SUMMARY
The primate APOBEC3 gene locus encodes a family of proteins (APOBEC3A-H) with various antiviral and anti-retroelement activities. Here, we trace the evolution of APOBEC3H activity in hominoids to identify a human-specific loss of APOBEC3H antiviral activity. Reconstruction of the predicted ancestral human APOBEC3H protein shows that human ancestors encoded a stable form of this protein with potent antiviral activity. Subsequently, the antiviral activity of APOBEC3H was lost via two polymorphisms that are each independently sufficient to destabilize the protein. Nonetheless, an APOBEC3H allele that encodes a stably expressed protein is still maintained at high frequency, primarily in African populations. This stable APOBEC3H protein has potent activity against retroviruses and retrotransposons, including HIV and LINE-1 elements. The surprising finding that APOBEC3H antiviral activity has been lost in the majority of humans may have important consequences for our susceptibility to retroviral infections as well as ongoing retroelement proliferation in the human genome.
INTRODUCTION
A substantial fraction of the human genome is comprised of active and extinct retroelements, a result of numerous germline insertions by both endogenous retroviruses and non-LTR retrotransposons (such as LINE and SINE elements) (Lander et al., 2001). Due to the potentially detrimental effects of the integration of mobile elements, such as insertional mutagenesis and an increase in the likelihood of ectopic recombination events (Boissinot et al., 2006), restricting the replication of mobile retroviral elements by host genes has likely been an ongoing source of genetic conflict during primate evolution. In particular, primates have evolved an expanded locus of genes, the APOBEC3 locus, that encodes seven genes in humans (APOBEC3A to APOBEC3H) that can inhibit retroviruses and retroelements through cytidine deamination and other mechanisms (Malim and Emerman, 2008). The role of APOBEC3 proteins in limiting the success of retroviral invaders in vivo has been demonstrated in mice where APOBEC3−/− mice have increased susceptibility to the replication of the mouse retroviral pathogen, mouse mammary tumor virus (MMTV) (Okeoma et al., 2007).
The human immunodeficiency virus (HIV) can successfully infect APOBEC3-expressing cells because the virally encoded Vif protein is able to cause destruction of APOBEC3 proteins by targeting APOBEC3 proteins to a proteaseome-mediated degradation pathway (reviewed in (Harris and Liddament, 2004; Malim and Emerman, 2008)). Despite efficient inactivation by HIV Vif, APOBEC3 proteins remain important in vivo because viruses that no longer encode a functional Vif protein are sensitive to APOBEC3-mediated restriction and bear hallmarks of hypermutation due to cytidine deamination (Simon et al., 2005).
Genetic conflict between host and pathogen drives rapid change in interacting host and pathogen proteins (rapid evolution) as they attempt to increase or decrease interactions with one another in the battle for dominance. Therefore, a signature of positive selection is often observed for host proteins that are directly involved in pathogen defense. APOBEC3G and APOBEC3H genes, in particular, demonstrate strong signatures of positive selection in primates that are likely the result of successive sweeps of APOBEC3 alleles with antiviral activity against an ever-changing array of rapidly evolving retroviruses and retroelements (OhAinle et al., 2006; Sawyer et al., 2004). These signals of adaptive evolution are present throughout primate phylogeny, suggesting that the conflict of primate APOBEC3 genes with exogenous retroviruses and endogenous retroelements is ancient and has taken place over many millions of years. This ancient conflict has shaped the evolution of a landscape of APOBEC3 genes with diverse antiviral functions and has important implications for the susceptibility of humans to current retroviral pathogens, such as HIV.
Despite the fact that the APOBEC3H gene is conserved in mammals and shows positive selection throughout primate evolution, previous work from our lab and others showed that whereas rhesus macaque APOBEC3H is an efficient retroviral inhibitor, human APOBEC3H shows little activity against retroviruses and LINE-1 elements (Dang et al., 2008; Dang et al., 2006; Kinomoto et al., 2007; Muckenfuss et al., 2006; OhAinle et al., 2006; Virgen and Hatziioannou, 2007) . The lack of antiviral activity of human APOBEC3H correlates with its low steady-state expression at the protein level although mRNA levels between human APOBEC3H and macaque APOBEC3H are similar (OhAinle et al., 2006). Moreover, the protein is stable and enzymatically active in bacteria (OhAinle et al., 2006). Thus, it appears that humans have lost a functional APOBEC3 gene that has potent antiviral activity in other primates.
Here, we investigated the evolutionary history of human APOBEC3H. We found that the stability of the human APOBEC3H protein was independently lost twice during recent human evolution through the acquisition of two different amino acid mutations that decrease the half-life of the protein. Moreover, we reconstructed the sequence of the human ancestral APOBEC3H protein and showed that it is well expressed and is very active against retroviruses and the prolific primate non-LTR retroelement, LINE-1. Despite the presence of two inactivating mutations in APOBEC3H alleles in humans, a sequence survey of the APOBEC3H alleles from several different human populations reveals the existence of a stable APOBEC3H allele currently circulating in the human population. This allele lacks either destabilizing mutation, is active against LINE-1 elements and HIV, and is sensitive to the HIV Vif protein. This stable and active APOBEC3H allele is more prominent in certain world populations, particularly in African populations and may reflect local selective pressures on APOBEC3H alleles across world populations. Together, these data suggest that our recent human ancestors possessed an APOBEC3H allele that was active against modern retroviruses and retrotransposons, but whose function is now lost in the majority of the human population.
RESULTS
Human APOBEC3H Protein is Uniquely Unstable
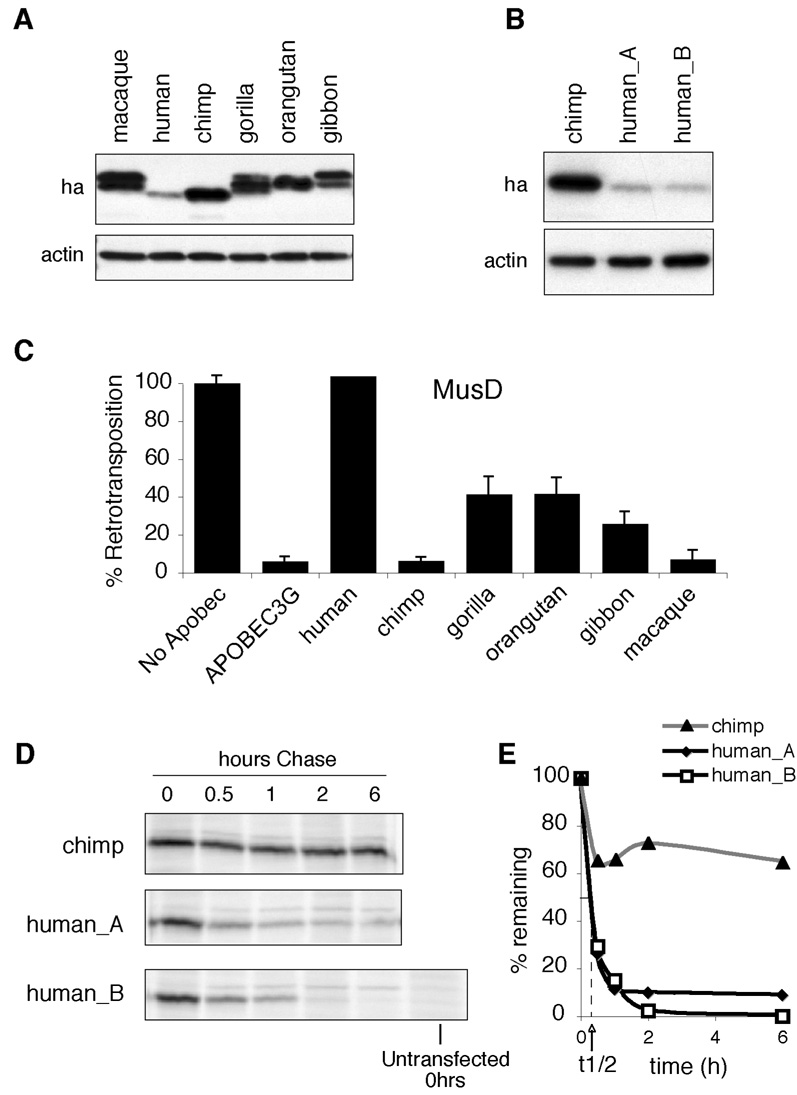
Rhesus macaque APOBEC3H is an efficient inhibitor of retroelements, but human APOBEC3H is not (OhAinle et al., 2006). We previously showed that the lack of antiviral activity of human APOBEC3H corresponds with poor steady-state protein expression (OhAinle et al., 2006). To track the origin of the instability of the APOBEC3H protein in primate evolution and, therefore, the likely cause of the loss of antiviral activity, we cloned and expressed a panel of hominoid APOBEC3H proteins by transient transfection and evaluated their steady-state expression levels by western blotting. All hominoid APOBEC3H proteins, except human, are expressed at levels similar to that of macaque APOBEC3H (Figure 1A). Importantly, chimpanzee APOBEC3H, the hominoid APOBEC3H protein with the highest amino acid identity to human APOBEC3H, is expressed at high levels suggesting that the stability of human APOBEC3H was lost sometime after human-chimpanzee speciation. We also cloned and tested the expression of a second human APOBEC3H allele that differs from the original human allele at four amino acid sites (R18L, G105R, K121D and delN15 - amino acid numbering is maintained based on the original clone). The expression of the second human APOBEC3H protein (human_B) is similar to that of the original human APOBEC3H protein (human_A) and is much less than that of chimpanzee APOBEC3H protein (Figure 1B).
