Abstract
Multiple tolerance to stressful environmental conditions of the black, yeast-like fungus Aureobasidium pullulans is achieved through different adaptations, among which there is the restructuring of the lipid composition of their membranes. Here, we describe three novel genes encoding fatty-acid-modifying enzymes in A. pullulans, along with the levels of their mRNAs under different salinity conditions. High levels of Δ9-desaturase and Δ12-desaturase mRNAs were seen at high salinities, which were consistent with an increased desaturation of the fatty acids in the cell membranes. Elevated levels of elongase mRNA were also detected. Surprisingly, increases in the levels of these mRNAs were also seen following hypo-osmotic shock, while hyperosmotic shock had exactly the opposite effect, demonstrating that data that are obtained from up-shift and down-shift salinity studies should be interpreted with caution.
Keywords: Aureobasidium pullulans, desaturase, elongase, extremotolerance, halotolerance, salt stress
INTRODUCTION
The salinisation of irrigated land has become a major agricultural problem, while attempts to breed crops with increased salt tolerance have still not yielded satisfactory results. Studies of the basic adaptation mechanisms of halophilic and halotolerant organisms, and particularly of eukaryotic ones, should help in the breeding of such crops in the future.
Aureobasidium pullulans (de Bary) G. Arnaud (Dothideaceae, Ascomycota) is a ubiquitous, saprophytic, black, yeast-like fungus. It is often found in the phyllosphere (Andrews et al. 1994), the air (Lugauskas et al. 2003) and many other, often extreme, environments, such as hypersaline water in solar salterns around the World (Gunde-Cimerman et al. 2000) and Arctic glacier ice (Gunde-Cimerman et al. 2003). A. pullulans is of great biotechnological interest because of its production of extracellular pullulan (Leathers 2003), and is also a well known pathogen that can cause a variety of localised, and rarely even, systemic infections (Hawkes et al. 2005). A. pullulans thrives under many different environmental conditions, and it can tolerate a variety of environmental stresses. It can grow from 4 °C up to 35 °C, and although it thrives best without NaCl, it can tolerate up to 17 % NaCl in its growth medium; it even shows an upward shift in its salinity optimum at high temperatures (Torzilli et al. 1985, Zalar et al. 1999, Gostinčar unpublished data).
Cell membranes have crucial roles in the adaptation of any organism to environmental extremes, such as low temperature and high salinity. The active restructuring of the membrane lipid composition that can occur in response to environmental changes preserves the suitable dynamic state of the membrane bilayer and restores membrane function following environmental insult (Hazel & Williams 1990). This restructuring of the membrane lipid composition in response to environmental cues can occur either through changes in the products of cell lipid biosynthesis, or through the selective degradation of lipid species with inappropriate properties (Hazel & Williams 1990).
Such cell membrane changes have been particularly well studied in salt-sensitive Saccharomyces cerevisiae. As in other eukaryotes, the de-novo biosynthesis of saturated fatty acids in these yeast requires acetyl-CoA carboxylase and the fatty-acid-synthase complex. Yeast are only able to synthesise mono-unsaturated fatty acids containing a Δ9 double bond, using a desaturase that is encoded by the OLE1 gene, the expression of which is severely repressed by unsaturated fatty acids (Trotter 2001). Regulation of this gene's expression occurs at the level of OLE1 transcription and via mRNA stability (Gonzalez & Martin 1996). In S. cerevisiae, several different enzyme systems have been described that can elongate the fatty-acyl-CoAs that are formed from de-novo synthesis or are acquired from the growth medium, some of which are essential for cell viability (Trotter 2001).
The halotolerant A. pullulans has so far been studied only at the level of its membrane composition and fluidity when exposed to hypersaline conditions (Turk et al. 2004, 2007). The levels of unsaturated fatty acids in the cell membranes of A. pullulans cells grown in 5 % and 10 % NaCl increase significantly due to their enrichment in C18:2Δ9,12 fatty acids. Significant changes in the lengths of the fatty acids were not seen, although such changes have been detected in another black, yeast-like fungus, the halophilic Hortaea werneckii (Horta) Nishim. & Miyaji (Turk et al. 2004). Although A. pullulans is more sensitive to salt than H. werneckii (but less so than S. cerevisiae), it is of interest because of its multiple tolerances to many other types of stress.
The aim of the present study was to find desaturase and elongase genes that are involved in these fatty-acid modifications in A. pullulans, to compare them to other known homologous enzymes from different fungi, and to study their differential expression under saline and non-saline conditions. In particular, we wanted to determine whether these changes in the fatty-acid saturation levels can be explained by the differential expression of the genes encoding desaturases, and whether the expression of the elongase gene changes in cells subjected to salt stress.
MATERIALS AND METHODS
Strain and culture conditions
The EX-F150 strain of A. pullulans was originally isolated from the Sečovlje solar salterns (Slovenia) (Gunde-Cimerman et al. 2000) and has been maintained in the Extremophilic Fungi (Ex) Culture Collection of the Department of Biology, Biotechnical Faculty, University of Ljubljana (Slovenia), and as MZKI B-802 in the Microbial Culture Collection of the National Institute of Chemistry (Slovenia). A. pullulans was grown using the standard Yeast Nitrogen Base (YNB) chemically defined liquid medium, both without NaCl and with different concentrations of NaCl (2.5 %, 5.0 %, 7.5 %, 10.0 % and 13.0 %; w/v). The liquid cultures were grown at 28 °C and under constant shaking (180 rpm).
DNA and RNA isolation
For DNA isolation, A. pullulans was grown in YNB, and the biomass was harvested by centrifugation in the mid-exponential growth phase (OD600 = 0.8–1.0). The cell pellet was frozen in liquid nitrogen and homogenised using a mortar and pestle. The DNA was then isolated according to the protocol described by Rozman & Komel (1994).
