Abstract
NAD+ is a co-enzyme for hydride transfer enzymes and an essential substrate of ADP-ribose transfer enzymes and sirtuins, the type III protein lysine deacetylases related to yeast Sir2. Supplementation of yeast cells with nicotinamide riboside extends replicative lifespan and increases Sir2-dependent gene silencing by virtue of increasing net NAD+ synthesis. Nicotinamide riboside elevates NAD+ levels via the nicotinamide riboside kinase pathway and by a pathway initiated by splitting the nucleoside into a nicotinamide base followed by nicotinamide salvage. Genetic evidence has established that uridine hydrolase, purine nucleoside phosphorylase, and methylthioadenosine phosphorylase are required for Nrk-independent utilization of nicotinamide riboside in yeast. Here we show that mammalian purine nucleoside phosphorylase but not methylthioadenosine phosphorylase is responsible for mammalian nicotinamide riboside kinase-independent nicotinamide riboside utilization. We demonstrate that so-called uridine hydrolase is 100-fold more active as a nicotinamide riboside hydrolase than as a uridine hydrolase and that uridine hydrolase and mammalian purine nucleoside phosphorylase cleave nicotinic acid riboside, whereas the yeast phosphorylase has little activity on nicotinic acid riboside. Finally, we show that yeast nicotinic acid riboside utilization largely depends on uridine hydrolase and nicotinamide riboside kinase and that nicotinic acid riboside bioavailability is increased by ester modification.
NAD+ and its phosphorylated and reduced derivatives are essential co-enzymes for hydride transfer enzymes central to intermediary metabolism. NAD+ is also a consumed substrate of three classes of enzymes, which produce ADP-ribosyl products plus nicotinamide (Nam)4 (1). Sirtuins utilize the ADP-ribose moiety of NAD+ to accept the acetyl modification of lysine, thereby producing a deacetylated protein plus Nam and a mixture of 2′- and 3′-acetylated ADP-ribose (2–4). Such reactions are important for chromatin silencing (5) and regulation of transcription factors and enzymes, thereby controlling a variety of genomic transactions (6), metabolic switches (7, 8), and lifespan (9–11). ADP-ribose transferases and polyADP-ribose polymerases utilize NAD+ to add ADP-ribose as a post-translational modification and/or to form ADP-ribose polymers (12, 13). Finally, cyclic ADP-ribose synthases produce and hydrolyze the calcium-mobilizing compound, cADP-ribose (14, 15). Thus, via pleiotropic ways and means, NAD+ is a central mediator of cellular and organismal metabolism and signaling.
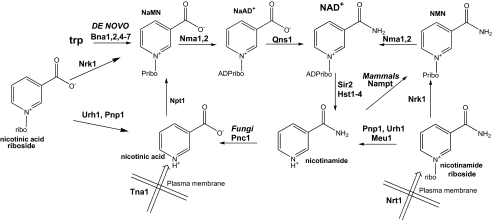
Although co-enzymatic NAD+ functions do not necessitate ongoing NAD+ synthesis, the activities of the NAD+-consuming enzymes mandate either ongoing de novo or salvage synthesis (see Fig. 1). In yeast, de novo synthesis from tryptophan maintains intracellular NAD+ at ∼0.8 mm, at which concentration cells grow well but perform Sir2-dependent gene silencing poorly and have relatively short replicative life spans (16). However, supplementation with 10 μm nicotinamide riboside (NR) more than doubles intracellular NAD+ and doubles replicative longevity(16). NR is imported into yeast cells by a specific, pH-dependent NR transporter, Nrt1, with a Km of 22 μm (17).
FIGURE 1.
NAD+ biosynthetic pathways in S. cerevisiae and mammals. NAD+ is synthesized from tryptophan by the de novo biosynthetic pathway and from vitamin precursors, NA, Nam, and NR, through salvage pathways (1, 44). Salvage pathways in mammals and yeast differ in the utilization of Nam. Yeast convert Nam to NA via the nicotinamidase, Pnc1, whereas mammals convert Nam to nicotinamide mononucleotide (NMN) through the activity of the nicotinamide phosphoribosyltransferase, Nampt. In yeast, NR is salvaged via the Nrk1 pathway and the Urh1/Pnp1 pathway with slight flux through Meu1 (16). In yeast, NaR is salvaged by Nrk1 and the Urh1/Pnp1 pathway (20). According to the data in this study, mammalian Nrk-independent NR salvage relies solely on Pnp and, in yeast, Nrk-independent usage of NaR relies principally on Urh1. Urh1 is now classified as an NR hydrolase, with lower activity on uridine and NaR. ribo, riboside; ADPribo, ADP-ribose.
NR is converted to NAD+ in two steps by nicotinamide riboside kinase (Nrk)-dependent phosphorylation and adenylylation by nicotinamide mononucleotide adenylyltransferase (18). Additionally, NR is split into Nam plus a ribosyl product for NAD+ synthesis through Nam salvage. Genetic analysis indicates that enzymes initially characterized for splitting other nucleosides, namely uridine hydrolase (Urh1), purine nucleoside phosphorylase (Pnp1), and methylthioadenosine phosphorylase (Meu1), are responsible for Nrk-independent NR utilization (16, 19). Nicotinic acid riboside (NaR), synthesized from nicotinic acid riboside ethyl ester, can function as an NAD+ precursor in yeast, through both an Nrk-dependent and an Nrk-independent pathway (20). Human Nrk1 and Nrk2 are closely related ATP- and GTP-dependent metabolite kinases, which can function in place of yeast Nrk1 (18). Both enzymes have approximately equal activity in phosphorylation of NaR and NR and exhibit a crystallographically defined nucleoside-binding site with spatial complementarity to both substrates (20).
