Abstract
A review of the work done in the Laboratory of Biodynamics of Tokyo Institute of Technology in the last decade has been summarized in this article in relation to the results reported from other laboratories. The emphasis here is the application of nanomechanics based on the force mode of atomic force microscopy (AFM) to proteins and protein-based biological structures. Globular proteins were stretched in various ways to detect the localized rigidity inside of the molecule. When studied by this method, bovine carbonic anhydrase II (BCA II), calmodulin and OspA protein all showed the presence of localized rigid structures inside the molecules. Protein compression experiments were done on BCA II to obtain an estimate of the Young modulus and its change in the process of denaturation. Then, the AFM probe method was turned on to cell membranes and cytoplasmic components. Force curves accompanying the extraction process of membrane proteins from intact cells were analysed in relation to their interaction with the cytoskeletal components. By pushing the AFM probe further into the cytoplasm, mRNAs were recovered from a live cell with minimal damage, and multiplied using PCR technology for their identification. Altogether, the work introduced here forms the basis of nanomechanics of protein and protein-based biostructures and application of the nanomechanical technology to cell biology.
Keywords: atomic force microscopy, force spectroscopy, protein stretching, receptor mapping, protein–ligand interaction, membrane protein extraction
1. Introduction
In terms of mechanical rigidity, the life-supporting materials are roughly a thousand times softer than most of the man-made materials developed for our daily use. Young's moduli of the former range, in a very approximate term, from 1 MPa to 1 GPa, whereas those of the latter from 1 GPa to 1 TPa. The physical reason for the softness of biological materials comes from the fact that most of them are built upon non-covalent interactions between covalently formed macromolecules (Howard 2001). For example, the covalent structure of a globular protein is a linear array of several hundreds of amino acid residues but its function comes from specific combinations of non-covalent segmental interactions in its folded conformation. Protein molecules in their native states have reasonably uniform and rigid conformations so that they can be crystallized, but they are flexible enough to change their conformations upon binding with specific ligands or substrates. Upon binding, protein molecules are thought to become more rigid but the actual values of parameters that represent their mechanical rigidity and its change upon ligand binding have not been well established. Among earlier reports on the measurement of the Young modulus of proteins at the single molecular level, those of Morozov & Morozova (1981), Kojima et al. (1994), Radmacher et al. (1994), Suda et al. (1995) and Tachibana et al. (2004) need to be mentioned.
Kojima et al. stretched a single actin fibre to a relative extension of a few per cent using a thin glass rod as a force transducer and calculated the Young modulus of a single actin molecule to be 2.5 GPa (Kojima et al. 1994). Tachibana et al. measured the velocity of the sound wave in a solution of lysozyme and converted it to the isothermal compressibility, κ, and obtained the Young modulus, Y, of 2 GPa for lysozyme. Suda et al. (1995) used the surface force apparatus to obtain two different values for Y for myosin head, 0.3 and 2 GPa for two different modes of compression. Morozov & Morozova (1981) published several papers on the measurement of resonance vibration of protein crystals and used the result of measurement to extract the Young modulus of the constituent protein. For lysozyme and albumin, they obtained values from 200 MPa to close to 1 GPa. Radmacher et al. (1994) applied the force mode of AFM to compress single molecules of lysozyme as adsorbed on a mica surface and analysed the data to obtain Y to be in the range of 500±200 MPa. We used the same method as that used by Radmacher et al. and obtained the lower limit estimate of Y of 75 MPa for bovine carbonic anhydrase in the native state and 2 MPa in its denatured state (Afrin et al. 2005).
The measurement of mechanical parameters of proteins and protein-based biostructures in our laboratory aims at, first, to understand the mechanical background of their biological activities, and second, to form the basis of mechanical manipulation of live cells for biomedical applications.
An enzyme reaction is a good example to illustrate the former interest. Before and after the reaction of converting the substrate S to the product P as catalysed by an enzyme E, both (E+S) and (E+P) states are equilibrated rapidly with little residual mechanical stresses. However, in the activated state of the reaction, both E and S are in strained conformations. An enzyme molecule must have sufficient rigidity to sustain the mechanical stress imposed on it during the activation process (Vanselow 2002), but there has not been an appropriate method to detect the presence or absence of rigid substructures within a single molecule of protein until the recent advent of the atomic force microscope (AFM; Sarid 1994).
The AFM has two operating modes, i.e. imaging and force mode. In this article, we focus on the results obtained from the force mode operation of AFM by pulling and pushing macromolecular samples. When the AFM is operated in the force mode, the piezo tube under the sample stage makes only a vertical (z) movement at a fixed lateral position so that it is possible to stretch a protein molecule from its two ends while constantly monitoring the tensile force as a function of chain extension (Mitsui et al. 1996; Rief et al. 1997; Wang & Ikai 1999). To ensure the correct analysis of force versus extension relationship of a protein molecule (F–E curve), it must be established, at least to a reasonable level, that the observation is made on a single chain of the sample protein and not on multiple chains. We and most other researchers in this field use the following criterion. The percentage of successful recording of chain elongation must be less than 10–20% of the total number of trials so that the probability of double or even triple capture events would be negligibly small, i.e. less than 0.1–0.4%.
In many cases, the affinity of protein–ligand interactions can be quantified by measuring the binding constant (K), but there are cases where measurement of K is not easy due to the exceedingly high value of the association constant. One such example is the anchoring of membrane proteins to the lipid bilayer membrane (Bell 1978), where the measurement of anchoring force is more practical than that of binding constant. We wanted to test Bell's estimate for the anchoring force of a membrane protein to the cell surface by using AFM.