For RNA isolation, A. pullulans was cultured in YNB with different concentrations of NaCl; the biomass was harvested by centrifugation in the mid-exponential growth phase (OD600 = 0.8–1.0). The cells that were subjected to osmotic shock were initially grown in YNB without NaCl or with 10 % NaCl (w/v). These cells were also harvested in the mid-exponential growth phase, and then subjected to osmotic shock by resuspension in YNB with 10 % NaCl or without NaCl, respectively (at the same pH and temperature) for specific times (5–120 min); they were then harvested by filtration. Adapted cells that were grown constantly at the final concentration of NaCl were used as an additional end-point reference. The biomass was frozen in liquid nitrogen and stored at –80 °C until further analysis, when they were homogenised in liquid nitrogen using a mortar and pestle. The RNA was isolated using TRI REAGENT™ (Sigma), according to the manufacturer instructions. Possible contaminating DNA was degraded with deoxyribonuclease I (Fermentas). The integrity and purity of the RNA was evaluated spectrometrically and with formaldehyde agarose electrophoresis.
Amplification and sequencing of genes
Partial sequences of the genes were amplified by polymerase chain reaction (PCR), which was performed as described by Lanišnik Rižner et al. (1999). In all cases, the annealing temperatures were 60 °C. Non-degenerate oligonucleotide primers were constructed on the basis of highly conserved domains in known fungal desaturases: 5'- TAC ACC GAT ACC GAC AAG GAC CCC TA-3' and 5'-GGA ACT CGT GGT GGA AGT TGT GGT A-3' for Δ9-desaturase, and 5'-CCA TCA AGG AGA TCC GTG ATG CCA T-3' and 5'-ATG TTA CCA GTG GCC TTG TGG TGC T-3' for Δ12-desaturase. Primers specific for part of the elongase gene were designed using the CODEHOP algorithm (Rose et al. 1998), from protein sequences of known fungal elongases: 5'-GTC ATC TAC TAC ATC ATC ATH TTY GGN GG-3' and 5'- CGG GCG GAC TGG AAG TAG TAV YAR TAC AT-3'. The amplified fragments were recovered from electrophoresis gels using Perfectprep Gel Cleanup (Eppendorf), and then sequenced. Fragments of the elongase gene were cloned into an Escherichia coli plasmid (pGEM®-T Vector System, Promega) prior to sequencing. The same regions were also amplified using cDNA as a template, to confirm the existence of introns indicated by alignment with sequences of homologous genes in the GenBank database. New non-degenerate primers were designed and later used in RT-PCR reactions: 5'-ATC TCC GAC CTC ACG ACG AC-3' and 5'-CTC ACC GAG AGT GAC GAT GG-3 for Δ9-desaturase gene; 5'-CCG AGA TAC ATT CCC TCG AC-3' and 5'-CCA TGA GAA GTA AGG GAC AAG G-3' for Δ12-desaturase gene; and 5'-TGG TAG GTG TGG AGG AAA GC-3' and 5'-CAT ATT TGG CGG CAG AGA GT-3' for the elongase gene.
Southern blotting was performed as described previously (Turk & Plemenitaš 2002). DNA fragments were amplified as described above (using non-degenerate primers in the case of the elongase gene), and then radioactively labelled and used as probes.
The upstream and downstream sequences of all three of these genes were obtained using the GenomeWalker™ Universal Kit (Clontech), according to the manufacturer instructions, using oligonucleotide primers that were specific for the adapters supplied in the kit, and oligonucleotide primers specific for known fragments of the genes. These were as follows: ApOLE1 upstream 5'-GGC AAC GAG GGT GGG GAA GAT CAA A-3', upstream nested 5'-TGA GGT AGT TCT TGT GCT GCC AGA CGA CAA-3', downstream 5'-ACT GGC TCG GTG ACC AGC CTT TCG AT-3', downstream nested 5'-CGT CAC TCT CGG TGA GGG CTA CCA CAA CT-3'; ApODE12 upstream 5'-GTG TCG TTG AGG GTC TTG GAG GGA GAG AA-3', upstream nested 5'-GGT GTA GAG AGC CCA GAG GCC AGC TCT AA-3', downstream 5'-TTC TCT CCC TCC AAG ACC CTC AAC GAC AC-3', downstream nested 5'-CTC CCA CGG CAA GCA CCA CAA CTT CT-3'; ApELO1 upstream 5'-CAG GGT CAG GTA GAG GTT GTG GAC CTT GAA-3', upstream nested 5'-GCA TGA ACT CTC TGC CGC CAA ATA TGA TG-3', downstream 5'-CAC CGC AGT CTC ATG GGT CCC CAT TA-3', downstream nested 5'-TGA ACT TGA CCG TCC ACG TCG TCA TGT ACT-3'.
Gene phylogeny reconstruction
Homologues of the Δ9-desaturase and Δ12-desaturase and the elongase were identified by BLAST searches (Altschul et al. 1997) against a GenBank non-redundant protein database. In addition to fungal homologues, sequences from other species were added. Protein sequences were aligned using ClustalX (Thompson et al. 1997) and edited in BioEdit software (Hall 1999). Gene trees were generated with MrBayes software applying Bayesian inference (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003). Runs were performed for two million generations with mixed amino-acid models, default temperature and numbers of chains. The trees were sampled every 100 generations. Trees sampled before the analysis reached stationarity of likelihood values and those sampled before the average standard deviation of the split frequencies lowered under 0.5 % were excluded from the final analysis. The stationarity of likelihood values was checked using the Tracer software (Rambaut & Drummond 2007). Enzymes from non-fungal organisms were used as outgroups: Vibrio fischeri (Δ9-desaturases) and Arabidopsis thaliana (Δ12-desaturases and elongases).
