Abstract
Rock surfaces are unique terrestrial habitats in which rapid changes in the intensity of radiation, temperature, water supply and nutrient availability challenge the survival of microbes. A specialised, but diverse group of free-living, melanised fungi are amongst the persistent settlers of bare rocks. Multigene phylogenetic analyses were used to study relationships of ascomycetes from a variety of substrates, with a dataset including a broad sampling of rock dwellers from different geographical locations. Rock-inhabiting fungi appear particularly diverse in the early diverging lineages of the orders Chaetothyriales and Verrucariales. Although these orders share a most recent common ancestor, their lifestyles are strikingly different. Verrucariales are mostly lichen-forming fungi, while Chaetothyriales, by contrast, are best known as opportunistic pathogens of vertebrates (e.g. Cladophialophora bantiana and Exophiala dermatitidis, both agents of fatal brain infections) and saprophytes. The rock-dwelling habit is shown here to be key to the evolution of these two ecologically disparate orders. The most recent common ancestor of Verrucariales and Chaetothyriales is reconstructed as a non-lichenised rock-inhabitant. Ancestral state reconstructions suggest Verrucariales as one of the independent ascomycetes group where lichenisation has evolved on a hostile rock surface that might have favored this shift to a symbiotic lifestyle. Rock-inhabiting fungi are also ancestral to opportunistic pathogens, as they are found in the early diverging lineages of Chaetothyriales. In Chaetothyriales and Verrucariales, specific morphological and physiological traits (here referred to as extremotolerance) evolved in response to stresses in extreme conditions prevailing on rock surfaces. These factors facilitated colonisation of various substrates including the brains of vertebrates by opportunistic fungal pathogens, as well as helped establishment of a stable lichen symbiosis.
Keywords: Evolution of rock-dwelling habit; evolution of lichenisation; multigene phylogeny; ancestral state reconstruction; Verrucariales and Chaetothyriales (Chaetothyriomycetidae, Eurotiomycetes)
INTRODUCTION
During the evolutionary history of the Fungi, associations with various substrates have evolved, together with a wide range of nutritional modes and numerous symbiotic interactions. As a consequence, fungi are often referred to as “ecological opportunists”, in order to emphasise their ecological diversity (Thompson 1994, Gargas et al. 1995). The evolution of ecological traits in fungi has been the focus of several studies, in particular for symbiotic fungi like lichens and mycorrhiza (Gargas et al. 1995, Hibbett et al. 2000, Lutzoni et al. 2001, Grube et al. 2004, Schmitt et al. 2005). Recent improvement in higher-level classification of Fungi has been reached by large-scale multigene analyses (Lutzoni et al. 2004, James et al. 2006, Schoch et al. in press). These studies have clarified phylogenetic relationships within and between main groups of fungi, and some clades were delimited with an increased certainty. Several of these newly defined groups comprise puzzling assemblages of ecologically diverse taxa, for which we need to develop a deeper understanding of the evolution of their lifestyles.
Verrucariales and Chaetothyriales, two closely related ascomycete orders from the class Eurotiomycetes, are a good example of one of these evolutionary puzzles. Strongly supported as sister groups in many studies (Lutzoni et al. 2001, 2004, Lumbsch et al. 2005), these two orders are extremely disparate ecologically. Verrucariales are mostly lichenised, i.e., forming stable symbiotic associations with one or two photosynthetic partners (mostly from the green algae), and have a preference for mineral substrates. Chaetothyriales, in the other hand, was originally best known for its animal and human opportunistic pathogens (Winka et al. 1998), often called black yeasts in reference to their melanisation and growth form. This order of mostly anamorphic species also includes teleomorphs that occur as saprophytes on decaying wood and mushrooms (Untereiner et al. 1995, Untereiner 1997, 2000). Previous studies of the evolution of lichenisation have shown that the ancestor of these two orders was likely lichenised, and that opportunistic pathogens and saprophytes within Chaetothyriales probably derived from this ancestor by a loss of lichenisation (Lutzoni et al. 2001, James et al. 2006). Since two additional non-lichenised orders, Coryneliales and Mycocaliciales, have been affirmatively placed within this class (Geiser et al. 2006, Spatafora et al. 2006), our comprehension of the phylogenetic relationships and hence of the class Eurotiomycetes was drastically changed. Association with different habitats and substrates has gained an important role while the evolution of the Chaetothyriales and a broader selection of fungi from bare rock surfaces came into consideration.
This peculiar guild of ascomycetes, which inhabit bare rock surfaces, has consistently been overlooked when considering the evolution of ecological traits in fungi. First discovered in extreme environments, in the hot deserts and in Antarctica (Krumbein & Jens 1981, Friedmann 1982, Staley et al. 1982, Henssen 1987, Danin 1993), these fungi were shown to persistently colonise rock surfaces under more temperate climates (Urzi et al. 1995, Sterflinger & Prillinger 2001). These rock-inhabitants are particularly diverse in semiarid and arid habitats, where they thrive largely due to the absence of competition, but also their extraordinary extremotolerance. A number of specific and universally present morphological and physiological characters enable them to tolerate surprisingly wide ranges of temperature, irradiation and osmotic stress (Palmer et al. 1990, Sterflinger 1998, Ruibal 2004, Gorbushina 2007). Melanisation protects the cells against UV and solar radiation, but also extremes of temperature and desiccation. The production of internal asexual spores and their typical isodiametrical (meristematic) growth form keep the volume-surface ratio optimal and, therefore, enable these fungi to survive extreme drought (Wollenzien et al. 1995). Finally, their ability to rely exclusively on sparse, airborne, low molecular weight nutrients (oligotrophism), contributes to the amazing survival capabilities that extremotolerance confers to rock-inhabiting fungi in these hostile habitats. The remarkable survival abilities of these fungi along with a capacity to penetrate minerals make this guild an attractive study object in microbial ecophysiology and applied research, such as biodeterioration of monuments and exobiology (Gorbushina et al. 1993, Diakumaku et al. 1995, Wollenzien et al. 1997, Gorbushina et al. 2002, Gorbushina 2003).