Figure 1. Instability and Inactivity of APOBEC3H is Unique to Humans and Shared Among Human APOBEC3H Alleles.
(A) Western blot analysis of primate APOBEC3H proteins expressed in transiently transfected 293T cells. Actin is shown as a loading control.
(B) Western blot analysis of chimpanzee and two versions of the human APOBEC3H protein cloned from two individuals (human_A and human_B).
(C) Inhibition of MusD replication by human APOBEC3G and primate APOBEC3H homologs. Values are shown as the % Retrotransposition relative to the assay without any APOBEC (No Apobec) expression plasmid. Averages of three replicates (+/− SEM) are shown.
(D) Pulse-chase analysis of chimpanzee and both human APOBEC3H proteins over a six-hour time-course (0, 0.5, 1, 2 and 6 hours). HA-tagged APOBEC3H proteins were immunoprecipitated and radiolabeled proteins detected at each time point by autoradiography following SDS-PAGE. An untransfected control was included to determine background. This is a representative experiment that was done three times with similar results.
(E) Autoradiography was used to quantitate protein levels at each time point. Quantities of radiolabeled proteins are shown as the percent of each protein remaining (% remaining) at that time point relative to time 0 (pulse only). The stability of the chimpanzee APOBEC3H is shown in the grey line and triangles; human_A is shown in the solid line and diamonds; human_B is shown in the solid line and squares. The half-life (t1/2) equal to the time in which 50% of the APOBEC3H protein remaining for human_A and human_B is marked on the graph.
To ask if stable expression correlates with antiviral activity, we tested all the APOBEC3H proteins for inhibition of the mouse retrotransposon, MusD. All primate APOBEC3H homologs, except for human, were found to inhibit the replication of MusD (shown in Figure 1C) as well as other retroviruses and retrotransposons (HIV vif-, SIVagm vif-, and the L1.3 LINE-1 elements; data not shown). Thus, the human APOBEC3H protein was uniquely incapable of inhibiting the retroviruses and retroelements tested.
We then performed a pulse-chase analysis to determine if the lack of steady-state expression is due to increased protein turnover. A 30-minute pulse with 35S-Cys/Met resulted in approximately equivalent levels of labeled chimpanzee and human APOBEC3H proteins, suggesting that equivalent amounts of both APOBEC3H proteins were synthesized (Figure 1D). However, both human APOBEC3H proteins showed a dramatic decrease during the 6-hour chase (Figure 1D). Quantification of the rate of degradation showed that both human APOBEC3H proteins have a half-life of less than 30 minutes, while the chimpanzee APOBEC3H protein has a half-life of over 6 hours (Figure 1D & 1E). Thus, we can attribute the lack of activity of human APOBEC3H protein to the evolution of a dramatically increased rate in protein turnover that occurred in the human lineage after speciation from a common ancestor with chimpanzees.
The C-terminal Tail of APOBEC3H Contributes to Subcellular Localization but is not the Major Determinant of Protein Stability
Macaque and human APOBEC3H proteins differ at 31 single amino acid sites. Further, macaque APOBEC3H encodes a 27 amino acid C-terminal tail that is missing in human and chimp APOBEC3H due to a premature termination codon (PTC) that arose in the APOBEC3H coding sequence in the common ancestor of human and chimpanzee (Figure 2A). Recently it was suggested that the PTC may have a role in determining relative levels of APOBEC3H protein (Dang et al., 2008). However, we found that while both human and chimpanzee APOBEC3H alleles encode the PTC, these alleles differ dramatically in the rate of protein turnover (see Figure 1D & 1E), leading us to deduce that the PTC is not a major determinant of turnover of the APOBEC3H protein. We specifically addressed the role of the C-terminal tail in protein stability and antiviral activity of APOBEC3H by first deleting the 27-residue C-terminal tail from the macaque APOBEC3H protein (Macaque−tail) and then by fusing the C-terminal tail to the human APOBEC3H protein (Human +tail). While the tail modifications did result in changes in expression levels of the proteins (Figure 2B; compare Macaque to Macaque −tail and Human to Human +tail), the relative increase or decrease was not nearly as dramatic as the tail-independent differences in expression levels of the human and chimpanzee proteins (see Figure 1A).
Figure 2. The C-terminal Tail Controls the Sub-cellular Localization but not the Stability or Antiviral Activity of APOBEC3H.
(A) Schematic of the primate APOBEC3H protein. Location of the HAE and PC--C motifs of APOBEC3H that are conserved in other APOBEC proteins (Conticello et al., 2005) are shown in black. A Premature Termination Codon (PTC) truncates human and chimpanzee APOBEC3H homologs by 30 amino acid residues (asterisk). The leucine residues making up the core of the putative Nuclear Export Sequence (NES) downstream of the PTC are highlighted in red in an alignment of human, chimpanzee, gorilla, orangutan, gibbon and rhesus macaque APOBEC3H C-terminal tail domains.
(B) Western blot analysis of human, macaque and chimeric human/macaque APOBEC3H proteins. Actin is shown as a loading control.
(C) Sub-cellular localization of human, macaque and chimeric human/macaque APOBEC3H proteins in HeLa cells. APOBEC3H proteins were detected with an anti-HA antibody (red) and DAPI staining was used to define the nucleus (blue).
(D) Inhibition of HIV-1 vif- by human, macaque and chimeric human/macaque APOBEC3H proteins in a single-cycle infectivity assay. Averages of triplicate assays (+/− SEM) are shown as the % Infectivity relative to the infectivity of HIV in the absence of an APOBEC expression plasmid (No Apobec).