As newly discovered eukaryotic NAD+ precursors, NR and NaR have the potential to be important mammalian nutritional supplements and/or drugs (19, 21). However, problems remain in understanding the nature of NR and NaR utilization in fungi and mammalian systems. Here we demonstrate that human Pnp but not human Mtap functions in two different yeast assays for NR utilization. Confirming the genetic results, we demonstrate that NR cleavage in mammalian systems is entirely phosphate-dependent and is sensitive to a specific inhibitor of Pnp. We purified yeast Pnp1 to conduct comparative enzymology with yeast and bovine enzymes. Although both enzymes convert NR to Nam with a specificity constant that is 8–13% as great as the corresponding inosine reaction, bovine but not yeast Pnp is also an NaR phosphorylase. Despite a recent claim that Urh1 is a uridine-specific nucleoside hydrolase (22), our kinetic analysis of recombinant Urh1 demonstrates a 100-fold greater specificity for NR than for uridine. Urh1 also functions in vitro and in vivo as an NaR hydrolase such that Nrk-independent NaR salvage in yeast principally depends on Urh1.
EXPERIMENTAL PROCEDURES
Strains, Media, and Plasmids—Construction of yeast strains has been described (16, 17). A complete strain and plasmid list is found in the supplemental data. Yeast media were prepared as described (16).
For yeast expression, human PNP and MTAP cDNAs were cloned under the control of the yeast PNP1 promoter. The yeast PNP1 promoter was amplified using primers 14093 and 14094, which appended HindIII and KpnI restriction sites. Human PNP and MTAP coding sequences were amplified from cDNAs purchased from ATCC and Origene using the primer pair 14091 and 14092 and the primer pair 14095 and 14096, respectively, which appended KpnI and EcoRI restriction sites. The promoter and cDNA products were digested with KpnI and then ligated, and the resulting linear products were gel-purified. Plasmid pRS327 and the linear products were then digested with HindIII and EcoRI and ligated to obtain pPAB011 and pPAB008 for yeast expression of PNP and MTAP, respectively.
For bacterial expression, Pnp1 and Urh1 were fused to maltose-binding protein (MBP). The PNP1 coding sequence was amplified from DNA from yeast strain BY4742 using primers 14083 and 14084, which appended EcoRI and BsaI restriction sites. The PCR product was digested with BsaI, filled in with Klenow fragment, and then digested with EcoRI. The pMAL-c2x vector (New England Biolabs) was digested with XmnI and EcoRI and ligated with the blunt to EcoRI insert to generate plasmid pPAB006. The pMAL-URH1 plasmid pPAB003 was generated similarly, initiated with primers 14082 and 14085. DNA primer sequences are provided in the supplemental data.
Enzyme Expression, Purification, and Kinetics—TB1 Escherichia coli transformants carrying pPAB003 or pPAB006 were induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside at room temperature for 8 h. Cells (1 liter at an A(600 nm) of 3.8) were lysed by sonication, and the MBP fusion proteins were purified by amylose affinity chromatography and eluted with maltose according to the New England Biolabs protocol. Maltose was removed by hydroxylapatite chromatography. MBP was cleaved from the enzymes of interest with factor Xa. Urh1 and Pnp1 were then recovered from the flow-through of a second amylose column. Bovine Pnp was purchased from Molecular Probes.
Hydrolysis and/or phosphorolysis of NR, uridine, NaR, and NaR methyl ester (meNaR) were measured spectroscopically at 269, 280, 260, and 260 nm, respectively, as described (23, 24). Phosphorolysis was measured in 50 mm sodium phosphate, pH 7.0, and hydrolysis was measured in 50 mm Tris-HCl, pH 7.0. Phosphorolysis of inosine was measure using the coupled xanthine oxidase procedure (23). Hydrolysis and phosphorolysis activity of crude extracts and phosphorolysis activity of NaR by Pnp1 were measured by strong anion exchange HPLC separation using a sodium phosphate gradient as mobile phase (20). Reactions at all substrate concentrations were performed alongside no-enzyme controls to account for nonenzymatic nucleoside degradation. All reactions were conducted at room temperature. Data were fit and analyzed using Sigma Plot.
Nucleosides—Inosine was purchased from Sigma. NR was produced both enzymatically and chemically as described (16, 25). Immucilin-H was a generous gift of Vern L. Schramm. Syntheses of NaR and meNaR and their verification by NMR are described in the supplemental data. The identities of all nucleosides were determined by strong anion exchange HPLC and matrix-assisted laser desorption mass spectrometry.
RESULTS AND DISCUSSION
Human Pnp but Not Mtap Functions in NR Utilization—As Shown in Fig. 1, Qns1-independent NR utilization depends on Nrk1 (18). However, when Nrk1 is deleted, NR can be converted to NAD+ via nucleoside splitting and Nam salvage (16). By genetic criteria, the enzyme with the greatest apparent role in Nrk1-independent NR utilization is Urh1, which has homologs in a limited number of fungi, protista, and eubacteria but not animals. The two other enzymes in yeast, Pnp1 and Meu1, the yeast homologs of mammalian Pnp and Mtap (26, 27), play a moderate and minor role, respectively, in Nrk1-independent NR utilization (16). Pnp is an extensively studied enzyme (28) whose human mutation produces symptoms similar to that of severe combined immunodeficiency (29, 30). Indeed, because Pnp is critically important for T cell function, it is a target for immunosuppressive drugs (31). Mtap is required for the recycling of methylthioadenosine to maintain cellular S-adenosylmethionine (32), and its deletion is common in tumor cells (33).
