Abstract
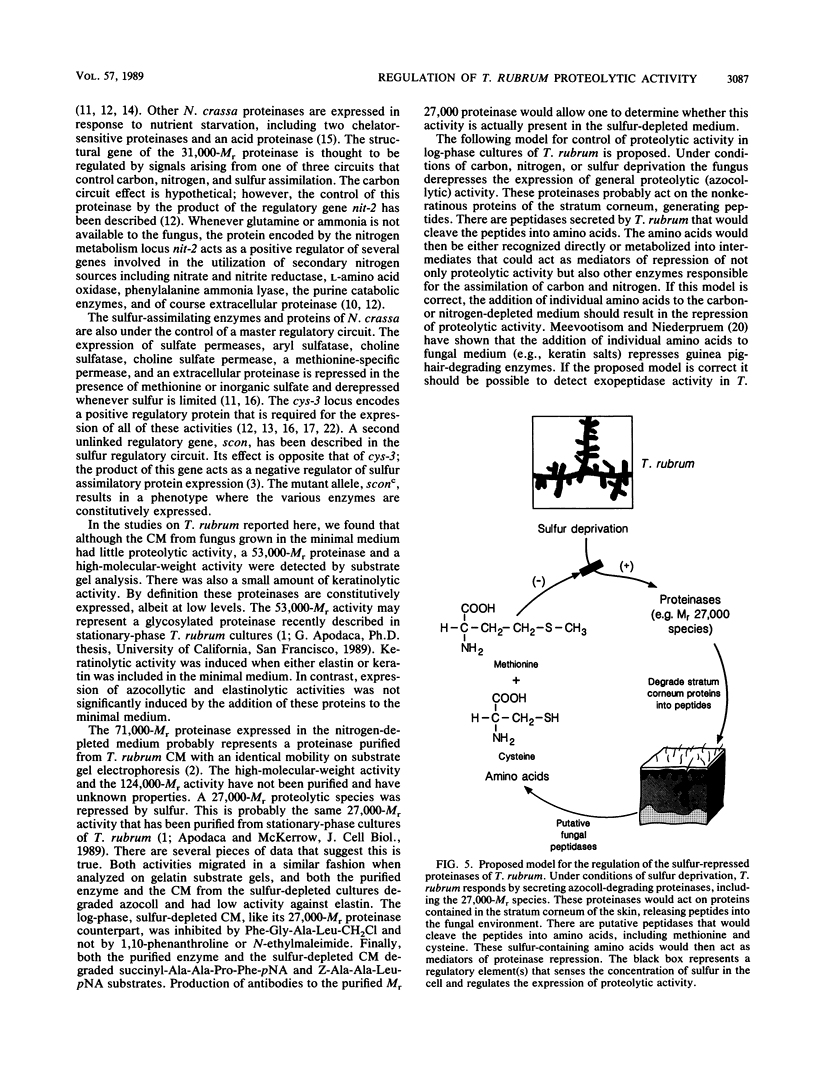
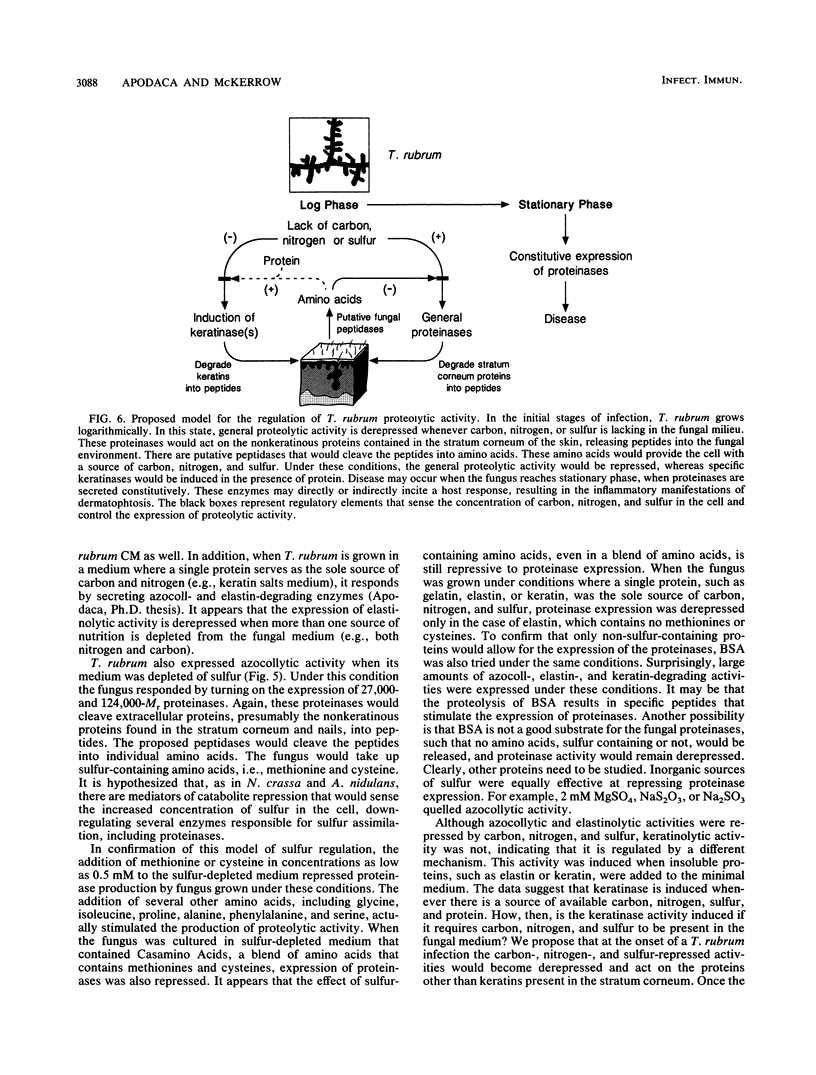
Trichophyton rubrum is the most common dermatophyte of humans and normally colonizes the superficial layers of the epidermis (stratum corneum). Several proteinases with a possible role in the metabolism of host proteins have been purified from this fungus. The regulation of these enzymes and their role in fungal metabolism were studied at the biochemical level. General proteolytic (azocollytic) activity was repressed when log-phase cultures of T. rubrum were grown in a minimal medium that contained readily metabolized sources of carbon, nitrogen, sulfur, and phosphorus. When either carbon, nitrogen, or sulfur was deleted from this minimal medium, azocollytic activity was derepressed. In all cases a high-molecular-weight activity (Mr, greater than 200,000) was expressed. A 71,000-Mr proteinase was observed in nitrogen-depleted cultures, and proteolytic species of Mr 124,000 and 27,000 were secreted in sulfur-depleted cultures. The addition of either inorganic (MgSO4, Na2SO3, NaS2O3) or organic (methionine, cysteine) sulfur to the sulfur-depleted medium repressed the expression of azocollytic activity. In contrast, keratinolytic activity was not repressed by carbon, nitrogen, or sulfur but instead was induced when a protein source was included in the minimal medium. Stationary-phase cultures of T. rubrum secreted all proteolytic activities constitutively. Unlike log-phase cultures, the stationary-phase cultures secreted azocollytic, elastinolytic, and keratinolytic activity in minimal medium. These activities fell in the carbon-, nitrogen-, and phosphorous-depleted media but remained high in sulfur-depleted medium. The following model is proposed for the regulation of T. rubrum proteolytic activity. In the initial stages of infection, T. rubrum grows logarithmically. In this state, proteolytic activity is derepressed whenever carbon, nitrogen, or sulfur is lacking in the fungal milieu. The general proteinases produced would act on the nonkeratinous proteins in the stratum corneum. There are probably peptidases, as yet unidentified, that would cleave the peptides generated by the initial proteolysis into amino acids. These amino acids would provide the cell with a source of carbon, nitrogen, and sulfur. Under these conditions, the expression of general proteinases would be repressed, whereas specific keratinases would be induced in this nutrient-rich environment. Disease may occur when