On the other hand, the C-terminal tail did dramatically alter the subcellular localization of APOBECH proteins. Truncation of the C-terminal tail from macaque APOBEC3H altered its localization from cytoplasmic to nuclear while addition of the C-terminal tail to human APOBEC3H altered its localization from mostly nuclear to mostly cytoplasmic (Figure 2C). It appears that cytoplasmic localization may be necessary for the antiviral activity of macaque APOBEC3H, since truncation of the C-terminal tail impairs its ability to efficiently restrict HIV-1 (Figure 2D; compare Macaque to Macaque−tail). In contrast, the native chimpanzee APOBEC3H protein lacks the C-terminal tail, yet retains robust antiviral activity (see Figure 1C). More importantly, the human APOBEC3H chimera with the macaque C-terminal tail does not restrict HIV-1 (Figure 2D), likely due to a lack of sufficient expression (see Figure 2B). Although not explored further, we note that there is a possible Nuclear Export Sequence (NES) in the C-terminal tail that is conserved in other APOBEC family members, such as APOBEC1 and AID (Ito et al., 2004) (Figure 2A). Thus, although the C-terminal tail of primate APOBEC3H appears to play a role in protein localization, we conclude that the major determinants of stability of primate APOBEC3H proteins do not lie within the C-terminal tail.
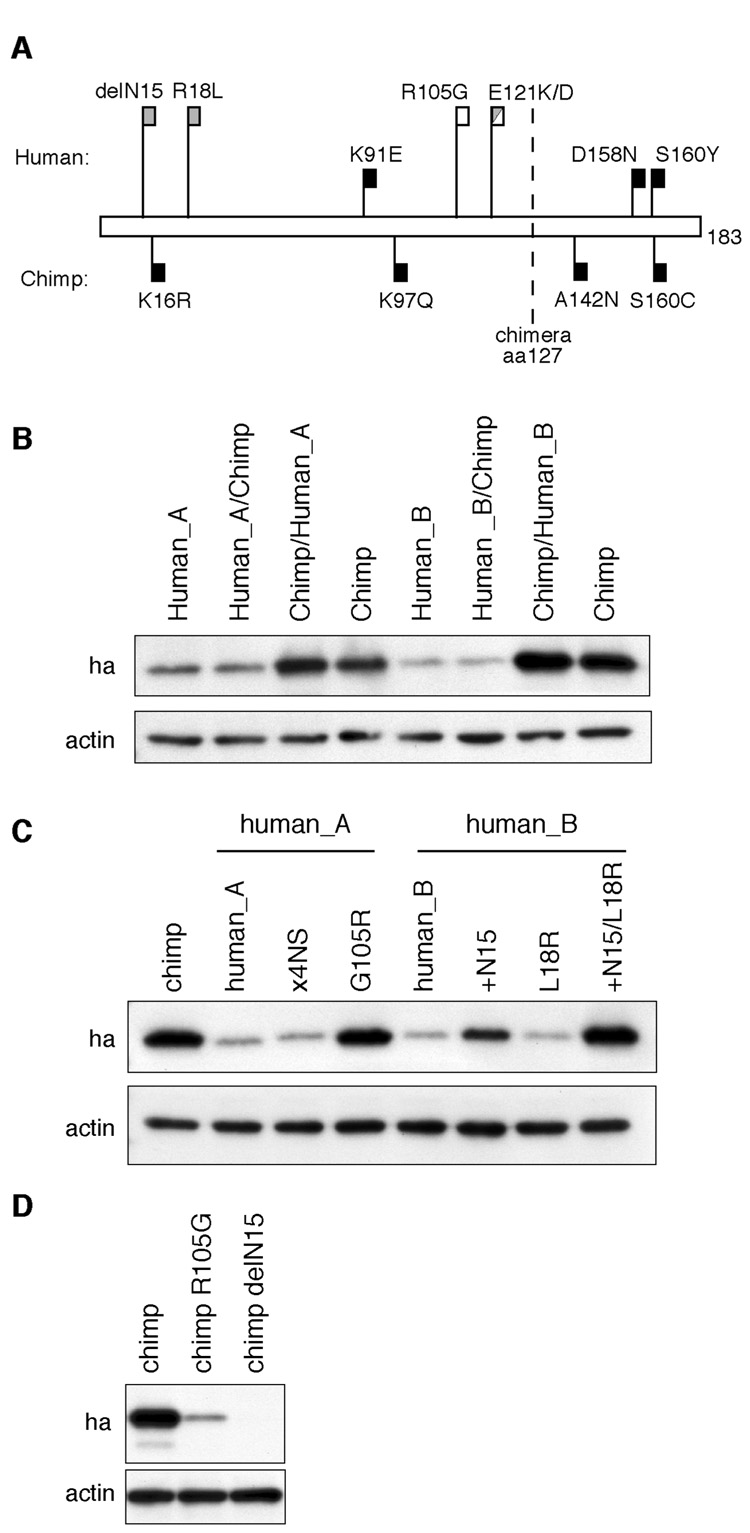
Loss of Stability of Human APOBEC3H Proteins Is Due to Two Independent Mutations
Since the instability and lack of antiviral activity of human APOBEC3H is not determined by the C-terminal tail, we hypothesized that changes in the APOBEC3H coding sequence must distinguish human and chimpanzee APOBEC3H orthologs. We used parsimony to infer those sites in the APOBEC3H coding sequence that have changed specifically in either the human or chimpanzee lineages, using the gorilla sequence as an outgroup. We identified four amino acid changes that have been fixed in the chimpanzee lineage (upside down black flags in Figure 3A) and three amino acid changes that have been fixed in humans (short black flags in Figure 3A). Based on polymorphisms currently segregating in the human population, the human_A protein differs from chimpanzee APOBEC3H at 2 additional sites (open flags in Figure 3A) and human_B differs at 3 additional sites (grey flags in Figure 3A). In sum, human and chimpanzee APOBEC3H proteins differ at 9 and 10 sites, respectively, with some changes shared in common between both human proteins and other changes specific to each allele. By creating chimeric proteins between the chimpanzee APOBEC3H and human APOBEC3H proteins, we mapped the determinants of the instability of both human proteins to the N-terminal two-thirds of APOBEC3H (Figure 3B). Next, we experimentally tested all remaining non-synonymous changes that distinguish the chimpanzee from the human APOBEC3H proteins. We found that only reversion of the R105G polymorphism in human_A could stabilize this protein (Figure 3C).
Figure 3. Two Independent Mutations Determine the Instability of Human APOBEC3H.
(A) Schematic of all non-synonymous differences (change the encoded amino acid) between both human (human_A and human_B) and chimpanzee APOBEC3H proteins relative to the ancestral amino acid at that position as determined using gorilla APOBEC3H as the outgroup. The four non-synonymous mutations specific to chimpanzee APOBEC3H are shown in upside down black flags (K16R, K97Q, A142N and S160C -note that only a single chimpanzee APOBEC3H was analyzed; therefore, these changes may not necessarily be fixed in the chimpanzee lineage). The three changes fixed in the human population are shown in as the short solid flags (K91E, D158N and S160Y). Human APOBEC3H polymorphisms are shown as long flags; open for for human_A (R105G and E121K) and grey for human_B (delN15, R18L and E121D). Position of the breakpoint of the chimeric human/chimpanzee APOBEC3H proteins analyzed in panel B is shown as the dotted line at position 127.
(B) Western blot analysis of native and chimeric human (human_A and human_B) and chimpanzee APOBEC3H proteins. APOBEC3H expression was detected with an HA antibody and actin is shown as a control.
