Summary
Catch bonds are biological interactions that are enhanced by mechanical force pulling a ligand-receptor complex apart. So far, the existence of catch bond-forming cellular adhesins has been ascertained for the most common Escherichia coli adhesin, FimH, and P-/L-selectins universally expressed by leukocytes and blood vessel walls. One compelling model for these force-enhanced interactions proposes that conformation of the ligand-binding pocket in the receptor protein is allosterically linked to the quaternary configuration of the receptor domains. The catch bond properties are likely widespread among adhesive proteins, calling for a detailed understanding of the underpinning mechanisms and physiological significance of this elusive phenomenon.
“Everything flows, nothing stands still” Heraclitus of Ephesus
Introduction
By definition, adhesive interactions must withstand tensile force to one extent or another. In biological systems, the mechanical force is derived from shear stress and/or active contraction powered by molecular motors.
Living organisms, unicellular or multicellular, consist mostly of water-based fluids and their flow along a surface creates a shear stress that drags along everything that is on it. Resisting removal by shear stress can be considered a critical characteristic of biological adhesion, because most adhesive interactions between bacterial and/or eukaryotic cells or between cells and non-cellular surfaces are initiated, and then, sustained under some flow conditions. The shear is created, for example, by flow from the heart pumping blood, saliva or tear secretion, eyelid blinking, intestinal peristalsis, emptying of the urinary bladder, lung mucus movement by ciliated epithelium or … gum chewing. Vascular endothelial cells are exposed to fluid shear stresses that are typically 1–2 dyn/cm2 on the venous side and 10–20 dyn/cm2 (up to 50 dyn/cm2) on the arterial side of the circulation [Davies, 1995; Guo, 1995]. Shear stress generated at the tooth surface by salivary flow is approximately 0.8 dyn/cm2 [Prakobphol, 1999]. Urine flow creates shear of 0.17 dyn/cm2 in the proximal renal tubule [Essig, 2003]. The dragging force on the adhesive bonds increases with an increase in fluid velocity and viscosity as well as with the diameter of the attaching cell and the angle between the receptor-ligand tether and adhesive surface [Thomas, 2008](Figure 1A).
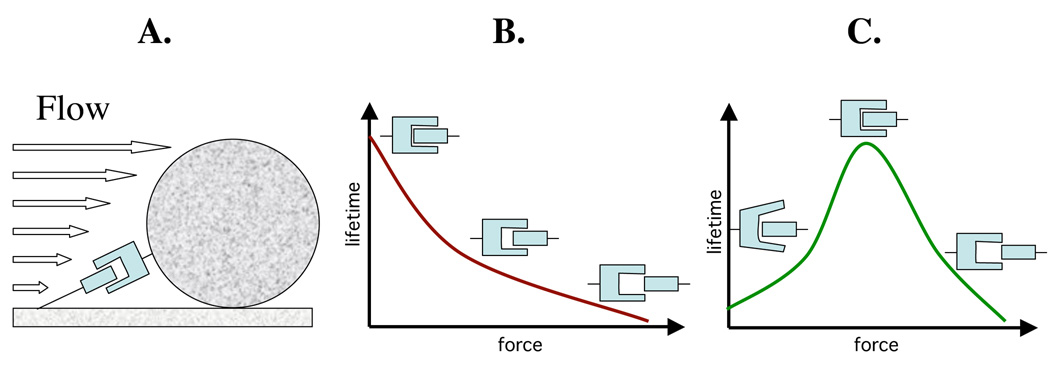
Figure 1. Dragging force and its effect on slip or catch bonds.
A. Schematic presentation of drag force on an adhering cell; B. Dependence of the lifetime of receptor-ligand interactions on the force level in slip bonds, and C. same but in catch bonds. Note that the full-in and half-in configurations of the ligand correspond to strong and weak binding, respectively, that in turn reflects low and high probability, respectively, of the bond dissociation.
Upon initiation of adhesion, the adhesive bonds must accommodate the need to sustain attachment and, at the same time, allow for the movement of migratory eukaryotic cells, or bacterial cells spreading along the colonizing surface. This involves the continuous formation and breakage of cell-surface or inter-cellular interactions (as reviewed in Vogel, 2006). In these processes, cytoskeletal rearrangements or retraction of adhesive organella can play the major role and provide an additional source of tensile stress on the adhesive receptor-ligand bonds.
Slip bonds vs. catch bonds
According to conventional wisdom, if interacting molecules are being pulled apart by tensile force, the probability that the bond will break increases with force [Bell, 1978; Dembo, 1988; Evans, 1997], i.e. the ligand should slip out of the binding pocket more rapidly under higher tensile force (Figure 1B). Thus, these types of force-inhibited interactions are called ‘slip’ bonds [Dembo, 1988]. When adhesive interactions are based on slip bonds, the strongest adhesion under flow is expected to occur at the lowest shear stress where force is the weakest. Indeed, for many bacteria that bind to tissues or other surfaces under fluid flow, it was shown that shear stress both prevents bacterial attachment and washes off bacteria that are already bound [Christersson, 1988; Shive, 1999; Prakobphol, 1995].
Several studies, however, indicated that in some instances shear stress seemed not to inhibit, but actually to promote adhesion. For example, it was known that in order for a platelet to bind to a surface coated with the plasma protein, von Willebrand factor, a certain level of shear stress was required [Savage, 1996; Kroll, 1996; Fredrickson, 1998]. Also, it became clear that without shear, leukocytes do not bind to an endothelial or PSGL-1 coated surface, but above a certain shear threshold, they are able to attach in a rolling fashion [Finger, 1996]. If the flow is reduced, the rolling cells completely detach from the surface. In bacteria, it was noticed that under shear stress induced by a rotational viscometer, the bacteria-induced agglutination of red blood cells occurred significantly more rapidly and that the clumps were significantly larger when shear was relatively high [Brooks, 1989, 1983a, 1983b]. It has also been reported that increased shear stress results in an increase in the number of S. aureus adhering to a collagen-coated surface [Li, 2000]. Different explanations have been advanced to explain the observations of shear-enhanced adhesion from a slip-bond perspective. The platelet binding requirement for shear has been explained by a need for von Willebrand factor to unravel its structure under high fluid flow, exposing a cryptic ligand-binding pocket [Siedlecki, 1996; Schneider, 2007]. For the shear threshold of leukocyte rolling, it has been proposed that increased flow rate results in faster formation of adhesive bonds. That is, increased shear increases the rate at which bonds form more than the rate at which they break [Dwir, 2000]. Also, deformation of the cellular shape under flow could increase the number of selectin molecules involved in the adhesive interactions. The shear-enhanced bacteria-erythrocyte clumping was proposed to be driven by the increased mobility and, thus, clustering of the receptors along the membrane when shear is applied [Brook, 1983b].
All these slip bond-based models are plausible and could have merit in explaining, at least in part, the described phenomena of shear-enhanced adhesion. However, another concept has been advanced that postulates the existence of receptor-ligand interactions that are activated by force (i.e. shear increase decreases the bonds dissociation rate) and therefore different from that seen for slip bonds, where bond dissociate faster under force [Dembo, 1988]. These counter-intuitive force-strengthened bonds were named ‘catch’ bonds to distinguish them from slip bonds (also named therein as such for the first time). While postulating the alternative concept of bond behavior under stress (and suggesting the quite ‘catchy’ bond nomenclature), this study was still purely theoretical in nature and mathematical in format, without any experimental evidence for the existence of catch bonds, or a structural model for their possible mechanism. It also assumed an infinite increase in bond strength under increasing tensile force, which is impossible, as any bond in nature would break under sufficient force. However, with a correction for the latter, the dependence of a catch bond’s lifetime on force could be illustrated as follows: at low force the bonds are weak and relatively short-lived; with force increase, the bond’s strength (i.e. life-time) would increase to a certain level, after which a further increase in force would eventually break the bond (Figure 1C).
