Abstract
The loading of oligomeric helicases onto replication origins marks an essential step in replisome assembly. In cells, dedicated AAA+ ATPases regulate loading, however, the mechanism by which these factors help recruit and deposit helicases has remained unclear. To better understand this process, we determined the structure of the ATPase region of the bacterial helicase loader DnaC from Aquifex aeolicus to 2.7 Å resolution. The structure shows that DnaC is a close paralog of the bacterial replication initiator, DnaA, that unexpectedly forms a right-handed helical assembly similar to the quaternary state adopted by ATP-bound DnaA. Complementation and ssDNA-binding assays validate the importance of homomeric DnaC interactions, while pull-down experiments show that the AAA+ domains of DnaC and DnaA interact in a nucleotide-dependent manner. These findings implicate DnaC as a molecular adaptor that uses ATP-activated DnaA as a docking site for regulating the recruitment and correct spatial deposition of the DnaB helicase onto origins.
INTRODUCTION
All cellular organisms use a multi-component molecular machine termed the replisome to couple rapid nucleic acid synthesis with DNA unwinding during replication (Baker and Bell, 1998; Johnson and O’Donnell, 2005). Cells rely on dedicated initiation factors to assist with replisome assembly and coordinate the establishment of competent replication forks with cell cycle timing signals (Kaguni, 2006; Mott and Berger, 2007; Stillman, 2005). Initiation proteins act at select regions of chromosomal DNA, termed replication origins, where they direct the recruitment and deposition of replicative helicases onto DNA strands. Although the specific mechanisms of initiation can vary across the three domains of life, a common theme is a reliance on proteins belonging to the ATPases Associated with various cellular Activities (AAA+) superfamily and ATP turnover for function (Davey et al., 2002b; Duderstadt and Berger, 2008; Lee and Bell, 2000; Neuwald et al., 1999)
In E. coli, the AAA+ proteins DnaC and DnaA help transform a single origin of replication (oriC) into a proper substrate for replisome assembly. Initiation begins with the assembly of the DnaA initiator onto a series of 9-base-pair (bp) sequences known as DnaA-boxes (Fuller et al., 1984; Matsui et al., 1985). Oligomerization of DnaA in the presence of ATP permits the engagement of secondary binding sites (I-sites and ATP-DnaA boxes), generating a large nucleoprotein complex that stimulates the unwinding of a nearby AT-rich DNA unwinding element (DUE) (Bramhill and Kornberg, 1988; Kowalski and Eddy, 1989; Leonard and Grimwade, 2005; Speck et al., 1999). Following melting, ATP-bound DnaC helps deposit two hexamers of the DnaB helicase onto the single-stranded DNA bubble, an event facilitated by a direct interaction between DnaB and DnaA (Kobori and Kornberg, 1982; Marszalek and Kaguni, 1994; Marszalek et al., 1996; Seitz et al., 2000; Wickner and Hurwitz, 1975). The presence of DnaB at the replication fork directs the assembly of other replisomal components, leading to the formation of two fully-equipped replication forks and progression into the elongation stage of DNA replication (Johnson and O’Donnell, 2005).
To date, relatively little is known as to how ATP controls the activity of DnaC and DnaA. The recent crystal structure of DnaA bound to the ATP analog AMPPCP has established that ATP binding causes a subtle conformational rearrangement in the initiator that allows the protein to self-associate (Erzberger et al., 2006). Surprisingly, oligomerized DnaA takes the form of a helical filament, rather than the more common closed-ring state adopted by the majority of AAA+ assemblies (Erzberger and Berger, 2006; Neuwald et al., 1999; Ogura and Wilkinson, 2001). How this complex subsequently remodels DNA and regulates the DnaC-dependent loading of DnaB onto oriC has remained unclear. Interestingly, phylogenetic analyses have suggested that the AAA+ domain of DnaC is most closely related to that of DnaA (Koonin, 1992). It has not been certain, however, to what extent the structural properties of DnaC mirror those of the DnaA, or how the ATPase domain of the loader coordinates its function with those of an ATP-activated initiator complex engaged with oriC.
To begin to address these issues and better understand the significance of the evolutionary and functional relationships between DnaC and DnaA, we determined the structure of the DnaC AAA+ domain from Aquifex aeolicus. The structure establishes that the loader and initiator are close structural paralogs, and identifies active site residues responsible for ATP binding in DnaC that were previously unclear from sequence alignments alone. The structure further reveals that DnaC can adopt a right-handed helical assembly similar to that of DnaA, and that this property is due to a conserved α-helical insertion shared with the initiator. Mutation of active-site residues and amino acids lining the subunit interface disrupt DnaC function in vivo and compromise the ability of the loader to productively associate with ssDNA in vitro, showing that DnaC requires bipartite ATPase interactions and self-assembly for function. Significantly, pull-down experiments show that the structural similarities between DnaC and DnaA extend to an ability of the two proteins to interact directly with each other in a nucleotide-dependent manner. Our findings suggest that DnaC may use an ATP-activated DnaA assembly as a docking site to assist with helicase recruitment, and provide a unifying model to account for the orientation-specific deposition of DnaB onto a melted DUE.