(C) Western blot analysis of native and mutant APOBEC3H proteins. Four of the five non-synonymous mutations in the N-terminal region that distinguish the human_A protein from the chimpanzee protein were mutated in human_A to match the chimpanzee sequence (x4NS). A single mutation was also analyzed (G105R). Two single mutations (+N15 and L18R) as well as the double-mutant (+N15/L18R) were analyzed for the human_B protein.
(D) Western blot analysis of native and single-mutant chimpanzee APOBEC3H proteins (chimp R105G/chimp delN15).
The fact that the R105G mutation was able to stabilize the human_A APOBEC3H protein was surprising because this change is not found in the human_B APOBEC3H protein, suggesting that an independent change is responsible for its high instability. Indeed, we found that by reverting the deletion of a single amino acid residue (delN15), we were able to restore stable expression of the human_B protein (Figure 3C). In addition, we found that either polymorphism (R105G or delN15) is independently sufficient to destabilize the otherwise stable chimpanzee APOBEC3H protein (Figure 3D). Thus, remarkably, the human species independently lost stable expression of APOBEC3H twice through different mutations (G105R and delN15) that are each independently capable of destabilizing the APOBEC3H protein.
A Stable Human APOBEC3H Protein Inhibits HIV-1 Replication and Is Neutralized by Vif
The mutations that cause the instability of human APOBEC3H proteins are both polymorphic in the human population. To determine whether or not stable and active alleles of APOBEC3H might still circulate in the human population, we sequenced the APOBEC3H coding exons from a panel of individuals from diverse world populations (Figure S1). The major haplotypes (set of polymorphisms that are inherited together on a single chromosome) were then inferred using PHASE 2.1.1 software (Table S1) (Stephens and Donnelly, 2003; Stephens et al., 2001). Four predicted haplotypes were found at frequencies > 10% in our panel (haplotypes I – IV in Figure 4A; and in bold in Table S1. The total counts for all haplotypes are listed in Tables S2). Amino acid polymorphisms in haplotypes I–IV are diagramed in Figure 4A.
Figure 4. A Stable and Active Human APOBEC3H Protein Blocks the Replication of vif-HIV.
(A) Schematic of the four dominant haplotypes of APOBEC3H (hap I, hap II, hap III and hap IV) found in the human population. Polymorphisms present in each haplotype are marked with a flag and shading is maintained based on the original human_A and human_B APOBEC3H alleles described in the legend to Figure 3. Of note, human_A corresponds to haplotype I while human_B corresponds to haplotype IV except that it lacks the E178D polymorphism.
(B) Western blot analysis of the four human APOBEC3H haplotypes and chimpanzee APOBEC3H.
(C) Inhibition of HIV vif-by human APOBEC3H proteins as well as by human APOBEC3G. Single-cycle infectivity assays were performed for both wild-type (HIV WT) and vif-deficient HIV (HIV vif-) in the presence or absence of APOBEC expression. Averages of triplicate assays (+/− SEM) are shown as % Infectivity of the control infection (No Apobec). Note the log scale on the Y-axis.
To ask if any of the major human APOBEC3H haplotypes identified in our panel might be stable and active, we reconstructed the four major haplotypes by site-directed mutagenesis of the human_A and human_B cDNAs. Three of the four haplotypes (hap I, hap III and hap IV) contain one of the destabilizing polymorphisms (either R105G or delN15) identified in Figure 3. As expected, all three haplotypes encoding either of the destabilizing mutations were found to encode unstable APOBEC3H proteins (Figure 4B). However, a single human APOBEC3H haplotype (hap II) encodes an APOBEC3H protein that is as stable as chimpanzee APOBEC3H (Figure 4B). Therefore, in spite of the two destabilizing polymorphisms that arose during recent human evolution, a stable APOBEC3H allele still persists in the human population.
To ask if the stable human APOBEC3H protein is capable of retroviral inhibition, APOBEC3H proteins were tested for activity against both wild type and vif-deficient HIV-1 (HIV vif-). Rhesus macaque APOBEC3H is a potent inhibitor of HIV-1 and is not neutralized by Vif from HIV-1, but is neutralized by Vif from SIV from african green monkeys (SIVagm) and rhesus macaques (SIVmac) (OhAinle et al., 2006; Virgen and Hatziioannou, 2007). Consistent with our observed correlation between stability and antiviral activity of APOBEC3H proteins, the human APOBEC3H haplotypes that encode unstable proteins have no effect on HIV replication (Figure 4C). In contrast, both chimpanzee APOBEC3H and the stable human APOBEC3H (hap II) proteins can inhibit HIV-1 (Figure 4C) but are both counteracted by HIV-1 Vif (although to a less extent than APOBEC3G) (Figure 4C; compare white to black bars). Thus, an active form of APOBEC3H does exist in the human population and, like APOBEC3F and APOBEC3G, it is sensitive to inactivation by HIV-1 Vif.
Human Ancestors Possessed APOBEC3H Alleles That Block Retroviruses and LINE-1 Replication
To demonstrate that the loss of stability and antiviral activity of APOBEC3H occurred specifically during recent human evolution, we reconstructed and tested the predicted ancestral states of human APOBEC3H since human-chimpanzee speciation. Using the gorilla and chimpanzee APOBEC3H sequences as outgroups to the human APOBEC3H alleles, we constructed both a hypothetical human ancestor sequence (referred to as human ancestor) as well as a hypothetical human/chimpanzee ancestor sequence (referred to as human/chimp ancestor) based on parsimony (Figure S2). Specifically, human APOBEC3H protein sequences were aligned with both chimpanzee and gorilla APOBEC3H. Sites that are polymorphic in humans were reverted to the ancestral state, as predicted based on the chimpanzee sequence, to create the human ancestor APOBEC3H protein. Similarly, amino acid sites that differ among humans and chimpanzees were identified and, using the gorilla APOBEC3H sequence as the outgroup, the ancestral state at each position was inferred by parsimony.
Next, we used this information to define the evolutionary path of changes that occurred in the APOBEC3H coding sequence during recent human evolution (Figure 5A). Four amino acid changes were fixed in the chimpanzee lineage (black flags in Figure 5A along the chimp lineage), three non-synonymous changes were fixed in the human lineage (black flags in Figure 5A above “human ancestor”), and six non-synonymous changes occurred in recent human evolution that have not been fixed in the human population and remain polymorphic in humans (open and grey flags in Figure 5A). The ancestral APOBEC3H cDNAs (circled nodes in Figure 5A) were then reconstructed in vitro by site-directed mutagenesis of human and human/chimpanzee APOBEC3H constructs. Both the human/chimp APOBEC3H ancestor and the human APOBEC3H ancestor encode stable APOBEC3H proteins that are expressed at levels similar to that of the native chimpanzee protein and much higher than the ‘unstable’ human APOBEC3H proteins (Figure 5B). Further, both the ancestral APOBEC3H proteins efficiently inhibit retroviral replication, as indicated by their potent activity against vif-deficient HIV (Figure 5C, open boxes).
Figure 5. Ancestral APOBEC3H Proteins Inhibit Retroviruses and LINE-1 Elements.
(A) The evolution of modern A3H alleles is presented as a phylogeny of the human/chimpanzee clade. Four changes (K16R, K97Q, A142N and S160C – purple) occurred during the evolution of modern chimpanzees. Three changes (K91E, D158N and S160Y – solid flags) are fixed among human APOBEC3H alleles. Haplotype I accumulated two further changes (R105G and E121K – open flags) while haplotypes II – IV are the result of sequential accumulation of four additional changes (E121D and E178D for haplotypes II; plus del15N for haplotype III; plus R18L for haplotype IV – grey flags). The nodes representing the human ancestor and the human/chimpanzee ancestor are circled.