RT-PCR and statistical analysis
Total cDNA was synthesised from the RNA samples isolated as described above, using the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas). Approximately 12.5 ng of the synthesised cDNA was used as a template for multiplex PCR with each pair of the oligonucleotide primers described above and oligonucleotide primers specific for the 28S rRNA (Lanišnik Rižner et al. 1999). The PCR consisted of 21, 24 and 22 cycles for the Δ9-desaturase and Δ12-desaturase and for the elongase, respectively. The primer dropping method was used (Wong et al. 1994), with 28S primers added to the reaction for the last 16 PCR cycles. The products of the reactions were separated on agarose gels and stained with ethidium bromide, with the relative abundance of each amplified fragment evaluated by measuring its luminescence with the UN-SCAN-IT software (v. 5.1, Silk Scientific Corporation). The values for the genes of interest were standardised with the amounts of the 28S rRNA fragments, the expression of which remained unchanged under the different environmental conditions. Transcription patterns of given genes were established through comparison of these values between different samples. RT-PCR analyses were carried out in triplicate.
Means and standard deviations were calculated, and the variance due to systematic error was subtracted. The data were analysed for statistically significant differences using a one-way ANOVA test (the variance was checked for homogeneity), followed by the Tukey (HSD) test.
RESULTS
Specific products were successfully amplified using PCR with the oligonucleotide primers specific for parts of the genes encoding a Δ9-desaturase, a Δ12-desaturase and a fatty-acid elongase. Sequencing of the fragments produced 306 bp, 234 bp and 548 bp partial sequences. Using the GenomeWalker™ Universal Kit, complete coding sequences of three new genes from A. pullulans were obtained. Searches with the BLAST programme (Altschul et al. 1997) showed similarities with several genes for Δ9-desaturases and Δ12-desaturases and for fatty acid elongases. The genes were named ApOLE1, ApODE12 and ApELO1, respectively. The sequences have been stored in GenBank under the accession numbers DQ901954 (ApOLE1), DQ901955 (ApODE12) and EF123104 (ApELO1).
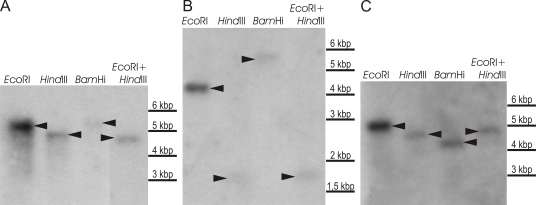
The coding regions were determined through the alignment of the sequences with known homologous genes obtained from the GenBank database and with cDNA sequences. The deduced ApOle1 protein is 458 amino acids long and the gene contains a 50-bp intron. ApODE12 contains no introns, while the putative ApOde12 protein is of 480 amino acids. Two 52 and 53 bp introns were discovered in the ApELO1 fragment, and their sequences were found to be 50 % identical. The hypothetical ApElo1 protein is of 343 amino acids. Southern blotting of the genomic DNA did not suggest the existence of more than one copy of any of the three genes in the genome of A. pullulans (Fig.1).
Fig. 1.
Determination of the ApOLE1 (A), ApODE12 (B) and ApELO1 (C) gene copies in A. pullulans. Southern blotting of genomic DNA digested with different restriction endonucleases (EcoRI, HindIII, BamHI, EcoRI+HindIII, as indicated). Southern blots of the gels with separately digested DNAs were probed with radiolabelled fragments, and amplified with oligonucleotide primers specific for parts of the genes in question.
Good convergence of the runs was reached when constructing all three of the gene trees with MrBayes. The likelihood values reached plateaus after approximately 10,000 (Δ9-desaturases), 8,000 (Δ12-desaturases) and 6,000 generations (elongases), while the average standard deviations of the split frequencies dropped below 0.5 % after approximately 400,000 (Δ9-desaturases) and 800,000 (Δ12-desaturases and elongases) generations. The first 4,000 (Δ9-desaturases) and 8,000 (Δ12-desaturases and elongases) trees were discarded as burn-in. The posterior probabilities for the amino-acid models were 1 for the Blosum62 model (Henikoff & Henikoff 1992) for Δ9-desaturases and Δ12-desaturases, and 1 for the WAG model (Whelan & Goldman 2001) for elongases. All three of the deduced proteins (ApOle1, ApOde12 and ApElo1) clustered with homologous enzymes from fungi belonging to Pezizomycotina.
The relative abundances of the ApOLE1, ApODE12 and ApELO1 genes were studied by RT-PCR. Their profiles for growth at different salinities, and under hyper- and hypo-osmotic shock were analysed separately. One-way ANOVA (α = 0.05) showed significant differences (3.13 ×10-07< P <0.003) in all cases. The results of the Tukey (HSD) post-hoc testing are shown in Figures 2, 3, 4.
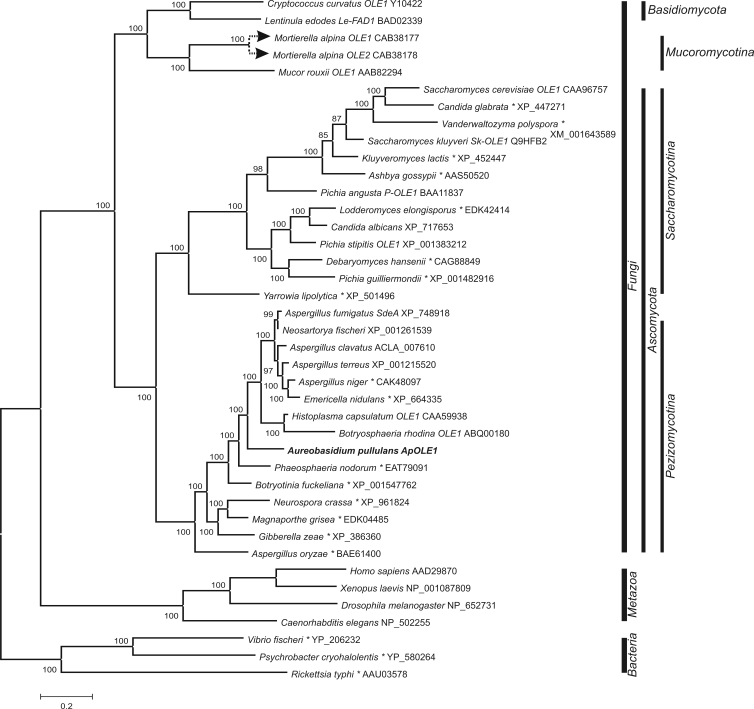
Fig. 2.