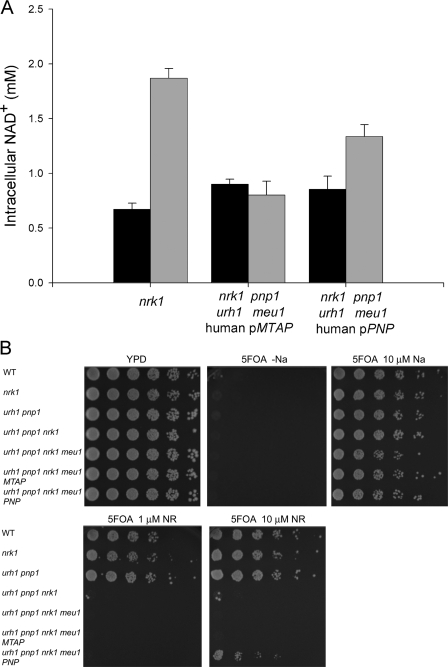
As an initial assay of whether Pnp or Mtap might function in NAD+ metabolism, human cDNAs encoding Pnp and Mtap were cloned into the pRS327 vector under the control of the yeast PNP1 promoter. We previously showed that the addition of 10 μm NR increases intracellular NAD+ from ∼0.8 to ∼2.0 mm in vitamin-free medium and that genetic deletion of nrk1, urh1, pnp1, and meu1 eliminates NR utilization (16). Although the expression of Mtap on a multicopy vector failed to restore NR utilization to an nrk1 urh1 pnp1 meu1 strain, human Pnp allowed the yeast strain to increase intracellular NAD+ from 0.85 to 1.34 mm with the addition of 10 μm NR. By comparison, a yeast strain with an intact Nrk-independent salvage pathway (genotype nrk1) increased NAD+ levels from 0.67 to 1.87 mm (Fig. 2A).
FIGURE 2.
Human Pnp but not Mtap functions in yeast NR utilization. A, intracellular NAD+ concentrations of the indicated genotypes were measured in NA-free SDC media (black bars) and in NA-free SDC media plus 10 μm NR (gray bars). pMTAP and pPNP indicate the addition of a pRS327 plasmid encoding a human cDNA fused to the yeast PNP1 promoter. Expression of human Pnp allows Nrk1-independent NR utilization. B, all strains are gene silencing-deficient on medium without NA and are gene silencing-proficient on medium with 10 μm NA. 10 μm NR restores gene silencing to wild-type (WT) yeast but not yeast strains lacking both NR salvaging pathways. Human Pnp but not Mtap provides a partial rescue of the gene silencing deficiency, indicating function in NR salvage.
We previously demonstrated that provision of NR improves Sir2-dependent telomeric gene silencing (16). This assay is more sensitive than a measurement of intracellular NAD+ because it detects the slight but reproducible effect of MEU1 on NR-promoted gene silencing, whereas no effect of meu1 deletion was observed in an assay of NAD+ levels (16). The gene silencing assay utilizes a yeast strain in which the URA3 gene is integrated at a Sir2-silenced telomeric locus. Because URA3 expression confers sensitivity to 5-fluoroorotic acid (5FOA), URA3 silencing can be scored by resistance to 5FOA (34). In Fig. 2B, all strains exhibited 5FOA-resistant growth when supplemented with 10 μm nicotinic acid (NA), indicating strong gene silencing, and showed complete 5FOA sensitivity on NA-free plates, indicating poor silencing. The wild-type strain and the strains lacking only one of the NR-salvaging pathways (i.e. nrk1 or urh1 pnp1 meu1) exhibited 5FOA-resistant growth with 1 or 10 μm NR. However, the strain with nrk1 deleted along with deletion of urh1 pnp1 and meu1 remained 5FOA-sensitive with the addition of NR to 10 μm. Although expression of human Pnp restored convincing 5FOA-resistant growth, the Mtap construct was incapable of contributing to gene silencing (Fig. 2B). Thus, of the two human enzymes homologous to yeast enzymes, which participate in Nrk-independent NR salvage, we demonstrate a role for Pnp but not Mtap as an NAD+ biosynthetic enzyme.