RESULTS
DnaC is a DnaA paralog
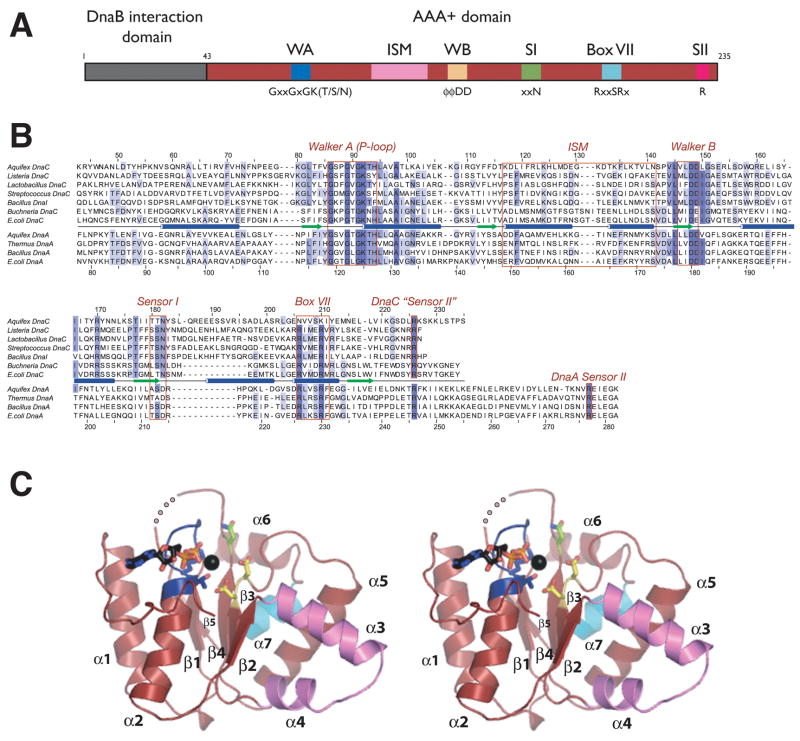
All DnaC orthologs identified to date consist of a small, N-terminal DnaB-binding domain fused to a larger, C-terminal AAA+ ATPase domain (Figure 1A) (Ludlam et al., 2001; Neuwald et al., 1999). Using sequence homology as a guide (Figure 1B), we cloned, expressed, and purified a monodisperse truncation variant of Aquifex aeolicus DnaC encompassing the C-terminal ATP-binding region of the protein (residues 43–235, termed DnaCAAA+). Consistent with biochemical studies on E. coli DnaC and its B. subtilis homolog DnaI (Galletto et al., 2000; Ioannou et al., 2006), fluorescence polarization assays using nucleotides labeled with BODIPY-fluorescein showed that A. aeolicus DnaC AAA+ is competent to bind both ADP and ATP analogs (Sup. Figure 1A). Rod-shaped crystals of selenomethionine-labeled DnaCAAA+ belonging to the space group P61 were grown by vapor diffusion in the presence of ADP·Mg2+ and used for structure determination by multiwavelength anomalous dispersion (MAD) (see Methods and Sup. Figure 2A). The final model, which includes ADP·Mg2+ and all residues except for amino acids 188–190 and the last 14 C-terminal residues, was refined to 2.7 Å resolution with an Rwork/Rfree of 23.3/27.8% and no amino acids in disallowed regions of Ramachandran space (Table 1).
Figure 1. Structure of DnaCAAA+.
(A) Domain representation of DnaC. The N-terminal helicase binding region is colored gray and the central AAA+ domain is shown in red. Numbers refer to amino acid positions. AAA+ motifs are highlighted. WA – Walker-A, WB – Walker-B, SI – sensorI, SII – sensor-II, ISM – Initiator Specific Motif.
(B) Sequence alignment of selected DnaC and DnaA homologs. Alignment was generated by ClustalX (Thompson et al., 1997).
(C) Stereo view of DnaCAAA+. Walker-A and -B motifs are blue and yellow, respectively. The sensor-I residue is green and the Box VII helix cyan. ADP and the coordinating magnesium ion (black) are shown within the ATP binding cleft. An internal disordered region is shown as a dotted line. This and all other molecular figures were generated with PyMOL (pymol.sourceforge.net).