Despite this presentation of the catch bond concept in 1988, none of the studies prior to 2002 that reported shear-enhanced adhesion invoked the idea of force-enhanced bond lifetime to explain the observed phenomena.
Experimental evidence for catch bonds
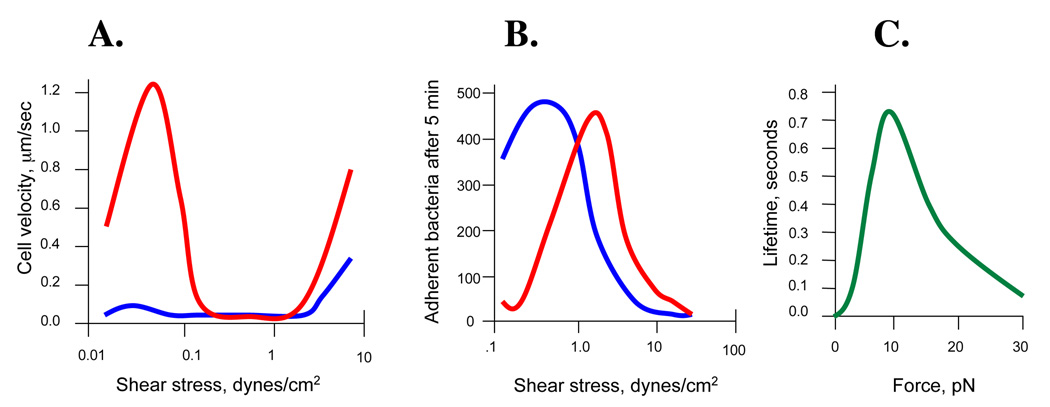
The first experimental study that claimed to identify a force-enhanced receptor-ligand interaction utilized Escherichia coli that possess type 1 fimbriae. Also called pili, these hair-like surface appendages present on their tip a mannose-binding adhesin, the 30 kDa protein FimH. In this study [Thomas, 2002], fimbriated bacteria were immobilized on the surface of a flow chamber. Then, guinea pig red blood cells (rich in mannosylated glycoproteins) were loaded on the bacterial carpet; the washing buffer was flowed at different rates and the behavior of the erythrocytes on the surface was monitored by video-microscopy. When the buffer was flown slowly (creating shear stress of 0.02 dynes/cm2) red blood cells that were attached to the bacterial carpet moved along the surface (Figure 2A). When the washing buffer was fed into the flow chamber at an increased velocity, the rolling velocity at first increased, but then suddenly dropped, with all red blood cells becoming stationary at a shear of 0.8 dynes/cm2. Thus, the binding strength of red blood cells and the mannose-specific fimbriae of E. coli was enhanced by shear.
Figure 2. Experimental evidence for catch bonds.
A. Rolling velocity of red blood cells over a carpet of type 1-fimbriated E. coli, dependent on shear level (red – shear-enhanced wild-type FimH; blue – shear-independent V156P FimH mutant) [adapted from Thomas, 2002]; B. Accumulation of type 1 fimbriated E. coli on a mannosylated surface dependent on shear level (red – wild-type FimH; blue – V156P FimH mutant) [adapted from Thomas, 2004], and C. Lifetime of P-selectin-PSGL-1 bonds dependent on the level of tensile force applied by AFM [adapted from Marshall, 2003].
Unlike the previous studies that reported shear-enhanced adhesion, special attempts were made in the Thomas et al. study to determine whether the phenomenon had a structural origin, i.e. whether it was dependent on the effect of tensile force on the intrinsic properties of individual bonds. A mutant form of the adhesive protein, FimH, was identified that mediated shear-independent binding, where the red blood cell binding was already strong under the lowest shear (Figure 2A). The FimH V156P mutation did not affect the number or morphology of the fimbriae nor did it increase the amount of adhesins per fimbria. Instead, steered molecular dynamics (SMD) simulations were used to predict that this mutation would ease a conformational change in FimH expected to be induced by shear-originated mechanical force - extension of a short linker chain connecting the lectin (mannose-binding) domain of FimH with its pilin (fimbria-incorporating) domain. These simulations were also used to predict point mutations that would suppress the linker extension, and bacteria expressing FimH with these mutations were even more dependent on shear stress for strong adhesion than were the wild-type FimH bacteria. Also, shear-enhanced adhesion was dependent on shear stress (i.e. force) but not flow rate arguing against a possible impact of the cell deformation on the increased adhesion strength. Thus, it was proposed that shear-enhanced FimH-mediated adhesion depends on a conformational change (linker extension) in FimH that, according to SMD, occurred under tensile force after the mannose binding, thus affecting (decreasing) the bonds OFF-rates. This was in contrast to prior studies on shear-enhanced cell adhesion that suggested that the phenomenon was caused by an increased ON-rates of the bonds, i.e. by kinetics factors that play roles before the bond is formed [Savage. 1996; Chang, 1999; Chen, 1999].
An independent follow-up minireview immediately called the FimH-mannose bond the long-sought counter-intuitive catch bond [Isberg, 2002]. The idea that catch bonds were due to intrinsic properties of the FimH-mannose bond was further supported in the next experimental study on FimH where the shear-enhanced binding of type 1 fimbriated E. coli was also shown using free floating bacteria and mannose-BSA coated plastic surfaces [Thomas, 2004]. Binding of the bacteria to the mannosylated surface was low (and in a loose rolling mode) at low shear, but increased more than 10-fold (and switched to strong stationary mode) with shear increase (Figure 2B). The shear-independent FimH V156P mutant exhibited high and stationary binding already under low shear (Figure 2B). The shear-enhanced binding of FimH was also later shown in a bacteria-free setting, by adhesion of mannose-coated microbeads to surface-immobilized fimbriae [Forero, 2004].
Though the original claims of discovering force-enhanced adhesion were based on a strong supportive evidence from the FimH binding, at that time some room for other interpretations could still be remaining. A direct proof for the existence of catch bonds had to come from single-molecule studies where the response of individual receptor-ligand interactions to tensile force could be measured. The first such study [Marshall, 2003] came soon after the original FimH report, where Atomic Force Microscopy (AFM) was used to stretch a bond between P-selectin and its ligand - P-selectin glycoprotein ligand-1 (PSGL-1). P-selectin are receptor proteins expressed by activated endothelial cells that recognize LewisX oligosaccharide of PSGL-1, expressed on the surface of leukocytes. As mentioned above, leukocyte binding to the endothelium (manifested in surface rolling) requires a certain shear threshold, i.e. is dependent on, or enhanced, by shear. This surface rolling allows cells to slow down in rapidly flowing blood enough to interact with other signals in the vessel wall and is involved in immune responses, inflammation, hemostasis and thrombosis. An AFM cantilever was coated with PSGL-1 and used to probe a supported lipid bilayer containing P-selectin. Upon formation of a single P-selectin-PSGL-1 bond, the cantilever was pulled away to create a certain level of constant force on the bond. The time it took for the bond to break (dissociate) under a defined force load was recorded. The lifetime of bonds steadily increased with an incremental rise in the level of force from 5 pN to 11 pN (Figure 2C). A further increase in tensile force resulted in the increase of dissociation rate as the bonds were overpowered by excessive force. The increase in bond strength with a moderate force could not be explained by a slip bond mechanism of the interaction, but corresponded well with the bonds behaving according to the catch bond concept.
The experimental studies on P-selectin-PSGL1 adhesive interactions provided solid evidence for the existence of catch bonds in nature, prompting a declaration that catch bonds were “finally caught” [Konstantopoulos, 2003]. In a recent study, single-molecule AFM experiments have also demonstrated that the FimH-mannose interaction is enhanced by pulling force [Yakovenko, 2008], affirming the accuracy of the original claim that E. coli FimH forms catch bonds.