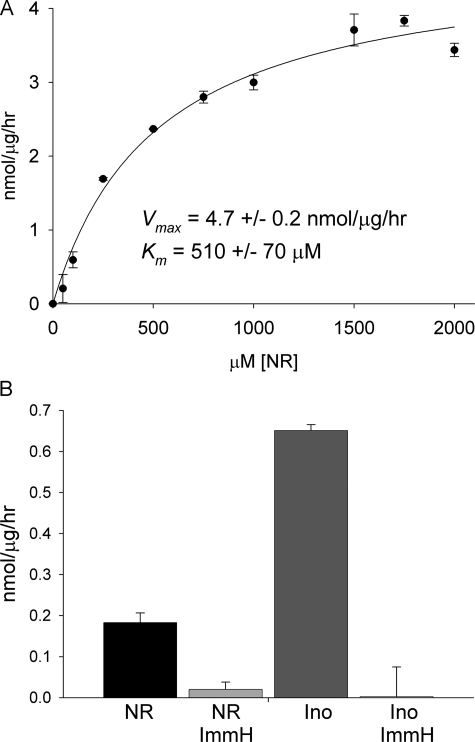
Mammalian NR to Nam Conversion Is Phosphorolytic and Sensitive to a Specific Inhibitor of Pnp—Phosphorolysis of NR has been reported to be a function of purine nucleoside phosphorylase (23, 35), although other investigators have claimed that NR phosphorolysis is a function of a different enzyme (36). Given our discovery of yeast Urh1 as an apparent NR (16) and NaR (20) hydrolytic enzyme, we wished to test whether mammalian NR-splitting extracts contain any hydrolase activity and whether the phosphorylase activity is sensitive to a specific inhibitor of Pnp. We prepared a cellular lysate from 13 mouse livers and recovered the majority of NR-splitting activity in the 60% ammonium sulfate pellet fraction. When this enzyme fraction was dialyzed against phosphate-free buffer (50 mm Tris-HCl, pH 7.0) and assayed in the same buffer, no NR to Nam conversion was observed despite increasing the amount of enzyme fraction by 100-fold and extending incubation to overnight. As shown in Fig. 3A, the addition of 50 mm phosphate to this reaction restored enzymatic activity to a Vmax of 4.7 ± 0.2 nmol/μg/h, indicating that all detectable NR to Nam cleavage in mouse liver is phosphorolytic. Despite the crude system, the Km of this reaction, 510 ± 70 μm, is within 2-fold of the Km of bovine Pnp (Table 1).
FIGURE 3.
Mammalian NR phosphorylase activity is Pnp. A, crude mouse liver NR phosphorylase activity. B, specific activity of CACO-2 cell lysate phosphorolysis of inosine (Ino) and NR without and with immucillin-H (ImmH) inhibition.
TABLE 1.
Kinetic parameters of Pnp1, bovine Pnp and Urh1
| Km | kcat | kcat/Km | |
|---|---|---|---|
| μm | s-1 | s-1m-1 | |
| Pnp1 | |||
| Inosine | 33.0 ± 5.2 | 3.8 ± 0.1 | 115,000 |
| NR | 1,020 ± 140 | 11. ± 0.6 | 11,000 |
| NaR | 3,000 ± 1,000 | 0.13 ± 0.02 | 40 |
| Bovine Pnp | |||
| Inosine | 51.0 ± 5.1 | 14 ± 3.8 | 275,000 |
| NR | 900 ± 100 | 31.2 ± 2.2 | 35,000 |
| NaR | 900 ± 230 | 4.4 ± 0.6 | 4,900 |
| Urh1 | |||
| Uridine | 1,600 ± 500 | 20.0 ± 4.4 | 125,000 |
| NR | 16.0 ± 2 | 23.4 ± 0.8 | 1,430,000 |
| NaR | 150.0 ± 40 | 1.8 ± 0.9 | 12,000 |
| MeNaR | 45.0 ± 15 | 2.3 ± 0.2 | 51,000 |
Nucleoside to nucleobase conversion was also tested in soluble extracts of human CACO-2 cells, an intestinal epithelial cell line. At 500 mm inosine, phosphorolytic conversion to hypoxanthine occurred with a specific activity of 650 ± 10 pmol/μg/h. NR to Nam conversion at 500 mm was ∼3.5 times slower with a specific activity of 180 ± 20 pmol/μg/h. Immucilin-H is a specific inhibitor of Pnp with a Ki of 56 pm for the human enzyme (31). As shown in Fig. 3B, inosine to hypoxanthine conversion and NR to Nam conversion were exquisitely sensitive to inhibition by 100 nm immucillin-H. This result, in combination with data from Figs. 2 and 3A, indicate that Pnp is the apparently unique Nrk1-independent NR salvage enzyme in mammalian cells.
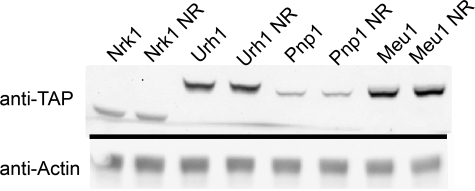
Urh1 Is Expressed at a Higher Level than Pnp1—Previously, we demonstrated that Urh1 makes a greater contribution to Nrk1-independent NR salvage than does Pnp1 or Meu1 (16). To determine whether this is due, in part, to increased protein levels, we measured the steady-state levels of endogenously expressed, C-terminally epitope-tagged Urh1, Pnp1, Meu1, and Nrk1 (37) by Western blot of cells grown in the presence and absence of 100 μm NR. As shown in Fig. 4, the addition of NR did not alter expression of any of the NR salvage enzymes. Earlier, we showed that in the Nrk1-independent NR salvage pathway, the rank order of contributions is Urh1 > Pnp1 > Meu1. However, the rank order of steady-state expression is Meu1 > Urh1 > Nrk1 > Pnp1. Thus, although the greater role of Urh1 than Pnp1 in cellular NR to nicotinamide salvage may be attributable, in part, to greater expression level, the lesser role of Meu1 in NR salvage is despite high expression.
FIGURE 4.