The first molecular phylogenetic studies to include some of these rock fungi showed that they belonged to two main groups of ascomycetes, the Dothideomycetes and the Chaetothyriales (Sterflinger et al. 1997, 1999, Ruibal 2004). These early studies did not allow a precise phylogenetic placement of most rock-inhabiting fungi as they either included many strains but a fast evolving phylogenetic marker (Ruibal 2004, Sert et al. 2007), or a slowly evolving marker but limited taxon sampling (Sterflinger et al. 1997, 1999). A recent detailed study of a broad sampling of rock-inhabiting fungi from Central Spain and Mallorca (Ruibal et al. 2005, 2008) revealed a large number of undiscovered rock-inhabiting strains and allowed us to infer the phylogenetic relationships of their Chaetothyrialean members in respect to other members of this order using a multigene analysis. By incorporating these unusual fungi within a phylogenetic study, we were able to obtain a better representation of the ecological diversity of this group. Taking into account recent contributions to fungal molecular phylogenetics and newly-discovered lineages of rock-inhabiting fungi, two main questions were addressed: (i) is the ancestor of the two orders Chaetothyriales and Verrucariales still reconstructed as lichenised, as in previous studies, and (ii) is the rock-dwelling habit an ancestral trait to both the lichenised Verrucariales and the pathogen-rich Chaetothyriales. To answer these questions, we explore the evolution of the lichenisation and the rock-dwelling habit using ancestral state reconstruction methods. Our results are discussed within the broader framework of the origin of lichenisation and pathogenicity in ascomycetes.
MATERIAL AND METHODS
Taxon sampling
Two different datasets were used. The evolution of lichenisation, which was shown in previous studies to be highly dependent on the relationships inferred between classes of ascomycetes (Lutzoni et al. 2001, James et al. 2006), was analysed with a dataset representative of almost all main groups of this phylum, except for some early diverging lineages. This dataset included a total of 92 accessions (Pezizomycotina dataset; Appendix 1 in Supplementary Information), sampled in order to represent the phylogenetic and ecological (lichenised vs. non-lichenised) diversity of this phylum. The study of the evolution of the rock-inhabiting habit was limited to our group of interest, the class Eurotiomycetes, and included a total of 183 accessions (Eurotiomycetes dataset; Appendix 2 in Supplementary Information). This dataset includes 25 isolates of rock-inhabiting fungi, most of which selected from a larger pool of strains (Ruibal 2004, Ruibal et al. 2008), to represent all groups separated by a 95 % ITS similarity criterion.
Molecular data
The Pezizomycotina dataset included three phylogenetic markers, the small and large subunits of the nuclear ribosomal RNA gene (nucSSU and nucLSU) and the largest subunit of the RNA polymerase II (RPB1). For this dataset, the gene sequences of 75 taxa were obtained from GenBank. The Eurotiomycetes dataset included the same three loci, and one additional phylogenetic marker, the small subunit of the mitochondrial ribosomal RNA gene (mtSSU). In total, this study generated 68 sequences of nucLSU, 64 of nucSSU, 129 of mtSSU and 61 of RPB1, most of them for taxa within the Eurotiomycetes (Appendices 1 and 2). The sequences of nucLSU, nucSSU and RPB1 were generated using protocols described in Gueidan et al. (2007), and sequences of mtSSU were obtained following the protocol published in Zoller et al. (1999).
Alignments and phylogenetic analyses
Sequence editing, alignment and congruence assessment were done as described in Gueidan et al. (2007). Phylogenetic relationships and confidence were inferred using a Bayesian approach. Additional support values were estimated using maximum likelihood (ML) bootstrap. For the Bayesian approach, the Akaike Information Criterion, as implemented in Modeltest 3.7 (Posada & Crandall 1998), was used to estimate models of molecular evolution. For both datasets, a GTR+I+G model was used for the different partitions (nucLSU, nucSSU, mtSSU, RPB1 first, second and third codon positions), except for the 3rd codon position of RPB1 region D-G, which was not concatenated with region A-D in the Eurotiomycetes dataset, and was subjected to a HKY+I+G model. For each dataset, two independent analyses of two parallel runs and four chains were carried out for 5,000,000 generations using MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003), and trees were sampled every 500 generations. A burn-in sample of 5000 trees was discarded from the first run. The remaining 5000 trees were used to estimate branch lengths and posterior probabilities (PPs) with the sumt command in MrBayes. The program RAxML-VI-HPC (Stamatakis et al. 2005) was used for the ML bootstrap analysis with 1000 replicates and a GTRMIX model of molecular evolution applied to the partitions previously defined.
Ancestral state reconstructions
A recent study showed that estimates of ancestral states may vary depending on the reconstruction methods applied (Ekman et al. 2008). For this reason, two methods were used (comparisons in Tables 1 and 2): a Bayesian approach using BayesMultiState (Pagel et al. 2004) and an ML approach applied to a sample of 5 000 Bayesian trees using MultiState (Pagel et al. 2004). Ancestral state reconstructions of lichenisation were carried out on 23 nodes of the Pezizomycotina dataset (Fig. 1, Table 1), and the rock-dwelling habit on 28 nodes of the Eurotiomycetes dataset (Fig. 2, Table 2). All nodes were present in all 5 000 Bayesian trees, except for nodes 5, 7, 10, and 12 of the Pezizomycotina dataset (Table 1). The program uses a continuous-time Markov model of trait evolution, allowing unequal rates of losses and gains (Lewis 2002). For the Bayesian approach, a uniform prior (0, 100) was chosen, and the analysis was allowed to run for 5 000 000 iterations, with a sample period of 100 and with the option AddMRCA. Output files from BayesMultiState were analysed using Tracer v.1.2.1 (Rambaut & Drummond), with a Burn-in of 500 000 iterations, and output files from MultiState were analysed with Excel v. 11.3.3 (Microsoft).
Table 1.