One promising application of nanomechanical measurement at the molecular level is the detection of specific receptor molecules on the live cell surface using an AFM probe modified with covalently immobilized specific ligand molecules. The probe makes a two-dimensional survey over the cell surface recording the interaction force between the probe and the surface as the indication of the presence of receptor molecules with biochemical specificity to the ligand on the probe (Ikai et al. 2002; Kim et al. 2003, 2004). In this respect, it is also important to establish the range of force required to extract membrane proteins from the live cell surface in comparison with the unbinding force of the ligand–receptor pairs of interest (Afrin et al. 2003, 2004; Ikai & Afrin 2003).
Recently, we and several other groups realized that the pulling mechanics of the intrinsic membrane protein is strongly influenced by the presence or absence of the non-covalent interactions between the targeted protein and the cytoskeletal structure via linker proteins. In this respect, we analysed the pulling force curve of glycophorin A and Band 3 protein from the surface of the red blood cell (Afrin & Ikai 2006).
If the AFM probe is allowed to penetrate into the cytoplasm, it is possible to extract intracellular macromolecules as adsorbed to the probe. Uehara et al. have shown that, by combining this procedure with RT-PCR and PCR methods, local concentrations of specific mRNAs could be calculated (Osada et al. 2003; Uehara et al. 2004).
2. Material and Methods
(a) Chemicals and proteins
The silanization reagent, 3-aminopropyltriethoxysilane (APTES), was obtained from Shin-Etsu Chemical Co. (Tokyo, Japan). Disuccinimidyl suberate (DSS) and N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) were purchased from Pierce (Rockford, IL). They were stored under a highly dehydrated and low temperature conditions and used without further purification. PBS+ (phosphate-buffered saline with Ca2+) as a medium for cells during AFM experiments was obtained from Gibco. The other chemicals were bought from Sigma (St Louis, MO). The proteins used in the stretching experiments were produced by the recombinant method with the insertion of cysteine residues immediately outside the N and C termini together with N-terminal histidine tags to facilitate purification by affinity chromatography (Alam et al. 2002).
(b) Cells
Mouse 3T3 fibroblast cells were grown in Dulbecco's modified minimum Eagle's medium for several days and the medium was changed to PBS in the final stage of AFM experiments. The red blood cells were prepared from fresh supplies of blood. A droplet of blood is diluted and washed with PBS five times by centrifugation at 2000g for 5 min.
(c) Atomic force microscope
Several commercially available AFM instruments were used including a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA), an NVA 100 AFM (Olympus, Tokyo, Japan) and a BioScope (Veeco Japan, Tokyo, Japan). Cantilevers were purchased from Olympus and Veeco. The cantilevers and crystalline silicon substrate were silanized with APTES and further modified with SPDP to make their surface reactive to -SH groups on the engineered proteins with cysteine residues. For immobilization of ligand proteins, AFM tips were modified with DSS so that the proteins could be immobilized through their amino groups to the tips. Gold-coated cantilevers (Olympus) were also used after direct modification with SPDP. For membrane protein extraction, silicon nitride cantilevers were modified with DSS and gold-coated ones with SPDP.
3. Results
(a) Protein stretching experiment
The probe and the sample stage are chemically functionalized so that macromolecules caught in between them are covalently cross-linked to the tip on the one end and to the substrate on the other. The sample molecule thus sandwiched between the tip and the substrate will then be put under a continuous tensile stress by increasing the tip-stage distance. The sandwiched sample will then be stretched until one or more of the weaker bonds in the system are broken. During the stretching process of the protein molecule, the mechanical response of the sample against the tensile stress produces a downward deflection of the cantilever which holds the probe at its free end (Mitsui et al. 1996). The extent of cantilever deflection, d, is detected by a bisected photodiode detector and, by knowing the cantilever force constant, the tensile force inflicted on the sample is calculated as F=−kd. The extension of the sample during the process of increasing the probe-stage distance (E) is calculated as (D−d), where D is the distance between the probe and the sample surface.
(i) Bovine carbonic anhydrase II (BCA II)
3.1.1.1 Molecular structure
BCA II has 259 amino acid residues with predominantly β-sheet protein, and especially the active site is built on the core β-sheet structure in the central part of the molecule. Three histidine residues are liganded to hold a zinc ion in the active centre. The terminal end of the protein is apparently folded to form a pseudoknot. From the number of amino acid residues and assuming the effective length per each residue is approximately 0.35–0.37 nm, the total contour length of the molecule was estimated to be 95–100 nm depending on the magnitude of the failure force of the cross-linking system.
3.1.1.2 Mechanical stretching from N and C termini
To stretch the protein from its two ends, two cysteine residues were inserted at both ends of the molecule, one for each end using the recombinant technology. The surface of crystalline silicon was chemically functionalized with the silanization reagent, APTES, as well as that of silicon nitride probes for AFM. Both functionalized surfaces were further modified with the covalent cross-linker, SPDP, which has a succinimidyl group to react with the amino end of APTES on the substrate, and a pyridyldithio group to form a covalent bond with the cysteine residue on BCA II.
