultra-thin sections of cryofixed, HM20-embedded cells (Figure 4). Although the tER sites and Golgi stacks were less defined than in Epon-embedded samples (compare Figure 4B with C and D), it was clear that Och1p-myc localized to the cis side of the Golgi stacks near the tER, whereas Sec7p-GFP localized to the opposite trans side of the stacks (Figure 4, C and D). To quantify this distribution, gold particles were counted on a total of 70 stacks labeled with either an anti-myc or an anti-GFP antibody. Figure 4A shows that Och1p-myc localized primarily to C1 and C2 cisternae, whereas Sec7p-GFP was concentrated on C3 and TGN cisternae. The background labeling in other parts of the cell was negligible. These data confirm the validity of using Och1p and Sec7p as cis and trans-Golgi markers in P. pastoris and demonstrate that the osmium staining gradient across the stacks is paralleled by a functional compartmentation of the P. pastoris Golgi.
Figure 4.
Localization of Och1p-myc and Sec7p-GFP by immunoelectron microscopy. (A) Summary of the immunolocalization studies. Och1p-myc labeling was quantified on 37 random stacks and Sec7p-GFP labeling was quantified on 33 random stacks. Och1p-myc localizes to the cis side and Sec7p-GFP to the trans side of the stacks. (B) Electron micrograph of an Epon section illustrating a tER site and a Golgi stack for reference. (C) Electron micrograph of an HM20 section showing a tER site and a Golgi stack with immunogold-labeled Och1p-myc on the cis side. (D) Electron micrograph of an HM20 section showing a Golgi stack with immunogold-labeled Sec7p-GFP at the trans side. Bar, 100 nm.
3D Tomographic Models of tER Sites, Golgi Stacks, and TGN Cisternae
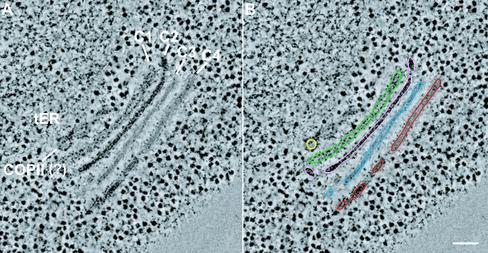
To explore the detailed 3D architecture of the tER–Golgi system in P. pastoris, we performed electron tomography. Twelve tomograms were produced. Several of the tomograms were from serial sections, which were combined to yield serial tomograms containing entire Golgi stacks. Figure 5A shows a tomographic slice from a dual-axis tomogram containing a tER site plus an almost complete Golgi stack and TGN. This single slice represents a thickness of ∼2.3 nm, and the full tomogram comprises 257 such slices, corresponding to a cell volume ∼0.6 μm in thickness. The resolution of the tomogram was ∼7 nm, which is 10–20 times better than can be achieved with serial thin section reconstruction methods. The preservation and staining characteristics of the structures were identical to those in ultra-thin sections (Figure 1), and were adequate for visualizing membrane bilayers and protein coats. One tomogram contained two tER sites and Golgi stacks, enabling us to compare variability between these structures within the same sample (our unpublished data).
Figure 5.
A tomographic slice from a dual-axis tomogram of a cell volume containing a tER site and a Golgi stack. The same 2.3-nm-thick slice is shown in A and B. The tomogram comprised a total of 257 optical slices, which corresponds to a 0.6-μm-thick cell volume. All of the tomographic slices of this tomogram are shown in Movie 1. (A) In the tomogram, the appearance of the Golgi cisternae and membrane bilayers is similar to that seen in conventional ultrathin sections (compare with Figure 1). (B) The tomographic data were analyzed by tracing membranes of interest in all of the slices of the tomogram. Bar, 100 nm.
Using the IMOD software, the tomographic data were interpreted by tracing all membranes in each slice of the tomograms. An example is given in Figure 5B where the membranes of a vesicle and a Golgi stack have been traced. Although it would be difficult to determine exactly where to draw membrane boundaries in a single slice, this task is made easier by studying the neighboring slices, and by combining five to 10 adjacent slices into a two-dimensional projection by using the slicer tool in IMOD. Movie 1 shows the tomographic slices of the tomogram illustrated in Figure 5. By scanning these slices, the distribution of fenestrae and tubules could be determined unambiguously. A 3D model was generated by rendering a surface fit to the contours (Figures 6 and 8).
Figure 6.
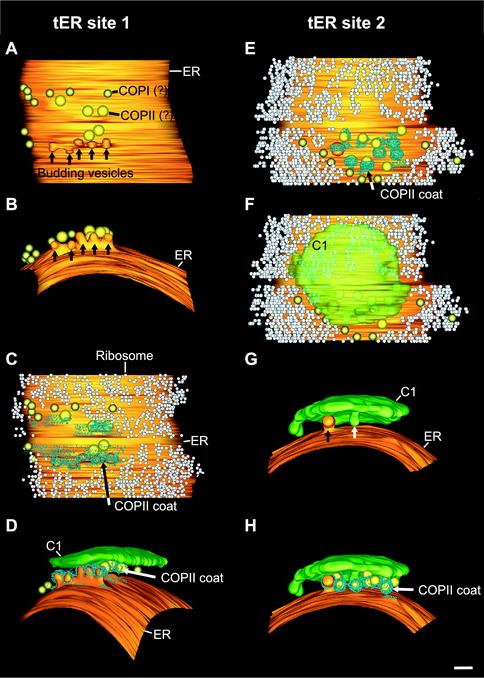
3D models of two tER sites. (A–D) Different views of a tER site with budding profiles and associated free vesicles. (E–H) A second tER site and its associated structures. (A and B) Two classes of vesicles are found between the tER site and the C1 cisterna. The larger vesicles have a diameter of ∼42 nm and a lightly staining lumen and are sometimes coated with a proteinaceous material similar to that on the budding tER profiles (Figure 7). We propose that these structures are anterograde vesicles and that the coat is composed of COPII components. The smaller vesicles have a diameter of ∼34 nm and a darkly staining lumen and are sometimes seen outside the tER-Golgi interface (Figures 7 and 8). We propose that these structures are retrograde COPI vesicles. (C and E) The tER sites are outlined by a ribosome-excluding zone. (D, F, G, and H) The C1 cisternae are very close to the tER sites, confining the movement of the free vesicles to this space. (G) Side view of a tER-associated budding vesicle and a budding/fusing structure on the C1 cisterna. Bar, 100 nm.
Figure 7.