Table 1.
Data collection and refinement statistics
| ADP | ADP·BeF3 | |||
|---|---|---|---|---|
| Data Collection | native | peak | remote | |
| Resolutiona (Å) | 50–2.25 | 50–2.5 | 30–2.5 | 73–2.12 |
| Rsyma (%) | 5.0 (38) | 5.0 (34) | 4.5 (33) | 5.7 (60) |
| I/σ | 8.0 (1.6) | 8.5 (2) | 10.0 (2) | 11.6 (1.8) |
| % Completeness | 99.5(100) | 99.5(100) | 99(98.4) | 96.4 (79.6) |
| Redundancy | 3.6(3.7) | 3.5(3.6) | 3.5(3.1) | 3.9 (2.7) |
| Phasing | ||||
| # of sites | 2 Se | |||
| FOMb | 0.27 | |||
| Refinement | ||||
| Resolution (Å) | 50–2.7 | 50–2.7 | ||
| Rwork/Rfreec % | 23.3/27.8 | 28.4/30.6 | ||
| Number of atoms | ||||
| Protein | 1434 | 1467 | ||
| Waters | 47 | 30 | ||
| Other | 28 | 32 | ||
| RMS deviation | ||||
| bond lengths, Å | 0.008 | 0.007 | ||
| Bond angles, ° | 1.01 | 0.924 | ||
| Ramachandran plot | ||||
| Most favored (%) | 86.2 | 89 | ||
| Additional (%) | 10.7 | 10.4 | ||
| Generous (%) | 3.1 | 0.6 | ||
| Disallowed (%) | 0.0 | 0.0 | ||
numbers in parentheses refer to highest resolution shell
where P(〈) is the probability that the phase angle 〈 is correct
Rwork=Σ||Fobs|−|Fcalc||/Σ|Fobs where Fobs and Fcalc are observed and model structure factors, respectively. Rfree was calculated by using a randomly selected set (5%) of reflections.
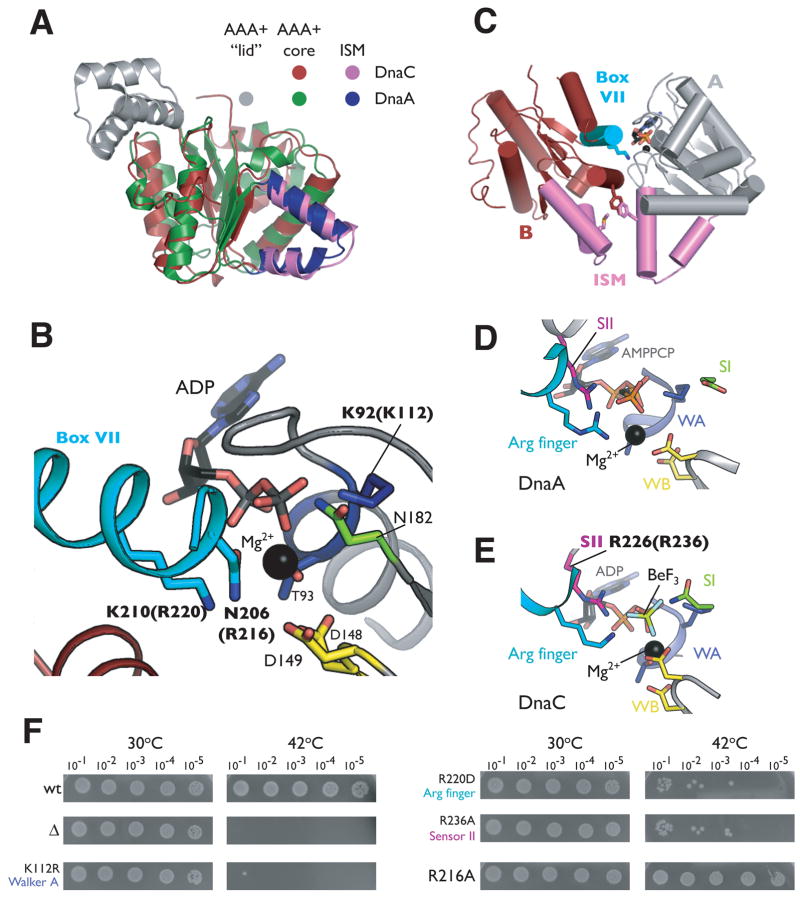
DnaC AAA+ adopts a compact αβα fold typical of the core AAA+ nucleotide-binding domain, in which five parallel β-strands (β1–β5) are sandwiched on both sides by α-helices (α1–α2 on one face and α3–α7 on the other, Figure 1C) (Iyer et al., 2004). Because the sequence identity between DnaC and DnaA orthologs is only 15–20%, a value comparable to the homology between the loader and certain other classes of AAA+ proteins, assignment of the evolutionary lineage of the loader has been not been definitive (e.g., DnaC is generally ~13–18% identical to archaeal Cdc6/Orc1 initiators). In support of a recently proposed classification scheme from Aravind and colleagues that places cellular replication initiators and helicase-loading factors into a distinct subgroup, or “clade”, of the AAA+ superfamily (Iyer et al., 2004), a search of the PDB using the DALI server (Holm and Sander, 1995) confirms that the closest structural paralog of DnaC is DnaA (Z-score 18.9). Cdc6/Orc1 is also a close structural match to DnaC (Z-score 11.0), although some nucleic acid dependent AAA+ proteins such as RuvB and the small subunit of archaeal Replication Factor C return similar Z-scores. Superposition of the AAA+ domains of DnaC and DnaA confirm that despite their sequence divergence, the two folds are highly homologous at the molecular level (1.9Å CαRMSD over 160 Cαpositions) (Figure 2A). The principle difference between the two proteins is that DnaC lacks a small, C-terminal α-helical subdomain (referred to hereafter as the “lid”) found in DnaA and most other AAA+ proteins.
Figure 2. Nucleotide binding by DnaCAAA+.