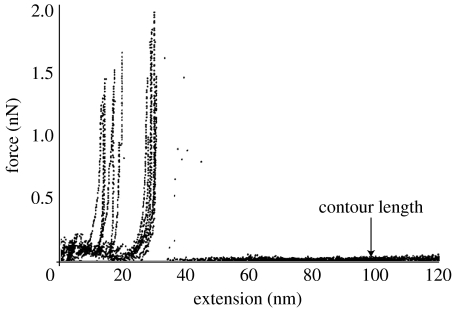
When the molecule was stretched by a tensile force applied from the AFM cantilever, it extended to approximately 20–30 nm with a small force less than 100 pN after which the force abruptly started to rise with little extension of the molecule (Wang & Ikai 1999; Afrin et al. 2005), which was taken as an indication of the knot-tightening effect. The force rose to approximately 2 nN and suddenly the force was reduced to zero, indicating a breakdown somewhere in the covalently linked system. The result suggested that the knot tightened without allowing a slippage of the still-folded main body of the protein. Only very rarely was a continuous stretching of the molecule after breakdown of the knot observed (figure 1).
Figure 1.
Stretching curve of native BCA II which had the intact knot structure. The protein was stretched up to approximately 20–30 nm with a low force but then the force rapidly increased to higher than 1.5 nN where covalent system was broken. Reproduced from Afrin et al. (2005) with permission.
3.1.1.3 Knot-free stretching and type I and II conformers
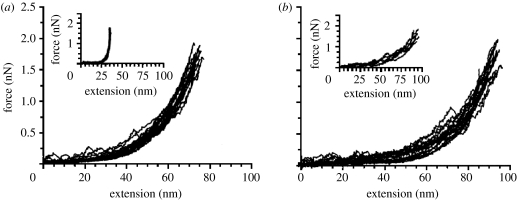
Alam et al. prepared a mutant protein with a replacement of the glutamine residue at the 253rd position instead of the C-terminal with a cysteine residue (Alam et al. 2002). Cysteine residue addition at the N terminus was kept as above. In this case, since the position of the 253rd residue is displaced from the knot-forming interaction at the C terminus, stretching the molecule from this position and the N terminus was expected to give knot-free stretching mechanics. Indeed, this time, stretching of BCA II was much easier than that for the native form of the protein, the extension reaching approximately 70–100 nm. Furthermore, by adding the known inhibitor of the enzyme, they successfully showed that the mutant protein actually contained two conformational isomers, one called type I with full enzymatic activity and a three-dimensional conformation almost exactly the same as that of the native protein (Saito et al. 2004), and the other conformer called type II which was enzymatically inactive and not crystallizable. Type II had a highly folded conformation as far as the optical properties such as CD or fluorescence spectra were concerned, suggesting that its secondary structure folding is almost complete but lacked some special packing of the secondary structures into the native three-dimensional structure. The pulling curves of type I and II conformers are given in figure 2 with the inset of pulling curves in the presence of an inhibitor.
Figure 2.
F–E curves of (a) type I and (b) type II conformers of BCA II with the replacement of Gln253 to Cys. The insets are curves obtained in the presence of an inhibitor. Reproduced from Alam et al. (2002) with permission.
Occasionally, a transition from type I F–E curve to that of type II was observed in the process of stretching of the mutant BCA II after approximately 60–70 nm in extension. The critical force was approximately 1 nN. The transition was interpreted as the cooperative breakdown of the final rigid core in the molecule showing a brittle fracture behaviour.
A partial digestion of the C-terminal residues by a carboxypeptidase treatment followed by MALDI-TOF analysis of the molecular weight of the protein indicated that the C-terminal region of type II was not folded as tightly as that in the native protein. It was suggested that, in type II, the knot structure was not formed leaving the last compacting step of folding unfinished.
3.1.1.4 Inhibitor binding
Inhibitor binding to the native as well as to type I conformers made them softer in the initial 30–40 nm of stretching but then the force went up to the level of covalent bond breaking. The result was in agreement with the previous observation of the change in the thermal factor in X-ray crystallography before and after inhibitor binding. After inhibitor binding, the thermal factor increased in the peripheral region of the molecule, but decreased in its central region.
3.1.1.5 Stretching of partially and completely denatured forms
Afrin et al. studied the stretching curves of the partially denatured forms of BCA II (Afrin et al. 2005) in the presence of 2M GdmCl. The protein was extensible to its full length most of the time indicating that the knot structure had been destroyed. The compression experiment showed that, with a height data of 3.5–5 nm, the protein was not expanded as much as the completely denatured form, which had a height of approximately 9 nm.
(ii) Stretching of OspA and calmodulin
When the globular protein, OspA, was sandwiched between the AFM tip and the substrate using bifunctional covalent cross-linkers and subsequently stretched from two ends as the distance between the tip and the substrate was increased, we observed F–E curves with two force peaks (Hertadi & Ikai 2002).β-sheet, it was natural to interpret that the two force peaks corresponded to the breakdown of the two domains and the low stress stretching in the initial 30–40 nm represented unzipping of the central β-sheet. To prove this intuitive interpretation was at least partly correct, Hertadi et al. constructed two amino acid replacements in the central region of the molecule. The first case was a single amino acid replacement and the second was the insertion of an extra β-sheet in the mid-region of the molecule. In both the cases, the original two-peak pattern was changed to single-peak patterns indicating that the central β-sheet region was responsible for at least one of the two original force peaks. The F–E curve in the presence of Ca2+ has a single force peak and the one without Ca2+ did not show any force peaks (Hertadi & Ikai 2002).
In all three cases of BCA II, OspA and calmodulin, the presence of locally rigid structures was detected by the AFM stretching experiments. It is most likely that each protein structure is characterized by folding with locally varying mechanical rigidity.