Several additional studies demonstrated that other receptor-ligand interactions exhibit catch bond properties. L-selectin were shown to have catch bond properties by using AFM pulls with PSGL1 and endoglycan ligands or by measuring lifetimes of the transient cell/microsphere tethers [Sarangapani, 2004], and catch bond were shown to be behind the L-selectin-mediated shear-dependent rolling of leukocytes [Yago, 2004]. In an optical tweezers study on interaction of the motor protein myosin and filamentous actin (which drives cellular contractility), catch bond behavior was shown as well [Guo, 2006]. Catch bonds have been proposed in the shear-enhanced interaction of FimH with another distinct ligand - tri-mannose, where interaction is much stronger than with single mannose ligand [Nilsson, 2006].
Models proposed for the catch bond mechanism
How does pulling apart make the interaction stronger? Currently, there are insurmountable technical difficulties in observing sufficient atomic level details of the structural changes occurring in proteins and their ligands under mechanical tension. Thus, most of the predictions for how catch bonds work have been made by fitting kinetic of adsorption and unbinding with mathematical models. Four major models have been advanced in recent years:
The two-pathway model, where the ligand can exit from the binding pocket of the receptor via two different ways (pathways with different energy barrier heights) – one relatively easy and another not as easy – and where a sufficiently strong force only allows the ligand to escape via the hard pathway [Pereversev, 2005a, 2005b]. This mechanism can be similar to two hooks locked together, and which can easily separate if they are not being pulled against each other, but ‘catch’ one another stably when tensile force is applied;
The deformation model, where tensile force directly causes a conformational change of the binding pocket and/or ligand, resulting in a tighter fit [Pereversev, 2006]. This model is somewhat analogous to the mechanism underlying the ‘finger-trap’ toy – a short mesh tube that narrows and tightens when stretched by fingers inserted in each end, effectively gripping and entrapping them inside the tube;
The sliding-rebinding model, where flexibility of the receptor protein under force leads to alignment of the ligand and binding pocket interface in parallel fashion, allowing new binding contacts to form and the original contacts to rebind [Lou, 2006, 2007], and
The allosteric model, where force-induced structural alterations in one part of the receptor protein are linked to a shift from low- to high-affinity conformation of the ligand-binding site located in another part of the protein [Thomas, 2006; Yakovenko, 2008]. The two-state model [Barsegov, 2005; Evans 2004] is mathematically similar in most respects to the allosteric one.
All four models have been more or less fitted quantitatively to the experimental catch-bond data. Unfortunately, quantitative fittings have only limited success in determining the actual structural mechanism involved. Another way is to use computer SMD simulations of available crystal structures to predict how the protein conformation might change under force conditions at an atomistic level, and how specific structural mutations might affect the dynamic binding properties. The two-pathway and deformation models have not been related to particular conformational changes in the atomistic protein structures and thus remain untested to date. In contrast, atomistic simulations have lead to the development and, importantly, some experimental testing of two other models - the allosteric model for FimH and sliding-rebinding model for P-selectin.
Mechanism how the FimH-mannose catch bond works
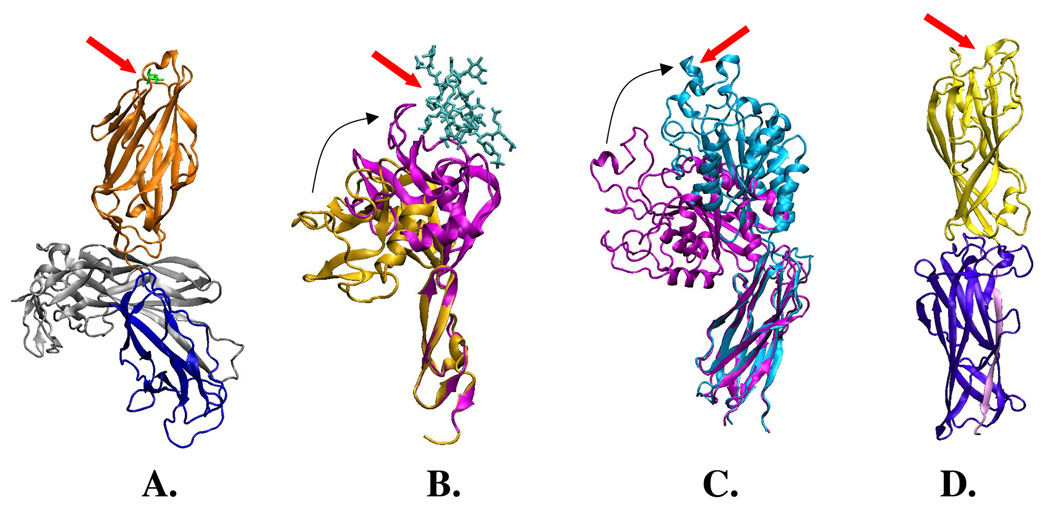
As mentioned above, FimH is located on the tip of the type 1 fimbrial rod (that mostly consists of the major (pilin) subunit, FimA) and, according to the crystal structure [Choudhury, 1999], contains the distally-located mannose-binding lectin domain that is connected via a linker chain to a pilin domain incorporating the FimH into the fimbria (Figure 3A). As also described above, when tensile force was applied in SMD simulations between the mannose-interacting pocket residues on the top of lectin domain and the end of the interdomain linker chain, the linker chain connecting the lectin and pilin domains was pulled out under force [Thomas, 2002]. Based on the functional effect of mutations predicted to facilitate or, alternatively, suppress the linker extension, it was proposed that the extension of the interdomain linker chain is associated with the increased binding strength of the pocket. It is important to remember that the mannose-binding pocket and interdomain linker are positioned on the opposite sides of the β-barrellike lectin domain.
Figure 3. Ribbon crystal structure of known and potential catch bond receptor proteins.
Ligand-binding pockets are indicated by the red arrow. A. FimH adhesin in complex with FimC chaperone (in grey), with pilin domain (blue), lectin domain (orange) and bound mannose (green); B. P-selectin EGF and lectin domains. The magenta structure was co-crystallized with PSGL-1 (cyan), while the gold structure is ligand-free. The two structures are aligned on the EGF domain to show the swing of the lectin domain (indicated by black arrow) that can occur upon binding to PSGL-1; C. Structure of beta-3 integrin I and ‘thigh’ domains in the low-affinity state (blue) and high-affinity state (magenta). The two are aligned by the ‘thigh’ domain; the I-domain binds ligand, and the swing of I-domain is indicated by black arrow (a ligand-mimetic present in the binding pocket of the swing-out conformation is omitted), and D. CfaE adhesin of enterotoxigenic E. coli, with lectin domain (yellow) and pilin domain (purple) with self-complementing β-strand (pink).
In a later study [Aprikian, 2007], it was determined that, in absence of the molecular chaperone that is wedged between the FimH domains (Figure 3A) in the crystal structure, the pilin domain is able to interact with the lectin domain. Structural mutations in the interdomain interface that weaken the domains’ interaction resulted in a dramatic increase in a) bacterial adhesion level under static or low shear conditions, and b) in affinity to mannose under equilibrium conditions (from KD 300 µM−1 to 1.2 µM−1). The highest affinity reached corresponded to the affinity of the purified lectin domain (i.e. in the absence of the pilin domain).
Thus, the FimH structure/function studies suggested that tight interaction between the lectin and pilin domains is associated with a low-affinity conformation of the pocket, while separation of the domains and linker extension, is associated with a high-affinity conformation of the pocket. This, in turn, implies that the interdomain interface might be allosterically linked to the binding pocket, i.e. a structural change in the interdomain region is structurally coupled to the binding site.
The most direct evidence for the allosteric properties of FimH came from the studies where a monoclonal antibody clone was identified that recognized the purified lectin domain and the high-affinity FimH mutants, but bound to the wild-type FimH only in the presence of soluble or surface-immobilized mannose, under no-force static conditions [Tchesnokova, 2008]. The ligand-induced binding site (LIBS) epitope was mapped to the interdomain region of the lectin domain, i.e. far away from the mannose-binding pocket. Moreover, binding of the antibodies (or Fab fragments) to the LIBS results in a 50-fold increase in affinity of the wild-type FimH towards mannose, effectively locking FimH in the high-affinity conformation. Thus, mannose binding increases antibody binding to the interdomain interface, while antibody binding between the domains increases mannose-binding. Such reciprocity of the structural events is a hallmark of allosteric proteins, proving that there is an allosteric link between the mannose-binding pocket and the interdomain configuration of FimH.