(A) Superposition of DnaCAAA+ and the AAA+ domain from Aquifex aeolicus DnaA (PDB ID 2HCB). (B) Close-up view of the ADP-DnaCAAA+ active site. Residues from two protomers of DnaCAAA+ are shown in gray and red, respectively. The Box VII helix of one molecule is cyan. Residues selected for mutation are labeled in bold; numbering for equivalent residues in E. coli is given in parentheses. (C) Cartoon representation of two associated ADP-DnaCAAA+ protomers, colored gray and red, respectively. The Box VII helix of monomer B is cyan and Lys210 is shown as sticks. The ISMs of both subunits are violet. Residues lining the ISM that were selected for mutational studies are indicated as sticks (see also Figure 3B). (D) The ATP-DnaA active site. AAA+ motifs of the initiator are colored according to the scheme presented for DnaCAAA+ in Figure 1A. (E) The DnaCAAA+ active site in the presence of the ATP mimetic ADP·BeF3. (F) Mutations in DnaC AAA+ motifs cause growth defects in vivo. Cultures of the E. coli dnaC2 strain transformed with a plasmid carrying the indicated DnaC mutant were serially diluted (1:10) and grown overnight on LB/Kan plates at the indicated temperature. wt= wild-type, Δ= empty vector.
Nucleotide binding properties of DnaC
Phylogenetic analyses had shown previously that DnaC possesses some of the canonical AAA+ motifs involved in nucleotide binding, most notably the Walker-A and -B signature sequence elements (Koonin, 1992). The identification of additional residues important for nucleotide binding and hydrolysis had been hampered, however, by the relatively infrequent occurrence of the helicase loader in the bacterial kingdom, and its significant divergence from other AAA+ proteins. The structure of DnaCAAA+ provides a detailed description as to how nucleotide is liganded within its active site and whether the general structural similarities observed between DnaC and DnaA extend to their nucleotide-binding properties as well.
As expected, the Walker-A motif amino acids of DnaC, located between the β1/α2 junction, create a phosphate-binding loop (P-loop) in which an invariant lysine (Lys92) directly contacts the β-phosphate of ADP (Figure 2B). A conserved threonine (Thr93) in the element further coordinates an associated Mg2+ ion. The Walker-B motif, located at the C-terminal tip of β3, consists of two conserved aspartates (Asp148 and Asp149), both of which are positioned to interact with the water coordination shell of Mg2+. A highly conserved asparagine residue (Asn182), found in the loop immediately following β4, fulfills the role of the sensor-I element, a motif associated with nucleotide hydrolysis and/or the detection of a γ-phosphate in the active site (Guenther et al., 1997).
Interestingly, examination of crystal packing contacts revealed extensive intermolecular contacts between individual DnaCAAA+ protomers (Figure 2C). Oligomerization is stabilized in part through the docking of helix α7 from one molecule into the ADP-occupied active site of an adjacent protomer. Helix α7 thus appears well positioned to serve as the BoxVII or SR(C/F) motif, an element known to play a critical role in the catalytic function of AAA+ proteins, notably due to the presence of an arginine that assists with the liganding and hydrolysis of nucleotide bound by a partner subunit (Neuwald et al., 1999; Ogura and Wilkinson, 2001). Consistent with this theme, a single amino acid, Lys210, projects from one molecule of DnaCAAA+. into the active site of its partner protomer. As Lys210 is an arginine in all bacterial DnaC orthologs except A. aeolicus, it is likely that this residue acts as the equivalent of the arginine finger for DnaC.