(iii) Molecular dynamics simulation
In the application of computer simulation to protein stretching, steered molecular dynamics (SMD) has been used in several cases with some success (Izrailev et al. 1997; Gao et al. 2002; Brockwell et al. 2003). In the SMD simulation, a spring was attached between the model AFM tip and the protein. Since the tip is moved at a constant velocity, the protein unfolding may not take place at the same velocity. The difference in the two velocities will stretch the hypothetical spring whose extension will be interpreted as the tensile force on the protein. We applied this method to the unfolding process of carbonic anhydrase and obtained results consistent with experimental observations (Ohta et al. 2004).
(b) Protein compression
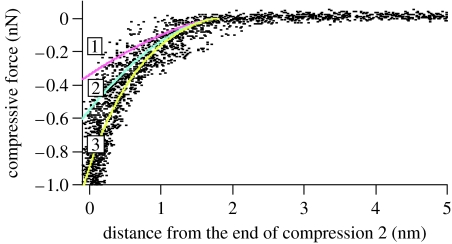
Since the sample deformation is so extensive and the shape change is not known, it is difficult to obtain mechanical parameters such as the Young modulus from the analysis of force curves. We made an attempt to do so in Ikai (2005) but the result is strongly model dependent. It is therefore better to analyse compression curves to obtain such parameters. Radmacher reported the result of analysing the compression curve of bound lysozyme on a solid surface obtained on the AFM according to the theoretical model of Hertz (Hertz 1882; Johnson 1985) and obtained an estimate of the Young modulus of 500±200 MPa (Radmacher et al. 1994). Afrin & Ikai (2006) applied a similar method to obtain the Young modulus of carbonic anhydrase II of bovine origin (BCA II). The protein was derivatized with two cysteine residues each at two ends of the molecule and cross-linked to a functionalized silicon surface with APTES and SPDP. After washing unbound BCA II from the surface, a similarly modified AFM probe was brought into contact with the surface of the immobilized protein so that another covalent bond was formed between the probe and the protein, thereby sandwiching BCA II molecules between the probe and the substrate. The probe was further closed in onto the substrate so that the sandwiched protein was compressed and the force–indentation relationship was obtained as shown in figure 3.
Figure 3.
Compression curve of BCA II. Experimental results are given in dots and theoretical fitting curves are (1) Hertz model, (2) Tatara model, and (3) exponential curve. Reproduced from Afrin & Ikai (2006) with permission.
An analysis of the compression curve was done according to the modified version of the Hertz model, i.e. the Tatara model which was developed in 1980s and 1990s to deal with the compression curves of macroscopic rubber spheres obtained under the condition of a large deformation regime (Tatara 1989, 1991; Tatara et al. 1991–2). The original Hertz model was constructed on the assumption that the deformation resulting from the forced contact of two spheres is small so that there is no lateral extension of the spheres, whereas the Tatara model allows a large deformation leading to lateral extension. The compression curve of BCA II was fitted for over 50% of the extent to one of the theoretical models of Tatara giving a constant value of 75 MPa as the Young modulus. A forced fitting of the Hertz model curve over the same range gave a two to three times larger value. The values of the Young modulus obtained so far represent the lower limits of BCA II under given conditions because the reversibility of compression might not have been complete. The compression process of BCA II was simulated by molecular dynamics simulations by Tagami et al. (2006). The compression curve of a completely denatured protein in 6 M guanidinium chloride (GdmCl) was fitted with a single value of 2 MPa for the Young modulus for approximately 80% of the entire range.
As the concentration of GdmCl was increased from 0 to 3 M, the Young modulus of BCA II decreased rapidly from 75 MPa to lower values but its height increased only slightly suggesting that the protein became much softer without increasing its diameter very much. The result confirmed that the denaturation process of BCA II proceeded via unfolding intermediates which could be characterized as keeping the compact globular structure of the native state but became much more loosened with regard to the native segmental interactions.
(c) Protein–protein interactions
(i) Biotin–avidin interaction
The measurement of protein–protein interaction is another popular area of application of force spectroscopy. As the first example of such experiment, the well-characterized ligand–receptor system of biotin–avidin (Florin et al. 1994) and antibody–antigen systems were investigated (Schwesinger et al. 2000).
(ii) Chaperonine system studied by non-compressive force spectroscopy method
Chaperonine is a protein known to help newly synthesized proteins correctly form biologically active, specific three-dimensional structures from the disordered conformation in the initial stage (Hartl & Hayer-Hartl 2002). It has been shown that chaperonine has a sevenfold rotational symmetry arising from the circular arrangement of seven subunits in one tire and each subunit has a single binding site for an unfolded polypeptide. After binding to such sites, the denatured protein is transferred into the molecular cavity of the chaperonine with the help of ATP where the protein-folding reaction can take place. Because unbinding of the denatured protein from the binding site on the subunit plays a crucial step in the internalization of unfolded protein into the chaperonine cavity, it is of interest to know the magnitude of unbinding force at this site.
The AFM tip modified with pepsin molecules that were denatured at an experimental pH of 7.4 was brought to bind with immobilized GroEL (a chaperonine) on the surface of the substrate (Sekiguchi et al. 2003). The tip and the substrate were kept at a non-contact distance during the force curve measurement to avoid unnecessary compression of GroEL.