Phylogeny of the Δ9-desaturase enzymes. The tree was constructed from the alignment of protein sequences found in the GenBank database with the BLAST programme. Analyses were performed with MrBayes software in two runs with two million generations each. Mixed amino-acid models were used, and the first 20 % of the trees were excluded from the final consensus tree. Full scientific names of source organisms are followed by gene names (if available) and GenBank accession numbers. Entries marked with an asterisk (*) are available in the database as hypothetical proteins without any assigned functions.
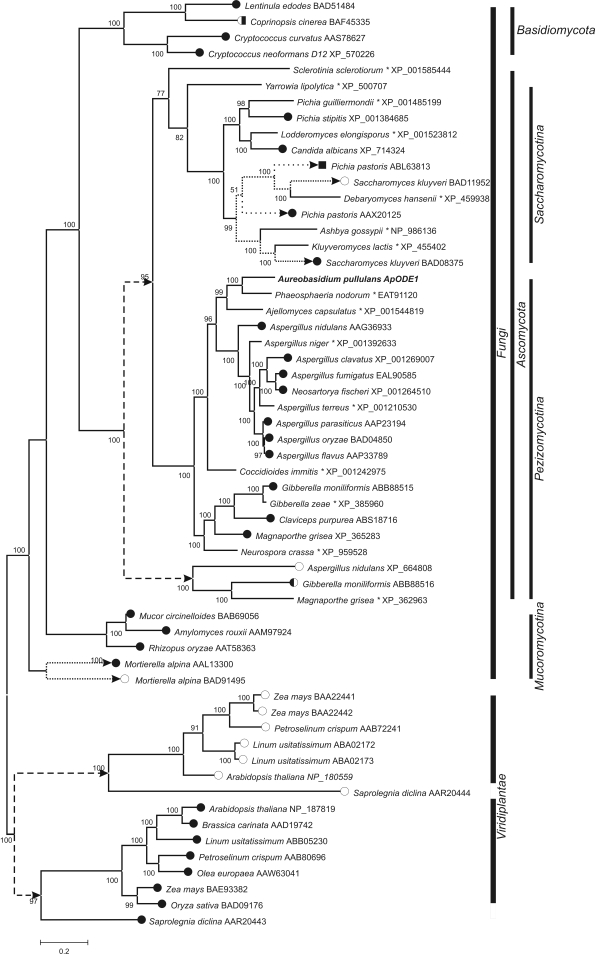
Fig. 3.
Phylogeny of the Δ12- Δ15- and ω3-desaturase enzymes. The tree was constructed from the alignment of the protein sequences found in the GenBank database with the BLAST programme. Analyses were performed with MrBayes software in two runs with two million generations each. Mixed amino-acid models were used, and the first 40 % of the trees were excluded from the final consensus tree. Full scientific names of source organisms are followed by the GenBank accession numbers. the presumed functions of the enzymes are marked with full circles • (Δ12 activity), empty circles ○ (ω3 activity), squares ▪ (Δ15 activity), or combinations thereof. Entries marked with an asterisk (*) are available in the database as hypothetical proteins without assigned function. Presumed large duplications of the ancestor genes are marked with arrows and dashed lines. Other gene duplicates are connected with dotted lines.
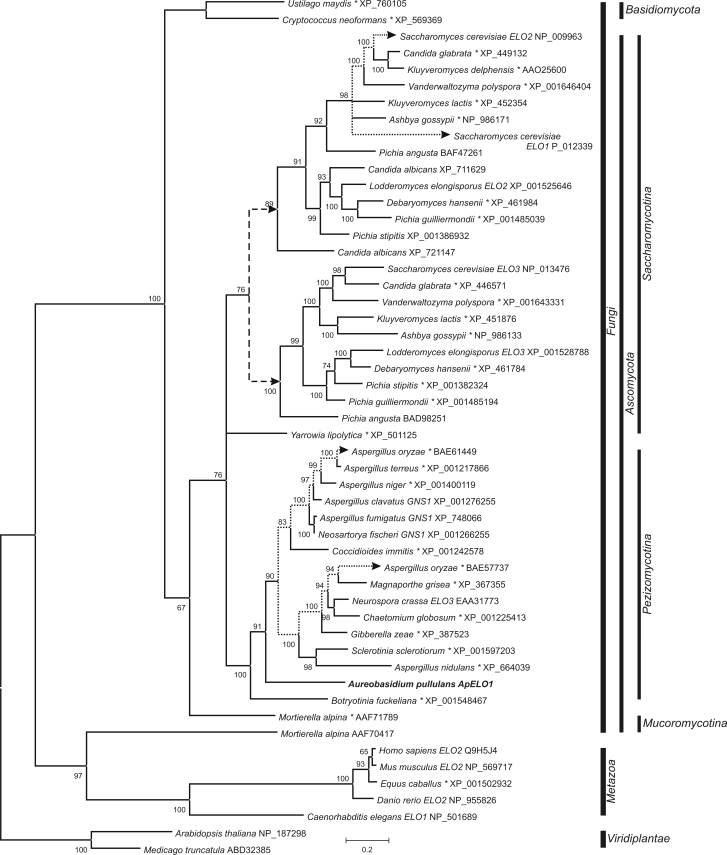
Fig. 4.