(B) Western blot analysis of human (haplotype I), human ancestor and human/chimpanzee ancestor APOBEC3H proteins. (C) Inhibition of HIV by chimpanzee and ancestral human APOBEC3H proteins. Averages of triplicate assays (+/− SEM) are shown as the % Infectivity relative to the control assay with no APOBEC expression (No Apobec). The open boxes are infections with HIV that contain a deletion in the Vif gene (HIV vif-) and the solid boxes are infections with wild-type HIV that contains a functional Vif gene (HIV WT). Note the log scale on the Y-axis.
(D) Inhibition of a human LINE-1 element (L1.3) by chimpanzee, ancestral human (human ancestor and human/chimp ancestor) and human APOBEC3H proteins. APOBEC3G and APOBEC3A are shown as a negative and positive control, respectively. Averages of triplicate assays (+/− SEM) are shown as the % Infectivity relative to the control assay with no APOBEC expression (No Apobec).
We consistently find that the ancestral versions of APOBEC3H are more active than either the extant human (Figure 4C) or chimpanzee versions of this protein (Figure 5C, open boxes). We also tested the Vif-sensitivity of the reconstructed human/chimp ancestor of APOBEC3H and the reconstructed human ancestor of APOBEC3H. We find that both are at least partially counteracted by a modern version of HIV-1 Vif (Figure 5C, solid boxes). These results suggest that while there has been some degradation of function of APOBEC3H in both the chimpanzee and the human lineage, the APOBEC3H protein has still retained a surface that is important for interaction with a modern day HIV-1 Vif.
Since APOBEC3H is conserved in eutherian mammals and appears to have evolved under constant positive selection in primates (OhAinle et al., 2006), it is likely to have evolved to counteract a more ancient and pervasive pathogen than lentiviruses. Endogenous retroelements, such as the LINE and SINE non-LTR retrotransposons, represent such a constant, pervasive threat in human evolution. Therefore, we tested both modern and ancestral human APOBEC3H proteins for the ability to block the replication of the human LINE-1 element. We find that all stably expressed APOBEC3H proteins inhibit the replication of LINE-1 (Figure 5D). In contrast, the unstable proteins have no impact on LINE-1 replication. Similar results were obtained with an Alu element (data not shown), consistent with previous findings that APOBEC3G potently blocks Alu transposition (Bogerd et al., 2006; Hulme et al., 2007). Significantly, the ancestral human APOBEC3H protein is as good or better than any of the extant hominoid versions of this gene in terms of its anti-LINE1 activity (Figure 6D). Thus, recent human ancestors encoded an APOBEC3H protein with potent activity against retroviruses and LINE-1 elements, but this activity now persists in only a fraction of the human population.
Figure 6. Frequency of the Active APOBEC3H Allele Varies Across World Populations.
(A) Genotypes were determined for African American (AA; N=22), European American (EA; N=23) and Han Chinese from Los Angeles (CA; N=23) for both the R105G (black bars) and N15 deletion (gray bars) polymorphisms. A list of all inferred haplotypes and their frequencies is found in Table S1, and a list of the number of individuals with each haplotype is found in Table S2.
(B) An FST statistic was calculated for both the delN15 and R105G polymorphisms in pairwise comparisons of each subpopulation from Panel A with the other two subpopulations (see Experimental Procedures for details). FST values near 0 are indicative of no population differentiation while large FST values suggest either local selection or demographic effects.
(C) Genotypes from HapMap populations (www.hapmap.org) from Yorubans in Ibadan, Nigeria (YRI; N=90), Utah residents with ancestry from northern and western Europe (CEU; N=90), and combined data from Japanese in Tokyo and Han Chinese in Beijing (JPT/CHB; N=92) for both the R105G (black bars) and delN15 (gray bars) polymorphisms.
(D) An FST statistic was calculated for both the delN15 and R105G polymorphisms in pairwise comparisons of each subpopulation from Panel C with the other two subpopulations.
(E) Predicted frequencies of APOBEC3H alleles predicted to encode stable proteins (black sector; that is, with R105 and without the deletion at N15; R105/N15) and unstable proteins (open sectors; that is either R105G or the deletion at N15; R105G/delN15) among the all six three subpopulations. The % of APOBEC3H alleles predicted to encode stable proteins is marked to the right of each bar graph.
Active APOBEC3H Alleles Exist at Higher Frequency in Individuals of African Descent
To determine the approximate frequencies of stable and active alleles across human populations, we genotyped both the inactivating polymorphisms of APOBEC3H (R105G and delN15) in a larger panel of unrelated individuals from three American populations with diverse ancestry, African Americans (AA), European Americans (EA) and Han Chinese from Los Angeles (CA) (Hinds et al., 2005). Both destabilizing human polymorphisms are found at significant frequencies in the populations we sampled (Figure 6A). Specifically, the N15 deletion is found at relatively similar frequencies in all three populations (0.27 in African Americans, 0.21 in European Americans and 0.30 in Han Chinese) (gray bars in Figure 6A), while R015G differs in frequency among populations (0.22 in African Americans, 0.61 in European Americans and 0.65 in Han Chinese) (black bars in Figure 6A). Pairwise comparisons of FST values, a measure of population differentiation that reflects either demographic effects or regional selection (Lewontin and Krakauer, 1973), indicate that there is little to no difference in frequencies of the N15 deletion polymorphism between the three populations (Figure 6B). In contrast, the R105G polymorphism is found at significantly higher frequencies in European Americans and Han Chinese as compared to African Americans (black bars in Figure 6A). The pairwise FST values for the APOBEC3H R105G SNP are 0.26 and 0.31 for comparisons between African American and the other two populations, respectively (Figure 6B); these values are higher than ~90% of human SNPs compared in the same populations in a genome-wide study (Hinds et al., 2005)
We were concerned about the potential confounding effects of admixture in the 3 populations we had thoroughly analyzed. Therefore, we examined the incidence of both destabilizing polymorphisms in the HapMap dataset that examined a larger panel of humans, that is relatively devoid of admixture. . These included the Yoruban (YRI) panel from Nigeria, the Caucasian (CEU) panel of Utah residents with ancestry from Northern and Western Europe and a combined East Asian (JPT/CHN) population that included Japanese individuals from Tokyo and Han Chinese individuals from Beijing (more information about these populations is available from www.hapmap.org). We tabulated the incidences of both the R105G polymorphism (already typed by the HapMap consortium) and the N15 deletion. Again, we found no significant discordance in the incidence of the N15 deletion allele frequencies across these three populations (Figure 6C). The N15 deletion frequencies ranged from 0.28 to 0.4 with no significant discordance in allele frequencies (low Fst values) between any two populations. In contrast, we found that the R105G polymorphism existed at a frequency of 0.689 and 0.608 in the HCB and CEU populations respectively, but only at an incidence of 0.075 in the YRI population (Figure 6C). This is an even more dramatic discordance in allele frequencies than what we have typed in our smaller dataset (Figure 6A, 6B). The Fst values that highlight this discordance between African (YRI) and non-African populations (CEU, HCB) are presented in Figure 6D.