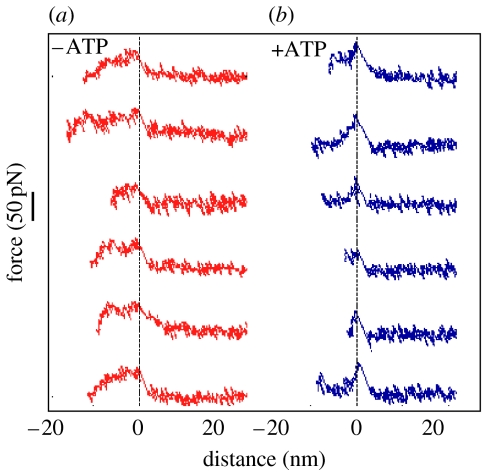
The F–E curves obtained in the absence of ATP showed a more or less flat plateau of approximately 11 nm and with a force of 42±17 pN, which was terminated by a jump to the horizontal position of the cantilever. The appearance of the plateau was interpreted as resulting from successive unbinding of pepsin from multiple binding sites on GroEL. The representative F–E curves are shown in figure 4.
Figure 4.
F–E curves of denatured pepsin detachment from GroEL in the (a) absence or (b) presence of ATP. Reproduced from Sekiguchi et al. (2003) with permission.
In the presence of ATP where the seven binding sites are widely spread out, F–E curves had a single sharp force peak without plateau. The result was interpreted as, because the distance between binding sites on a GroEL became large, pepsin could interact with only one or at the most two binding sites. The numerical value of the unbinding force was similar to that obtained in the absence of ATP confirming that the effect of ATP is restricted to the change in the geometry of GroEL without significant influence on the binding property of individual sites.
(d) Membrane protein mapping
The method to measure the force to unbind ligand–receptor pairs was applied to the cell surface receptors as a new method of mapping the presence of a particular receptor type on the surface (Ikai et al. 2002; Kim et al. 2003; Kim et al. 2004). In one of such applications, the AFM tip was covered with the protein, α2-macroglobulin, and scanned over the surface of a living cell in the culture medium point by point to find the presence of its specific receptor. Ideally, the force to separate the cantilever from the cell surface is zero where there is no receptor, and non-zero, finite value when the ligand–receptor interaction is established (Ikai et al. 2002).
This mapping method was later extended to several different membrane systems and the advantages and disadvantages of the AFM-based mapping method in contrast to conventional methods based on receptor labelling with fluorescent dyes were discussed. A problem in the force mapping of receptors was the lack of confirmation that the force measured at the end of the force response was actually that of the unbinding of ligand–receptor pairs. If the force was due to some other events such as the uprooting of receptor molecules from the cell surface, the receptor mapping method loses ground. If the unbinding force is comparable or even larger than the force required to extract receptors from the cell membrane, receptors would have been extracted with a finite probability that obscures the result of force mapping. It was necessary therefore for us to devise a new experiment in which membrane proteins were to be extracted with certainty and as a result establish the force range of protein extraction separately from ligand–receptor unbinding events.
(e) Membrane protein extraction
The cell membrane is a composite structure of phospholipids and proteins. Among cell surface-associated proteins, the intrinsic proteins are those that span the lipid membrane from one side to the other having a central hydrophobic segment connecting its extracellular domain to the cytoplasmic one and securely anchored to the membrane. In general, they are not displaced from the membrane unless the membrane is destroyed by, for example, detergents.
It is of fundamental and practical interest to measure the force to dislodge the intrinsic membrane proteins from the cell surface by forcibly pulling them out by AFM. For this purpose, we have to make sure that the weakest bond in the force spectroscopy system between the tip and the substrate is the targeted bond or interaction (Afrin et al. 2003, 2004). We therefore used the modified AFM tip with the amino-reactive bifunctional covalent cross-linkers, such as SPDP, sulfo-SPDP and DSS. Once covalent bonds are formed between the cross-linkers on the tip and the amino bearing molecules on the cell surface, the subsequent separation of the tip from the cell surface should disrupt non-covalent interactions between the membrane and the intrinsic protein, provided such interactions could be ruptured with a force significantly less than the force needed to sever a covalent bond (Grandbois et al. 1999; Afrin et al. 2004).
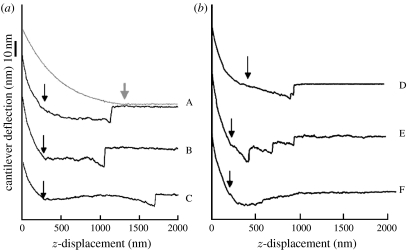
Some of the experimentally observed force curves obtained by pulling membrane proteins by using covalent cross-linking system under such an experimental set-up are shown in figure 5.
Figure 5.
Various pulling force curve of membrane proteins from the cell surface showing long extension on the retraction regime and stepwise release of the AFM probe from the cell surface. (a,b) Curve A has both approach (light grey line with an arrow showing the contact to the cell surface) and retraction curves (dark lines with an arrow showing detachment point from the cell surface). For B to F, only retraction parts are given. The AFM probes were modified with bifunctional covalent cross-linkers. Reproduced from Afrin et al. (2004) with permission.
When a modified tip with covalent cross-linkers was used, a prolonged downward deflection corresponding to the tether pulling from the cell membrane in the retraction regime was terminated abruptly. Sometimes, two or three such jumps were observed before the final one.
When the force difference associated in the jump events is collected as a histogram, the majority of the force value clustered between 200 and 700 pN with a mean of 450 pN and standard deviation of 220 pN. It was shown that the AFM tip used in this experiment actually carried the tensile material on its surface which showed extensive elongation that was easily removed by the treatment with proteolytic enzymes, proving that the tensile material on the tip was protein. The identification of the extracted proteins was attempted by the immunofluorescence method based on which integrins and fibronectin were identified (Afrin et al. 2003).