Support values for the phylogeny of the Pezizomycotina dataset (PP and ML bootstrap) and for the ancestral state reconstruction of lichenisation. The state “non lichenised” was coded as 0, and the state “lichenised” as 1. For the Bayesian reconstructions (MCMC), the PPs of each state are indicated [P(0) and P(1)]. For the ML reconstructions on the Bayesian tree sample (ML), the number of trees supporting each state [Nt(0) and Nt(1)] or ambiguous [Nt(a)] are indicated, as well as the mean of the probabilities for each state across the 5000 trees [Pm(0) and Pm(1)]. Node numbers in this table refer to node numbers in Figure 1. Bold numbers indicate PP greater than 95 %. Results with AddNode option: 1 P(0)=1.00 for node 5; 2 P(0)=0.98 for node 7; 3 P(0)=0.83 for node 10; 4 P(1)=1.00 for node 12. Results using RAxML BS replicates: 5 Pm(0)=0.81 for node 13; 6 Pm(0)=0.89 for node 17; 7 Pm(0)=0.68 for node 18; 8 Pm(0)=0.78 for node 21.
|
Ancestral state reconstruction of lichenisation (0=non lichenised,
1=lichenised)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
MCMC
|
ML
|
|||||||||
| Nodes | Taxonomical groups | PP (%) | ML BS (%) | P(0) | P(1) | Nt(0) | Nt(a) | Nt(1) | Pm(0) | Pm(1) |
| 1 | Pezizomycotina | 100 | 88 | 0.98 | 0.02 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 2 | Pezizomycotina without Pezizomycetes | 100 | 100 | 0.91 | 0.09 | 4983 | 17 | 0 | 0.99 | 0.01 |
| 3 | - | 100 | 84 | 0.71 | 0.29 | 270 | 4730 | 0 | 0.88 | 0.12 |
| 4 | Leotiomycetes + Sordariomycetes | 100 | 99 | 0.99 | 0.01 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 5 | Leotiomycetes* | 65.38 | 67 | 1.001 | 0.00 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 6 | Sordariomycetes | 100 | 100 | 0.99 | 0.01 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 7 | Pezizomycetes* | 99.66 | 74 | 1.002 | 0.02 | 5000 | 0 | 0 | 0.99 | 0.01 |
| 8 | Arthoniomycetes | 100 | 100 | 0.01 | 0.99 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 9 | Trypetheliaceae | 100 | 100 | 0.00 | 1.00 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 10 | Dothideomycetes* | 50.34 | <50 | 0.583 | 0.42 | 14 | 4986 | 0 | 0.63 | 0.37 |
| 11 | Dothideomycetes without Trypetheliaceae | 100 | 91 | 1.00 | 0.00 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 12 | Lichinomycetes + Lecanoromycetes* | 99.68 | <50 | 0.00 | 1.004 | 0 | 14 | 4986 | 0.00 | 1.00 |
| 13 | Eurotiomycetes | 100 | 88 | 0.98 | 0.02 | 5000 | 0 | 0 | 1.005 | 0.00 |
| 14 | Mycocaliciales | 100 | 100 | 1.00 | 0.00 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 15 | Eurotiomycetidae | 100 | 100 | 0.99 | 0.01 | 5000 | 0 | 0 | 1.00 | 0.01 |
| 16 | Eurotiales/Onygenales | 100 | 100 | 1.00 | 0.00 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 17 | Eurotiomycetidae + Chaetothyriomycetidae | 100 | 99 | 0.91 | 0.09 | 4969 | 31 | 0 | 0.986 | 0.03 |
| 18 | Chaetothyriomycetidae | 100 | 100 | 0.55 | 0.45 | 0 | 5000 | 0 | 0.647 | 0.36 |
| 19 | Pyrenulales | 100 | 100 | 0.05 | 0.95 | 0 | 555 | 4445 | 0.04 | 0.96 |
| 20 | Chaetothyriales | 100 | 100 | 0.99 | 0.01 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 21 | Chaetothyriales + Verrucariales | 100 | 100 | 0.93 | 0.07 | 4985 | 15 | 0 | 0.988 | 0.02 |
| 22 | Rock Fungi/Verrucariales | 100 | 75 | 0.59 | 0.41 | 0 | 5000 | 0 | 0.65 | 0.35 |
| 23 | Verrucariales | 100 | 100 | 0.00 | 1.00 | 0 | 0 | 5000 | 0.00 | 1.00 |
Table 2.