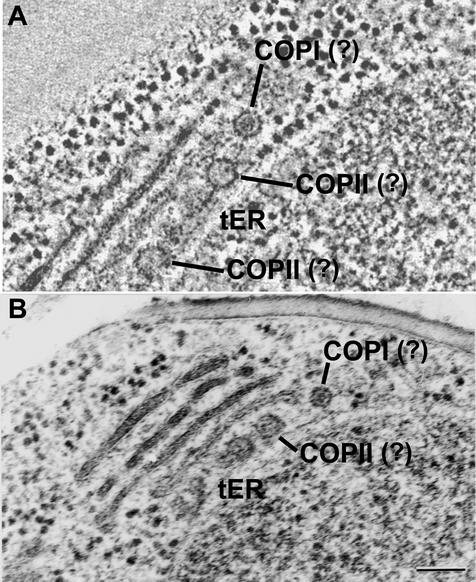
Tomographic and electron micrographic images of tER sites. (A) A tomographic slice from a dual-axis tomogram showing a tER site. (B) Electron micrograph showing a tER site similar to that in A. Both A and B show the presence of two different vesicles types at the tER-Golgi interface. The putative COPII vesicles have lightly stained lumens, whereas the putative COPI vesicles have darkly stained lumens. Bar, 100 nm.
Vesicles and Coated Buds Fill the Space between a tER Site and the Adjacent cis Cisterna
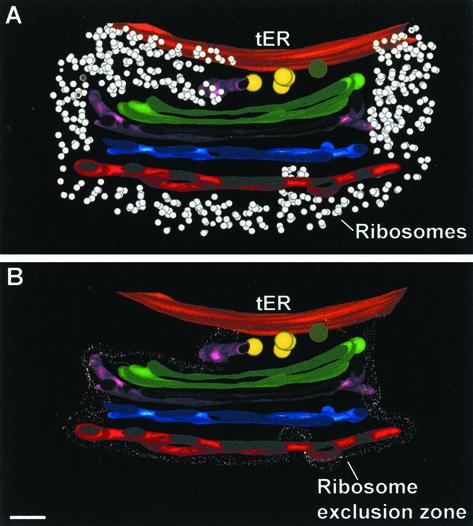
Three tER sites were modeled along with their Golgi stacks, and two of these models are shown in Figure 6. tER sites were present in flat, cisterna-like domains of the ER. Each tER site was clearly delineated by a ribosome exclusion zone. Unexpectedly, the cis most cisterna was not perfectly aligned with this zone (Figure 6F). tER site 1 in Figure 6 had five budding or fusing structures (Figure 6A), whereas tER site 2 had only one such structure (Figure 6G). Because all of the tER-associated structures had proteinaceous coats (Figure 6, C, D, E, and H), similar to the coats seen on a subset of the adjacent vesicles (Figure 6, C, E, and H), we postulate that these structures correspond to budding COPII vesicles. Consistent with this view, previous immunoelectron microscopy of P. pastoris indicated that COPII components are concentrated at the tER-Golgi interface (Rossanese et al., 1999). Interestingly, the five budding structures at tER site 1 looked aligned. The space between the tER site and the cis most cisterna was always very narrow, confining all the budding, fusing, and free vesicles in this zone to a single plane (Figure 4B). This planar organization was also seen in the tomographic models (Figure 6, D and H).
The vesicles between the tER sites and the C1 cisternae were of two size classes, with inner diameters of ∼42 and 34 nm (Figure 7). The larger vesicles had a lightly and uniformly stained lumen resembling the lumen of the budding structures at the tER sites, and virtually all of the larger vesicles were covered by a proteinaceous coat resembling the coat on the budding structures at the tER sites. Based on these criteria, we propose that the 42-nm vesicles are anterograde COPII vesicles (Figure 6), and we will refer to them in the text as putative COPII vesicles. The smaller 34-nm vesicles could be distinguished from the larger vesicles by their densely stained lumens (Figure 7). These smaller vesicles were occasionally seen outside the tER-Golgi interface, mostly along the margins of the stacks (Figure 8). Based on these observations, the 34-nm vesicles are probably retrograde COPI vesicles, and we will refer to them in the text as putative COPI vesicles. Our vesicle diameters are slightly smaller than those previously reported for COPI and COPII vesicles (Rothman and Wieland, 1996; Schekman and Orci, 1996), presumably because the swelling artifacts that can be induced by chemical fixation and dehydration were eliminated here by using high-pressure freezing and freeze substitution.
Figure 8.
3D model of a P. pastoris Golgi stack and associated vesicles. A and B are two different views of the same Golgi stack. The model represents the Golgi stack shown in Figure 5 and was generated by rendering a surface fit to the traced contours. This stack has four cisternae. An interesting feature is the tubule from the margin of C2 that reaches around the C1 cisterna. Bar, 100 nm.
One budding or fusing vesicle profile was seen at the center of the C1 cisterna of tER site 2 (Figure 6G). Judging from the size and staining characteristics of this structure, it was potentially an anterograde COPII vesicle fusing with the C1 cisterna.
We never observed a tER site without an adjacent Golgi stack, or vice versa. This result is consistent with previous fluorescence microscopy studies, which indicated that tER sites and Golgi stacks in P. pastoris are closely associated with one another (Rossanese et al., 1999; Bevis et al., 2002).
P. pastoris Golgi Cisternae Exhibit Systematic Morphological Changes across the Stacks
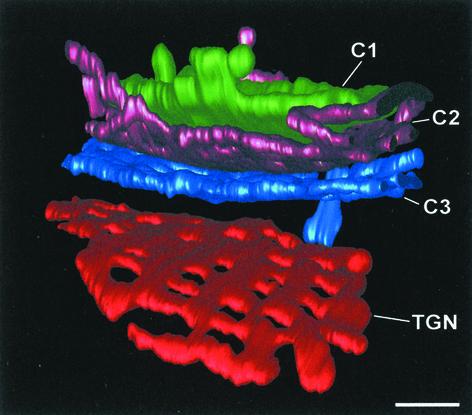
Three complete Golgi stacks with four cisternae each were modeled from serial tomograms. With this method, ∼30 nm of information is lost between adjacent sections, and in our 3D models these gaps have been filled by interpolation, manifesting as smoother domains. The edges of some cisternae could not be reconstructed because they fell into the gaps between adjacent sections. Slices of Golgi stacks were also modeled from single tomograms of 250- to 300-nm-thick cell volumes. By using the IMOD software (Kremer et al., 1996), the individual components of the models were studied from multiple angles. In Figure 9, the individual cisternae of two modeled Golgi stacks are depicted in face-on views, as seen from the cis side of the stack (Stack 1 is the same one that is shown in Figure 8). In all of the models, the cisternae exhibited several characteristic morphological features, and each of the stacks seemed to possess a cis (C1), a medial (C2), a trans (C3), and a TGN cisterna. These structural differences complemented the differences in cisternal staining and biochemical composition (Figures 1 and 4).