Overall, the bipartite nucleotide-binding pocket of DnaCAAA+ is highly organized, with its signature residues coordinating nucleotide in a manner similar to that seen in other functional AAA+ assemblies (Erzberger and Berger, 2006; Ogura and Wilkinson, 2001). The active site configuration of DnaCAAA+ is particularly similar to that of DnaA, whereby the arginine finger of each protein (Lys210 for DnaC, Arg230 for DnaA) occupy analogous positions with respect to bound nucleotide, despite the presence of ADP in DnaCAAA+ and an ATP analog associated with DnaA (Figure 2B and D). Engagement of the AMPPCP γ-phosphate by the arginine finger in DnaA previously had been observed to require a conformational change between the core AAA+ element and its lid subdomain (Erzberger et al., 2006). The absence of the C-terminal lid in DnaC eliminates this steric constraint, and may have allowed us to capture inter-protomer associations in the presence of ADP due to the high protein concentrations used during crystallization.
DnaC associates with ATP-mimetics
The comparable arrangement of the bipartite ADP-DnaCAAA+ active site with that of ATP-DnaA suggested that the loader might accommodate ATP as well. To determine what changes, if any, would occur upon ATP binding, we first tried to co-crystallize DnaCAAA+ with ATP or other nucleotide analogs such as AMPPNP. These attempts routinely gave rise to small and poorly diffracting crystals, and further induced the protein to rapidly precipitate out of solution. To overcome these solubility issues, we instead soaked crystals of ADP-DnaCAAA+ overnight in a solution of 2mM BeF3, a widely used substitute for the γ-phosphate of ATP (Petsko, 2000). Crystals treated in this manner diffracted similarly to those obtained for ADP-DnaCAAA+, and permitted determination of the structure by molecular replacement to 2.7Å resolution (Table 1). Examination of difference density maps confirmed the presence of BeF3 liganded to ADP·Mg2+ in the active site (Sup. Figure 2B).
Overall, the spatial positioning of residues previously identified to be important for nucleotide binding in ADP-DnaCAAA+ changes very little in the ADP·BeF3-DnaCAAA+ structure (hereafter referred to as ATP-DnaCAAA+) (Figure 2B and E). As anticipated, the conserved lysine from the BoxVII motif now makes direct contact to the BeF3moiety. Unexpectedly, however, extra density for five additional residues at the extreme C-terminus of DnaCAAA+ also became evident in difference density maps from BeF3-soaked crystals. Surprisingly, this structural rearrangement results in the formation of a short loop immediately following strand β5 of the AAA+ core, and the extension of a single arginine (Arg226) into the active site where it engages BeF3. Sequence alignments indicate that this arginine is invariant among DnaC orthologs (Figure 1B). This degree of conservation, together with the spatial placement of the residue within the active site of DnaCAAA+, suggests that Arg226 may serve as the equivalent of the sensor-II arginine, an amino acid that generally resides in the lid subdomain of AAA+ proteins and is important for catalytic function (Guenther et al., 1997; Ogura et al., 2004). Although it cannot be ruled out that ATP may invoke still other conformations than that imaged here, the properly organized bipartite active site around the γ-phosphate mimetic shows that the helical oligomeric form of DnaC observed in the crystal is nevertheless compatible with binding to an ATP analog.
DnaC active site residues are required for function in vivo
Kaguni and coworkers have identified a number of mutations falling in the N-terminal DnaB-binding region of DnaC that impair activity in vivo (Ludlam et al., 2001). The only specific AAA+ mutation of DnaC biochemically characterized to date, however, is that of a lysine to arginine alteration in the Walker-A motif of the E. coli helicase loader (Davey et al., 2002a). Specifically, the K112R mutant fails to bind nucleotide in vitro, and is defective for helicase loading in oriC-dependent replication assays. The relative importance of other nucleotide-binding residues to DnaC function has remained unestablished.