the fungus reaches stationary phase, when proteinases are secreted constitutively. These enzymes may directly or indirectly incite a host response, resulting in the inflammatory manifestations of dermatophytosis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apodaca G., McKerrow J. H. Purification and characterization of a 27,000-Mr extracellular proteinase from Trichophyton rubrum. Infect Immun. 1989 Oct;57(10):3072–3080. doi: 10.1128/iai.57.10.3072-3080.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahi M., Lindquist R., Fukuyama K., Apodaca G., Epstein W. L., McKerrow J. H. Purification and characterization of major extracellular proteinases from Trichophyton rubrum. Biochem J. 1985 Nov 15;232(1):139–144. doi: 10.1042/bj2320139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton E. G., Metzenberg R. L. Novel mutation causing derepression of several enzymes of sulfur metabolism in Neurospora crassa. J Bacteriol. 1972 Jan;109(1):140–151. doi: 10.1128/jb.109.1.140-151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. L. Regulation of intracellular and extracellular neutral and alkaline proteases in Aspergillus nidulans. J Gen Microbiol. 1973 Dec;79(2):311–320. doi: 10.1099/00221287-79-2-311. [DOI] [PubMed] [Google Scholar]
- Cohen B. L. The neutral and alkaline proteases of Aspergillus nidulans. J Gen Microbiol. 1973 Aug;77(2):521–528. doi: 10.1099/00221287-77-2-521. [DOI] [PubMed] [Google Scholar]
- Drucker H. Regulation of exocellular proteases in Neurospora crassa: induction and repression of enzyme synthesis. J Bacteriol. 1972 Jun;110(3):1041–1049. doi: 10.1128/jb.110.3.1041-1049.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker H. Regulation of exocellular proteases in Neurospora crassa: metabolic requirements of the process. J Bacteriol. 1975 Jun;122(3):1117–1125. doi: 10.1128/jb.122.3.1117-1125.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker H. Regulation of exocellular proteases in Neurospora crassa: role of Neurospora proteases in induction. J Bacteriol. 1973 Nov;116(2):593–599. doi: 10.1128/jb.116.2.593-599.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 May;7(5):1691–1696. doi: 10.1128/mcb.7.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. A., Marzluf G. A. Control of the synthesis of a single enzyme by multiple regulatory circuits in Neurospora crassa. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1240–1244. doi: 10.1073/pnas.72.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. A., Marzluf G. A. Regulation of a sulfur-controlled protease in Neurospora crassa. J Bacteriol. 