Figure 9.
Face-on view of the individual cisternae of two P. pastoris Golgi stacks, seen from the cis side. Stack 1 is the same as in Figure 8. Protein coats are shown in bright yellow. The C1 cisternae are smaller in diameter than the other cisternae. Each of the C1 cisternae shown here has one fenestra, whereas the C2 to TGN cisternae have multiple fenestrae. The fenestrae seem to form a narrow ring in the C2 cisternae and a wider ring in the C3 cisternae and are found throughout the TGN cisternae, which resemble networks. Tubules protrude from the margins of C2 and to a lesser extent from C3. The TGN has a protein coat, perhaps Golgi matrix proteins, distributed over its surface. Arrows, budding profiles. Bar, 100 nm.
When the cisternae were compared across the stack, several features were seen to be shared by two or more types of cisternae. Surface area and volume were very similar for all of the cisternal types (Figure 2, B and C). The C1, C2, and C3 cisternae possessed budding/fusing structures along their margins, and some of these structures were covered by a densely staining coat (Figure 9, C1 and C3 of stack 1). The C1 and TGN cisternae lacked tubules, whereas all of the C2 and C3 cisternae had tubules projecting from their rims. None of the tubules seemed to be fused to other cisternae, indicating that each cisterna in a Golgi stack was a physically separate compartment.
Superimposed on these shared features was a clear morphological change across the stacks (Figure 9). The most notable change was the number and location of fenestrae. C1 cisternae were disk shaped, had slightly bulging margins, and were basically devoid of fenestrae (Figures 8 and 9). In contrast, the C2 cisternae exhibited numerous fenestrae of fairly uniform size. These fenestrae were concentrated near the rims of the C2 cisternae. The C3 cisternae had a similar number of fenestrae as the C2 cisternae (Figure 2D), but the individual C3 fenestrae tended to be larger. Many of the C3 fenestrae were distant from the cisternal rims, but they were still largely absent from the central portions of the cisternae (Figure 9). The TGN cisternae had larger and more evenly distributed fenestrae than the C3 cisternae, giving the TGN cisternae a network-like architecture. In some places the TGN networks seemed to be in the process of breaking up along the margins.
Intact TGN Cisternae Apparently Dissociate from Golgi Stacks
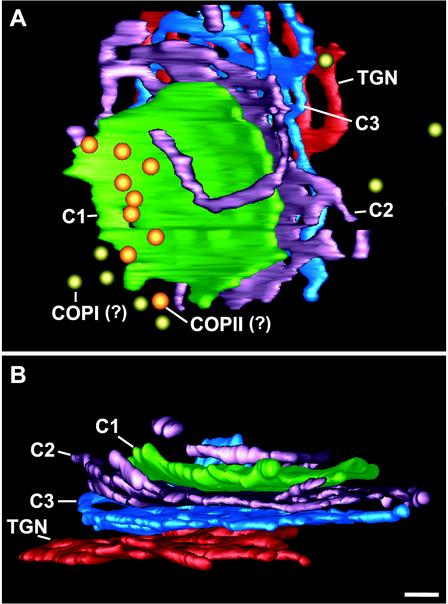
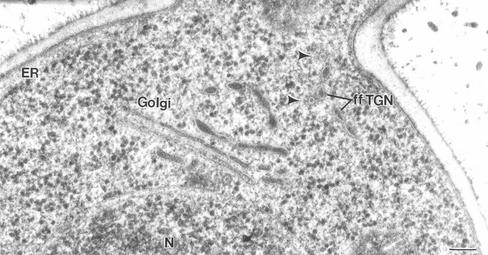
The coherence of P. pastoris Golgi stacks was underscored by the exclusion of ribosomes from the regions between the C1 to C3 cisternae (Figures 1, 5, 7, 11, and 14). These three cisternae were closely juxtaposed and evenly spaced. However, ribosomes were frequently observed between the C3 and TGN cisternae (Figures 1 and 11), suggesting that the TGN cisternae were less tightly associated with the stacks. TGN-like structures were often seen to be completely separated from a stack by ribosome-containing cytoplasm (Figure 11). Such independent structures were named “free-floating” TGN cisternae (ff-TGNs). The separation of a TGN cisterna from a parental stack is illustrated in the 3D model in Figure 10, and Figure 11, A and B, show what we interpret as two different stages of TGN release from parental stacks. To verify that the structures identified as ff-TGNs were indeed TGN cisternae, we used immunoelectron microscopy to test whether they contained the trans/TGN marker Sec7p-GFP. In Figure 11C, two immunogold-labeled Sec7p-GFP–containing cisternae seem to be in the process of separating from a stack. Figure 11D shows an ff-TGN cisterna with labeling of the Sec7p-GFP marker.
Figure 11.
Electron micrographs of ultrathin sections showing Golgi stacks or cisternae. (A and B) Sections of Epon samples showing two different stages of TGN dissociation from the parental stack. (C and D) HM20 sections with immunogold labeling of the trans/TGN marker protein Sec7p-GFP. In C the labeled cisternae seem to be peeling off from the stack, equivalent to the situation in A. (D) ff-TGN structure with Sec7p-GFP labeling, indicating that these structures can carry resident Golgi proteins after dissociating from the stack. Bar, 100 nm.
Figure 14.
View showing a slice of a reconstructed P. pastoris Golgi stack. (A) Ribosomes are excluded from the spaces between the cisternae and the tER-Golgi interface. (B) Same stack as in A, but with the border of the ribosome-excluding zone outlined. Note how the ribosomes penetrate a large fenestra in the red TGN cisterna. Bar, 100 nm.
Figure 10.
Side view of a P. pastoris Golgi stack. The TGN cisterna seems to be in the process of dissociating from the stack. Bar, 100 nm.