To determine whether residues such as the Box VII and sensor-II arginines identified here are necessary for DnaC activity, we employed genetic complementation assays using an E. coli strain carrying a thermosensitive dnaC allele (dnaC2) (Carl, 1970). This mutant blocks the initiation of DNA replication at 42°C and has been used extensively for synchronizing bacterial cell cultures (Withers and Bernander, 1998). Expression of a wild-type copy of the dnaC+ gene from a plasmid under conditions that allow leaky expression from an arabinose-inducible promoter permits robust cell growth in the dnaC2 strain at both 30° and 42°C (Figure 2F). By contrast, transformation of an empty vector is unable to support growth at the non-permissive temperature. A control assay employing a dnaC mutant in which the conserved catalytic lysine residue of the Walker-A motif is substituted for an arginine also fails to grow at 42°C, consistent with its deficient biochemical properties in vitro (Davey et al., 2002a).
As shown in Figure 2F, an arginine to aspartate mutation for the E. coli equivalent of the Box VII arginine finger (R220D, see Sup. Table 1) is greatly reduced in its ability to complement the growth of the temperature sensitive strain at 42°C. Similarly, an arginine to alanine substitution for the sensor-II residue (R236A) also is compromised severely for viability at the non-permissive temperature. Both results confirm that the prospective catalytic AAA+ motifs identified in our structure are important for DnaC function. In contrast, mutation of a second arginine that resides near the Box VII motif, R216A (Asn206 in A. aeolicus), had no affect on growth at 42°C as compared to the wild-type control. Although highly conserved, this residue does not make any noticeable contacts to the nucleotide-binding pocket of either ADP- or ATP-DnaCAAA+. Interestingly, DnaA contains an arginine at the same position as Arg216 of DnaC that is conserved between orthologs of the initiator. Studies have shown that mutation of this amino acid does not affect the ability of DnaA to oligomerize and unwind the DUE in vitro, but does interfere with the DnaC-dependent loading of DnaB (Felczak and Kaguni, 2004).
DnaC oligomerization
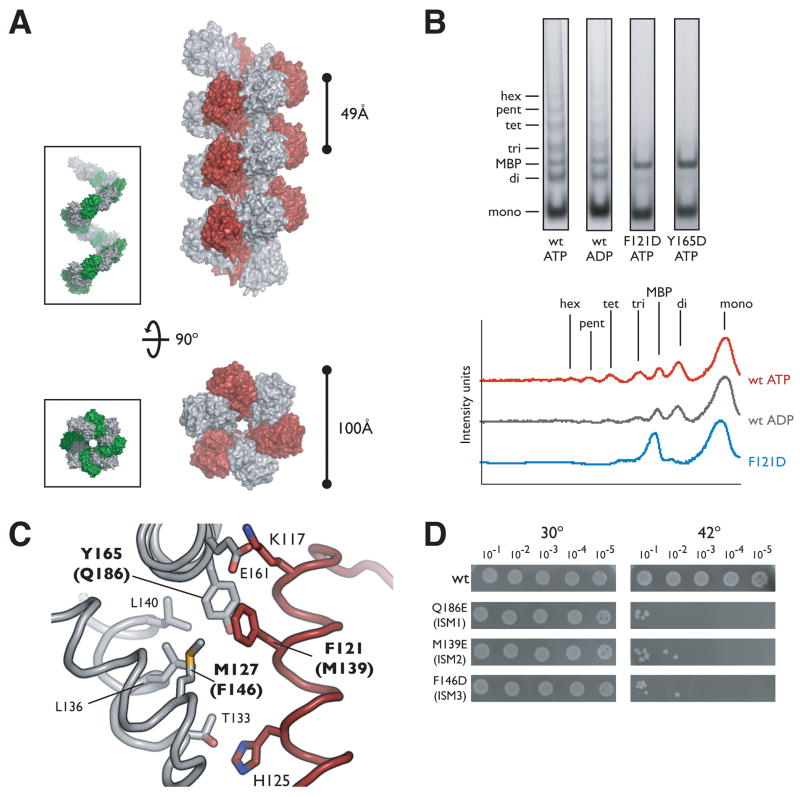
One of the hallmarks underlying the phylogenetic grouping of cellular helicase loaders such as DnaC with replication initiator proteins in the AAA+ superfamily is the presence of an extra α-helix inserted into one edge of the AAA+ fold (Iyer et al., 2004). Our structure of DnaCAAA+ confirms the existence of this distinguishing helix, hereafter referred to as an ISM (Initiator/loader specific motif) (Dueber et al., 2007), and shows that this element, together with the α-helix that follows, forms a distinctive V-shaped insertion that connects strands β2 and β3. The spatial positioning of this region in DnaCAAA+ is highly similar to that seen in the DnaA protomer (Figure 2A). In ATP-oligomerized DnaA, the protrusion of these two helices into the subunit interface causes a relative lateral displacement of adjoining initiator subunits, promoting the formation of a helical filament. In DnaCAAA+, the ISM performs a similar function, forcing associated protomers into a right-handed spiral assembly that winds around the crystallographic 61axis (Figure 3A). Thus, DnaC is capable not only of higher-order assembly, but also adopts a structural state which parallels that observed for the ATP-bound form of DnaA.