Almost a flat dependence of the extraction force on the loading rate was apparent and it confirmed the theoretical framework of Bell's proposition that the activation distance of protein extraction from the lipid bilayer would be as long as a few nanometres thus reducing the slope of the mean force versus log[loading rate] plot as low as a few piconewtons. Under a similar loading rate condition, the force of uprooting intrinsic membrane protein is at least five times greater than the unbinding force of the ligand–receptor interaction involving integrins and fibronectin, and it is safe to conduct receptor mapping experiment based on the force spectroscopic method.
(f) Membrane protein pulling
We have recently tried to pull out intrinsic membrane proteins from the surface of red blood cells using modified AFM probes with several species of lectins which have specific binding affinities towards some membrane proteins (Afrin et al. 2006). For example, wheat germ agglutinin (WGA) and Psathyrella velutina lectin (PVL) are specific for glycophorin A, whereas concanavalin A binds Band 3 otherwise known as the anion exchanger protein.
When WGA-modified probes were used to pull glycophorin A on the red blood cell surface, Afrin et al. (2006) obtained force curves with a long plateau force extending up to 1–2 μm and followed by a sudden release of the cantilever from the cell surface with a single- or two-step jumps.
The force curves obtained using AFM probes modified with concanavalin A were classified into types I and II. Type I curves were similar to those obtained with WGA on the tip, but type II curves were different as they were characterized with the appearance of sharp force peaks in addition to long plateau forces.
After deglycosylation, the force curves were collected by using AFM probes modified with the covalent bifunctional cross-linker, SPDP, reactive towards amino groups on the cell surface. Approximately two-thirds to three-fourths of the force curves showing non-zero force in the retraction regime could be classified as type II according to the previous criteria. Almost all force curves obtained on deglycosylated and heat-treated red blood cells, where cytoskeletal structures were disrupted, were reduced to type I, strongly suggesting that the force peaks observed prior to deglycosylation were indeed due to interaction between membrane proteins and the cytoskeletal structure.
(g) mRNA extraction
Previously, Uehara et al. demonstrated that the AFM-based method of mRNA extraction was useful to reveal the localization of β-actin mRNA inside of the cell (Osada et al. 2003; Uehara et al. 2004). In the resting state of cultured fibroblasts, β-actin mRNAs were shown to be localized in proximal regions to the nucleus but, when the cells were activated by the addition of foetal calf serum into the culture medium, mRNAs were redistributed to a distal region from the nucleus especially to the front side of cell movement.
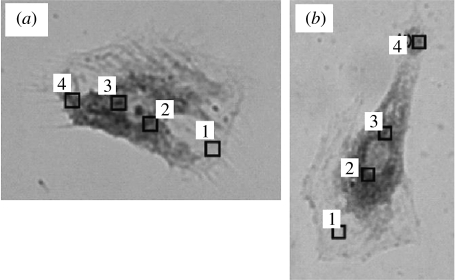
A new result was obtained in regard to the agreement between the AFM-based mRNA detection and the conventional FISH method (Uehara et al. 2007). First, AFM probes were used to recover β-actin mRNA from several loci of live cells and used for RT-PCR conversion and amplification of the recovered mRNA. The same cells were later fixed using glutaraldehyde and stained with fluorescently labelled complementary RNAs against β-actin mRNAs. The result of the two methods are shown in figure 6 in the form of FISH results as fluorescence microscopic photographs with overlaid site maps of AFM probe insertion loci.
Figure 6.
The result of matching map of mRNA distribution as probed by AFM-based recovery method and that of the traditional FISH method. The dark staining inside of the cell represents the concentrated presence of mRNA of β-actin. The number of similar mRNAs as recovered on AFM tips from numbered loci. The results on two different cells are given in (a) mRNA (fg) (1) 0, (2) 44, (3) 471, (4) 2 and (b) mRNA (fg) (1) 0, (2) 65, (3) 300, (4) 0. Reproduced from Uehara et al. (2007).
In general, the agreement between the two methods was excellent, meaning that from the strongly stained area by FISH method, large numbers of mRNAs were recovered by the AFM-based method. At a few locations where the AFM probe did not reveal the recovery of mRNAs despite fairly strong staining by FISH method, the result of the AFM-based method was taken as correct, emphasizing the high lateral resolution of the AFM-based method and possible change in the mRNA location during fixing procedure for FISH method.
This method was also used for detecting time-dependent change of several species of mRNA in the fibroblast cell after stimulation of cellular activity from the resting stage by the addition of foetal calf serum to the culture medium (Uehara et al. 2007).
4. Discussion
The measurement of force to disrupt biostructures based on protein–protein and protein–lipid interactions is the focus of this article. The results of recent measurements are expected to reveal the mechanical principles of the internal architecture of these structures which cannot be explicitly measured by the conventional technology and therefore have not been investigated in the past.
The F–E curves obtained during the process of forced unfolding of several globular proteins gave a rare insight into the presence of rigidly folded local substructures inside the native conformation of such proteins. It is noteworthy that the classical question of how can proteins be soft and yet sustain the activated state of covalently bonded molecules has been partially answered by the new evidence for the presence of locally rigid substructures. Owing to the presence of such substructures, the internal mechanics of protein is rather more complex than hitherto imagined. It is also true, as has been amply demonstrated, that proteins cannot be just rigid. The rigid parts of a protein molecule must coordinate with the more flexible and soft parts of the same molecule for the whole molecule to perform its biological roles in unison.
5. Conclusions
We have established the method to investigate hitherto unexplored physical properties of globular proteins using atomic force spectroscopy based on the force curve mode of the AFM. Globular proteins have been shown to possess mechanically rigid substructures which, when under tensile stress, showed fracture mechanics similar to brittle materials. Membrane proteins can be extracted from the cell surface by a force lesser than that required to break a covalent bond but significantly greater than that needed to separate ligand–receptor pairs on the cell surface. Receptor mapping and cell membrane harvest can, therefore, be done in distinct force ranges. The application of AFM to explore cytoplasmic localization of specific mRNAs will prove to be a useful biotechnological methodology.