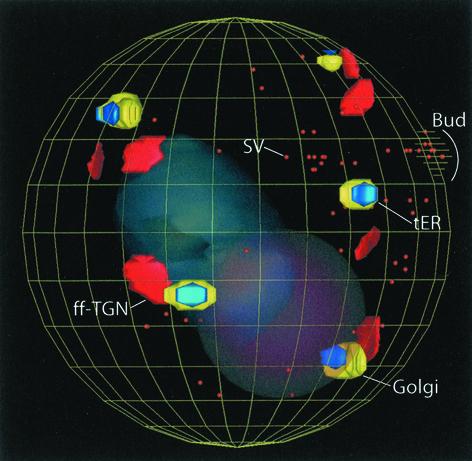
To learn more about the ff-TGNs, we analyzed 16 P. pastoris cells by serial thin-section analysis. The IMOD software was used to build 3D models of several of these cells (Figure 12). This approach has a lower resolution than tomographic analysis, but it was sufficient to identify ff-TGN cisternae and to determine their number and distribution with respect to Golgi stacks and tER sites. Most of the ff-TGNs seemed to be in the general vicinity of their putative parental stacks rather than randomly distributed throughout the cytosol (Figure 12). This result suggests that the ff-TGNs fragment before moving too far from the parental stacks, or that moving further away is a rare and/or rapid event that would seldom be observed.
Figure 12.
3D model of an entire P. pastoris cell. The model is based on serial thin-section reconstruction and shows Golgi stacks, tER sites, ff-TGNs, secretory vesicles (SV), the nucleus (faint red), and the vacuole (faint blue). The ff-TGNs are all near Golgi stacks rather than being randomly distributed throughout the cytoplasm. Note that the secretory vesicles are concentrated toward the emerging bud.
We did not investigate the detailed 3D structure of the ff-TGNs by tomographic analysis. However, in thin-section electron micrographs, structures that exhibited the morphological features of clathrin-coated vesicles (Staehelin et al., 1990) were occasionally seen adjacent to ff-TGN cisternae (Figure 13). These putative clathrin-coated vesicles were never seen next to TGN cisternae that were associated with a stack, although due to the limited number of observations we cannot rule out that such vesicles might also arise from stack-associated TGN cisternae. As seen in Figure 13, the putative clathrin-coated vesicles had a densely staining lumen similar to that of the ff-TGNs.
Figure 13.
Electron micrograph showing the apparent association of clathrin-coated vesicles with a ff-TGN. N, nucleus. Bar, 100 nm.
A Ribosome-excluding Matrix Encompasses Each tER-Golgi Unit
The coherence of the P. pastoris Golgi stacks may be due to matrix proteins that glue the cisternae together. In support of this idea, we found that each tER-Golgi unit seemed to be embedded in a “matrix” that excluded ribosomes (Figure 14). This zone of ribosome exclusion extended from the tER site to the cis most cisterna and around the rest of the adjacent Golgi stack. The putative COPII vesicles were always seen inside the exclusion zone between the tER site and the Golgi stack, whereas most of the putative COPI vesicles were located near the edge of this exclusion zone. Interestingly, ribosomes were sometimes seen to penetrate the larger fenestrae of the TGN cisternae (Figure 14). These findings support the idea that the matrix is anchored to the tER and Golgi membranes and that it has a defined thickness. Moreover, the frequent presence of ribosomes in the region between the C3 and TGN cisternae suggests that release of a TGN cisterna from a Golgi stack involves disruption of the matrix.
DISCUSSION
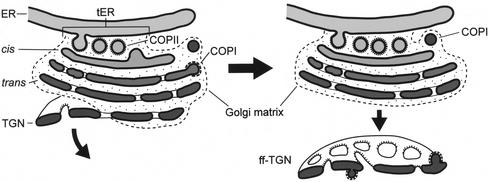
In this study, we have used state-of-the-art structural, immunocytochemical, and fluorescence microscopic techniques to analyze the early secretory pathway in P. pastoris. The principal findings include 1) the existence of a ribosome-excluding matrix that encompasses each Golgi stack and connects it to a tER site; 2) the confinement of putative COPII vesicles to the center of the tER-Golgi interface, and of putative COPI vesicles to the periphery of this interface; 3) the lack of fenestrae in cis Golgi cisternae; 4) the apparent addition of fenestrae to medial cisternae, and the relatively constant number of fenestrae in medial, trans, and TGN cisternae; 5) the presence of at least one ff-TGN cisterna in the vicinity of each Golgi stack; and 6) the apparent formation of clathrin-coated vesicles from ff-TGN cisternae. A diagrammatic interpretation of these and other results is shown in Figure 15.
Figure 15.
Interpretive model of the structural organization of P. pastoris tER sites and Golgi stacks. See the text for details.
COPII and COPI Vesicles Accumulate at the tER-Golgi Interface
The data presented herein confirm and extend the previous observation of Rossanese et al. (1999) that P. pastoris Golgi stacks are closely associated with tER sites. Indeed, each of the more than 80 Golgi stacks that we examined by serial thin-section analysis and dual-axis tomography was located adjacent to a tER site.
According to the cisternal maturation model, a new cis cisterna forms when tER-derived COPII vesicles fuse with one another and with Golgi-derived retrograde COPI vesicles (Glick and Malhotra, 1998). A predicted early intermediate in this process is a group of COPII and COPI vesicles. Such an intermediate has not previously been observed, but seems to exist in P. pastoris. As shown in Figures 1, 5, 6, and 8, the interface between a tER site and the adjacent cis cisterna contains tER-associated buds and free vesicles that are presumably generated by the COPII pathway (Schekman and Orci, 1996). Interestingly, protein coats are observed not only on all of the tER-associated buds, but also on most of the tER-derived vesicles. Smaller vesicles, presumably generated by the COPI pathway (Rothman and Wieland, 1996), are relegated to the edges of the tER-Golgi interface. In the 12 tomograms produced and analyzed in this study, we did not encounter a structure that was clearly an intermediate in the assembly of a cisterna from vesicles. However, several samples did possess cis cisternae with smaller diameters than the adjacent medial cisternae. We speculate that the initial fusion of COPII and COPI vesicles to form a new cis cisterna is a rapid, synchronous event, and that the resulting cisterna quickly adopts a mature architecture. If this interpretation is correct, our data set was not large enough to capture the short-lived intermediates in cisternal formation. Because the assembly of cis cisternae is a key prediction of the cisternal maturation model, it will be important in future studies to determine whether intermediates in cisternal assembly can be identified in P. pastoris.
In mammalian cells, presumptive intermediates in cisternal assembly are readily observed and are known as elements of the ER-Golgi intermediate compartment (ERGIC) or vesicular-tubular clusters (Hauri and Schweizer, 1992; Saraste and Kuismanen, 1992; Bannykh and Balch, 1997; Ladinsky et al., 1999). These ERGIC elements act as transport carriers by moving along microtubules from peripheral tER sites to the juxtanuclear Golgi ribbon (Presley et al., 1997; Scales et al., 1997). In P. pastoris, the Golgi stacks are directly apposed to tER sites, so there is apparently no need for long-lived ERGIC-type transport intermediates.