Next we asked if the stable APOBEC3H alleles are found at a significant frequency in any of the six subpopulations that we had sampled in Figure 6A and 6C. Based on our phasing analysis, the loci encoding the two destabilizing mutations (R105G and delN15) are tightly linked and, therefore, are rarely found on the same chromosome (only 2 of 86 chromosomes analyzed in our panel are predicted to encode both polymorphisms -- see Table S2). Therefore, summing the frequencies of both polymorphisms within a population gives a robust estimation of the frequency of active (stable) versus inactive (unstable) APOBEC3H alleles in each subpopulation. The relative frequencies of active (gray) and inactive (black in Figure 6C) APOBEC3H alleles show that the frequency of active APOBEC3H alleles varies significantly between populations, from 51–52% in African Americans (AA) and Yorubans (YRI) respectively, 10–18% in Europeans (CEU) and European Americans (EUR) respectively and only 3–4 in East Asian (JPT/CHN) and Chinese-American populations respectively (Figure 6E). Thus, a gradient of active APOBEC3H alleles exists with increasing distance from Africa, from nearly 53% for the African population (YRI), to only 3% for the East Asian population (JPT/CHN) (Figure 6). Therefore, the ancestral active APOBEC3H allele has been maintained at higher frequency in Africa and has been lost with increasing distance from Africa.
Remarkably, the trend of loss of APOBEC3H activity in human populations is similar to that observed for another APOBEC3 gene, APOBEC3B. APOBEC3B is found at a frequency of nearly 100% in African populations but is present in only 40% of East Asians due to a large structural deletion (Kidd et al., 2007). Since APOBEC3B and APOBEC3H are present in the same genetic location, we wished to ascertain whether there was any correlation between the unstable APOBEC3H alleles and the loss of APOBEC3B. Therefore, we typed the presence or absence of the APOBEC3B gene in the same individuals from all three subpopulations (Table S3). We did not find a significant correlation between non-functional alleles of these two genes within individuals (2 by 2 contingency χ2 test; p =0.08). This either means that the loss of APOBEC3B and APOBEC3H were independent events or that our test was not powerful enough to identify any linkage between the two loci. Nevertheless, whether independent events or not, populations that lack the APOBEC3B gene, in particular East Asian populations, are also more likely to encode inactive APOBEC3H proteins.
DISCUSSION
We present evidence of the evolutionary history of APOBEC3H in humans in which two independent mutations arose during recent human evolution and gave rise to an unstable APOBEC3H protein with no detectable antiviral activity. This is in contrast to our finding of stable expression and antiviral activity of other hominoid APOBEC3H proteins. Both of the inactivating mutations occur in distinct regions of APOBEC3H, but similarly act to decrease the half-life of the APOBEC3H protein. Reconstruction of the ancestral human APOBEC3H gene suggests that recent human ancestors encoded a very powerful anti-retroviral and anti-retroelement version of this gene. While most humans possess unstable APOBEC3H alleles, we found that a minor fraction of humans still encode a stable allele of APOBEC3H that still retains activity and can be targeted by the HIV Vif protein.
Our finding of an APOBEC3H allele with variable activity among individuals may have important consequences for the interaction of the human APOBEC3 locus with HIV. First, in those individuals homozygous or heterozygous for the active APOBEC3H haplotype, there may be an additional APOBEC-mediated block to infection of HIV target cells in addition to the activities of APOBEC3F and APOBEC3G. However, we must point out that although APOBEC3H mRNA is present in the appropriate cell types (T cells), (OhAinle et al., 2006)) it is still unclear if the APOBEC3H protein is present. Second, our finding that HIV Vif can inhibit the active APOBEC3H allele argues that HIV Vif may have specifically evolved to counteract the active APOBEC3H protein in addition to the other active APOBEC3 proteins. The mechanism by which HIV Vif recognizes and inhibits APOBEC3H may or may not overlap with the mechanisms of inhibition of other APOBEC3 proteins. Therefore, successful replication of HIV in individuals with active APOBEC3H alleles may further challenge Vif evolution even though HIV-1 Vif may be sufficient to counteract any potential benefit of active APOBEC3H in humans. Significant differences in HIV-1 progression have been shown to be correlated with APOBEC3G variation (An et al., 2004), however, we do not yet know if individuals heterozygous or homozygous for active APOBEC3H are more resistant to successful HIV replication. On the other hand, since the stable APOBEC3H proteins appear to be able to inhibit multiple retroelements (against both LINE-1, Alu elements and retroviruses), it is also possible that APOBEC3H served an ancient role and has no modern impact on HIV-1.
In light of the multiple protective functions served by human APOBEC3H (against both LINE-1, Alu elements and retroviruses), it is puzzling that a majority of humans have lost this activity and even more puzzling that loss of activity happened twice in recent human evolution. This suggests the interesting possibility that non-functional APOBEC3H alleles have been actively selected for in humans. The “less-is-more” hypothesis suggests that loss of function mutations can be maintained at high frequency during a shift of environmental pressures under which they may be selectively advantageous, particularly if the functional allele presents a cost to the host (Olson, 1999). For example, there may be a significant cost associated with maintaining active APOBEC3 proteins at high levels in the cell as APOBEC3 proteins present a risk to the host if they were to deaminate the wrong targets, such as cellular mRNA or genomic DNA. Consistent with this hypothesis, APOBEC3H was recently shown to hypermutate human papillomavirus, a double-stranded DNA virus thought to closely resemble nuclear DNA (Vartanian et al., 2008). We speculate that APOBEC3H may be particularly toxic relative to other APOBEC3 family members as no duplications of the APOBEC3H-like domain have been tolerated in any mammalian genome, in striking contrast to the multiple duplications of the non-APOBEC3H domain (Conticello et al., 2005; OhAinle et al., 2006). Further, the expression of APOBEC3H in germline tissues (OhAinle 2006) poses a mutagenic threat with serious evolutionary consequences and, therefore, could be particularly costly to the host. In this context, APOBEC3H may be especially 'costly' in humans and chimps, compared to other primates, because nuclear localization of the APOBEC3H protein occurred only recently during the evolution of the protein in hominoids, corresponding with loss of the C-terminal domain in the human/chimpanzee ancestor. Therefore, the nuclear localization and potential risk of increased genomic DNA editing are greater for humans and chimpanzees than for other primates. The benefits of maintaining stable and active APOBEC3H alleles (due to significant selective pressure from an active exogenous viral infection or from active endogenous retroviruses or retrotransposons) might outweigh the costs associated with the stable APOBEC3H protein in chimpanzees but not in humans. This is plausible given that retroviral infections and retroelement amplifications are known to be distinct among even these two closely-related species (Jern et al., 2006; Polavarapu et al., 2006; Yohn et al., 2005).
Taken together, our findings suggest that there may be significant evolutionary pressure to downregulate levels of APOBEC3H protein in the cell in times of decreased pathogenic pressure. Under this scenario, maintaining a stable and active APOBEC3H protein capable of retroelement inhibition would have to be balanced with the costs associated with maintaining high doses of this protein in the cell. Other APOBEC3 proteins that are localized to the nucleus (and therefore may also contribute to higher genomic mutation) might also display evidence of this increased cost. Consistent with this model, APOBEC3B has also been lost in a significant fraction of human populations (Kidd et al., 2007), while the start codon of the APOBEC3A gene is missing from chimpanzees (unpublished data). These debilitating events appear specific to the APOBEC3 genes that have a nuclear localization, supporting the 'costly defense' selective scenario for the acquisition of destabilizing mutations in human APOBEC3H.