Phylogeny of elongase enzymes. The tree was constructed from the alignment of the protein sequences found in the GenBank database with the Blast programme. Analyses were performed with MrBayes software in two runs with two millions generations each. A mixed amino-acid model was used, with the first 40 % of trees were excluded from the final consensus tree. Full scientific names of source organisms are followed by gene names (if available) and GenBank accession numbers. Entries marked with an asterisk (*) are available in the database as hypothetical proteins without assigned functions. Presumable large duplication of the ancestor gene is marked with arrows and a dashed line. Other gene duplicates are connected with dotted lines.
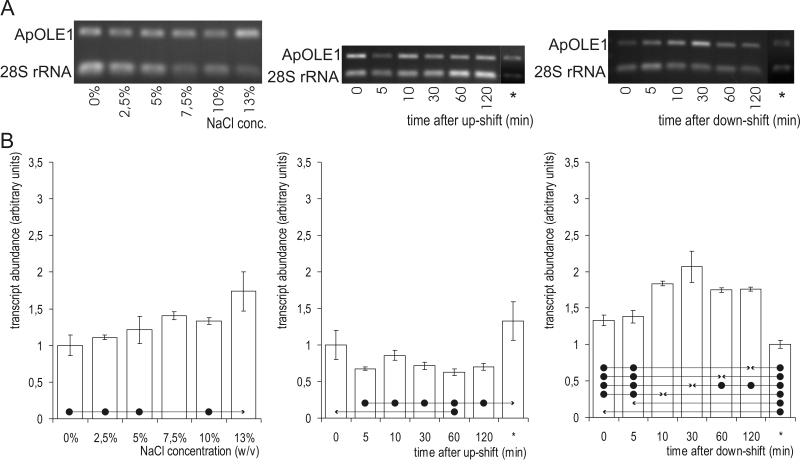
The abundance of ApOLE1 mRNA (Fig. 5) was significantly higher at 13 % NaCl than at the lower salinities. When A. pullulans cells were subjected to osmotic shock, the responses differed considerably. Surprisingly, after hyperosmotic shock, the ApOLE1 mRNA level was reduced, although a significant decrease was detected in only one sample (1 h after the up-shift). All other levels, except that initial prior to up-shift, were significantly lower than the final steady-state levels at 10 % NaCl. In contrast, hypo-osmotic shock led to increased levels of ApOLE1 mRNA. A significant difference was first detected 10 min after the down-shift, and it reached its peak after 30 min. One hour after the down-shift, the ApOLE1 mRNA decreased again, but two hours after the down-shift it was still significantly higher than prior to the osmotic shock. Nevertheless, the steady-state mRNA levels in the cells not exposed to NaCl was significantly lower than in all of the samples obtained after the hypo-osmotic shock.
Fig. 5.
ApOLE1 mRNA abundance. A. Semi-quantitative transcriptional analysis by RT-PCR. Samples were prepared from cells grown at different salinities (first column) or subjected to a salinity shift from 0 % to 10 % NaCl (second column) and 10 % to 0 % NaCl (third column). mRNA abundances in cells completely adapted to the final salinity (10 % in the case of an up-shift, and 0 % in the case of a down-shift) are marked with an asterisk (*). Amplified fragments of ApOLE1 and 28S rRNA genes were run on 1 % agarose gel and stained with ethidium bromide. B. ApOLE1 gene expression as detected by RT-PCR. Data represent means ±SD of three independent experiments. Significant differences (Tukey's HSD) were seen between the value marked with an arrow-head and each of the samples marked with the dot.
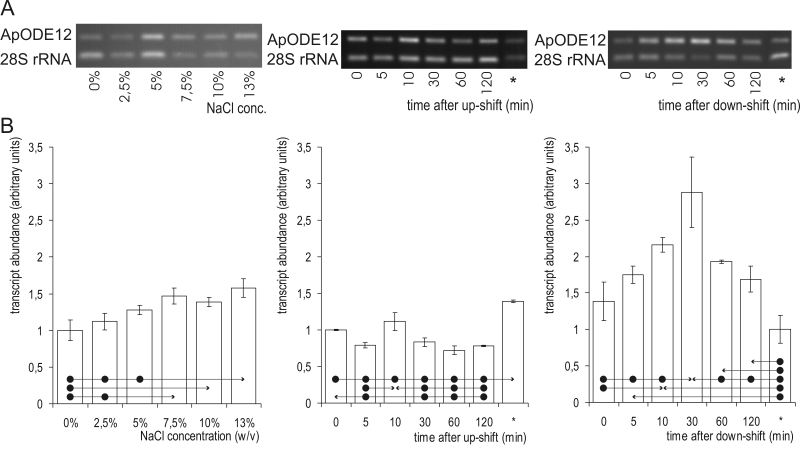
The levels of ApODE12 mRNA were generally higher at higher salinities (Fig. 6): the levels were significantly higher at 13 % NaCl as compared to 0.0 %, 2.5 % and 5.0 % NaCl. As seen for the ApOLE1 gene, the levels of ApODE12 mRNA decreased significantly after exposing the cells to hyperosmotic shock. A transient increase in ApODE12 mRNA levels was seen 10 min after the up-shift, which was significantly higher than after 30 min, and 1 h and 2 h later. Hypo-osmotic shock resulted in transiently increased ApODE12 mRNA levels. The peak here was reached 30 min after the down-shift. A significant increase was first seen 10 min after exposing the cells to the shock and 2 h later it could not be detected any more.
Fig. 6.