In addition to participating in the assembly of new cisternae, COPI vesicles have been proposed to function within the Golgi stack to carry anterograde secretory cargo (Rothman and Wieland, 1996; Pelham and Rothman, 2000; Cosson et al., 2002) and to recycle resident Golgi proteins (Glick et al., 1997; Martínez-Menárguez et al., 2001; Storrie and Nilsson, 2002). Consistent with these models, coated budding profiles were seen on P. pastoris Golgi cisternae (Figure 9). However, only relatively few vesicles were seen in the vicinity of the Golgi stacks (Figure 8). The situation is different in mammalian cells, where numerous vesicles are tethered to the Golgi cisternae (Orci et al., 1998; Ladinsky et al., 1999; Marsh et al., 2001). This difference becomes less striking when considering that a P. pastoris Golgi stack has <5% of the surface area of a mammalian Golgi stack. We have calculated that the vesicle density per unit surface area for P. pastoris Golgi stacks is ∼30% of the vesicle density per unit surface area for normal rat kidney (NRK) cell Golgi stacks (Ladinsky et al., 1999). It remains to be tested whether the vesicles around the P. pastoris Golgi stacks are indeed COPI vesicles, as we propose based on our structural criteria, and whether they carry resident Golgi proteins as predicted by the cisternal maturation model (Glick et al., 1997; Martínez-Menárguez et al., 2001; Storrie and Nilsson, 2002).
A Ribosome-excluding Matrix Links a tER Site and a Golgi Stack into a Functional Unit
Typically, budded P. pastoris cells possess four to eight tER-Golgi units, depending on the stage of the cell cycle (Figure 12; Rossanese et al., 1999). Live-cell fluorescence imaging indicated that tER-Golgi units form de novo (Bevis et al., 2002). This result was interpreted to mean that a new tER site gives rise to a new Golgi stack, but we have not been able to detect such an event in our electron micrographs. The few Golgi stacks that had only two cisternae (Figure 2) may have been recently formed, but we have never observed a tER site associated with a single cisterna or with only a cluster of COPII vesicles. Of course, any such intermediates that are very transient would be difficult to capture by electron microscopy.
It has been suggested that the formation of coherent Golgi stacks in P. pastoris may be a kinetic phenomenon caused by restriction of the COPII vesicle machinery to discrete tER sites (Rossanese et al., 1999; Glick, 2000). If tER sites are long lived then repeated rounds of COPII vesicle budding should allow new cisternae to be assembled repeatedly at the same locations. Videomicroscopy confirmed that tER sites in P. pastoris persist for at least 30 min (Bevis et al., 2002). Thus, multiple rounds of cisternal assembly next to a tER site could potentially generate an ordered Golgi stack, provided that diffusion of the cisternae is slow relative to cisternal maturation. This kinetic model can explain the observation that in S. cerevisiae, COPII vesicle budding occurs throughout the ER and the Golgi cisternae are scattered throughout the cytoplasm (Rossanese et al., 1999; Rambourg et al., 2001). However, this model does not account for the close association and regular spacing between tER sites and cis, medial, and trans cisternae in P. pastoris. We therefore propose that additional physical constraints help to generate coherent tER-Golgi units.
Each P. pastoris tER-Golgi unit is embedded in a ribosome-free zone (Figures 14 and 15) that is probably created by a dense proteinaceous network. Similarly, plant Golgi stacks are completely surrounded by a ribosome-excluding Golgi matrix (Staehelin and Moore, 1995), which may protect the stacks from shearing as they are propelled through the cytoplasm (Nebenfuhr and Staehelin, 2001). In mammalian cells, a similar Golgi matrix excludes ribosomes (Lucocq et al., 1987) and is thought to contain structural proteins such as spectrin and ankyrin as well as the Golgin family of coiled coil proteins (DeMatteis and Morrow, 2000; Barr et al., 2001). This matrix has been implicated in generating the higher order architecture of the Golgi apparatus and in Golgi inheritance during mitosis (Seemann et al., 2000, 2002). P. pastoris is a promising experimental system for defining the composition and function of the Golgi matrix.
Membrane Continuities between Golgi Cisternae Are Absent in P. pastoris
Despite the close apposition of Golgi cisternae, we found no evidence for membrane continuities that would link the lumenal spaces of different cisternae. Unlike mammalian Golgi stacks, which are laterally interconnected to form a ribbon (Rambourg and Clermont, 1997), P. pastoris Golgi stacks are separate from one another. Golgi tubules are seen on the C2 and C3 cisternae (Figures 8 and 9), but these tubules tend to be relatively short, and they do not provide lumenal connections between either homologous or heterologous cisternae. These results argue against models for intra-Golgi transport that invoke membrane continuities between cisternae (Beznoussenko and Mironov, 2002).
P. pastoris Golgi Cisternae Show Properties Suggestive of Cisternal Maturation
Each cisterna of a P. pastoris Golgi stack exhibits distinct characteristics, and these characteristics change progressively across the stack. Such a pattern fits with multiple models for intra-Golgi trafficking. However, the cisternal maturation model makes specific predictions that can be tested by electron microscopy. One prediction is that many resident Golgi proteins should not be confined to individual cisternae, but should show gradients of distribution across the stack (Glick et al., 1997; Weiss and Nilsson, 2000). A second prediction is that the morphological variations between cisternae should be consistent with a precursor–product relationship.
Resident Golgi protein distributions in P. pastoris were analyzed by immunolocalizing the membrane-anchored mannosyltransferase Och1p and the peripheral membrane protein Sec7p. Och1p and Sec7p have previously been reported to be cis and trans-Golgi markers, respectively, based on fluorescence microscopy and biochemical data (Franzusoff et al., 1991; Nakayama et al., 1992; Gaynor et al., 1994; Harris and Waters, 1996; Rossanese et al., 1999). Herein, we show by electron microscopy that Och1p localizes predominantly to C1 and C2 cisternae, whereas Sec7p localizes predominantly to C3 and TGN cisternae (Figures 4 and 11, C and D). This compartmentation of the P. pastoris Golgi is similar to that described for the Golgi of S. cerevisiae, mammalian and plant cells (Staehelin et al., 1990; Farquhar and Palade, 1998; Brigance et al., 2000). As predicted by the cisternal maturation model, the marker proteins are not confined to single cisternae, but rather are distributed across the entire Golgi with concentration peaks at opposite sides of the stacks (Figure 4). This pattern resembles the distribution profiles of Golgi glycosyltransferases in mammalian cells (Nilsson et al., 1993; Rabouille et al., 1995).