Acknowledgments
This work was supported by grants-in-aid to A.I. from the JSPS Research for the Future Program (99R16701) and Scientific Research on Priority Areas (B).
Footnotes
One contribution of 17 to a Theme Issue ‘Japan: its tradition and hot topics in biological sciences’.
References
- Afrin R, Ikai A. Force profiles of protein pulling with or without cytoskeletal links studied by AFM. Biochem. Biophys. Res. Commun. 2006;348:238–244. doi: 10.1016/j.bbrc.2006.07.050. doi:10.1016/j.bbrc.2006.07.050 [DOI] [PubMed] [Google Scholar]
- Afrin, R., Takahashi, I., Ohta, S. & Ikai, A. 2003 Mechanical unfolding of alanine based helical polypeptide: experiment vs. simulation. Presented at the 47th Annual Meeting of the American Biophysical Society, San Antonio, Texas, USA, March 1–5 2003
- Afrin R, Yamada T, Ikai A. Analysis of force curves obtained on the live cell membrane using chemically modified AFM probes. Ultramicroscopy. 2004;100:187–195. doi: 10.1016/j.ultramic.2004.01.013. doi:10.1016/j.ultramic.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Afrin R, Alam M.T, Ikai A. Pretransition and progressive softening of bovine carbonic anhydrase II as probed by single molecule atomic force microscopy. Protein Sci. 2005;14:1447–1457. doi: 10.1110/ps.041282305. doi:10.1110/ps.041282305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M.T, Yamada T, Carlsson U, Ikai A. The importance of being knotted: effects of the C-terminal knot structure on enzymatic and mechanical properties of bovine carbonic anhydrase II. FEBS Lett. 2002;519:35–40. doi: 10.1016/s0014-5793(02)02693-5. doi:10.1016/S0014-5793(02)02693-5 [DOI] [PubMed] [Google Scholar]
- Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. doi:10.1126/science.347575 [DOI] [PubMed] [Google Scholar]
- Brockwell D.J, Paci E, Zinober R.C, Beddard G.S, Olmsted P.D, Smith D.A, Perham R.N, Radford S.E. Pulling geometry defines the mechanical resistance of a beta-sheet protein. Nat. Struct. Biol. 2003;10:731–737. doi: 10.1038/nsb968. doi:10.1038/nsb968 [DOI] [PubMed] [Google Scholar]
- Florin E.L, Moy V.T, Gaub H.E. Adhesion forces between individual ligand–receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. doi:10.1126/science.8153628 [DOI] [PubMed] [Google Scholar]
- Gao M, Craig D, Vogel V, Schulten K. Identifying unfolding intermediates of FN-III10 by steered molecular dynamics. J. Mol. Biol. 2002;232:939–950. doi: 10.1016/s0022-2836(02)01001-x. doi:10.1016/S0022-2836(02)01001-X [DOI] [PubMed] [Google Scholar]
- Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub H.E. How strong is a covalent bond? Science. 1999;265:1727–1730. doi: 10.1126/science.283.5408.1727. doi:10.1126/science.283.5408.1727 [DOI] [PubMed] [Google Scholar]
- Hartl F.U, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. doi:10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- Hertadi R, Ikai A. Unfolding mechanics of holo- and apocalmodulin studied by the atomic force microscope. Protein Sci. 2002;11:1532–1538. doi: 10.1110/ps.3600102. doi:10.1110/ps.3600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz H. Über die Berührung festen elastische Körper. J. Reine und Angewandte Mathematik. 1882;92:152–171. [Google Scholar]
- Howard J. Sinauer Associates, Inc; Sunderland, MA: 2001. Mechanics of motor proteins and the cytoskeleton. [Google Scholar]
- Ikai A. Local rigidity of a protein molecule. Biophys. Chem. 2005;116:187–191. doi: 10.1016/j.bpc.2005.04.003. doi:10.1016/j.bpc.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Ikai A, Afrin R. Toward mechanical manipulations of cell membranes and membrane proteins using an atomic force microscope: an invited review. Cell Biochem. Biophys. 2003;39:257–277. doi: 10.1385/CBB:39:3:257. doi:10.1385/CBB:39:3:257 [DOI] [PubMed] [Google Scholar]
- Ikai A, Afrin R, Itoh A, Thogersen H.C, Hayashi Y, Osada T. Force measurements for membrane protein manipulation. Colloids Surf. B: Biointerfaces. 2002;23:165–171. doi:10.1016/S0927-7765(01)00230-2 [Google Scholar]
- Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin–biotin complex. Biophys. J. 1997;59:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.L. Contact Mechanics. Cambridge University Press; Cambridge UK: 1985. p. 56. [Google Scholar]
- Kim H, Arakawa H, Osada T, Ikai A. Quantification of cell adhesion force with AFM: distribution of vitronectin receptors on a living MC3T3-E1 cell. Ultramicroscopy. 2003;97:359–363. doi: 10.1016/S0304-3991(03)00061-5. doi:10.1016/S0304-3991(03)00061-5 [DOI] [PubMed] [Google Scholar]
- Kim H, Tsuruta S, Arakawa H, Osada T, Ikai A. Quantitative analysis of the number of antigens immobilized on a glass surface by AFM. Ultramicroscopy. 2004;100:203–210. doi: 10.1016/j.ultramic.2004.01.015. doi:10.1016/j.ultramic.2004.01.015 [DOI] [PubMed] [Google Scholar]