Nonetheless, there are important differences between the loss of APOBEC3B and APOBEC3H activities in humans. While APOBEC3B has been lost due to a genomic deletion, APOBEC3H still appears to be expressed but encodes an unstable protein. We cannot rule out the possibility that instability, rather than loss of APOBEC3H function, is what has been selected. Even the unstable APOBEC3H alleles may still be sufficient to carry out some antiviral function that we have not assayed. A simple ‘loss-of-function’ hypothesis might be insufficient to explain the high prevalence of the unstable alleles (~50%) even in the African populations, where APOBEC3H activity still appears to be selectively retained. Instead, our findings suggest that the two unstable alleles appear to be independently under selection, based on the discordance in their allele frequencies in different human populations.
Independent of the true nature of selective pressures shaping the human APOBEC3 locus, the consequences of these pressures on the antiviral repertoire are significant. Combining the presence of two 'loss of stability' mutations in APOBEC3H and the loss of the APOBEC3B gene in humans, we can conclude that at least two APOBEC3 antiretroviral proteins are maintained at higher frequencies in African populations and have subsequently been lost to varying degrees in European and Asian populations. This is consistent with a model in which a lower pathogenic burden outside Africa may allow for the accumulation of inactivating mutations in antiviral genes outside of Africa (either due to genetic drift or due to selection). This implies both that a relatively recent (and perhaps ongoing) pathogenic pressure is maintaining APOBEC3H and APOBEC3B activity in African populations, and further that these populations might have an associated higher risk of genomic mutations. Regardless of the cause, it is clear that much of the redundancy of an expanded APOBEC3 locus that built up over primate evolution to defend the genome against the deleterious effects of retrotransposition or unknown retroviruses has been lost from a substantial fraction of the human species.
Experimental Procedures
APOBEC3H cDNA Cloning
APOBEC3H cDNAs were cloned by RT-PCR or nested RT-PCR (QIAGEN One-Step RT PCR Kit) from RNA derived from chimpanzee (Pan troglodytes verus – Coriell GM03448), gorilla (gorilla gorilla – Coriell AG05251), orangutan (Pongo pygmaeus pygmaeus - Coriell AG05252) and gibbon (Nomascus leucogenys leucogenys - GM051496). Primers used were as follows: chimpanzee APOBEC3H was amplified using an initial round of RT-PCR (50 cycles) with primers JAK012/JAK015 followed by a second round of 30 cycles (Pfu Turbo – Stratagene) with primers JAK012/MO136. Gorilla APOBEC3H was amplified with an initial round of RT-PCR using primers JAK012/JAK015 followed by a second round of 30 cycles with JAK012/MO143. Orangutan APOBEC3H was amplified in a single round of RT-PCR (50 cycles) using primers JAK012/MO173. Gibbon APOBEC3H was amplified in a single round of 50 cycles using primers JAK012/JAK015. RT-PCR products were gel purified, TOPO cloned (Invitrogen) and sequenced. Cloning of human (= human_A) and macaque APOBEC3H cDNAs were described previously (OhAinle 2006). The “human_B” APOBEC3H cDNA was amplified by RT-PCR from a Druze individual (Coriell #11521) using primers JAK012/JAK015. GenBank accession numbers for the primate APOBEC3H sequences described here are EU861357 through EY861361.
Expression Constructs & Plasmids
All APOBEC3H cDNAs were cloned into the XhoI/EcoRI sites of pcDNA3.1 (Invitrogen) with a primer-encoded 5’ HA tag (MYPYDVPDYA). Chimeric human/macaque APOBEC3H plasmids were constructed by PCR using primers that match both the macaque and human APOBEC3H sequences. The Human +tail plasmid was constructed by PCR using overlapping primers specific for both the human APOBEC3H C-terminus and the macaque APOBEC3H C-terminal tail domain. First, these primers were used with external primers to create human APOBEC3H and macaque tail fragments, followed by amplification with external primers to create the chimeric Human +tail plasmid. The truncated macaque APOBEC3H plasmid (Macaque −tail) was constructed by amplification of macaque APOBEC3H with the human APOBEC3H 3’ primer, resulting in truncation of macaque APOBEC3H at the STOP found in the human sequence. Chimeric human/chimpanzee constructs were created by digesting chimpanzee and human APOBEC3H constructs with EcoRI/BamHI and BamHI/XhoI, purification of plasmid backbone and both fragments and triple-ligation. All constructs were confirmed by sequencing. Point mutants were constructed by site-directed mutatgenesis of plasmids using the Quickchange kit (Stratagene). Primers used in this study are described in Table S4.
Cell Lines, Transfections and Western Blot Analysis
HEK293T and HeLa cell lines (ATCC) were maintained in DMEM/5% Pen/Strep/10% FBS at 37°C in a CO2 incubator. Transfections were performed with TransIT-LT1 transfection reagent (Mirus Bio) at a reagent:plasmid DNA ratio of 3:1. For Western blot analysis, cells were lysed in ice-cold NP40doc buffer (1% NP40, 0.2% Sodiumdeoxycholate, 0.12M NaCl, 20mM Tris pH 8.0) with Protease Inhibitors (Roche Complete Mini, EDTA-free tablets), resolved by 12% SDS-PAGE, transferred to PVDF and probed with anti-HA (Santa Cruz Biotech) and actin (Sigma) antibodies.
Viral Infectivity Assays
Single-round HIV-1 infectivity assays were performed as previously described (OhAinle 2006)(Yamashita and Emerman, 2004). All assays were performed by transfection of 1.25 × 105 293T cells in 24-well plates with a 1:1 ratio of pcDNA/APOBEC plasmid (250ng) to BruLuc2 env- proviral plasmid (250ng). MusD and LINE-1 retroelement assays were performed essentially as previously described (Esnault et al., 2002; Ribet et al., 2004). For MusD assays, HeLa’s were plated at 2.5 × 105 cells/mL in 2 mLs in 6-well plates and triplicate wells transfected with 0.75 µg MusD plasmid and 0.75 µg APOBEC expression plasmid. At Day 3 post-transfection, cells were replated @ 1.5 × 105 cells/mL in 2 mLs of a 6-well dish. At Day 4, cells were selected in G418+ media (0.4 mg/mL) and fresh G418+ media added every 2 days until single colonies remained. For LINE-1 assays, HeLa’s were plated at 5 × 104 cells/mL in 2 mLs in a 6-well plate and transfected with 0.3 µg LINE-1 plasmid and 0.6 µg APOBEC expression plasmid.