To analyze morphological variations in P. pastoris Golgi cisternae, we focused on fenestration patterns. Golgi fenestrae are generated by an unknown mechanism that has been postulated to involve either “periplasmic fusion” of the lumenal surfaces of cisternal membranes (Rothman and Warren, 1994) or fusion of Golgi tubules with their parental cisternae (Weidman et al., 1993). In P. pastoris, fenestrae are absent from the C1 cisternae but are present in the C2, C3, and TGN cisternae (Figures 2 and 9). The fenestrae remain roughly constant in number from the C2 through TGN cisternae, but their distribution progressively changes. In the C2 cisternae, fenestrae are found in a narrow ring near the cisternal rims; in the C3 cisternae, this ring is wider but the cisternal centers remain largely devoid of fenestrae; and in the TGN cisternae, fenestrae are evenly distributed (Figure 9). This result suggests that Golgi fenestrae in P. pastoris form at the rims of the cisternae and then migrate inward as the cisternae mature.
P. pastoris Golgi Stacks Seem to Shed TGN Cisternae
Some of our most striking electron micrographs seem to capture different stages in the release of intact TGN cisternae from P. pastoris Golgi stacks (Figures 1, 10, and 11). We propose that after a TGN cisterna is released from a Golgi stack, it becomes a ff-TGN in the cytoplasm (Figures 11, 12, 13). The shedding of TGN cisternae was recently confirmed in a videomicroscopy study of living P. pastoris cells expressing a Sec7p-DsRed fusion protein (Bevis et al., 2002). TGN shedding is consistent with the cisternal maturation model, which posits that a TGN cisterna must be removed for every new cisterna that is added to the cis side of the stack (Glick et al., 1997). Images consistent with TGN shedding have previously been obtained with other cell types (Mollenhauer and Morré, 1991), but our work goes further by showing that Sec7p is present both on Golgi stack-associated TGN cisternae and on ff-TGNs, strongly suggesting that Golgi stacks give rise to ff-TGNs.
One or two ff-TGN cisternae are found in the vicinity of each tER-Golgi unit, implying that ff-TGNs persist for at least as long as it takes to create a new cis cisterna. After their release from the Golgi stacks, ff-TGNs presumably mature into secretory carriers. This event may trigger active translocation of the ff-TGNs toward regions of polarized growth (Bevis et al., 2002). As they mature, ff-TGNs produce vesicles that seem to be clathrin-coated (Figure 13). To our knowledge, this result is the first reported visualization of an organellar site for clathrin-coated vesicle budding in yeast. We have observed clathrin-coated vesicles budding only from ff-TGNs, never from Golgi stack-associated TGN cisternae (Figure 13). This specialization is reminiscent of the NRK cell Golgi, in which clathrin-coated vesicles bud exclusively from the trans-most C7 cisternae and not from the adjacent C6 cisternae (Ladinsky et al., 1999). Based on these findings, we postulate that ff-TGNs are major sites for cargo sorting and packaging in P. pastoris. Our work is consistent with genetic data from S. cerevisiae showing that clathrin functions in export from the TGN and/or recycling from a post-TGN compartment (Costaguta et al., 2001; Deloche et al., 2001; Pelham, 2002; Valdivia et al., 2002;).
When TGN cisternae are released from the stacks, they must “escape” from the ribosome-excluding Golgi matrix. This release seems to involve a breakdown of the matrix at the trans side of the stacks, as evidenced by the penetration of ribosomes into the space between the C3 and TGN cisternae (Figures 1, 11, and 15). Disassembly of the Golgi matrix may therefore control the release of TGN cisternae.
Cross-Species Comparisons Highlight Conserved and Nonconserved Features of the tER-Golgi System
Comparisons between species have frequently offered insights into Golgi organization and dynamics (Mollenhauer and Morré, 1991; Munro, 2002). The data reported herein for P. pastoris can be combined with previous work, particularly tomographic studies of NRK cells (Ladinsky et al., 1999), to distinguish between conserved and nonconserved features of the Golgi.
Golgi stacks show a compositional polarity in every species examined. Some aspects of this polarity are statistical in nature, because resident Golgi proteins tend to be distributed in overlapping gradients of concentration across the stack rather than being restricted to specific cisternae (Rabouille et al., 1995). However, sharper boundaries are seen for the distribution of coat proteins. In both NRK cells and P. pastoris, budding COPI vesicles are restricted to the cis through trans cisternae of the Golgi stack, whereas budding clathrin-coated vesicles are restricted to the most distal TGN compartment (Ladinsky et al., 1999). Within the framework of the cisternal maturation model, this observation suggests that a rapid functional switch occurs during the conversion of a trans to a TGN cisterna.
Certain structural properties of the Golgi are also conserved. An example is the fenestration of Golgi cisternae. Unlike the Golgi fenestrae in P. pastoris, the Golgi fenestrae in NRK cells are more prevalent in cis and trans than in medial cisternae, and they are frequently aligned from one cisterna to the next (Ladinsky et al., 1999), suggesting that the processes controlling Golgi fenestration are somewhat different in the two species. Regardless of their origin, the widespread occurrence of Golgi fenestrae suggests that they are important for Golgi function. We have previously suggested that fenestrae may serve as “spot welds” to prevent the cisternae from swelling (Ladinsky et al., 1999).
A comparison of Golgi structures in P. pastoris and mammalian cells also reveals key differences. For example, the Golgi stacks in mammalian cells are laterally interconnected to form a ribbon (Rambourg et al., 1997; Ladinsky et al., 1999), whereas P. pastoris Golgi stacks are individual units. Moreover, ER membranes in mammalian cells are intimately associated with Golgi cisternae at the trans side of the stack (Rambourg et al., 1997; Ladinsky et al., 1999; Marsh et al., 2001), but no such association is seen in P. pastoris. These results suggest that interstack connections and associations between the ER and the trans-Golgi are cell type-specific phenomena that are not essential for basic Golgi function. Other differences between the mammalian Golgi and the P. pastoris Golgi are quantitative rather than qualitative. Most notably, NRK cell Golgi stacks have seven cisternae (Ladinsky et al., 1999), whereas P. pastoris Golgi stacks have three to four cisternae. It is attractive to speculate that in all eukaryotes, Golgi cisternae pass through four biochemically distinct stages of maturation that can be defined as cis, medial, trans, and TGN. In this scenario, P. pastoris contains a “minimal” Golgi that has all of the standard machinery but few of the optional features.