Relative protein abundance of NR salvage enzymes. Yeast strains, containing tandem affinity purification-tagged NR salvage enzymes, were grown in synthetic media without or with the addition of 100μm NR. The relative abundance of the proteins, Meu1 > Urh1 > Nrk1 > Pnp1, was determined by Western blotting with an anti-tandem affinity purification antibody CAB1001 (Open Biosystems). Half of the blot was probed with anti-actin antibody as a loading control.
Urh1 Prefers NR to Uridine by 100-fold—Prior to the identification of the URH1 gene in 2001, a uridine nucleosidase activity purified from baker's yeast was described. This activity possessed a Km for uridine of 0.9 mm and showed strong discrimination against other nucleosides including thymidine, cytidine, adenosine, inosine, and guanosine (24). URH1 was subsequently cloned, and its biochemical activity was ascribed to that of the uridine nucleosidase without any characterization of the recombinant enzyme (38, 39). Urh1 was recently classified as a uridine-preferring pyrimidine nucleoside hydrolase in a study that investigated pyrimidine versus purine nucleoside specificity (22). However, given our genetic assignment of Urh1 as an NR (16) and NaR (20) hydrolase, it was important to characterize pyrimidine versus pyridine nucleoside specificity.
Urh1 activity was unaffected by EDTA or the addition of MgCl2. Urh1 hydrolyzed uridine to uracil at high substrate concentrations. As shown in Table 1, the Kmvalue for uridine was 1.6 mm, with a kcat of 20 s–1. However, despite a gene and enzyme name suggesting that uridine is the preferred substrate, Urh1 prefers NR by greater than 100-fold by depression of the Km to 16 μm and maintaining kcat at 23 s–1. The actual contributions of Urh1 to uridine and NR flux remain unknown because the concentrations of these substrates are unknown. Thus, it is conceivable that uridine hydrolysis at high cellular concentrations of uridine is an unavoidable consequence of NR hydrolysis at low cellular concentrations of NR. It is also conceivable that Urh1 is tuned to the cellular concentrations of NR and uridine.
Yeast and Bovine Pnp Prefer Inosine to NR by 10-fold—Although terms such as “uridine hydrolase” and “purine nucleoside phosphorylase” suggest a simplified set of reactions in which single enzymes have only one or two substrates, in fact, multiple enzymes, expressed at different levels, participate in the consumption of multiple competitive substrates. Bovine and human Pnp are enzymes with well characterized specificity for guanosine and inosine (40). Bovine and bacterial Pnp have been reported to exhibit a 50-fold preference for inosine over NR (23). We purified yeast Pnp1 to measure the phosphorylase activity of recombinant Pnp1 and bovine Pnp on both inosine and NR and to place the kinetic constants of these enzymes on a common scale with Urh1 and the human Nrk enzymes (20).
As shown in Table 1, yeast Pnp1 exhibited a specificity constant for NR greater than 9% of the corresponding value for inosine with the difference mediated by a 31-fold disadvantage in Km offset by a 2.9-fold advantage in kcat. Bovine Pnp exhibited similar discrimination. NR was phosphorylized at 13% of the inosine specificity constant with the difference mediated by an 18-fold disadvantage in Km offset by a 2.2-fold advantage in kcat.
The absolute specificity constants (Table 1) and relative protein expression levels (Fig. 4) of Urh1 and Pnp1 can also be compared. Urh1 possesses greater than 100-fold more activity on NR than does Pnp1 and is expressed at a higher level. Thus, the fact that Pnp1 appears to be responsible for 25–35% of Nrk1-independent NR salvage suggests that in vivo Urh1 activity might be attenuated by competition for other substrates.
NaR Is a Hydrolysis Substrate of Urh1 but a Poor Substrate of Yeast Pnp1—Our previously work showed that NaR, synthesized from NaR ethyl ester, can serve as an NAD+ precursor in a manner that depends on the Nrk1 and the Urh1/Pnp1 pathways (20). However, the increase in NAD+ levels provided by this precursor supplied at 10 μm was only 34% of the increase in NAD+ levels with provision of 10 μm NR. We also demonstrated that human Nrk1 and Nrk2 phosphorylate NaR and NR with equal efficiency (20). These data indicated that NaR is a substrate of Pnp1 and/or Urh1. Moreover, there is a suggestion in the data that NaR is either transported less efficiently than NR or split less efficiently by the enzymes of the Urh1/Pnp1 pathway.
As shown in Table 1, Urh1 hydrolyzes NaR to NA with a Km of 150 μm and a kcat of 1.8 s–1. The resulting specificity constant of 11,000 s–1m–1 is 130-fold lower than that for NR and on a par with the activity of Urh1 for uridine.
Yeast Pnp1 showed no NaR phosphorolysis activity in 30-min continuous spectroscopic assays. However, HPLC runs of the endpoints of the assays indicated low level enzymatic consumption of NaR. To measure this activity, we ran 18-h end point assays and analyzed products by HPLC. As shown in Table 1, the measured Km for NaR is 3 mm, and the kcat is 0.13 s–1. The resulting specificity constant for yeast Pnp1 for NaR phosphorolysis, 40 s–1m–1, compares poorly with that of Urh1 for NaR hydrolysis. These data suggest that Pnp1 may play a negligible role in NaR utilization.
Bovine Pnp proved to be over 120-fold more active on NaR than yeast Pnp1. Moreover, as shown in Table 1, the Km of bovine Pnp for NaR, 900 μm, is nearly identical to its Km for NR, such that discrimination in favor of NR is driven by the kcat term. Although the expression levels of Pnp and Nrk isozymes in mammalian systems are likely to be cell type-specific and regulated, our data support the prediction that NR and NaR utilization in mammals depends on both enzyme systems.