Support values for the phylogeny of the Eurotiomycetes dataset (PP and ML bootstrap) and for the ancestral state reconstruction of the rock-dwelling habit (PP). The state “non rock-inhabitant” was coded as 0, and the state “rock-inhabitant” as 1. For the Bayesian reconstructions (MCMC), PPs of each state are indicated [P(0) and P(1)]. For the ML reconstructions on the Bayesian tree sample (ML), the number of trees supporting each state [Nt(0) and Nt(1)] or ambiguous [Nt(a)] are indicated, as well as the mean of the probabilities for each state across the 5000 trees [Pm(0) and Pm(1)]. Node numbers in this table refer to node number in Figure 1. Bold numbers indicate PP greater than 95 %.
|
Ancestral state reconstruction of the rock dwelling habit (0=non
rock-inhabitant, 1=rock-inhabitant)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
MCMC
|
ML
|
|||||||||
| Nodes | Taxonomic groups | PP (%) | ML BS (%) | P(0) | P(1) | Nt(0) | Nt(a) | Nt(1) | Pm(0) | Pm(1) |
| 1 | Eurotiomycetes | 100 | 100 | 0.80 | 0.20 | 0 | 5000 | 0 | 0.83 | 0.17 |
| 2 | Eurotiomycetidae | 100 | 98 | 0.80 | 0.20 | 0 | 5000 | 0 | 0.83 | 0.17 |
| 3 | Eurotiales/Onygenales/Arachnomycetales | 100 | 100 | 0.95 | 0.05 | 4765 | 235 | 0 | 0.96 | 0.04 |
| 4 | Onygenales | 100 | 80 | 0.94 | 0.06 | 4973 | 24 | 0 | 0.97 | 0.03 |
| 5 | Eurotiales | 100 | 100 | 0.99 | 0.01 | 5000 | 0 | 0 | 1.00 | 0.00 |
| 6 | Eurotiomycetidae/Chaetothyriomycetidae | 100 | 100 | 0.59 | 0.41 | 0 | 5000 | 0 | 0.59 | 0.42 |
| 7 | Chaetothyriomycetidae | 100 | 100 | 0.22 | 0.78 | 0 | 5000 | 0 | 0.18 | 0.82 |
| 8 | Pyrenulales | 100 | 100 | 0.85 | 0.15 | 0 | 5000 | 0 | 0.88 | 0.12 |
| 9 | Chaetothyriales | 100 | 100 | 0.14 | 0.86 | 0 | 4774 | 226 | 0.08 | 0.92 |
| 10 | - | 100 | 93 | 0.27 | 0.73 | 0 | 5000 | 0 | 0.21 | 0.79 |
| 11 | - | 100 | 100 | 0.05 | 0.95 | 0 | 4 | 4996 | 0.03 | 0.97 |
| 12 | Coniosporium/Sarcinomyces | 100 | 100 | 0.03 | 0.97 | 0 | 0 | 5000 | 0.02 | 0.98 |
| 13 | - | 100 | 81 | 0.58 | 0.42 | 0 | 5000 | 0 | 0.58 | 0.42 |
| 14 | - | 100 | 100 | 0.92 | 0.09 | 717 | 4283 | 0 | 0.94 | 0.07 |
| 15 | Cladophialophora/Fonsecaea | 100 | 78 | 0.88 | 0.12 | 0 | 5000 | 0 | 0.90 | 0.10 |
| 16 | - | 100 | 94 | 0.34 | 0.66 | 0 | 5000 | 0 | 0.30 | 0.70 |
| 17 | - | 100 | 86 | 0.98 | 0.03 | 5000 | 0 | 0 | 0.99 | 0.01 |
| 18 | Verrucariales/Chaetothyriales | 100 | 100 | 0.05 | 0.95 | 0 | 0 | 5000 | 0.02 | 0.98 |
| 19 | Verrucariales | 100 | 100 | 0.01 | 0.99 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 20 | Staurothele/Catapyrenium/Placidiopsis | 100 | 96 | 0.00 | 1.00 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 21 | Polyblastia/Thelidium/Verrucaria | 100 | 99 | 0.00 | 1.00 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 22 | Placopyrenium/Verruculopsis | 100 | 97 | 0.92 | 0.08 | 578 | 4422 | 0 | 0.94 | 0.08 |
| 23 | Endocarpon/Neocatapyrenium | 100 | 95 | 0.00 | 1.00 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 24 | Hydropunctaria | 100 | 99 | 0.02 | 0.99 | 0 | 0 | 5000 | 0.01 | 0.99 |
| 25 | Verrucula/Placocarpus/Norrlinia | 100 | 82 | 0.53 | 0.47 | 0 | 5000 | 0 | 0.53 | 0.47 |
| 26 | Placidium/Heteroplacidium | 100 | 100 | 0.84 | 0.16 | 0 | 5000 | 0 | 0.87 | 0.13 |
| 27 | Bagliettoa/Parabagliettoa | 100 | 88 | 0.00 | 1.00 | 0 | 0 | 5000 | 0.00 | 1.00 |
| 28 | Mycocaliciales | 100 | 100 | 0.93 | 0.07 | 1725 | 3265 | 0 | 0.95 | 0.06 |
Fig. 1.
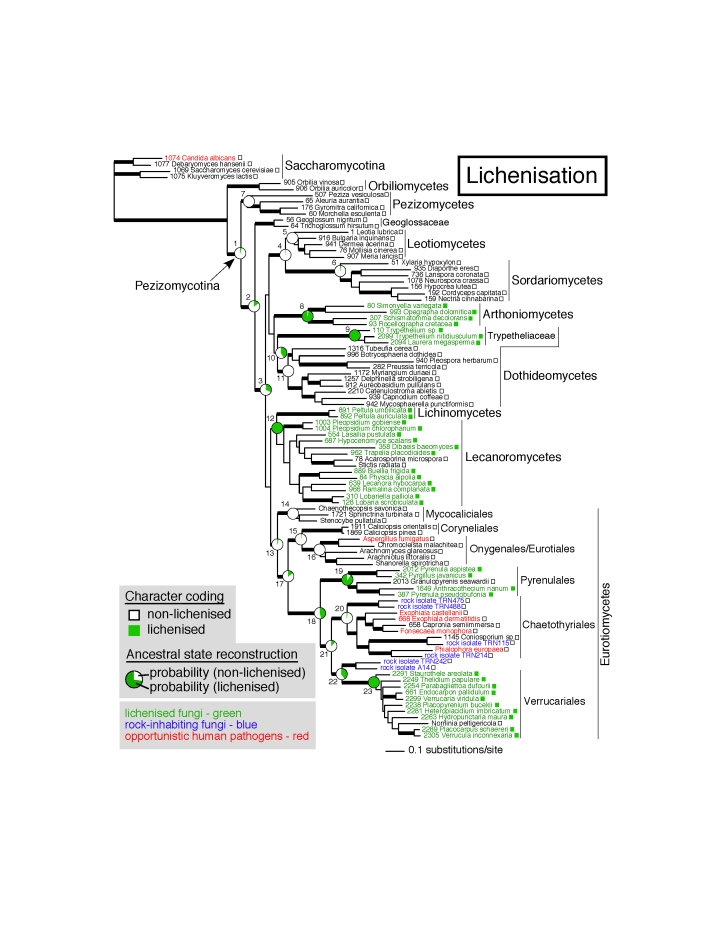
Three-locus phylogeny of the Pezizomycotina obtained from a Bayesian MCMC analysis depicting ancestral state reconstruction of lichenisation. Thick branches represent nodes supported by PP ≥ 95 % and ML bootstrap ≥ 70 % (see Table 1 for numerical values). Boxes after each name indicate the state for each extant taxon (white box = non-lichenised, green box = lichenised). Posterior probabilities for each of the two states are represented in pie charts at each reconstructed node (numbered from 1 to 23). Additional information for these 23 nodes is provided in Table 1.
Fig. 2.