Supplementary Material
Acknowledgments
We thank Drs. David Mastronarde and Tom Giddings for technical advice, and Dr. Pam Silver for providing the anti-GFP antibody. We are grateful to members of the Glick and Staehelin laboratories for advice. This work was supported by National Institutes of Health grants GM-61156 (to B.S.G.) and GM-61306 (to L.A.S.), and by a Danish Natural Science Research Council grant (to S.M.).
Abbreviations used: 3D, three-dimensional; ERGIC, endoplasmic reticulum-Golgi intermediate compartment; ff-TGN, free-floating trans-Golgi network; NRK, normal rat kidney; tER, transitional endoplasmic reticulum. Movie 1. The movie shows all the tomographic slices of the tomogram illustrated in Figure 5. Visualized this way, it is easier to define structures such as tubules and fenestrae.
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
References
- Bannykh, S.I., and Balch, W.E. (1997). Membrane dynamics at the endoplasmic reticulum Golgi interface. J. Cell Biol. 138, 1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh, S.I., Rowe, T., and Balch, W.E. (1996). The organization of endoplasmic reticulum export complexes. J. Cell Biol. 135, 19-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C. (2002). COPII-dependent transport from the endoplasmic reticulum. Curr. Opin. Cell Biol. 14, 417-422. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., Preisinger, C., Kopajtich, R., and Korner, R. (2001). Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 155, 885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, E.G., and Roth, J.P.D.D. (1997). The Golgi Apparatus. Boston: Birkhäuser Verlag.
- Bevis, B.J., Hammond, A.T., Reinke, C.A., and Glick, B.S. (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4, 750-756. [DOI] [PubMed] [Google Scholar]
- Beznoussenko, G.V., and Mironov, A.A. (2002). Models of intracellular transport and evolution of the Golgi complex. Anat. Rec. 268, 226-238. [DOI] [PubMed] [Google Scholar]
- Brigance, W.T., Barlowe, C., and Graham, T.R. (2000). Organization of the yeast Golgi complex into at least four functionally distinct compartments. Mol. Biol. Cell 11, 171-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, P., Amherdt, M., Rothman, J.E., and Orci, L. (2002). A resident Golgi protein is excluded from peri-Golgi vesicles in NRK cells. Proc. Natl. Acad. Sci. USA 99, 12831-12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta, G., Stefan, C.J., Bensen, E.S., Emr, S.D., and Payne, G.S. (2001). Yeast GGA coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell 12, 1885-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMatteis, M.A., and Morrow, J.S. (2000). Spectrin tethers and mesh in the biosynthetic pathway. J. Cell Sci. 113, 2331-2343. [DOI] [PubMed] [Google Scholar]
- Deloche, O., Yeung, B.G., Payne, G.S., and Schekman, R. (2001). Vps10p transport from the trans-Golgi network to the endosome is mediated by clathrin-coated vesicles. Mol. Biol. Cell 12, 475-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar, M.G., and Hauri, H.P. (1997). Protein sorting and vesicular traffic in the Golgi apparatus. In: The Golgi Apparatus, ed. E.G. Berger and J.P. Roth, Boston: Birkhäuser Verlag, 63-129.
- Farquhar, M.G., and Palade, G.E. (1998). The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 8, 2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff, A., Redding, K., Crosby, J., Fuller, R.S., and Schekman, R. (1991). Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 112, 27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, E.C., te Heesen, S., Graham, T.R., Aebi, M., and Emr, S.D. (1994). Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J. Cell Biol. 127, 653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, P.F.C. (1972). Reconstruction of a 3-dimensional structure from projections and its application to electron-microscopy. 2. Direct Methods. Proc. R. Soc. Lond. B Biol. Sci. 182, 89-102. [DOI] [PubMed] [Google Scholar]
- Gilkey, J.C., and Staehelin, L.A. (1986). Advances in ultra-rapid freezing for the preservation of cellular ultrastructure. J. Electron Microsc. Tech. 3, 177-210. [Google Scholar]
- Glick, B.S. (1996). Cell biology: alternatives to baker's yeast. Curr. Biol. 6, 1570-1572. [DOI] [PubMed] [Google Scholar]
- Glick, B.S. (2000). Organization of the Golgi apparatus. Curr. Opin. Cell Biol. 12, 450-456. [DOI] [PubMed] [Google Scholar]
- Glick, B.S., Elston, T., and Oster, G. (1997). A cisternal maturation mechanism can explain the asymmetry of the Golgi stack. FEBS Lett. 414, 177-181. [DOI] [PubMed] [Google Scholar]
- Glick, B.S., and Malhotra, V. (1998). The curious status of the Golgi apparatus. [letter; comment]. Cell 95, 883-889. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., McCollum, D., Spong, A.P., Heyman, J.A., and Subramani, S. (1992). Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast 8, 613-628. [DOI] [PubMed] [Google Scholar]
- Hammond, A.T., and Glick, B.S. (2000). Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11, 3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.L., and Waters, M.G. (1996). Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J. Cell Biol. 132, 985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri, H.P., and Schweizer, A. (1992). The endoplasmic reticulum-Golgi intermediate compartment. Curr. Opin. Cell Biol. 4, 600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde, D.N., and McIntosh, J.R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- Ladinsky, M.S., Mastronarde, D.N., McIntosh, J.R., Howell, K.E., and Staehelin, L.A. (1999). Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 144, 1135-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq, J.M., Pryde, J.G., Berger, E.G., and Warren, G. (1987). A mitotic form of the Golgi-apparatus in HeLa cells. J. Cell Biol. 104, 865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, B.J., and Howell, K.E. (2002). The mammalian Golgi-complex debates. Nat. Rev. Mol. Cell Biol. 3, 789-795. [DOI] [PubMed] [Google Scholar]
- Marsh, B.J., Mastronarde, D.N., Buttle, K.F., Howell, K.E., and McIntosh, J.R. (2001). Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl. Acad. Sci. USA 98, 2399-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Menarguez, J.A., Prekeris, R., Oorschot, V.M., Scheller, R., Slot, J.W., Geuze, H.J., and Klumperman, J. (2001). Peri-Golgi vesicles contain retrograde but not anterograde proteins consistent with the cisternal progression model of intra-Golgi transport. J. Cell Biol. 155, 1213-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343-352. [DOI] [PubMed] [Google Scholar]