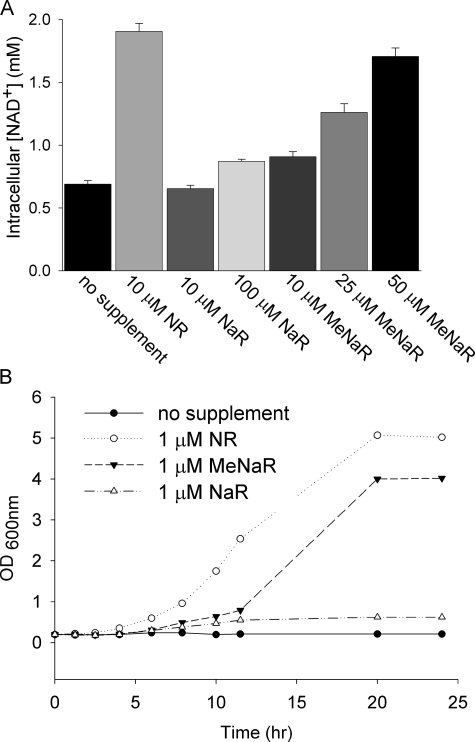
meNaR Is a Better NAD+ Precursor than NaR—We prepared NaR enzymatically from nicotinic acid mononucleotide (16) and chemically from meNaR (see supplemental data). We found that the pure, chemically or enzymatically synthesized NaR provided at 10 μm failed to increase intracellular NAD+, whereas 100 μm NaR increased intracellular NAD+ only 16% as much as did 10 μm NR (Fig. 5A). Surprisingly, 10 μm meNAR increased intracellular NAD+ to a greater degree than did 10 μm NaR. meNAR also produced a dose-dependent increase in intracellular NAD+. Moreover, as shown in Fig. 5B, although 1 μm NR and 1 μm meNAR supported the vigorous growth of a bna1 mutant, which is deficient in de novo NAD+ biosynthesis, 1 μm NaR did not. These data suggest that the methyl ester modification of NaR facilitates nucleoside transport and/or enzymatic cleavage of the methyl ester base from the ribose.
FIGURE 5.
NaR utilization is facilitated by methyl ester modification. A, intracellular NAD+ concentrations of wild-type yeast supplemented with NR, NaR, or meNaR at the indicated concentrations. B, de novo mutant bna1 was grown for 18 h in SDC and then grown to exhaustion for 18 h in NA-free SDC. It was then diluted to a starting A(600 nm) of 0.2 in the specified media. Although 1 μm NR was the best at promoting bna1 growth, meNaR supported bna1 growth after a lag period and 1 μm NaR did not.
meNaR Is a Better Urh1 Substrate than NaR—Data indicating that meNaR is a better NAD+ precursor than NaR prompted us to test whether the major Nrk-independent NR/NaR salvage enzyme, Urh1, prefers meNaR to NaR. Remarkably, as shown in Table 1, the methyl esterified pyridine nucleoside is preferred by Urh1 by a factor of 5.4-fold. On the other hand, Pnp1 had little detectable activity on meNaR. We were unable to measure specific kinetic constants for the cleavage of meNaR by Pnp1 because the nonenzymatic conversion of meNaR to NaR was ∼100 times faster than the enzymatic cleavage of the glycosidic bond. The specific enzymatic activity of Pnp1 on meNaR is <100 pmol/min/μg, which is ∼25% less than the specific activity of Pnp1 on NaR.
Although NR makes use of a specific nucleoside transporter with a Km for NR of 22 μm (17) and has the ability to support yeast cell growth at a supplement concentration of 1 μm, NaR is a poor supplement, which depends on synthetic ester modifications to improve availability to yeast. These data suggest that a distinction be drawn between NR and NaR as yeast NAD+ precursors. Although both compounds function as salvageable metabolites, NR has the characteristics of a yeast vitamin, whereas meNaR has the characteristics of a low potency provitamin, which can be utilized once it gains entry to cells.
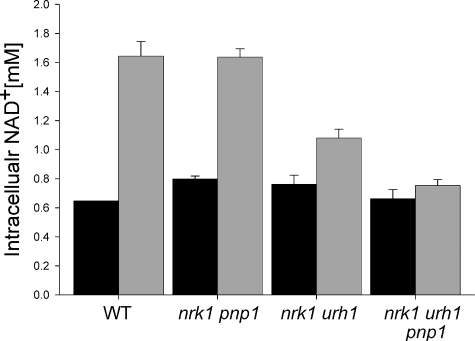
Nrk1 and Urh1 Are Responsible for in Vivo meNaR Utilization—Based on the protein expression data in Fig. 4 and the kinetic data in Table 1, we hypothesized that Urh1 is responsible for the majority of Nrk-independent NaR/meNaR utilization. We tested this hypothesis by measuring the meNaR-dependent NAD+ increase in wild-type yeast and in strains deleted for specific components of NaR salvage (Fig. 6). As suspected, deleting pnp1 in the nrk1 background had no detectable effect on meNaR utilization, indicating that Pnp1 does not play a major role in meNaR utilization. On the other hand, deleting urh1 in the nrk1 background lowered the meNaR-dependent NAD+ increase from 1.2 to 0.21 mm. This result indicates that Urh1 is responsible for the majority of Nrk-independent meNaR utilization. Finally, the triple deletion of nrk1 urh1 pnp1 abolished the NAD+ increase from meNaR, demonstrating that Pnp1 has a minor role in meNaR utilization in the absence of Urh1, consistent with the kinetic data.