ApODE12 mRNA abundance. A. Semi-quantitative transcriptional analysis by RT-PCR. Samples were prepared from cells grown at different salinities (first column) or subjected to salinity shifts from 0 % to 10 % NaCl (second column) and 10 % to 0 % NaCl (third column). mRNA abundances in cells completely adapted to the final salinity (10 % in case of up-shift and 0 % in case of down-shift) are marked with an asterisk (*). Amplified fragments of ApOLE1 and the 28S rRNA genes were run on 1 % agarose gels and stained with ethidium bromide. B. ApODE12 gene expression as detected by RT-PCR. Data represent means ±SD of three independent experiments. Significant differences (Tukey's HSD) were seen between the value marked with an arrow-head and each of the samples marked with dots.
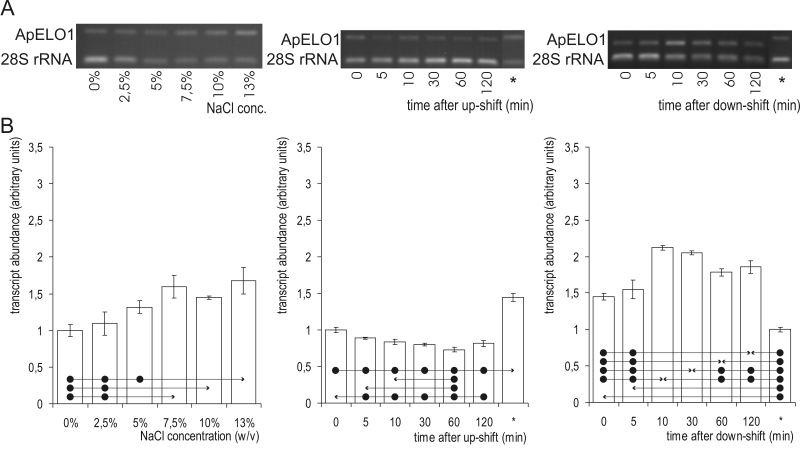
The levels of ApELO1 mRNA were highest at 13.0 % NaCl (Fig. 7), and at 7.5 % and 10.0 % NaCl they were also significantly higher than at both 0.0 % and 2.5 % NaCl. After exposing the cells to hyperosmotic shock, the ApELO1 mRNA levels decreased after 5 min, and the decrease remained significant throughout the shock period, even though the steady-state ApELO1 mRNA expression levels at 10.0 % NaCl were significantly higher than all others seen during the up-shift. Hypo-osmotic shock resulted in a significant increase in ApELO1 mRNA levels after 10 min, which then decreased slowly up to 1 h after the shock, when it was still significantly higher than prior to the down-shift.
Fig. 7.
ApELO1 mRNA abundance. A. Semi-quantitative transcriptional analysis by RT-PCR. The samples were prepared from cells grown at different salinities (first column) or subjected to salinity shifts from 0 % to 10 % NaCl (second column) and 10 % to 0 % NaCl (third column). mRNA abundances in the cells were completely adapted to the final salinities (10 % in the case of the up-shift, and 0 % in the case of then down-shift) are marked with an asterisk (*). Amplified fragments of ApOLE1 and the 28S rRNA genes were run on 1 % agarose gels and stained with ethidium bromide. B. ApELO1 gene expression as detected by RT-PCR. Data represent means ±SD of three independent experiments. Significant differences (Tukey's HSD) were seen between the value marked with an arrow-head and each of the samples marked with the dot.
DISCUSSION
The cell membranes of A. pullulans contain long-chain fatty acids with double bonds in positions 9 and 12 (Δ9, Δ12;Turk et al. 2004). We thus assumed that the A. pullulans genome must encode the corresponding fatty-acid desaturases as well as a fatty-acid elongase. We also wanted to know if changes in the mRNAs levels of these desaturases and elongases correlate with alterations in fatty-acid composition that have previously been seen at different salinities of the surrounding medium (Turk et al. 2004).
The proteins coded by the new genes that are involved in fatty-acid modifications in A. pullulans did not differ significantly from their homologues in other fungi. Analysis of the ApOLE1 sequence revealed the presence of an intron with atypical splice acceptor bases at its 3' end (TG instead of AG). The high degree of identity between two introns in the ApELO1 gene indicated that they might originate from a relatively recent duplication event and could thus be useful in studies of the evolutionary history of this species.
The essential OLE1 gene was first found in S. cerevisiae, and later also in several other fungi (Watanabe et al. 2004). A homologue in A. pullulans was therefore expected. The Δ12-desaturases are, on the other hand, less common, and have not been extensively studied (Watanabe et al. 2004). They introduce a cis double bond into fatty acids that already have a cis double bond at the Δ9 position. This additional double bond further increases the membrane fluidity, although its effect is less pronounced than the effect of the first double bond (Los & Murata 1998). Therefore, this Δ12-desaturase in A. pullulans may enable the cell to fine-tune its cell-membrane fluidity and may be one of the reasons for its halotolerance, in contrast to the salt-sensitive S. cerevisiae which is not capable of Δ12-desaturation. Like the Δ9-desaturases, elongases are essential for cell growth (Dittrich et al. 1998). Three different elongases exist in S. cerevisiae, each with a different substrate specificity (Trotter 2001). So far, only one elongase homologue has been identified in A. pullulans, using degenerate PCR and Southern blotting.
The evolutionary origins of all three of these enzymes from A. pullulans in the Pezizomycotina branch as shown by our phylogenetic analysis are not surprising.
Evolution of Δ9-desaturases also did not show any unusual characteristics with the exception of desaturases from Mucoromycotina which clustered with homologous enzymes from Basidiomycota.