- McEwen, B.F., and Marko, M. (1999). Three-dimensional transmission electron microscopy and its application to mitosis research. Method Cell Biol. 61, 81-111. [DOI] [PubMed] [Google Scholar]
- Mellman, I., and Simons, K. (1992). The Golgi complex: in vitro veritas? Cell 68, 829-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer, H.H., and Morré, D.J. (1991). Perspectives on Golgi-apparatus form and function. J. Electron Microsc. Tech. 17, 2-14. [DOI] [PubMed] [Google Scholar]
- Munro, S. (2001). What can yeast tell us about N-linked glycosylation in the Golgi apparatus? FEBS Lett. 498, 223-227. [DOI] [PubMed] [Google Scholar]
- Munro, S. (2002). More than one way to replicate the Golgi apparatus. Nat. Cell Biol. 4, E223-E224. [DOI] [PubMed] [Google Scholar]
- Nakayama, K., Nagasu, T., Shimma, Y., Kuromitsu, J., and Jigami, Y. (1992). OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 11, 2511-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr, A., and Staehelin, L.A. (2001). Mobile factories: Golgi dynamics in plant cells. Trends Plant Sci. 6, 160-167. [DOI] [PubMed] [Google Scholar]
- Nilsson, T., Pypaert, M., Hoe, M.H., Slusarewicz, P., Berger, E.G., and Warren, G. (1993). Overlapping distribution of two glycosyltransferases in the Golgi apparatus of HeLa cells. J. Cell Biol. 120, 5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci, L., Perrelet, A., and Rothman, J.E. (1998). Vesicles on strings: morphological evidence for processive transport within the Golgi stack. Proc. Natl. Acad. Sci. USA 95, 2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Mastronarde, D.N., Kang, B.H., Bednarek, S.Y., and Staehelin, L.A. (2001). Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13, 2033-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade, G. (1975). Intracellular aspects of the process of protein synthesis. Science 189, 347-358. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R. (2001). Traffic through the Golgi apparatus. J. Cell Biol. 155, 1099-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H.R.B. (2002). Insights from yeast endosomes. Curr. Opin. Cell Biol. 14, 454-462. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R.B., and Rothman, J.E. (2000). The debate about transport in the Golgi: two sides of the same coin? Cell 102, 713-719. [DOI] [PubMed] [Google Scholar]
- Presley, J.F., Cole, N.B., Schroer, T.A., Hirschberg, K., Zaal, K.J., and Lippincott-Schwartz, J. (1997). ER-to-Golgi transport visualized in living cells. [see comments]. Nature 389, 81-85. [DOI] [PubMed] [Google Scholar]
- Rabouille, C., Hui, N., Hunte, F., Kieckbusch, R., Berger, E.G., Warren, G., and Nilsson, T. (1995). Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J. Cell Sci. 108, 1617-1627. [DOI] [PubMed] [Google Scholar]
- Rambourg, A., and Clermont, Y. (1997). Three-dimensional structure of the Golgi apparatus in mammalian cells. In: The Golgi Apparatus, ed. E.G. Berger and J.P. Roth, Boston: Birkhäuser Verlag, 37-61.
- Rambourg, A., Clermont, Y., and Kepes, F. (1993). Modulation of the Golgi-apparatus in Saccharomyces cerevisiae Sec7 mutants as seen by 3-dimensional electron-microscopy. Anat. Rec. 237, 441-452. [DOI] [PubMed] [Google Scholar]
- Rambourg, A., Jackson, C.L., and Clermont, Y. (2001). Three dimensional configuration of the secretory pathway and segregation of secretion granules in the yeast Saccharomyces cerevisiae. J. Cell Sci. 114, 2231-2239. [DOI] [PubMed] [Google Scholar]
- Rossanese, O.W., Soderholm, J., Bevis, B.J., Sears, I.B., O'Connor, J., Williamson, E.K., and Glick, B.S. (1999). Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 145, 69-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J.E., and Warren, G. (1994). Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr. Biol. 4, 220-233. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E., and Wieland, F.T. (1996). Protein sorting by transport vesicles. Science 272, 227-234. [DOI] [PubMed] [Google Scholar]
- Saraste, J., and Kuismanen, E. (1992). Pathways of protein sorting and membrane traffic between the rough endoplasmic reticulum and the Golgi complex. Semin. Cell Biol. 3, 343-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales, S.J., Pepperkok, R., and Kreis, T.E. (1997). Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell 90, 1137-1148. [DOI] [PubMed] [Google Scholar]
- Schekman, R., and Orci, L. (1996). Coat proteins and vesicle budding. Science 271, 1526-1533. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., Damelin, M., Kahana, J., Taura, T., and Silver, P.A. (1999). Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19, 1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, 1022-1026. [DOI] [PubMed] [Google Scholar]
- Seemann, J., Pypaert, M., Taguchi, T., Malsam, J., and Warren, G. (2002). Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, 848-851. [DOI] [PubMed] [Google Scholar]
- Soderholm, J., Bevis, B.J., and Glick, B.S. (2001). Vector for pop-in/pop-out gene replacement in Pichia pastoris. Biotechniques 31, 306-310. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., Giddings, T.H., Jr., Kiss, J.Z., and Sack, F.D. (1990). Macromolecular differentiation of Golgi stacks in root tips of Arabidopsis and Nicotiana seedlings as visualized in high pressure frozen and freeze-substituted samples. Protoplasma 157, 75-91. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Moore, I. (1995). The plant Golgi-apparatus: structure, functional-organization and trafficking mechanisms. Annu. Rev. Plant Physiol. 46, 261-288. [Google Scholar]
- Storrie, B., and Nilsson, T. (2002). The Golgi apparatus: balancing new with old. Traffic 3, 521-529. [DOI] [PubMed] [Google Scholar]
- Valdivia, R.H., Baggott, D., Chuang, J.S., and Schekman, R.W. (2002). The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell 2, 283-294. [DOI] [PubMed] [Google Scholar]
- Weidman, P., Roth, R., and Heuser, J. (1993). Golgi membrane dynamics imaged by freeze-etch electron microscopy: views of different membrane coatings involved in tubulation versus vesiculation. Cell 75, 123-133. [PubMed] [Google Scholar]
- Weiss, M., and Nilsson, T. (2000). Protein sorting in the Golgi apparatus: a consequence of maturation and triggered sorting. FEBS Lett. 486, 2-9. [DOI] [PubMed] [Google Scholar]
- Zhang, G.F., and Staehelin, L.A. (1992). Functional compartmentation of the Golgi-apparatus of plant-cells: immunocytochemical analysis of high-pressure frozen-substituted and freeze-substituted sycamore maple suspension-culture cells. Plant Physiol. 99, 1070-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.