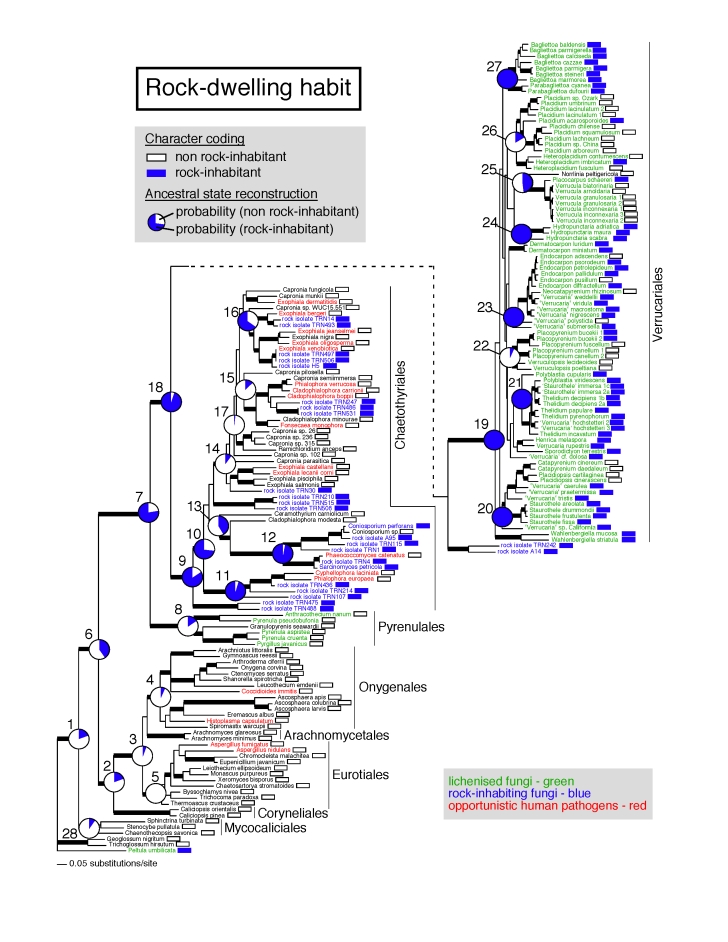
Four-locus phylogeny of the Eurotiomycetes obtained from a Bayesian MCMC analysis depicting ancestral state reconstruction of the rock-dwelling habit. Thick branches represent nodes supported by PP ≥ 95 % and ML bootstrap ≥ 70 % (see Table 2 for numerical values). Boxes after each name indicate the state for each extant taxon (white box = non rock substrate, blue box = rock substrate). Posterior probabilities for each of the two states are represented in pie charts at each reconstructed node (numbered from 1 to 28). Additional information for these 28 nodes is provided in Table 2.
RESULTS
Ancestral state reconstructions
Ancestral state reconstructions gave similar results with maximum likelihood (ML) and Bayesian methods (MCMC), except for some nodes supported in more than 95 % of the Bayesian trees with ML (nodes 2 and 17), but with only moderate supports with MCMC (90 % < posterior probability [PP] < 95 %).
Analyses were done using the option AddMRCA, which allows the reconstruction to be carried out on all trees, even if they do not have the node of interest, by finding, for these particular trees, a node that contains at least all the species otherwise present in the node of interest. For the four nodes that were not present in all Bayesian trees (nodes 5, 7, 10 and 12 of the Pezizomycotina dataset), the option AddNode, restricting the reconstructions to the trees having the node of interest, was also used for comparison (see legend of Table 1). For these four nodes, whether all trees were used (option AddMRCA) or only the trees with the node of interest (option AddNode), did not have a major effect on most of the reconstructions, at the exception of node 3, for which the posterior probability to be non-lichenised increased from 0.58 to 0.83 when using the option AddNode.
Bayesian and ML ancestral state reconstructions on Bayesian trees allow taking into account phylogenetic uncertainties (Pagel et al. 2004). However, because the Bayesian approach as implemented in MrBayes was shown to sometime overestimate posterior probabilities on short internodes (Alfaro et al. 2003), phylogenetic uncertainty linked to nodes with 100 % PP but low or moderate bootstrap (BS) values is not accounted for in our analyses. Therefore, the effect of the underestimation of the phylogenetic uncertainty linked to PPs was further explored for four nodes of interest of the Pezizomycotina dataset (nodes 13, 17, 18, and 21). Ancestral state reconstructions were carried out on these four nodes using a ML approach on 500 trees obtained with branch lengths from bootstrap replicates in RAxML (see legend of Table 2). The results show that the probability for the state “non-lichenised” for nodes 13, 17 and 21 (all superior to 98 % when using the Bayesian tree sample) gave only moderate support when using the bootstrap tree sample (PPs ranging from 78 to 89 %).
Evolution of lichenisation
A first phylogenetic analysis was conducted on a dataset including representatives from almost all main groups of ascomycetes (Fig. 1). The topology inferred is similar to those recovered in two recent large-scale phylogenetic analyses (James et al. 2006, Spatafora et al. 2006), except for the placement of the Geoglossaceae, which is here strongly supported as an early diverging lineage in the Pezizomycotina, the largest subphylum of ascomycetes (see also Wang et al. 2006, Schoch et al. in press). Lichenisation is shown to have evolved independently three to five times in the Pezizomycotina: once in the lineage including Lecanoromycetes and Lichinomycetes (node 12; PPMCMC= 1.00), once or twice in the lineage including Pyrenulales (node 19; PPMCMC= 0.95) and Verrucariales (node 23; PPMCMC= 1.00), and once or twice in the lineage including Arthoniomycetes (node 8; PPMCMC= 0.99) and Dothideomycetes (node 9; PPMCMC= 1.00 for Trypetheliaceae). In contrast to previous studies (Lutzoni et al. 2001, James et al. 2006), the ancestor of Eurotiomycetes is reconstructed as non-lichenised (node 13; PPMCMC= 0.98), as well as the ancestor of both Verrucariales and Chaetothyriales, although with lower support (node 21; PPMCMC= 0.93).
Evolution of the rock-dwelling habit
A second phylogenetic analysis was conducted on a dataset including representatives from all orders within Eurotiomycetes (Fig. 2). Relationships between orders were strongly supported and congruent with previous analyses (Lutzoni et al. 2004, Geiser et al. 2006). Most of the rock isolates are shown to belong to the Chaetothyriales, except for two strains (rock isolates TRN242 and A14), which are here resolved (but not supported) as sister to the lichen order Verrucariales. Within the Chaetothyriales, some rock-isolates are closely related to previously described species (e.g., rock isolate TRN4 and Phaeococcomyces catenatus), and are nested within a large group including most of the saprophytes and opportunistic pathogens. Other strains consist in early diverging lineages, some of which are novel (e.g., lineage including rock isolates TRN475 and TRN488). When reconstructing the rock-dwelling habit, the ancestor of the monophyletic group including Verrucariales and Chaetothyriales (node 18; PPMCMC= 0.95), as well as the ancestor of the Verrucariales (node 19; PPMCMC= 0.99), are supported as inhabiting rock surfaces. The ancestor of the Chaetothyriales is also reconstructed as rock inhabitant, although with lower support (node 9; PPMCMC= 0.86).