Taken together, these results show that FimH is an allosterically regulated protein that exists in two interchangeable conformational states – a low and a high-binding state to mannose. In the low-binding state, the domains interact with each other and the mannose-binding pocket is in the low-affinity conformation (Figure 4A). In the high-binding state, the domains are separated and the binding pocket is in the high-affinity conformation (Figure 4B). Any external factor that would keep the domains in the separated conformation will keep the binding pocket in high-affinity conformation. This could be structural mutations in or antibodies against interdomain interface. More importantly, such a factor could also be tensile force which is applied via the bound mannose thereby keeping the domains separated and thus, keeping the mannose-binding strong (Figure 4C). This provides a structural explanation for the shear-enhanced FimH-mannose catch bond that explains all available data. One needs to note that despite the strong evidence for the allosteric mechanism of FimH catch bonds, no high resolution structural details of the conformational link between interdomain interface and binding pocket configurations have been elucidated to date. SMD simulations, however, suggest that two binding pocket conformations exist which are distinct in the force-bearing hydrogen bond networks that the mannose ring can form with the base of the binding pocket (Nilsson, 2008).
Figure 4. Schematic representation of putative allosteric states of FimH.
The pilin domain is the lower part of the blue-filled structure, the lectin domain is above, with the mannose-binding site being an indent on the top and mannose ligand represented by the black-filled oval. A. low-binding state; B. high-binding state (transient), and C. high-binding state sustained by tensile forces (block arrows). Note: the structural details of the conformational link between the interdomain interface and mannose-binding pocket are currently unknown. While the wide and narrow configurations of the binding pocket correspond to low- and high-affinities, respectively, of the mannose-binding, alternative conformational associations have been proposed [Nilsson, 2008].
Here, it is also important to mention that the structural properties of the fimbrial rod could have been evolutionary adapted to optimize the catch bond properties of FimH [Forero, 2006]. The body of type 1 fimbriae consists of the 18 kDa FimA subunit, which forms the spiral-shaped rod with 3.4 subunits per coil. Under a certain level of tensile force, the quaternary structure of the rod is uncoiled and the fimbrial length can be stretched 8-fold [Forero, 2006; Miller, 2006]. Upon a drop in the force, the rod recoils back. Uncoiling/recoiling of the fimbria is expected to happen under variable shear conditions. Uncoiling of the fimbria is likely to dampen the mannose bond-breaking effect of high shear forces even when the adhesin is shifted to the high-affinity state. At the same time, if the shear drops to low level, the fimbrial recoiling would maintain a residual level of mechanical tension on the bond, preventing its fast backshift from the high- to the low-binding state. Thus, though the shear-enhanced binding can be accomplished by non-fimbrial forms of FimH (by using FimA-free fimbrial tips, for example [Aprikian, 2007]), the mechanical properties of the fimbrial rod appears to be tuned to sustain bacterial adhesion under pulsatile flow conditions.
Proposed models of selectin catch bonds
Like FimH, selectins have a lectin domain that binds the LewisX carbohydrate ligand (in PSGL-1) and a neighboring domain, the EGF domain, which serves to anchor the receptor to the endothelium or leukocytes. According to the crystal structures, the domains interact with each other and assume either a bent or a straightened configuration (Figure 3B) [Somers, 2000]. It was hypothesized that the hinge angle between the two domains acts to regulate the binding strength [Somers, 2000]. Later, it was proposed that the domains’ straightening by tensile force could explain catch bond formation by selectins [Konstantopoulos, 2003]. Indeed, a point mutation introduced in L-selectin that was predicted to eliminate a key hydrogen bond favoring the bent conformation, reduced the shear threshold for adhesion under flow and, in biomembrane force probe experiments, increased binding at low force [Lou, 2006]. Also, SMD simulations have predicted that mechanical force would straighten an interdomain hinge angle in selectins [Lou, 2007].
The apparent force-induced inter-domain conformational change in selectins from bent to straightened configuration is analogous to the force-induced switch in FimH from the interacting to separated conformation. Indeed, it remains possible that the putative domain-domain interaction in FimH also results in a bent conformation that straightens and separates under force.
Thus, selectin catch bonds may also be allosterically regulated, where the straightening of the interdomain region is structurally coupled to the PSLG-1 pocket thus regulating the shift from low to high-affinity conformation. Indeed, the idea of allosteric regulation by the hinge angle is supported by the fact that when an oligosaccharide moiety was introduced into P-selectin to wedge the hinge angle open, an increase in the affinity and lifetime of selectin-PSGL-1 bonds was observed in cell adhesion assays and, in surface plasmon resonance kinetic studies of isolated receptors, which do not generate significant drag force [Phan, 2006]. However, the affect of this mutation on catch bond behavior was not determined. Moreover, no monoclonal antibody studies or other means have been pursued so far that would differentially recognize and lock-in the open interdomain configurations of this system in solution. Thus, reciprocity between these configurations and low- and high-affinity conformation of the binding pocket has not yet been established which leaves it open whether the selectin-PSGL-1 catch bond is allosterically regulated.
The importance of the interdomain hinge straightening in enhancing selectin binding under force has prompted the advancement of an alternative, sliding-rebinding model of the selectin catch bond [Lou, 2006, 2007]. According to the model, at low force, the ligand bound to the bent receptor would be pulled directly away from the pocket until complete unbinding (without the interdomain straightening), and so the lifetime would reflect the initial binding state, which is short-lived. However, higher forces would cause the opening of the hinge angle that, in turn, would align the ligand-binding pocket to be parallel to the direction of force. The ligand would then be pulled so that it slides along the binding face of the straightened receptor, allowing new binding contacts to be formed in the pocket, thus prolonging bond lifetime. This concept was supported by SMD simulations that showed that when the hinge angle between the domains had been opened, the ligand slid within the binding pocket and formed new contacts [Lou, 2007]. The authors also hypothesize that, in the course of hinge opening, reversible unbinding and rebinding of the original or newly formed interactions would occur, further contributing to the increased bond lifetime. This model was suggested to be experimentally supported by a demonstration that a mutation predicted to straighten the hinge angle had the effect of increasing bond lifetime at forces too low to straighten the hinge angle in the native selectin [Lou, 2006]. One needs to note, however, that these experimental data do not contradict the allosteric model for selectin catch bonds since, while it correlates the hinge angle to increased binding, the underlying mechanism by which the hinge angle enhances binding has not yet been derived‥ Incidentally, it was stated that the sliding-rebinding model also contains an allosteric element based on the fact that force acting on a remote site (hinge) leads to a novel ligand-binding interface. One can argue, however, that no conformational change of the binding pocket structure is postulated by the sliding-rebinding model that implies that such a mechanism cannot be called allosteric according to the classical definition of the latter.
Thus, both the sliding-rebinding model and the allosteric model place importance to a structural change in conformation of the interdomain region, but differ significantly as the sliding-rebinding model does not require that a conformational change in the binding pocket will occur in response to the hinge straightening. Future studies are needed to clarify the mechanism by which selectins catch their ligands.
Possibilities of allosteric catch bond properties of other adhesins
While cofactor-regulated allosteric proteins are common, mechanical perturbations in a remote site could play an analogous role to cofactors. This prompts the question whether other adhesive systems might potentially act in a similar way. The basic requirements for the FimH-like allosteric model are: (i) a multi-domain structure of the receptor protein that includes minimally a binding and an anchoring domain; (ii) the ability of the binding domain to interact with the anchoring domain; (iii) the location of the ligand-binding pocket away from the interdomain interface, and most importantly (iv) the ability of the binding pocket to change its conformation from a low- to a high-bond strength when the domains separate and/or switch to a straightened orientation, both of which would be favored by tensile force.