FIGURE 6.
Urh1 is principally responsible for Nrk1-independent NaR/meNaR utilization. Intracellular NAD+ concentration of yeast strains was supplemented with 50 μm meNaR. The urh1 deletion from nrk1 greatly reduces meNaR utilization, which is eliminated by deletion of nrk1, urh1, and pnp1. WT, wild type.
Conclusions—We have undertaken to discover and dissect all components of eukaryotic NAD+ salvage pathways and have discovered unanticipated features and complexity (16–18, 20, 41–43). Here we clarified several features of fungal and mammalian salvage of NR and NaR. First, we provide evidence that Pnp but not Mtap has the characteristics of the mammalian NR phosphorylase, and we see no evidence of a mammalian NR hydrolytic activity. Second, we establish that the greater role in yeast of Urh1 than Pnp1 in NR and NaR metabolism is dually driven by higher expression and higher intrinsic activity. Third, we discover that yeast Urh1 is a highly specific NR hydrolase with lower activity on uridine and NaR. This has broad implications on the function of homologous enzymes, which have been termed pyrimidine nucleoside hydrolases (22). Fourth, we extend knowledge on mammalian Pnp activity, showing that both NR and NaR are alternative substrates for this enzyme. Finally, we reveal that the yeast NAD+ precursor activity of NaR depends largely on ester modification of the acid group, which promotes salvage by Urh1. In the future, we plan to determine to what degree NR utilization by particular mammalian cells depends on the Nrk and the Pnp pathways and to dissect the cellular basis for NR and NaR as intracellular metabolites.
Supplementary Material
This work was supported in part by National Institutes of Health Training Grant T32GM008704 (to P. B.). This work was also supported in part by a grant from the Shaklee Corp. and by National Science Foundation Grant MCB-0822581 (to C. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains DNA primer sequences, a complete strain and plasmid list, supplemental methods, and supplemental Fig. 1.
Footnotes
The abbreviations used are: Nam, nicotinamide; Nrk, nicotinamide riboside kinase; NR, nicotinamide riboside; NA, nicotinic acid; NaR, nicotinic acid riboside; meNaR, nicotinic acid riboside methyl ester; Urh1, uridine hydrolase; Pnp, purine nucleoside phosphorylase; Meu1, yeast methylthioadenosine phosphorylase; Mtap, human methylthioadenosine phosphorylase; 5FOA, 5-fluoroorotic acid; HPLC, high pressure liquid chromatography.
References
- 1.Belenky, P., Bogan, K. L., and Brenner, C. (2007) Trends Biochem. Sci. 32 12–19 [DOI] [PubMed] [Google Scholar]
- 2.Tanny, J. C., Dowd, G. J., Huang, J., Hilz, H., and Moazed, D. (1999) Cell 99 735–745 [DOI] [PubMed] [Google Scholar]
- 3.Tanner, K. G., Landry, J., Sternglanz, R., and Denu, J. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauve, A. A., Celic, I., Avalos, J., Deng, H., Boeke, J. D., and Schramm, V. L. (2001) Biochemistry 40 15456–15463 [DOI] [PubMed] [Google Scholar]
- 5.Guarente, L. (2000) Genes Dev. 14 1021–1026 [PubMed] [Google Scholar]
- 6.Michan, S., and Sinclair, D. (2007) Biochem. J. 404 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guarente, L., and Picard, F. (2005) Cell 120 473–482 [DOI] [PubMed] [Google Scholar]
- 8.North, B. J., and Sinclair, D. A. (2007) Trends Biochem. Sci. 32 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longo, V. D., and Kennedy, B. K. (2006) Cell 126 257–268 [DOI] [PubMed] [Google Scholar]
- 10.Dali-Youcef, N., Lagouge, M., Froelich, S., Koehl, C., Schoonjans, K., and Auwerx, J. (2007) Ann. Med. 39 335–345 [DOI] [PubMed] [Google Scholar]
- 11.Guarente, L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72 483–488 [DOI] [PubMed] [Google Scholar]
- 12.Burkle, A. (2005) FEBS J. 272 4576–4589 [DOI] [PubMed] [Google Scholar]
- 13.Schreiber, V., Dantzer, F., Ame, J. C., and de Murcia, G. (2006) Nat. Rev. Mol. Cell. Biol. 7 517–528 [DOI] [PubMed] [Google Scholar]
- 14.Lee, H. C. (2000) Chem. Immunol. 75 39–59 [DOI] [PubMed] [Google Scholar]
- 15.Ying, W. (2007) Front Biosci. 12 1863–1888 [DOI] [PubMed] [Google Scholar]