Figure 3.
DnaCAAA+ oligomerization
(A) DnaCAAA+ forms a right-handed filament. Side and axial views of 21 symmetry-related DnaCAAA+ monomers are shown in surface representation with alternating subunits colored gray and red. The inset depicts similar views for the AAA+ domains of the ATP-DnaA filament (PDB ID H2HCB) with alternating monomers in gray and green. (B) DnaCAAA+ forms multimers in solution. Crosslinking was performed by the addition of 1.25mM glutaraldehyde to wild-type (wt) DnaC, or either ISM mutant (F121D and Y165D) in the presence of the indicated nucleotide. The graph on the right compares the intensity traces for wt ATP, wt ADP, and the ISM mutant F121D. mono – monomer, di –dimer, tri – trimer, tet – tetramer, pent – pentamer, hex – hexamer, MBP – maltose binding protein. (C) Close-up view of the interface created by the packing of ISMs from adjacent DnaC AAA+ protomers in the filament. Residues selected for mutation are indicated in bold, the numbering for equivalent residues in E. coli is given in parentheses. (D) Mutation of residues involved in intermolecular DnaCAAA+ contacts result in significant growth defects in vivo. Experiments were performed similarly to those described for Figure 2F.
To determine if DnaCAAA+ could similarly oligomerize in the absence of crystallizing agents, we performed crosslinking assays using glutaraldehyde. DnaCAAA+ was incubated in the presence of ATP prior to addition of the crosslinking reagent, following which, reactions were stopped and analyzed by SDS PAGE. As shown in Figure 3B, DnaCAAA+ forms a ladder of oligomeric species under these conditions that run at the expected sizes for dimers, trimers, and higher-order multimers. Maltose binding protein (MBP) was included in the reaction mixture to ensure that the crosslinking observed was specific to DnaCAAA+. When the reactions included only ADP, oligomers were again observed, although the bands corresponding to particular multimers were markedly less intense or not present as when ATP (or ADP-BeF3) was present (Figure 3B and data not shown). This result is consistent with our finding that DnaC crystallized as an oligomer in the presence of ADP, but suggests that ATP further facilitates the formation of this state, likely due to the presence of extra contacts within the active site.
To test specifically whether the oligomeric interactions observed in the crystal were responsible for the self-association of DnaCAAA+, we next mutated hydrophobic residues emanating from the ISM that would be otherwise solvent exposed in the context of the monomer. The interface surrounding the ISM constitutes approximately 35% of the total buried surface area per protomer (1,197 Å2), and is composed of a complimentary combination of hydrophobic and polar residues. The most notable set of interactions comes from the docking of Phe121 from one molecule into a compact hydrophobic pocket created by residues Met127, Leu136, Leu140, and Tyr165 on the other (Figure 3C). We mutated both Phe121 and Tyr165 to aspartate residues and tested for their ability to promote oligomerization in the crosslinking assays. As shown in Figure 3B, neither mutation supports oligomerization, consistent with the idea that gluteraldehyde is capturing quaternary interactions similar to those seen the crystal, and that these amino acids play an important role in assembly.
Based on in vitro studies, DnaC is believed to form a 6:6 complex with DnaB (Kobori and Kornberg, 1982). The concentration of DnaC appears to be important in vivo as well, since an excess of the loader can inhibit replication (Allen and Kornberg, 1991). Given these observations, and the finding that the DnaB-binding domain of B. subtilis DnaI inhibits ATPase activity (Ioannou et al., 2006), is seemed likely that we captured a filament of DnaC because the N-terminal region (and the helicase) was absent during crystallization. We therefore set out to test whether residues that contribute to the filament interface are important for the function of the loader in the cell. Using full-length E. coli DnaC, we changed several hydrophobic residues along the ISM to an acidic amino acid (F146D, M139E, Q186E), and examined the mutants for their ability to complement the temperature sensitive dnaC2 strain. The results show that each substitution severely compromises growth at the non-permissive temperature as compared to the wild-type control (Figure 3D), indicating that residues involved in forming the DnaCAAA+ oligomer are also required for function in vivo.