One large class of potential catch bond adhesive receptors are integrins, the allosteric properties of which have been characterized in great detail. The integrins family of proteins is involved in cell adhesion and cell-to-cell signaling processes. Integrins are dimeric molecules with more than a dozen domains, multi-ligand binding and complex bidirectional regulatory properties. Despite the complexity of integrins relative to FimH, there are some stunning similarities in the allosteric properties of the two adhesive proteins, specifically in their LIBS-associated characteristics. Actually, LIBS were originally described and thoroughly studied in integrins [Frelinder, 1991; Khaspekova, 1996; Luo, 2004] where they are positioned far from the binding pockets and are found in both binding and non-binding domains, usually close to the interface between domains. Integrin LIBS become exposed upon binding to fibrinogen, fibronectin, collagen, or Mg++/Mn++ to the α-I/A and β propeller domains in the α-subunit or the β-I/A domain in the β-subunit [Frelinder, 1991; Khaspekova, 1996; Luo, 2004]. The binding domains are distally-positioned, forming the integrin’s ‘head’ (Figure 3C). The ligand (or cation) binding results in integrin activation - a ‘jack-knife’-like switch from a bent conformation of integrins with low-affinity towards the ligands, to a straightened high-affinity conformation (Figure 3C) [Mould, 2002; Arnaout, 2005]. As in FimH, antibody binding to the exposed LIBS sustains the high-affinity conformation of integrins [Humphries, 2000]. Because the bent-to-open quaternary change in the integrins would also be facilitated by a tensile mechanical force, it has recently been suggested that mechanical regulation can activate integrins, making them catch bonds [Chigaev, 2003]. Indeed, some recent studies provided experimental support for force-enhanced integrin function [Katsumi, 2005; Astrof, 2006; Alon, 2007; Woolf, 2007]. Also, SMD simulations have suggested specific hypotheses for how force-activation of integrins might occur [Jin, 2004; Puklin-Faucher, 2006]. Still, no direct evidence has been obtained yet that integrins do indeed form catch bonds.
Based on its allosteric similarities with FimH (and integrins), another cell adhesion protein - galactose/N-acetylgalactoseamine-specific calcium-dependent lectin (mMGL) on macrophages and lymphocytes – might form catch bonds. Indeed, in mMGL, a so-called LOM-11 epitope becomes accessible once the adhesin is activated, by either calcium-binding or ligand-binding [Kimura, 1995]. Antibody binding to this LIBS stabilizes the lectin in an “active” conformation so that the latter is able to bind ligands even under low-calcium conditions. Like FimH, and indeed the majority of cell-adhesion receptors, mMGL has separate carbohydrate-binding, and cell-anchoring, domains [Sato, 1992].
A similar allosteric mechanism could be involved in platelet adhesion via glycoprotein Ib to the multi-domain protein von Willebrand factor, which occurs only under high shear stress. Binding of the glycoprotein Ib to von Willebrand factor has also been shown to be enhanced by antibodies that bind to a neighboring domain [Ulrichts, 2004; 2006], removal of the neighboring domain [Ulrichts, 2006], and many point mutations that are near the anchoring termini and distal from the major glycoprotein Ib-binding site (Huizinga, Tsuji et al. 2002). The interaction of T-cells with antigen-presenting cells, where stabilization of the receptor sub-domain conformation is associated with increased longevity of inter-cellular binding [Qi, 2006], can also involve allosteric catch bond formation.
Shear-enhanced bacterial adhesion was shown to be mediated by the di-galactose specific P-fimbriae of E. coli, involving a 32 kDa PapG adhesin [Nilsson, 2006]. Like FimH, PapG is positioned on the fimbrial tip and has two domains – a ligand-binding and a fimbriae-anchoring one. Though only the structure of the former has been resolved [Dodson, 2001], it is clear that the galactose(1–4)galactose binding pocket is positioned away from the putative interdomain interface. Thus, there is good reason to postulate that the shear-enhanced binding might involve allosteric catch bond properties of PapG, but this needs to be investigated. Interestingly, the uncoiling/recoiling properties of the P-fimbriae have been demonstrated as well [Fallman, 2005; Jass, 2004].
Finally, we expect that, beside FimH and PapG, many additional bacterial adhesins mediate adhesion via catch bond mechanism. For example, a major structural category of fimbriae (class 1) is similar to the type 1 fimbriae of E. coli, and characterized by a spiral fimbrial rod, with the adhesive function carried by the adhesive protein positioned on the very tip of the fimbriae. These tip adhesins are usually similar to FimH in size, and are either known or predicted to have two domains – one ligand-binding and another fimbria-incorporating. Recently, a first chaperone-free structure was obtained for a class 1 fimbrial adhesin - CfaE adhesin of CFA fimbriae of enteropathogenic E. coli [Li, 2006]. Unlike FimH, CfaE was also crystallized without a bound ligand, the exact nature of which is yet unknown. In this structure, two domains have been identified, which tightly interact with one another (Figure 3D). The putative binding pocket for the ligand is mapped on the top of the binding domain. Thus, if CfaE is able to form catch bonds via a mechanism proposed for the FimH two-state allosteric model, the ligand-free crystal structure of CfaE could correspond to the low-binding state, with a closed interdomain configuration and low-affinity conformation of the binding pocket. Investigations are currently underway to determine whether CfaE mediates shear-enhanced adhesion to the target cells. In addition to fimbrial adhesins, afimbrial adhesins in bacteria are also usually of a multi-domain structure, with separate binding and cell-anchoring domains [Walsh, 2004]. As such, some of these might exhibit allosteric catch bond properties as well.
Thus, it appears that a great number of adhesive proteins, on the surface of both bacterial and human cells, have at least the potential to act as FimH-like mechanically regulated allosteric catch bond adhesins. In some adhesins, interaction between the binding and anchoring/neighboring domains or, even an allosteric-like connection between the binding pocket affinity and the interdomain configuration has already been established. We propose, therefore, that adhesion mediated by an allosteric catch bond mechanisms represents a more common phenomenon in nature, which likely govern many types of bacterial or cell adhesions under shear-derived or other types of mechanical tension.
Among the reasons for a rather slow identification of novel catch bonds could be the only recent appreciation for the existence force-enhanced adhesive interactions as well as certain technical challenges in studying adhesion dynamics under shear conditions or, especially, in single-molecule force measurements. Another significant obstacle in identifying catch bond adhesins could arise from using natural or lab-induced structural variants of the receptor protein that do not adequately support shear-enhanced adhesion. The latter could be a significant problem as it takes only a single point mutation to convert a shear-dependent catch bond-forming receptor variant into a slip bond-like receptor that mediates strong binding already under static conditions. Such recombinant variants are mentioned above for the FimH adhesin and selectins. For FimH, also, many naturally occurring variants have been described which arise among uropathogenic strains of E. coli and increase many-fold the binding to mannose and uroepithelial cells under static conditions [Sokurenko, 1998, 2004]. Such mutations appear to be adaptive for colonization of the urinary tract, where shear is estimated to be low (e.g. 0.17 dyn/cm2 in the renal tubule), with the exception of urethra in the course of urination, where shear is likely to be around 3–5 dyn/cm2 . Also, mutations that confer shear-independent properties might be selected under laboratory conditions, because they provide an adhesive phenotype that is easily observable by the investigator. To avoid this potential problem with natural or lab-induced mutations, comparative analyses of the function of several different natural variants of a particular receptor need to be undertaken.
Physiological significance of catch bonds
Catch bonds (independent of their mechanism of operation) have certain physiological advantages over slip bonds:
Catch bonds could provide a mechanism for binding to surface-anchored ligands much stronger than to soluble ligands, thus allowing adhesion to surface-bound ligands even in the presence of soluble ligands in solution. Under flow conditions, dragging force will induce and sustain the high-binding state of the receptor in the course of interaction with immobilized ligands. Interaction with soluble ligands, however, will mostly be via a low-binding state, because soluble ligands are too small to generate significant drag force. In contrast, slip bond affinity is similar to both soluble and immobilized forms of the ligand and adhesion inhibition can be avoided only by an increased avidity (multiplicity) of the surface interaction. Pharmacologically relevant, soluble inhibitors that prevent bacterial adhesion under low flow are ineffective washing off bacteria under high flow conditions, while they are effective detaching bacteria expressing a slip bond-forming mutant FimH [Thomas, 2002; Nilsson, 2006b].