Comparisons of the deduced amino-acid sequences of the Δ12-desaturases and ω3-desaturases indicate several independent duplications of their genes, leading to separate Δ12 and ω3 branches; this was previously reported by Damude et al. (2006). According to our analysis, the major duplication occurred before the separation of Pezizomycotina from Saccharomycotina, resulting in a separate ω3 branch with very few representatives. At least one duplication event could also be detected in Saccharomycotina and Mucoromycotina.
The phylogenetic analysis of the elongase genes indicates a duplication event early in the evolutionary history of Saccharomycotina, which led to two groups of elongase genes, each represented with (at least) one enzyme in most species. Surprisingly, Elo1 and Elo2 from S. cerevisiae belong to the same group, and Elo3 to the other one, although Elo2 and Elo3 are more functionally related. Two hypothetical proteins from Aspergillus oryzae indicate a possible duplication event in Pezizomycotina as well. Another interesting elongase is a long-chain polyunsaturated fatty acid elongation enzyme from Mortierella alpina described by Parker-Barnes et al. (2000), which proved to be quite different from all of the other elongases, and in our analysis it clustered with animal elongase enzymes.
The structural and functional integrity of biological membranes requires the presence of water. Changes in cellular water activities can have profound influences on membrane stability. Cations from salts interact with membrane constituents and can also affect their conformation. Furthermore, abrupt changes in salinity can expose cell membranes to physical stress because of the changes in osmotic pressure (Hazel & Williams 1990). Phenotypic adjustments in the cell-membrane lipid compositions as a response to altered salinity have been observed, among which there are increases in the relative proportions of anionic lipids. Increased levels of unsaturated acyl chains and longer fatty acids have been seen in yeasts and cyanobacteria at higher salinities (Hazel & Williams 1990, Turk et al. 2004), and there is evidence that these changes serve to increase the membrane fluidity at high salt concentrations (Russell 1989, Hazel & Williams 1990).
The high abundance of the desaturase mRNAs seen at high salinities in A. pullulans was expected, and it is consistent with an enrichment in the C18:2Δ9,12 fatty acids in the A. pullulans cell membranes at 5 % and 10 % NaCl (Turk et al. 2004). These changes may help to sustain sufficient levels of membrane fluidity at high salt concentrations (Russell 1989, Hazel & Williams 1990). Increased proportions of long chain fatty acids would oppose this effect, but when the A. pullulans cell membranes were analysed, no shifts in fatty-acid lengths were detected (Turk et al. 2004). A higher abundance of ApELO1 mRNA at high salinities might not therefore be associated with alterations in the fatty-acid composition. The reason for this discrepancy might lie in a faster turnover of the long-chain fatty-acid pool at high salinities, a lower efficiency of ApELO1 due to the high concentrations of compatible solutes, additional undetected elongases with complementary roles, or the posttranscriptional control of expression.
The changes in the mRNA levels following osmotic shock were contrary to expectations, considering the observations of the mRNA abundances at different salinities. A similar transient decrease in mRNA abundance after exposure of the cells to hyperosmotic shock was seen previously for the S. cerevisiae OLE1 gene. Expression of the OLE1 gene diminished 45 min after a shift to high osmolarity (Rep et al. 2000), and increased again after 90 min (Yale & Bohnert 2001). In both cases, the changes can probably be associated with the temporary growth arrest caused by the shock, a response that is similar to the previously reported expression of ribosomal proteins (Rep et al. 2000).
Another possibility is that these reactions act as part of a stress-response system in A. pullulans. By introducing double bonds into these phospholipid fatty-acyl chains and by decreasing the fatty-acyl chain length in artificial membranes, glycerol permeability is increased (Blomberg & Adler 1992). The immediate changes following the osmotic shock could thus be an emergency response, which in the case of hyperosmotic shock protects the cells from compatible solute leakage until other – possibly slower or energetically more demanding – mechanisms can take over, such as synthesis of other compatible solutes and membrane glycerol transporters, or cell wall melanisation. Indeed, in salt-sensitive S. cerevisiae, the ORF's encoding many known yeast salinity stress response proteins are either unaffected or down-regulated immediately following the up-shift, and gradually induced only later (Yale & Bohnert 2001). Similarly, a decrease in the saturation levels of the fatty acids, and therefore an increase in the permeability of the membranes for molecules such as glycerol could facilitate export of compatible solutes when they are no longer needed (Tamas et al. 1999), explaining the transient increases in the levels of the mRNAs encoding fatty acid desaturases.
Finally, maybe cells deal with abrupt changes in osmotic pressure caused by changes in salinity in a very different way than when they are grown in an environment with constant water activity with this “turgor shock” response somehow having priority over the “salinity shock” response.
The opposite response patterns in mRNA levels following hyper- and hypo-osmotic shock compared to the slow adaptation to growth at different salinities indicates that these two types of stress represent a fundamentally different challenge to cells.
Acknowledgments
This work was supported by the Ministry of Higher Education and Technology of the Republic of Slovenia in the form of Young Researcher's grant to C. Gostinčar and grant no. J1-6715.