- 16.Belenky, P., Racette, F. G., Bogan, K. L., McClure, J. M., Smith, J. S., and Brenner, C. (2007) Cell 129 473–484 [DOI] [PubMed] [Google Scholar]
- 17.Belenky, P. A., Moga, T. G., and Brenner, C. (2008) J. Biol. Chem. 283 8075–8079 [DOI] [PubMed] [Google Scholar]
- 18.Bieganowski, P., and Brenner, C. (2004) Cell 117 495–502 [DOI] [PubMed] [Google Scholar]
- 19.Ma, B., Pan, S. J., Zupancic, M. L., and Cormack, B. P. (2007) Mol. Microbiol. 66 14–25 [DOI] [PubMed] [Google Scholar]
- 20.Tempel, W., Rabeh, W. M., Bogan, K. L., Belenky, P., Wojcik, M., Seidle, H. F., Nedyalkova, L., Yang, T., Sauve, A. A., Park, H. W., and Brenner, C. (2007) PLoS Biol. 5 e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogan, K. L., and Brenner, C. (2008) Annu. Rev. Nutr. 28 115–130 [DOI] [PubMed] [Google Scholar]
- 22.Iovane, E., Giabbai, B., Muzzolini, L., Matafora, V., Fornili, A., Minici, C., Giannese, F., and Degano, M. (2008) Biochemistry 47 4418–4426 [DOI] [PubMed] [Google Scholar]
- 23.Wielgus-Kutrowska, B., Kulikowska, E., Wierzchowski, J., Bzowska, A., and Shugar, D. (1997) Eur. J. Biochem. 243 408–414 [DOI] [PubMed] [Google Scholar]
- 24.Magni, G., Fioretti, E., Ipata, P. L., and Natalini, P. (1975) J. Biol. Chem. 250 9–13 [PubMed] [Google Scholar]
- 25.Yang, T., Chan, N. Y., and Sauve, A. A. (2007) J. Med. Chem. 50 6458–6461 [DOI] [PubMed] [Google Scholar]
- 26.Lecoq, K., Belloc, I., Desgranges, C., Konrad, M., and Daignan-Fornier, B. (2001) J. Bacteriol. 183 4910–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chattopadhyay, M. K., Tabor, C. W., and Tabor, H. (2006) Biochem. Biophys. Res. Commun. 343 203–207 [DOI] [PubMed] [Google Scholar]
- 28.Mao, C., Cook, W. J., Zhou, M., Federov, A. A., Almo, S. C., and Ealick, S. E. (1998) Biochemistry 37 7135–7146 [DOI] [PubMed] [Google Scholar]
- 29.Markert, M. L. (1991) Immunodefic. Rev. 3 45–81 [PubMed] [Google Scholar]
- 30.Markert, M. L., Finkel, B. D., McLaughlin, T. M., Watson, T. J., Collard, H. R., McMahon, C. P., Andrews, L. G., Barrett, M. J., and Ward, F. E. (1997) Hum. Mutat. 9 118–121 [DOI] [PubMed] [Google Scholar]
- 31.Lewandowicz, A., Tyler, P. C., Evans, G. B., Furneaux, R. H., and Schramm, V. L. (2003) J. Biol. Chem. 278 31465–31468 [DOI] [PubMed] [Google Scholar]
- 32.Schramm, V. L., Gutierrez, J. A., Cordovano, G., Basu, I., Guha, C., Belbin, T. J., Evans, G. B., Tyler, P. C., and Furneaux, R. H. (2008) Nucleic Acids Symp. Ser. (Oxf.) 75–76 [DOI] [PMC free article] [PubMed]
- 33.Fernandez-Irigoyen, J., Santamaria, M., Sanchez-Quiles, V., Latasa, M. U., Santamaria, E., Munoz, J., Sanchez Del Pino, M. M., Valero, M. L., Prieto, J., Avila, M. A., and Corrales, F. J. (2008) Biochem. J. 411 457–465 [DOI] [PubMed] [Google Scholar]
- 34.Smith, J. S., Brachmann, C. B., Celic, I., Kenna, M. A., Muhammad, S., Starai, V. J., Avalos, J. L., Escalante-Semerena, J. C., Grubmeyer, C., Wolberger, C., and Boeke, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowen, J. W., and Kornberg, A. (1951) J. Biol. Chem. 193 497–507 [PubMed] [Google Scholar]
- 36.Imai, T., and Anderson, B. M. (1987) Arch. Biochem. Biophys. 254 253–262 [DOI] [PubMed] [Google Scholar]
- 37.Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S. (2003) Nature 425 737–741 [DOI] [PubMed] [Google Scholar]
- 38.Mitterbauer, R., Karl, T., and Adam, G. (2002) Appl. Environ. Microbiol. 68 1336–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtz, J. E., Exinger, F., Erbs, P., and Jund, R. (2002) Curr. Genet 41 132–141 [DOI] [PubMed] [Google Scholar]
- 40.Stoeckler, J. D., Poirot, A. F., Smith, R. M., Parks, R. E., Jr., Ealick, S. E., Takabayashi, K., and Erion, M. D. (1997) Biochemistry 36 11749–11756 [DOI] [PubMed] [Google Scholar]
- 41.Bieganowski, P., Pace, H. C., and Brenner, C. (2003) J. Biol. Chem. 278 33049–33055 [DOI] [PubMed] [Google Scholar]
- 42.Bieganowski, P., Seidle, H. F., Wojcik, M., and Brenner, C. (2006) J. Biol. Chem. 281 22439–22445 [DOI] [PubMed] [Google Scholar]
- 43.Wojcik, M., Seidle, H. F., Bieganowski, P., and Brenner, C. (2006) J. Biol. Chem. 281 33395–33402 [DOI] [PubMed] [Google Scholar]
- 44.Magni, G., Orsomando, G., Raffelli, N., and Ruggieri, S. (2008) Front Biosci. 13 6135–6154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.