Evolving catch bonds could provide a competitive advantage to help cells resist a wash-off under high flow, but to bind loosely and, thus, spread along the surface under low flow. In contrast, binding via slip bonds leads to slower or even stationary movement at reduce flow, and increased rolling and detachment at high flow, as demonstrated by simulations of cell adhesion in flow (Chang, Tees et al. 2000). This stronger adhesion at high flow presumably allows leukocytes to not become permanently ‘stuck’ on the endothelium surface in areas of low blood velocity, like capillaries, small arterioles or venules, and at the same time to stay attached and patrol the endothelium by rolling in the vessels where the flow is relatively high. Also, biofilm spread under flow occurs much more quickly in bacteria expressing wild-type FimH that able to mediate surface rolling, rather than shear-independent mutant FimH that always mediates firm adhesion [Anderson, 2007].
Catch bonds could provide a means for migrating or interacting eukaryotic cells to modulate the strength of interaction via changing mechanical tension in the foci of adhesive contacts. Cell migration involves a continuous process of membrane protrusions, traction and retraction, driven by rearrangement of actin cytoskeleton inside the cell. The assembly/disassembly of the cytoskeleton and the power strokes of myosins create variable mechanical tension in the adhesive interactions. Catch bonds should allow rapid dynamics in the attachment and detachment of cell tethers by merely modulating the level of tensile force on the adhesive bonds. Here, the known catch bond properties of actin-myosin interactions could be coupled with the proposed catch bond properties of integrins.
Catch bonds could provide a means to increase on-rates but decrease off-rates of binding. Indeed, if a receptor protein can assume weak- and strong-binding states, they could be associated with a relatively loose and tight conformation, respectively, of the binding pocket. In addition to effects on the binding strength, binding pocket conformation could affect kinetics. Specifically, if the pocket is in a loose conformation, this would allow fast entry and exit of the ligand (resulting in high on- and off-rates), while a tight pocket would slow entry and exit (resulting in low on- and off-rates). Indeed, recent kinetic studies support this notion for FimH (Tchesnokova, Aprikian et al. 2008). Because initial interaction of the receptor with the ligand occurs via the weak-binding state, which is able then to convert into the strong-binding state, this would ensure high on- but low off-rates of attachment, making the attachment initiation more efficient.
It is as yet unclear, however, to what extent the proposed or any other properties of catch bond adhesion are physiologically relevant.
Conclusion
The phenomenon of tensile force-enhanced, catch-bond adhesion is only beginning to be unraveled. We would like this review to stimulate the discovery of additional catch bond interactions as well as to facilitate further studies into the detailed structural mechanisms of how the various catch bonds described really work. For example, for the allosteric mechanism we need to define the currently unknown structural pathway of the conformational link between the binding pocket and the remote interdomain interface. To accomplish this, additional, more focused crystallography, NMR and computer simulation studies must be undertaken. However, what we already know about allosteric catch bonds allows for a shift in the paradigm as to how we can best exploit our understanding of these adhesive interactions. In particular:
Specific attention should be devoted to the design of adhesion inhibitors that act allosterically and that can either prevent a shift from a low- to high-binding state, or to induce reversion of the later to the former (i.e. to destroy bonds already formed). Such inhibitors would target, for example, the interdomain interface or conformational pathway between the interface and binding pocket. By the same token, allosteric compounds could be designed to enhance the adhesion, if desirable, by promoting and sustaining the switch from the low- to high-binding state.
The use of bacterial adhesins or adhesive domains for vaccines needs to be re-evaluated taking into account the possibility that potential LIBS epitopes might elicit antibodies that will enhance rather than inhibit the binding properties of the adhesin. While it will increase the sensitivity of the adhesin to inhibition by soluble ligands, paradoxically, this type of anti-adhesin antibodies will likely increase significantly the adhesive strength.
Acknowledgements
We gratefully acknowledge the many scientific contributions from the current and former members of our research teams, especially Lina Nilsson and Manu Forero, to whom we also grateful for critical reading of the manuscript. Funding for these studies has primarily been provided by NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon R, Chen S, et al. The kinetics and shear threshold of transient and rolling interactions of L-selectin with its ligand on leukocytes. Proc Natl Acad Sci U S A. 1998;95(20):11631–11636. doi: 10.1073/pnas.95.20.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R, Chen S, et al. The kinetics of L-selectin tethers and the mechanics of selectin-mediated rolling. J Cell Biol. 1997;138(5):1169–1180. doi: 10.1083/jcb.138.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon R, Hammer DA, et al. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 1995;374(6522):539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- Alon R, Dustin ML. Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels andantigen-presenting cells. Immunity. 2007;26:17–27. doi: 10.1016/j.immuni.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Anderson BN, Ding AM, Nilsson LM, Kusuma K, Tchesnokova V, Vogel V, Sokurenko EV, Thomas WE. Weak rolling adhesion enhances bacterial surface colonization. J Bacteriol. 2007;189:1794–1802. doi: 10.1128/JB.00899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RK, Lopez JA, et al. Molecular mechanisms of platelet adhesion and activation. Int J Biochem Cell Biol. 1997;29(1):91–105. doi: 10.1016/s1357-2725(96)00122-7. [DOI] [PubMed] [Google Scholar]
- Aprikian P, Tchesnokova V, Kidd B, Yakovenko O, Yarov-Yarovoy V, Trinchina E, Vogel V, Thomas W, Sokurenko E. Interdomain Interaction in the FimH Adhesin of Escherichia coli Regulates the Affinity to Mannose. J Biol Chem. 2007;282:23437–23446. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- Astrof NS, Salas A, et al. Importance of Force Linkage in Mechanochemistry of Adhesion Receptors. Biochemistry. 2006;45(50):15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsegov V, Thirumalai D. Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc Natl Acad Sci U S A. 2005 Feb 8;102(6):1835–1839. doi: 10.1073/pnas.0406938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Bouckaert J, Berglund J, et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol Microbiol. 2005;55(2):441–455. doi: 10.1111/j.1365-2958.2004.04415.x. [DOI] [PubMed] [Google Scholar]
- Brooks DE, Cavanagh J, Jayroe D, Janzen J, Snoek R, Trust TJ. Involvement of the MN blood group antigen in shear-enhanced hemagglutination induced by the Escherichia coli F41 adhesin. Infect Immun. 1989 Feb;57(2):377–383. doi: 10.1128/iai.57.2.377-383.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DE, Trust TJ. Enhancement of bacterial adhesion by shear forces: characterization of the haemagglutination induced by Aeromonas salmonicida strain 438. J Gen Microbiol. 1983a Dec;129(Pt 12):3661–3669. doi: 10.1099/00221287-129-12-3661. [DOI] [PubMed] [Google Scholar]
- Brooks DE, Trust TJ. Interactions of erythrocytes with bacteria under shear. Ann N Y Acad Sci. 1983b;416:319–331. doi: 10.1111/j.1749-6632.1983.tb35196.x. [DOI] [PubMed] [Google Scholar]