References
- Altschul SF, Madden TL, Shaffer AA, Zhang Z, Miller W, Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JH, Harris RF, Spear RN, Lau GW, Nordheim EV (1994). Morphogenesis and adhesion of Aureobasidium pullulans. Canadian Journal of Microbiology 40: 6–17. [DOI] [PubMed] [Google Scholar]
- Blomberg A, Adler L (1992). Physiology of osmotolerance in fungi. Advances in Microbial Physiology 33: 145–212. [DOI] [PubMed] [Google Scholar]
- Damude HG, Zhang H, Farrall L, Ripp KG, Tomb JF, Hollerbach D, Yadav NS (2006). Identification of bifunctional delta12/omega3 fatty acid desaturases for improving the ratio of omega3 to omega6 fatty acids in microbes and plants. Proceedings of the National Academy of Sciences of the United States of America 103: 9446–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich F, Zajonc D, Huhne K, Hoja U, Ekici A, Greiner E, Klein H, Hofmann J, Bessoule JJ, Sperling P, Schweizer E (1998). Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. European Journal of Biochemistry 252: 477–485. [DOI] [PubMed] [Google Scholar]
- Gonzalez CI, Martin CE (1996). Fatty acid-responsive control of mRNA stability. Unsaturated fatty acid-induced degradation of the Saccharomyces OLE1 transcript. Journal of Biological Chemistry 271: 25801–25809. [DOI] [PubMed] [Google Scholar]
- Gunde-Cimerman N, Sonjak S, Zalar P, Frisvad JC, Diderichsen B, Plemenitaš A (2003). Extremophilic fungi in Arctic ice: a relationship between adaptation to low temperature and water activity. Physics and Chemistry of the Earth 28: 1273–1278. [Google Scholar]
- Gunde-Cimerman N, Zalar P, Hoog GS de, Plemenitaš A (2000). Hypersaline waters in salterns - natural ecological niches for halophilic black yeasts. FEMS Microbiology, Ecology 32: 235–240. [DOI] [PubMed] [Google Scholar]
- Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hawkes M, Rennie R, Sand C, Vaudry W (2005). Aureobasidium pullulans infection: fungemia in an infant and a review of human cases. Diagnostic Microbiology and Infectious Disease 51: 209–213. [DOI] [PubMed] [Google Scholar]
- Hazel JR, Williams EE (1990). The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Progress in Lipid Research 29: 167–227. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG (1992). Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences of the United States of America 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Lanišnik Rižner T, Moeller G, Thole HH, Žakelj-Mavrič M, Adamski J (1999). A novel 17beta-hydroxysteroid dehydrogenase in the fungus Cochliobolus lunatus: new insights into the evolution of steroid-hormone signalling. Biochemical Journal 337: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathers TD (2003). Biotechnological production and applications of pullulan. Applied Microbiology and Biotechnology 62: 468–473. [DOI] [PubMed] [Google Scholar]
- Los DA, Murata N (1998). Structure and expression of fatty acid desaturases. Biochimica et Biophysica Acta 1394: 3–15. [DOI] [PubMed] [Google Scholar]
- Lugauskas A, Sveistyte L, Ulevicius V (2003). Concentration and species diversity of airborne fungi near busy streets in Lithuanian urban areas. Annals of Agricultural and Environmental Medicine 10: 233–239. [PubMed] [Google Scholar]
- Parker-Barnes JM, Das T, Bobik E, Leonard AE, Thurmond JM, Chaung LT, Huang YS, Mukerji P (2000). Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proceedings of the National Academy of Sciences of the United States of America 97: 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ (2007). Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer.
- Rep M, Krantz M, Thevelein JM, Hohmann S (2000). The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. Journal of Biological Chemistry 275: 8290–8300. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998). Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Research 26: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman D, Komel R (1994). Isolation of genomic DNA from filamentous fungi with high glucan level. BioTechniques 16: 382–383. [PubMed] [Google Scholar]
- Russell NJ (1989). Adaptive modifications in membranes of halotolerant and halophilic microorganisms. Journal of Bioenergetics and Biomembranes 21: 93–113. [DOI] [PubMed] [Google Scholar]
- Tamas MJ, Luyten K, Sutherland FC, Hernandez A, Albertyn J, Valadi H, Li H, Prior BA, Kilian SG, Ramos J, Gustafsson L, Thevelein JM, Hohmann S (1999). Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Molecular Microbiology 31: 1087–1104. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torzilli A, Vinroot S, West C (1985). Interactive effect of temperature and salinity on growth and activity of a salt marsh isolate of Aureobasidium pullulans. Mycologia 77: 278–284. [Google Scholar]
- Trotter PJ (2001). The genetics of fatty acid metabolism in Saccharomyces cerevisiae. Annual Review of Nutrition 21: 97–119. [DOI] [PubMed] [Google Scholar]
- Turk M, Abramovic Z, Plemenitaš A, Gunde-Cimerman N (2007). Salt stress and plasma-membrane fluidity in selected extremophilic yeasts and yeast-like fungi. FEMS Yeast Research 7: 550–557. [DOI] [PubMed] [Google Scholar]
- Turk M, Mejanelle L, Šentjurc M, Grimalt JO, Gunde-Cimerman N, Plemenitaš A (2004). Salt-induced changes in lipid composition and membrane fluidity of halophilic yeast-like melanized fungi. Extremophiles 8: 53–61. [DOI] [PubMed] [Google Scholar]
- Turk M, Plemenitaš A (2002). The HOG pathway in the halophilic black yeast Hortaea werneckii: isolation of the HOG1 homolog gene and activation of HwHog1p. FEMS Microbiology Letters 216: 193–199. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Oura T, Sakai H, Kajiwara S (2004). Yeast Delta 12 fatty acid desaturase: gene cloning, expression, and function. Bioscience, Biotechnology, and Biochemistry 68: 721–727. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular Biology and Evolution 18: 691–699. [DOI] [PubMed] [Google Scholar]
- Wong H, Anderson WD, Cheng T, Riabowol KT (1994). Monitoring mRNA expression by polymerase chain reaction: the »primer-dropping« method. Anals in Biochemistry 223: 251–258. [DOI] [PubMed] [Google Scholar]
- Yale J, Bohnert HJ (2001). Transcript expression in Saccharomyces cerevisiae at high salinity. Journal of Biological Chemistry 276: 15996–16007. [DOI] [PubMed] [Google Scholar]
- Zalar P, Hoog GS de, Gunde-Cimerman N (1999). Ecology of halotolerant dothideaceous black yeasts. Studies in Mycology 43: 38–48. [Google Scholar]