DISCUSSION
During the evolutionary history of the Fungi, many transitions in lifestyles and habitats have occurred, giving rise to an amazing ecological diversity. In order to better understand their evolution, ancestral state reconstructions methods have been used to characterise these ecological shifts (Hibbett et al. 2000, Lutzoni et al. 2001). However, the discovery of new fungal lineages and the recent contributions to fungal molecular phylogenetics have greatly changed our perception of the evolution of some group of fungi, as, for example, the two ascomycetes orders Verrucariales and Chaetothyriales. In our study, the inclusion of the newly discovered rock-inhabiting fungi and other groups recently shown to belong to the Eurotiomycetes is reshaping our understanding of the evolution of this group. Reconstructions prove the key-role of primordial habitats like rock surfaces by suggesting that the ancestor of both lichenised Verrucariales and pathogen-rich Chaetothyriales was most likely non-lichenised and dwelled on rock surfaces. Similar to present-day rock-inhabiting fungi, this ancestor most probably possessed characters universally effective against different stresses (temperature, desiccation, pH, radiation), such as melanisation, and meristematic growth. Our results further imply that existing views on the evolutionary history of lichenisation within ascomycetes should be revised. Rock surfaces are proposed as the ancestral habitat for the lichenised Verrucariales. The results also support the hypothesis that, in Chaetothyriales, factors involved in pathogenicity evolved from characters enabling survival to physical stresses and life in extreme environments.
Evolution of lichenisation
Previous studies have suggested that the lichen symbiosis has evolved multiple times in fungi (Gargas et al. 1995, Lutzoni et al. 2001). In ascomycetes, where the great majority of lichen species belong, the evolution of lichenisation was shown to be limited to only one-three origins, and three-four losses (Lutzoni et al. 2001). These results supported the hypothesis that lichenisation results from complex and specialised interactions between symbiotic partners, unlikely to have evolved frequently and independently in different lineages, but more conceivably to have been lost multiple times. Our present work, which aimed at reassessing these previous results based on a taxon sampling more representative of the ecological diversity of these fungi (lichenised versus non-lichenised), suggests a higher number of origins of lichenisation in the ascomycetes (three-five) than previously estimated (Lutzoni et al. 2001, James et al. 2006). In contrast to Lutzoni et al. (2001), the ancestor of the Eurotiales and Onygenales, two orders comprising medically and commercially important fungi (such as the genera Aspergillus and Penicillium), is here supported as being non-lichenised. The addition of non-lichenised lineages previously overlooked (Mycocaliciales, Coryneliales and the rock-inhabiting fungi) seems to account for most of the differences observed in ancestral state reconstructions (Lutzoni et al. 2001, James et al. 2006 and this study). A recent work (Schoch et al. in press), including the most exhaustive taxon and gene sampling currently available for ascomycetes (434 taxa and 6 genes), also suggests a higher number of lichenisation events (four-seven), supporting that, lichen symbiosis, although resulting from complex interactions, might have evolved independently many more times than previously inferred.
Rock surfaces as a cradle to lichen symbiosis
Based on our reconstructions, the Verrucariales likely resulted from an independent lichenisation event, and the ancestor of Verrucariales and Chaetothyriales inhabited rock surfaces. This rock-dwelling ancestor most likely relied on sparse nutrients provided by the winds or rainfalls. It is possible that, where unicellular free-living algae could survive on rock surfaces (mainly the northerly outcrops, which are less exposed to the sun), ancestral lineages of rock-inhabiting fungi were able to evolve symbiotic relationships with photosynthetic microorganisms in order to ensure a more constant source of carbohydrates. Recent studies have shown that, when cultured in vitro together with pure isolates of lichen algal symbionts, some rock-inhabiting fungi and one melanised lichen-colonising fungus could develop structures allowing a cellular contact with the algal cells (Gorbushina et al. 2005, Brunauer et al. 2007). This ability to develop symbiotic interactions with unicellular free-living algae might have allowed some rock-inhabiting fungal lineages to evolve lichenisation, such as the Verrucariales. Strikingly, preliminary data suggest that rock-inhabiting fungi also constitute early diverging lineages in Arthoniomycetes, another independently derived lichen lineage nested within a primarily non-lichenised group (data not shown).
From extremotolerance to pathogenicity
The order Chaetothyriales is particularly well known for its animal and human opportunistic pathogens (de Hoog et al. 2000). They infect their hosts either by inhalation, ingestion or traumatic inoculation, provoking a variety of illnesses, ranging from skin to fatal brain infections, both in immunocompromised and immunocompetent patients. Non-pathogenic asexual strains of Chaetothyriales can be isolated from the environment using selective culture methods (Prenafeta-Boldú et al. 2006), and closely related sexual saprophytes are found as secondary decomposers on decaying wood and mushrooms (Haase et al. 1999, Untereiner & Naveau 1999). Primary habitats of opportunistic pathogens and factors involved in pathogenicity are still not entirely understood (Prenafeta-Boldú et al. 2006). Factors conferring invasive capability include the production of melanin and the meristematic growth (Schnitzler et al. 1999, Feng et al. 2001). It was previously suggested that these factors might have primarily evolved in response to extreme environments (de Hoog 1993, Haase et al. 1999, Prenafeta-Boldú et al. 2006). This hypothesis is now supported by our results, as they show that lineages diverging early in the evolution of the Chaetothyriales comprise a particularly high diversity in rock-inhabiting fungi, and that the ancestor of this order is a rock-inhabitant. Therefore, specific traits evolved by a rock-inhabiting ancestor for life in extreme habitats and potentially shared by all descendants within this lineage, can explain the numerous independent shifts to pathogenicity that have occurred in this group of fungi. Because clinical strains have to survive and multiply at body temperature, relying only on nutrients provided by the hosts, tolerance to high temperatures and oligotrophism might also be prerequisites to the evolution of animal and human pathogens (Prenafeta-Boldú et al. 2006).
This new insight in the evolution of the Eurotiomycetes accentuates the role that traits enabling survival in extreme conditions might have had on lifestyle transitions often observed in this class. Other opportunistic fungal pathogens in Eurotiomycetes have also been shown to primarily colonise extreme habitats, such as Coccidioides immitis, the agent of the respiratory disease known as valley fever, which occurs in desert regions in the Americas (Fisher et al. 2001). Unfortunately, as for many other environmental opportunistic fungal pathogens, limited knowledge on primary niches, ecological preferences and factors involved in pathogenicity, is available for the Eurotiomycetes. Because opportunistic fungal pathogens have become an important concern in Public Health, further work on their ecology and evolution is of highest importance.