- Chang KC, Hammer DA. The forward rate of binding of surface-tethered reactants: effect of relative motion between two surfaces. Biophys J. 1999;76(3):1280–1292. doi: 10.1016/S0006-3495(99)77291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KC, Tees DF, et al. The state diagram for cell adhesion under flow: leukocyte rolling and firm adhesion. Proc Natl Acad Sci U S A. 2000;97(21):11262–11267. doi: 10.1073/pnas.200240897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Springer TA. An automatic braking system that stabilizes leukocyte rolling by an increase in selectin bond number with shear. J Cell Biol. 1999;144(1):185–200. doi: 10.1083/jcb.144.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular alpha4-integrin conformational activation. Biophys. J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- Christersson CE, Glantz PO, Baier RE. Role of temperature and shear forces on microbial detachment. Scand J Dent Res. 1988 Apr;96(2):91–98. doi: 10.1111/j.1600-0722.1988.tb01413.x. [DOI] [PubMed] [Google Scholar]
- Davies PF. Flow-Mediated Endothelial Mechanotransduction. Physiological Reviews. 1995 July;Vol. 75(No 3) doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Dodson KW, Pinkner JS, Rose T, Magnusson G, Hultgren SJ, Waksman G. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001 Jun 15;105(6):733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- Doggett TA, Girdhar G, et al. Selectin-like kinetics and biomechanics promote rapid platelet adhesion in flow: the GPIb(alpha)-vWF tether bond. Biophys J. 2002;83(1):194–205. doi: 10.1016/S0006-3495(02)75161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwir O, Kansas GS, Alon R. An activated L-selectin mutant with conserved equilibrium binding properties but enhanced ligand recognition under shear flow. J Biol Chem. 2000 Jun 23;275(25):18682–18691. doi: 10.1074/jbc.M001103200. [DOI] [PubMed] [Google Scholar]
- Essig M, Friedlander G. Tubular shear stress and phenotype of renal proximal tubular cells. J Am Soc Nephrol. 2003;14:s33–s35. doi: 10.1097/01.asn.0000067650.43083.df. [DOI] [PubMed] [Google Scholar]
- Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci U S A. 2004 Aug 3;101(31):11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fällman E, Schedin S, Jass J, Uhlin BE, Axner O. The unfolding of the P pili quaternary structure by stretching is reversible, not plastic. EMBO Rep. 2005 Jan;6(1):52–56. doi: 10.1038/sj.embor.7400310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger EB, Puri KD, Alon R, Lawrence MB, von Andrian UH, Springer TA. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature. 1996;379:266–269. doi: 10.1038/379266a0. [DOI] [PubMed] [Google Scholar]
- Forero M, Yakovenko O, Sokurenko EV, Thomas WE, Vogel V. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 2006 Sep;4(9):e298. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero M, Thomas W, Bland C, Nilsson L, Sokurenko EV, Vogel V. Applications of Catch-Bond based Nano-Adhesives to direct the attachment of particles to specific shear regions in a fluidic environment. Nanotechnology Letters. 2004;Vol. 4(No 9):1593–1597. [Google Scholar]
- Fredrickson BJ, Dong JF, McIntire LV, López JA. Shear-dependent rolling on von Willebrand factor of mammalian cells expressing the platelet glycoprotein Ib-IX-V complex. Blood. 1998;92:3684–3693. [PubMed] [Google Scholar]
- Frelinger ALd, Du XP, Plow EF, Ginsberg MH. Monoclonal antibodies to ligand-occupied conformers of integrin alpha IIb beta 3 (glycoprotein IIb-IIIa) alter receptor affinity, specificity, and function. J Biol Chem. 1991;266:17106–17111. [PubMed] [Google Scholar]
- Guo Z, Moreau M, Rickey DW, Picot PA, Fenster A. Quantitative investigation of in vitro flow using three-dimensional colour Doppler ultrasound. Ultrasound Med Biol. 1995;21(6):807–816. doi: 10.1016/0301-5629(95)00007-e. [DOI] [PubMed] [Google Scholar]
- Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci U S A. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MS, Hempel J, Brinton CC., Jr Purification of the Escherichia coli type 1 pilin and minor pilus proteins and partial characterization of the adhesin protein. J. Bact. 1988;170:3350–3358. doi: 10.1128/jb.170.8.3350-3358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Schroder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos. Trans. R. Soc. London. B Biol. Sci. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- Hung CS, Bouckaert J, et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. 2002;44(4):903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- Humphries MJ. Integrin structure. Biochem. Soc. Trans. 2000;28:311–339. [PubMed] [Google Scholar]
- Isberg RR, Barnes P. Dancing with the host; flow-dependent bacterial adhesion. Cell. 2002 Jul 12;110(1):1–4. doi: 10.1016/s0092-8674(02)00821-8. J Biol Chem. 2006 Jun 16;281(24):16656-63. [DOI] [PubMed] [Google Scholar]
- Jass J, Schedin S, Fällman E, Ohlsson J, Nilsson UJ, Uhlin BE, Axner O. Physical properties of Escherichia coli P pili measured by optical tweezers. Biophys J. 2004 Dec;87(6):4271–4283. doi: 10.1529/biophysj.104.044867. Epub 2004 Sep 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Andricioaei I, et al. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure. 2004;12(12):2137–2147. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Katsumi A, Naoe T, Matsushita T, Kaibuchi K, Schwartz MA. Integrin activation and matrix binding mediate cellular responses to mechanical stretch. J Biol Chem. 2005 Apr 29;280(17):16546–16549. doi: 10.1074/jbc.C400455200. [DOI] [PubMed] [Google Scholar]
- Khaspekova SG, et al. Conformational changes of the platelet membrane glycoprotein IIb-IIIa complex stimulated by a monoclonal antibody to the N-terminal segment of glycoprotein IIIa. Biokhimiia. 1996;61:412–428. [PubMed] [Google Scholar]
- Kimura T, Imai Y, Irimura T. Calcium-dependent conformation of a mouse macrophage calcium-type lectin. Carbohydrate binding activity is stabilized by an antibody specific for a calcium-dependent epitope. J Biol Chem. 1995;270:16056–16062. doi: 10.1074/jbc.270.27.16056. [DOI] [PubMed] [Google Scholar]
- Konstantopoulos K, Hanley WD, Wirtz D. Receptor-ligand binding: 'catch' bonds finally caught. Curr Biol. 2003;13:R611–R613. doi: 10.1016/s0960-9822(03)00529-3. [DOI] [PubMed] [Google Scholar]
- Kroll MH, Hellums JD, McIntire LV, Schafer AI, Moake JL. Platelets and shear stress. Blood. 1996;88:1525–1541. [PubMed] [Google Scholar]
- Lawrence MB, Kansas GS, et al. Threshold levels of fluid shear promote leukocyte adhesion through selectins (CD62L,P,E) J Cell Biol. 1997;136(3):717–727. doi: 10.1083/jcb.136.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Poole S, Rasulova F, McVeigh AL, Savarino SJ, Xia D. A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli. J Biol Chem. 2007 Aug 17;282(33):23970–23980. doi: 10.1074/jbc.M700921200. [DOI] [PubMed] [Google Scholar]
- Li ZJ, Mohamed N, Ross JM. Shear stress affects the kinetics of staphylococcus aureus adhesion to collagen. Biotechnol Prog. 2000 Nov;16(6):1086–1090. doi: 10.1021/bp000117r. [DOI] [PubMed] [Google Scholar]
- Liddington RC, Ginsberg MH. J. Cell Biol. 2002;158:833–839. doi: 10.1083/jcb.200206011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J. 2007;92:1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Yago T, Klopocki AG, Mehta P, Chen W, Zarnitsyna VI, Bovin NV, Zhu C, McEver RP. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174:1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Strokovich K, Walz T, Springer TA, Takagi J. Allosteric beta1 integrin anti-bodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J Biol Chem. 2004;279:27466–27471. doi: 10.1074/jbc.M404354200. [DOI] [PubMed] [Google Scholar]
- Marchese P, Saldivar E, et al. Adhesive properties of the isolated amino-terminal domain of platelet glycoprotein Ibalpha in a flow field. Proc Natl Acad Sci U S A. 1999;96(14):7837–7842. doi: 10.1073/pnas.96.14.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003 May 8;423(6936):190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Miller E, Garcia T, Hultgren S, Oberhauser AF. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys J. 2006 Nov 15;91(10):3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AP, Askari JA, Barton S, Kline AD, McEwan PA, Craig SE, Humphries MJ. J. Biol. Chem. 2002;277:19800–19805. doi: 10.1074/jbc.M201571200. [DOI] [PubMed] [Google Scholar]