Pulse-Chase Analysis
HEK293T cells were plated at 2 × 105 cells/mL in 6 cm poly-L-lysine plates (Becton Dickinson) and were transfected one day later with 5 µg pcDNA/APOBEC plasmids. One day post-transfection, the pulse was initiated by incubating cells in 1mL Starvation media (Met/Cys-free DMEM (Invitrogen)/5% Dialyzed FCS + L-Glutamine). After 30 minutes, starvation media was removed and 750 µL Pulse media was added. Pulse media is Starvation media plus 440µCi EasyTag EXPRE35S35S Protein Label (PerkinElmer) per plate. Following a 30-minute pulse, pulse media was removed and 1mL warm Chase media (Met/Cys-free DMEM + 5%FCS + 2mM Met + 2mM Cys) was added to all plates. Cells were harvested at 0, 30 min, 1 hr, 2 hr and 6 hr timepoints by washing 1x in PBS and lysing cells directly in 500µL ice-cold NP40doc buffer (1% NP40, 0.2% Sodiumdeoxycholate, 0.12M NaCl, 20mM Tris pH 8.0) with Protease Inhibitors (Roche Complete Mini, EDTA-free tablets). Debris was removed by spinning 5 minutes at 16,000g @ 4°C and supernatants were frozen @ −80°C until use. HA-tagged APOBEC3H proteins were Immunoprecipitated using Protein G-Sepharose beads (Zymed) conjugated to RAW HA.11 antibody. Lysates were pre-cleared by incubating lysates with unconjugated beads for 1 hr @ 4°C with rocking and antibody was pre-conjugated with beads for 1 hr @ 4°C with rocking. Beads were washed twice in NP40doc and spun out for 2 min @ 4000g @ 4°C. 0.01% BSA was added to antibody-conjugated beads which were incubated with pre-cleared lysates for 2 hours @ 4°C with rocking. Beads were spun out (2 min @ 4000g) and washed in a series of wash buffers with protease inhibitors (Roche Mini, EDTA-free tablets) as follows: RIPA (10mM Tris pH7.5, 150mM NaCl, 1% NP40, 1% sodiumdeoxycholate, 0.1% SDS), High Salt Buffer (2M NaCl, 10mM Tris pH7.4, 1% NP40, 0.5% sodiumdeoxycholate), Low Salt Buffer (0.5% NP40, 0.1% SDS in PBS) and RIPA. Bead slurries were resuspended in 20µL 2X Protein Loading Buffer (125mM Tris pH6.8, 4% SDS, 20% Glycerol and 10% β-mercaptoethanol), boiled for 10 minutes, beads spun out @ 16,000g for 1 min and supernatants used for 12% SDS-PAGE. Radioactivity was detected using a Typhoon Trio imager (Amersham Biosciences) and quantitated using ImageQuantTL software (Amersham Biosciences).
Immunofluoresence
HeLa cells plated onto coverslips in a 6-well dish at 2 × 105 cells/mL were transfected the next day with 1.5 µg pcDNA/HA-APOBEC expression vectors. 24 hours post-transfection slips were fixed in 4% Paraformaldehyde, permeabilized in 0.3% Triton-X/PBS and blocked in 10% FBS/PBS. HA-tagged proteins were detected using RAWHA.11 antibody (Santa Cruz Biotech) at a 1:100 dilution followed by anti-mouse-TexasRed conjugate (Invitrogen). Nuclei were stained with DAPI in mounting media (Vectashield; Vector Labs)and images were collected on an Olympus DeltaVision microscope.
Ancestor Reconstruction
The human/chimpanzee ancestor APOBEC3H protein sequence was inferred using the gorilla sequence as an outgroup. The human/chimpanzee ancestor APOBEC3H cDNA was then constructed in vitro by site-directed mutagenesis (QuikChange Site Directed Mutagenesis Kit – Stratagene) using the chimpanzee/human_A chimera as the template. The human ancestor APOBEC3H protein was defined by determining all changes that are fixed in the human population and reconstructed in vitro using the human hap I cDNA as a template by site-directed mutagenesis.
Human APOBEC3H Genotyping
The human APOBEC3H coding sequence was amplified/sequenced from genomic DNA samples with the following primer pairs (see Table S4 for primer sequences): Exon 1- Ex1_For/Ex1_Rev, Exons 2/3 -Ex2&3_For/Ex2&3_Rev and Exon 4 - Ex4_For/Ex4_Rev. A second round of sequencing was performed to obtain missing data with the following primer pairs: Exon 2 - MO276/MO271, Exon 3 - MO272/MO273 and Exon 4 - MO274/MO275. Human genomic DNA samples (Coriell) sequenced were: African Pygmy (GM10469 – 10473/GM10492 – 10494), South American (GM10965 – 10969), Central American (GM10975/6/8 & 9), Caucasian (GM893, 946, 1310, 1805, 1806, 1814, 1953, 8428, 9948 and 14492), African South of the Sahara (NA17341 – 17349) and African North of the Sahara (NA17378 – 17384). Extended genotyping of the delN15 and R105G polymorphisms was performed from the following Coriell samples: Han Chinese from Los Angeles (NA17733–17747, 17749, 17752 – 17757, 17759, 17761), African American (NA17102 – 17116, 17125, 17133 – 17140) and European American (NA06990, 07019, 07348, 07349, 10830, 10831, 10842 – 10845, 10848, 10850 – 10854, 10857, 10858, 10860, 10861, 12547, 12548, 12560, 17201). For the R105G polymorphism, genomic DNA was amplified (Supermix; Invitrogen) with primers JAK08 and JAK10 for 30 cycles (94°C – 30 sec.; 58°C – 30 sec; 72°C – 1.5 min.) and PCR product diluted and directly sequenced. For the delN15 polymorphism, genomic DNA was amplified with primers MO285 and MO286 with Taq for an initial denaturing for 5 min at 94°C , followed by 30 cycles (94°C – 15 sec; 58°C – 1 min) and a final 5 min extension at 72°C. DNA Fragment Analysis was performed on an Applied Biosystems 3730xl DNA Analyzer with a 36 cm capillary array and analyzed by GeneMapper 4.0 software (Applied Biosystems) to detect the presence or absence of the 3 bp deletion. Calculation of Hardy-Weinburg equilibrium (HWE) was used to assess genotyping accuracy; the data did not result in a significant rejection of HWE for both the delN15 and R105G polymorphisms in each population.
Population Genetic Analysis
SNP data was input to PHASE 2.1.1 software (http://stephenslab.uchicago.edu/software.html) and was used to infer phasing of haplotypes over five iterations. Human haplotypes found at > 10% frequency in the panel were reconstructed by site-directed mutagenesis (Quickchange; Stratagene) of human_A APOBEC3H and human_B APOBEC3H plasmids. Complete primer sequences used in this study are found in Table S4. FST values were calculated for each pairwise comparison using the following method of calculation: (MSP − MSG)/[MSP + {nc −1)MSG] (Akey et al., 2002).
Supplementary Material
Acknowledgments
We thank John Moran for the LINE-1 retrotransposition plasmid, Thierry Heidmann for the MusD and Alu retrotransposition plasmids, Rasmus Nielsen for advice on the FST analyses, Adam Geballe and Morgan Hakki for assistance with the pulse-chase analysis, Evan Eichler and Jason Kidd for access to the HapMap samples, the FHCRC Genomics and, Scientific Imaging Shared Resources, and Jaisri Lingappa, Evan Eichler, and Rasmus Nielsen for comments on the manuscript. This work was supported by NIH grant R37 AI30937 (M.E.), and Searle Scholar and Burroughs-Wellcome Investigator Awards (H.S.M). J.A.K. was supported by an NIH Postdoctoral Training Grant and M.O. was supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, Alobwede I, Trono D, Vlahov D, Donfield S, et al. J Virol. 2004. 78, APOBEC3G genetic variants and their influence on the progression to AIDS; pp. 11070–11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. Human Cytidine Deaminase APOBEC3H Restricts HIV-1 Replication. J Biol Chem. 2008;283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Casella JF, Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30:e49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Sperber GO, Blomberg J. Divergent patterns of recent retroviral integrations in the human and chimpanzee genomes: probable transmissions between other primates and chimpanzees. J Virol. 2006;80:1367–1375. doi: 10.1128/JVI.80.3.1367-1375.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile enviroment. Cell Host and Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polavarapu N, Bowen NJ, McDonald JF. Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol. 2006;7:R51. doi: 10.1186/gb-2006-7-6-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Dewannieux M, Heidmann T. An active murine transposon family pair: retrotransposition of "master" MusD copies and ETn trans-mobilization. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A New Statistical Method for Haplotype Reconstruction from Population Data. The American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J Virol. 2007;81:13932–13937. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CT, Jiang Z, McGrath SD, Hayden KE, Khaitovich P, Johnson ME, Eichler MY, McPherson JD, Zhao S, Paabo S, Eichler EE. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.