Acknowledgments
We thank G. Burleigh and H. O'Brien for their help with the phylogenetic analyses, C. Schoch, E. Fraker and K. Molnar for providing sequences, and the technical staff of the CBS for their assistance with the cultures. Thanks also to W.J. Broughton, S. Joneson, L. Margulis, N. Nagalingum, H. O'Brien and for valuable suggestions on the manuscript. This work was supported by an NSF DDIG grant (DEB-0508567) to F.L. and C.G., as well as an NSF AToL grant (AFTOL, DEB-0228725), and an NSF CAREER grant (DEB-0133891) to F.L.
References
- Alfaro ME, Zoller S, Lutzoni F (2003). Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Molecular Biology Evolution 20: 255–266. [DOI] [PubMed] [Google Scholar]
- Brunauer G, Blaha J, Hager A, Türk R, Stocker-Wörgötter E, Grube M (2007). An isolated lichenicolous fungus forms lichenoid structures when co-cultured with various coccoid algae. Symbiosis 44: 127–136. [Google Scholar]
- Danin A (1993). Pitting of calcareous rocks by organisms under terrestrial conditions. Israel Journal of Earth Sciences 41: 201–207. [Google Scholar]
- Diakumaku E, Gorbushina AA, Krumbein WE, Panina L, Soukharjeski S (1995). Black fungi in marble and limestones – an aesthetical, chemical and physical problem for the conservation of monuments. The Science of the Total Environment 167: 295–304. [Google Scholar]
- Ekman S, Andersen HL, Wedin M (2008). The limitations of ancestral state reconstruction and the evolution of the ascus in the Lecanorales (lichenised Ascomycota). Systematic Biology 57: 141–156. [DOI] [PubMed] [Google Scholar]
- Feng B, Wang X, Hauser M, Kaufmann S, Jentsch S, Haase G, Becker JN, Szaniszlo PJ (2001). Molecular cloning and characterization of WdPKS1, a gene involved in dyhydroxynaphtalene melanin biosynthesis and virulence in Wangiella (Exophiala) dermatitidis. Infection and Immunity 69: 1781–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Koenig GL, White TJ, San-Blas G, Negroni R, Gutiérrez Alvarez I, Wanke B, Taylor JW (2001). Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proceedings of the National Academy of Sciences of the United States of America 98: 4558–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann EI (1982). Endolithic microorganisms in the Antarctic cold desert. Science 215: 1045–1053. [DOI] [PubMed] [Google Scholar]
- Gargas A, DePriest PT, Grube M, Tehler A (1995). Multiple origins of lichen symbioses in Fungi suggested by SSU rDNA phylogeny. Science 268: 1492–1495. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, Hofstetter V, Fraker E, Schoch C, Tibell L, Untereiner WA, Aptroot A (2006). Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia 98: 1054–1065. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA (2003). Methodologies and techniques for detecting extraterrestrial (microbial) life. Microcolonial fungi: survival potential of terrestrial vegetative structures. Astrobiology 3: 543–554. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA (2007). Life on the rocks. Environmental Microbiology 9: 1613–1631. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Beck A, Schulte A (2005). Microcolonial rock inhabiting fungi and lichen photobionts: evidence for mutualistic interactions. Mycological Research 109: 1288–1296. [DOI] [PubMed] [Google Scholar]
- Gorbushina AA, Krumbein WE, Hamman CH, Panina L, Soukharjevski S, Wollensien U (1993). Role of black fungi in color-change and biodeterioration of antique marbles. Geomicrobiology Journal 11: 205–221. [Google Scholar]
- Gorbushina AA, Krumbein WE, Volkmann M (2002). Rock surfaces as life indicators: new ways to demonstrate life and traces of former life. Astrobiology 2: 203–213. [DOI] [PubMed] [Google Scholar]
- Gueidan C, Roux C, Lutzoni F (2007). Using a multigene analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota). Mycological Research 111: 1147–1170. [DOI] [PubMed] [Google Scholar]
- Grube M, Baloch E, Lumbsch HT (2004). The phylogeny of Porinaceae (Ostropomycetidae) suggests a neotenic origin of perithecia in Lecanoromycetes. Mycological Research 108: 1111–1118. [DOI] [PubMed] [Google Scholar]
- Haase G, Sonntag L, Melzer-Krick B, Hoog GS de (1999). Phylogenetic inference by SSU-gene analysis of members of the Herpotrichiellaceae with special reference to human pathogenic species. Studies in Mycology 43: 80–97. [Google Scholar]
- Henssen A (1987). Lichenothelia, a genus of microfungi on rocks. In: Progress and problems in lichenology in the eighties (Peveling E, ed.), Bibliotheca Lichenologica 25, J. Cramer, Berlin-Stuttgart: 257–293. [Google Scholar]
- Hibbett DS, Gilbert L-B, Donoghue MJ (2000). Evolutionary instability of ectomycorrhizal symbioses in basidiomycetes. Nature 407: 506–508. [DOI] [PubMed] [Google Scholar]
- Hoog GS, de (1993). Evolution of black yeasts: possible adaptation to the human host. Antonie van Leeuwenhoek 63: 105–109. [DOI] [PubMed] [Google Scholar]
- Hoog GS, de Guarro J, Gené J, Figueras MJ (2000). Atlas of clinical fungi. 2nd edition. CBS, Utrecht.
- James TY, Kauff F, Schoch C, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch T, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung G-H, Johnson D, O'Rourke B, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüßler A, Longcore JE, O'Donnell K, Mozley-Standridge K, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Sugiyama J, Rossman, AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Buck WR, Cole MS, Diederich P, Printzen C, Schmitt I, Schultz M, Yahr R, Zavarzin A, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006). Reconstructing the early evolution of the fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Krumbein WE, Jens K (1981). Biogenic rock varnishes of the Negev desert (Israel), an ecological study of iron and manganese transformation by cyanobacteria and fungi. Oecologia 50: 25–38. [DOI] [PubMed] [Google Scholar]
- Lewis PO (2002). A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Lumbsch HT, Schmitt I, Lindemuth R, Miller A, Mangold A, Fernandez F, Huhndorf S (2005). Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomycota). Molecular Phylogenetics and Evolution 34: 512–524. [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Kauff F, Cox C, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett D, James T, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Shoemaker R, Sung GH, Lücking R, Lumbsch T, O'Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hansen K, Harris RC, Hosaka K, Lim YW, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R (2004). Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American Journal of Botany 91: 1446–1480. [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Pagel M, Reeb V (2001). Major fungal lineages are derived from lichen symbiotic ancestors. Nature 41: 937–940. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D (2004). Bayesian estimation of ancestral character states on phylogenies. Systematic Biology 53: 673–684. [DOI] [PubMed] [Google Scholar]
- Palmer FE, Staley JT, Ryan B (1990). Ecophysiology of microcolonial fungi and lichens on rocks in Northeastern Oregon. New Phytologist 116: 613–620. [Google Scholar]
- Posada D, Crandall KA (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Prenafeta-Boldú FX, Summerbell R, Hoog GS de (2006). Fungi growing on aromatic hydrocarbons: biotechnology's unexpected encounter with biohazard? FEMS Microbiol Reviews 30: 109–130. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003). MrBayes v3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ruibal CV (2004). Isolation and characterization of melanized, slow-growing fungi from semiarid rock surfaces of central Spain and Mallorca. Ph.D. dissertation. Universidad Autónoma de Madrid/Merck, Sharp & Dohme de España, Madrid, Spain.