- Nilsson LM, Thomas WE, Sokurenko EV, Vogel V. Elevated shear stress protects Escherichia coli cells adhering to surfaces via catch bonds from detachment by soluble inhibitors. Appl Environ Microbiol. 2006b Apr;72(4):3005–3010. doi: 10.1128/AEM.72.4.3005-3010.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson LM, Thomas WE, Trintchina E, Vogel V, Sokurenko EV. Catch bond-mediated adhesion without a shear threshold: trimannose versus monomannose interactions with the FimH adhesin of Escherichia coli. J Biol Chem. 2006;281:16656–16663. doi: 10.1074/jbc.M511496200. [DOI] [PubMed] [Google Scholar]
- Nilsson LM, Thomas WE, Sokurenko EV, Vogel V. Beyond Induced-Fit Receptor-Ligand Interactions: Structural Changes that Can Significantly Extend Bond Lifetimes. Structure. 2008 doi: 10.1016/j.str.2008.03.012. (In Press) [DOI] [PubMed] [Google Scholar]
- Pereverzev YV, Prezhdo OV, Thomas WE, Sokurenko EV. Distinctive features of the biological catch bond in the jump-ramp force regime predicted by the two-pathway model. Phys Rev E. 2005b;72 doi: 10.1103/PhysRevE.72.010903. 010903. [DOI] [PubMed] [Google Scholar]
- Pereverzev Y, Prezhdo O. Force-Induced Deformations and Stability of Biological Bonds. Phys Rev E. 2006;73 doi: 10.1103/PhysRevE.73.050902. 050902. [DOI] [PubMed] [Google Scholar]
- Pereverzev Y, Prezhdo OV, Forero M, Sokurenko E, Thomas W. The Two-Pathway Model for the Catch-Slip Transition in Biological Adhesion. Biophys J. 2005a;89:1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Tsang J, et al. Regulation of endothelial cell function by GRGDSP peptide grafted on interpenetrating polymers. J Biomed Mater Res A. 2007 Nov;83(2):423–433. doi: 10.1002/jbm.a.31320. [DOI] [PubMed] [Google Scholar]
- Phan UT, Waldron TT, Springer TA. Remodeling of the lectin-EGF-like domain interface in P- and L-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat. Immunol. 2006;7:883. doi: 10.1038/ni1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakobphol A, Burdsal CA, Fisher SJ. Quantifying the strength of bacterial adhesive interactions with salivary glycoproteins. J Dent Res. 1995 May;74(5):1212–1218. doi: 10.1177/00220345950740051101. [DOI] [PubMed] [Google Scholar]
- Prakobphol A, Tangemann K, Rosen SD, Hoover CI, Leffler H, Fisher SJ. Separate oligosaccharide determinants mediate interactions of the low-molecular-weight salivary mucin with neutrophils and bacteria. Biochemistry. 1999 May 25;38(21):6817–6825. doi: 10.1021/bi990145m. [DOI] [PubMed] [Google Scholar]
- Puklin-Faucher E, Gao M, Schulten K, Vogel V. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J Cell Biol. 2006;175:349–360. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Krogsgaard M, Davis MM, Chakraborty AK. Molecular flexibility can influence the stimulatory ability of receptor-ligand interactions at cell-cell junctions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4416–4421. doi: 10.1073/pnas.0510991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Yago T, Klopocki AG, Lawrence MB, Fieger CB, Rosen SD, McEver RP, Zhu C. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- Sato M, Kawakami K, Osawa T, Toyoshima S. Molecular cloning and expression of cDNA encoding a galactose/N-acetylgalactosamine-specific lectin on mouse tumoricidal macrophages. J Biochem (Tokyo) 1992;111:331–336. doi: 10.1093/oxfordjournals.jbchem.a123758. [DOI] [PubMed] [Google Scholar]
- Savage B, Saldivar E, Ruggeri ZM. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84:289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]
- Schneider SW, Nuschele S, et al. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci U S A. 2007;104(19):7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shive MS, Hasan SM, Anderson JM. Shear stress effects on bacterial adhesion, leukocyte adhesion, and leukocyte oxidative capacity on a polyetherurethane. J Biomed Mater Res. 1999;46:511–519. doi: 10.1002/(sici)1097-4636(19990915)46:4<511::aid-jbm9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Siedlecki CA, Lestini BJ, et al. Shear-dependent changes in the three-dimensional structure of human von Willebrand factor. Blood. 1996;88(8):2939–2950. [PubMed] [Google Scholar]
- Smith MJ, Berg EL, et al. A direct comparison of selectin-mediated transient, adhesive events using high temporal resolution. Biophys J. 1999;77(6):3371–3383. doi: 10.1016/S0006-3495(99)77169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt KA, Struve C, Schembri MA, Hasty DL. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A. 1998 Jul 21;95(15):8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, Johnson JR, Dykhuizen DE. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004 Jul;21(7):1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Tsuji S, et al. Shear-dependent functions of the interaction between soluble von Willebrand factor and platelet glycoprotein Ib in mural thrombus formation on a collagen surface. Int J Hematol. 1999;69(1):48–53. [PubMed] [Google Scholar]
- Tchesnokova V, Aprikian P, Yakovenko O, Larock C, Kidd B, Vogel V, Thomas W, Sokurenko E. Integrin-like allosteric properties of the catch-bond forming FimH adhesin of E. coli. J Biol Chem. 2008 Mar 21;283(12):7823–7833. doi: 10.1074/jbc.M707804200. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V. Shear-dependent 'stick-and-roll' adhesion of type 1 fimbriated Escherichia coli. Mol Microbiol. 2004 Sep;53(5):1545–1557. doi: 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- Thomas W, Forero M, Yakovenko O, Nilsson L, Vicini P, Sokurenko E, Vogel V. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys J. 2006 Feb 1;90(3):753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WE. Catch Bonds in Adhesion. Ann Rev. Biomed Eng. 2008 doi: 10.1146/annurev.bioeng.10.061807.160427. In press. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Forero M, Yakovenko O, Nilsson L, Vicini P, Sokurenko EV, Vogel V. Catch Bond Model Derived from Allostery Explains Force-Activated Bacterial Adhesion. Biophys J. 2006;90:753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- Ulrichts H, Harsfalvi J, Bene L, Matko J, Vermylen J, Ajzenberg N, Baruch D, Deckmyn H, Tornai I. A monoclonal antibody directed against human von Willebrand factor induces type 2B-like alterations. J Thromb Haemost. 2004;2:1622–1628. doi: 10.1111/j.1538-7836.2004.00865.x. [DOI] [PubMed] [Google Scholar]
- Ulrichts H, Udvardy M, Lenting PJ, Pareyn I, Vandeputte N, Vanhoorelbeke K, Deckmyn H. Shielding of the A1 domain by the D'D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J Biol Chem. 2006;281:4699–4707. doi: 10.1074/jbc.M513314200. [DOI] [PubMed] [Google Scholar]
- Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Walsh EJ, O'Brien LM, Liang X, Hook M, Foster TJ. Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J Biol Chem. 2004 Dec 3;279(49):50691–50699. doi: 10.1074/jbc.M408713200. [DOI] [PubMed] [Google Scholar]
- Wellens A, Garofalo C, et al. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS ONE. 2008;3(4):e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf E, Grigorova I, Sagiv A, Grabovsky V, Feigelson SW, Shulman Z, Hartmann T, Sixt M, Cyster JG, Alon R. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nat Immunol. 2007 Oct;8(10):1076–1085. doi: 10.1038/ni1499. [DOI] [PubMed] [Google Scholar]
- Yago T, Wu J, Wey CD, Klopocki AG, Zhu C, McEver RP. Catch bonds govern adhesion through L-selectin at threshold shear. J Cell Biol. 2004 Sep 13;166(6):913–923. doi: 10.1083/jcb.200403144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas W. Fimh forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008 Apr 25;283(17):11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]