1973 Nov;116(2):785–789. doi: 10.1128/jb.116.2.785-789.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketter J. S., Marzluf G. A. Molecular cloning and analysis of the regulation of cys-14+, a structural gene of the sulfur regulatory circuit of Neurospora crassa. Mol Cell Biol. 1988 Apr;8(4):1504–1508. doi: 10.1128/mcb.8.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg R. A., Eirich L. D., Price J. S., Wolfinbarger L., Jr, Drucker H. Alkaline protease from Neurospora crassa. Purification and partial characterization. J Biol Chem. 1981 Jan 25;256(2):811–814. [PubMed] [Google Scholar]
- Lindberg R. A., Rhodes W. G., Eirich L. D., Drucker H. Extracellular acid proteases from Neurospora crassa. J Bacteriol. 1982 Jun;150(3):1103–1108. doi: 10.1128/jb.150.3.1103-1108.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATILE P. INTRAZELLULAERE LOKALISATION PROTEOLYTISCHER ENZYME VON NEUROSPORA CRASSA. I. FUNKTION UND SUBZELLULAERE VERTEILUNG PROTEOLYTISCHER ENZYME. Z Zellforsch Mikrosk Anat. 1965 Mar 16;65:884–896. [PubMed] [Google Scholar]
- Marzluf G. A., Metzenberg R. L. Positive control by the cys-3 locus in regulation of sulfur metabolism in Neurospora. J Mol Biol. 1968 Apr 28;33(2):423–437. doi: 10.1016/0022-2836(68)90199-x. [DOI] [PubMed] [Google Scholar]
- Meevootisom V., Niederpruem D. J. Control of exocellular proteases in dermatophytes and especially Trichophyton rubrum. Sabouraudia. 1979 Jun;17(2):91–106. doi: 10.1080/00362177985380141. [DOI] [PubMed] [Google Scholar]
- Minocha Y., Pasricha J. S., Mohapatra L. N., Kandhari K. C. Proteolytic activity of dermatophytes and its role in the pathogenesis of skin lesions. Sabouraudia. 1972 Mar;10(1):79–85. doi: 10.1080/00362177285190161. [DOI] [PubMed] [Google Scholar]
- Paietta J. V., Akins R. A., Lambowitz A. M., Marzluf G. A. Molecular cloning and characterization of the cys-3 regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 Jul;7(7):2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippon J. W., Garber E. D. Dermatophyte pathogenicity as a function of mating type and associated enzymes. J Invest Dermatol. 1969 Dec;53(6):445–448. doi: 10.1038/jid.1969.173. [DOI] [PubMed] [Google Scholar]
- Sanyal A. K., Das S. K., Banerjee A. B. Purification and partial characterization of an exocellular proteinase from Trichophyton rubrum. Sabouraudia. 1985 Jun;23(3):165–178. doi: 10.1080/00362178585380271. [DOI] [PubMed] [Google Scholar]