- Ruibal C, Platas G, Bills GF (2005). Isolation and characterization of melanized fungi from limestone formations in Mallorca. Mycological Progress 4: 23–38. [Google Scholar]
- Ruibal C, Platas G, Bills GF (2008). High diversity and morphological convergence among melanised fungi from rock formations in the Central Mountain System of Spain. Persoonia 21: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I, Mueller G, Lumbsch HT (2005). Ascoma morphology is homoplaseous and phylogenetically misleading in some pyrenocarpous lichens. Mycologia 97: 362–374. [DOI] [PubMed] [Google Scholar]
- Schnitzler N, Peltroche-Llacsahuanga H, Bestier N, Zündorf J, Lütticken R, Haase G (1999). Effect of melanin and carotenoids of Exophiala (Wangiella) dermatitidis on phagocytosis, oxidative burst, and killing by human neutrophils. Infection and Immunity 67: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Emily Fraker, Hodkinson BP, Bonito G, Yahr R, Groenewald JZ, Arzanlou M, Hoog GS de, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh S-O, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O'Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner WA, Johnston P, Stenroos S, Zuccaro A, Dyer P, Crittenden P, Cole MS, Hansen K, Trappe JM, Lutzoni F, Spatafora JW (2008). The Ascomycota Tree of Life: a phylum wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Systematic Biology (in press). [DOI] [PubMed]
- Sert HB, Sümbül H, Sterflinger K (2007). Microcolonial fungi from antique marbles in Perge/Side/Termessos (Antalya/Turkey). Antonie van Leeuwenhoek 91: 217–227. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, O'Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, Reeb V, Gueidan C, Fraker E, Lumbsch T, Lücking R, Schmitt I, Hosaka K, Aptroot A, Roux C, Miller A, Geiser D, Hafellner J, Hestmark G, Arnold AE, Büdel B, Rauhut A, Hewitt D, Untereiner WA, Cole MS, Scheidegger C, Schultz M, Sipman H, Schoch CL (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Staley JT, Palmer F, Adams JB (1982). Microcolonial fungi: common inhabitants on desert rocks? Science 215: 1093–1095. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H (2005). Raxml-iii: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463. [DOI] [PubMed] [Google Scholar]
- Sterflinger K (1998). Temperature and NaCl-tolerance of rock-inhabiting meristematic fungi. Antonie van Leeuwenhoek 74: 271–281. [DOI] [PubMed] [Google Scholar]
- Sterflinger K, De Baere R, Hoog GS de, De Wachter R, Krumbein WE, Haase G (1997). Coniosporium perforans and C. apollinis, two new rock-inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece). Antonie van Leeuwenhoek 72: 349–363. [DOI] [PubMed] [Google Scholar]
- Sterflinger K, Hoog GS de, Haase G (1999). Phylogeny and ecology of meristematic ascomycetes. Studies in Mycology 43: 5–22. [Google Scholar]
- Sterflinger K, Prillinger H (2001). Molecular taxonomy and biodiversity of rock fungal communities in an urban environment (Vienna, Austria). Antonie van Leeuwenhoek 80: 275-286. [DOI] [PubMed] [Google Scholar]
- Thompson JN (1994). The co-evolutionary process. University of Chicago Press, Chicago IL.
- Untereiner WA (1997). Taxonomy of selected members of the ascomycetes genus Capronia with notes on anamorph-teleomorph connections. Mycologia 89: 210–131. [Google Scholar]
- Untereiner WA (2000). Capronia and its anamorphs: exploring the value of morphological and molecular characters in the systematics of the Herpotrichellaceae. Studies in Mycology 45: 141–149. [Google Scholar]
- Untereiner WA, Naveau FA (1999). Molecular systematics of the Herpotrichiellaceae with an assessment of the phylogenetic positions of Exophiala dermatitidis and Phialophora americana. Mycologia 91: 67–83. [Google Scholar]
- Untereiner WA, Straus NA, Malloch D (1995). A molecular-morphotaxonomic approach to the systematics of the Herpotrichiellaceae and allied black yeasts. Mycological Research 99: 897–913. [Google Scholar]
- Urzi C, Wollenzien U, Criseo G, Krumbein WE (1995). Biodiversity of the rock inhabiting microbiota with special reference to Black Fungi and Black Yeasts. In: Microbial diversity and ecosystem function (Allsopp D, Colwell RR, Hawksworth DL, eds). CAB International, Wallingford: 289–302.
- Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS (2006). Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98: 1065–1075. [DOI] [PubMed] [Google Scholar]
- Winka K, Eriksson OE, Bång Å (1998). Molecular evidence for recognizing the Chaetothyriales. Mycologia 90: 822–830. [Google Scholar]
- Wollenzien U, Hoog GS de, Krumbein WE, Uijthof JMJ (1997). Sarcinomyces petricola, a new microcolonial fungus from marble in the Mediterranean basin. Antonie van Leeuwenhoek 71: 281–288. [DOI] [PubMed] [Google Scholar]
- Wollenzien U, Hoog GS de, Krumbein WE, Urzì C (1995). On the isolation of microcolonial fungi occurring on and in marble and other calcareous rocks. The Science of the Total Environment 167: 287–294. [Google Scholar]
- Zoller S, Scheidegger C, Sperisen C (1999). PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. The Lichenologist 31: 511–516. [Google Scholar]