- Kojima H, Ishijima A, Yanagida T. Direct measurement of stiffness of single actin filaments with and without tropomyosin by in vitro nanomanipulation. Proc. Natl Acad. Sci. USA. 1994;91:12 962–12 966. doi: 10.1073/pnas.91.26.12962. doi:10.1073/pnas.91.26.12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Hara M, Ikai A. Mechanical unfolding of alpha2-macroglobulin molecules with atomic force microscope. FEBS Lett. 1996;385:29–33. doi: 10.1016/0014-5793(96)00319-5. doi:10.1016/0014-5793(96)00319-5 [DOI] [PubMed] [Google Scholar]
- Morozov V.N, Morozova T.Ya. Viscoelastic properties of protein crystals: triclinic crystals of hen egg white lysozyme in different conditions. Biopolymers. 1981;20:451–467. doi: 10.1002/bip.1981.360200304. doi:10.1002/bip.1981.360200304 [DOI] [PubMed] [Google Scholar]
- Ohta S, Alam M.T, Arakawa H, Ikai A. Origin of mechanical strength of bovine carbonic anhydrase studied by molecular dynamics simulation. Biophys. J. 2004;87:4007–4020. doi: 10.1529/biophysj.104.045138. doi:10.1529/biophysj.104.045138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada T, Uehara H, Kim H, Ikai A. mRNA analysis of single living cells. J. Nanobiotechnol. 2003;1:2–7. doi: 10.1186/1477-3155-1-2. doi:10.1186/1477-3155-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmacher M, Fritz M, Clevel J.P, Walters D.A, Hansma P.K. Imaging adhesion forces and elasticity of lysozyme adsorbed on mica with the atomic force microscope. Langmuir. 1994;10:3809–3814. doi:10.1021/la00022a068 [Google Scholar]
- Rief M, Gautel M, Oesterhelt F, Fernandez J.M, Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. doi:10.1126/science.276.5315.1109 [DOI] [PubMed] [Google Scholar]
- Saito R, Sato T, Ikai A, Tanaka N. Structure of bovine carbonic anhydrase II at 1.95 A resolution. Acta Crystallogr. D Biol. Crystallogr. 2004;70:792–795. doi: 10.1107/S0907444904003166. doi:10.1107/S0907444904003166 [DOI] [PubMed] [Google Scholar]
- Sarid D. Oxford University Press; Oxford, UK: 1994. Scanning force microscopy. [Google Scholar]
- Schwesinger F, Ros R, Strunz T, Anselmetti D, Güntherodt H.J, Honegger A, Jermutus L, Tiefenauer L, Plückthun A. Unbinding forces of single antibody–antigen complexes correlate with their thermal dissociation rates. Proc. Natl Sci. Acad. USA. 2000;97:9972–9977. doi: 10.1073/pnas.97.18.9972. doi:10.1073/pnas.97.18.9972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi H, Arakawa H, Taguchi H, Ito T, Kokawa R, Ikai A. Specific interaction between GroEL and denatured protein measured by compression-free force spectroscopy. Biophys. J. 2003;85:484–490. doi: 10.1016/S0006-3495(03)74493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda H, Sugimoto M, Chiba M, Uemura C. Direct measurement for elasticity of myosin head. Biochem. Biophys. Res. Commun. 1995;211:219–225. doi: 10.1006/bbrc.1995.1799. doi:10.1006/bbrc.1995.1799 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Koizumi H, Kojima K. Temperature dependence of microhardness of tetragonal hen-egg-white lysozyme single crystals. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;84:051 921–051 924. [Google Scholar]
- Tagami K, Tsukada M, Afrin R, Sekiguchi H, Atsushi Ikai A. Discontinuous force compression curve of single bovine carbonic anhydrase molecule originated from atomistic slip. J. Surf. Sci. Nanotechnol. 2006;4:552–558. doi:10.1380/ejssnt.2006.552 [Google Scholar]
- Tatara Y. Extensive theory of force–approach relations of elastic spheres in compression and impact. J. Eng. Mater. Technol. 1989;111:163–168. [Google Scholar]
- Tatara Y. On compression of rubber elastic sphere over a large range of displacements—part 1: theoretical study. J. Eng. Mater. Tech. 1991;113:285–291. [Google Scholar]
- Tatara Y, Shima S, Lucero J.C. On compression of rubber elastic sphere over a large range of displacements—part 2: comparison of theory and experiment. J. Eng. Mater. Tech. 1991–2;113:292–295. [Google Scholar]
- Uehara H, Osada T, Ikai A. Quantitative measurement of mRNA at different loci within an individual living cell. Ultramicroscopy. 2004;100:197–201. doi: 10.1016/j.ultramic.2004.01.014. doi:10.1016/j.ultramic.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Uehara H, Kunitomi Y, Ikai A, Osada T. mRNA detection of individual cells with the single cell nanoprobe method compared with in situ hybridization. J. Nanobiotech. 2007;5:7. doi: 10.1186/1477-3155-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow D.G. Role of constraint in catalysis and high-affinity binding by proteins. Biophys. J. 2002;82:2293–2303. doi: 10.1016/S0006-3495(02)75575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ikai A. Protein stretching III: force-extension curves of tethered bovine carbonic anhydrase B to the silicon substrate under native, intermediate and denaturing conditions. Jpn J. Appl. Phys. 1999;38:3912–3917. doi:10.1143/JJAP.38.3912 [Google Scholar]