DnaC and ssDNA binding
Several studies have demonstrated that DnaC binds cooperatively to single-stranded DNA (ssDNA) in the presence of ATP, and that the AAA+ domain of DnaC alone is sufficient for this activity (Biswas et al., 2004; Davey et al., 2002a; Ioannou et al., 2006; Learn et al., 1997). The mechanism by which the helicase loader engages nucleic acid substrates has remained unknown, as has the molecular means by which ATP specifically stimulates ssDNA binding. Interestingly, in addition to binding duplex DNA sites at oriC, DnaA also associates with ssDNA in an ATP-dependent manner (Ozaki et al., 2008; Speck and Messer, 2001; Weigel and Seitz, 2002). Recent studies have suggested that residues located along the central axis of the DnaA oligomer are critical for this function (Ozaki et al., 2008).
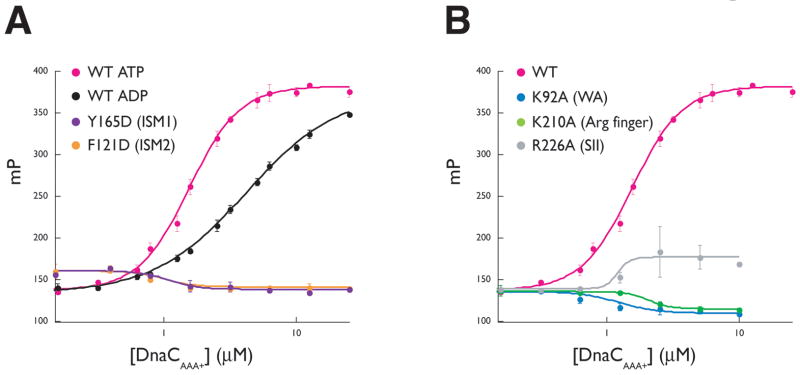
The similar higher-order structures we observed for oligomerized DnaC and DnaA suggested to us that the nucleotide-dependency of the DnaC:ssDNA interaction might be related to the ability of the protein to self-assemble. To test this idea, we used mutagenesis and fluorescence polarization assays to determine if residues that contribute to the formation of the DnaCAAA+ interface also were required for ssDNA binding. We first incubated wild-type A. aeolicus DnaCAAA+ with a fluorescein-labeled 25-mer dT oligonucleotide and monitored the change in fluorescence polarization of this substrate as a function of increasing DnaCAAA+ concentration. As shown in Figure 4A, ATP stimulates wild-type DnaCAAA+ to bind ssDNA compared to ADP, with an apparent affinity and cooperativity of association similar to that observed previously for full-length E. coli DnaC (Sup. Table 2) (Biswas et al., 2004). By contrast, mutations to the oligomeric interface of DnaCAAA+ (F121D and Y165D) abolish ssDNA binding (Figure 4A). Notably, these substitutions do not affect the ability of DnaCAAA+ to associate with ATP (Sup. Figure 1B). This observation indicates that at least some of the oligomeric contacts seen in the crystal structure are necessary to support the ATP-stimulated binding of single-stranded nucleic acid substrates.
Figure 4. Single-stranded DNA binding by DnaCAAA+.
(A) The effect of nucleotide and interface mutants on DnaC-ssDNA interactions. Binding reactions contained 10nM fluorescein-labeled dT25 oligonucleotide titrated against either wild-type DnaCAAA+ or either interface mutant. All assays were performed in triplicate the presence of 2mM ATP except where indicated. Error bars represent the standard deviation between measurements. (B) AAA+ motif mutations disrupt ssDNA binding by DnaCAAA+. Experiments were performed similarly as for (A) in the presence of 2mM ATP. Data points for wild-type DnaCAAA+ are the same as those indicated in (A), and shown again for comparison.
Our ssDNA-binding model for DnaC also predicts that the mutation of residues involved in the coordination of ATP between individual monomers may negatively affect DNA affinity, potentially by destabilizing oligomeric DnaC interactions. To test this assumption, we made alanine mutations of the A. aeolicus DnaCAAA+ arginine finger and sensor II residues (K210A and R226A). Both mutations were defective for ssDNA binding, with the K210A mutation displaying a more severe defect of the two, even though neither mutation affected the ability of DnaCAAA+ to bind ATP (Figure 4B and Sup. Figure 1C). The Walker A K92R mutant also showed no measurable binding, either for DNA or ATP consistent with the reported inability of this substitution in E. coli DnaC to bind nucleotide (Davey et al., 2002a), and our finding that this mutation is unable to support cellular growth. These results are consistent with both a role for ATP and its coordinating residues in stabilizing inter-DnaC contacts, as well as a need for DnaC self-assembly, in creating a properly configured oligomer competent to bind nucleic acid.
DnaA and DnaC interact in a nucleotide-dependent manner
In our analysis of DnaC, we were struck by its degree of similarity to DnaA, particularly with regard to the respective active site configurations and oligomeric states of the two proteins (Figures 2 and 3). In particular, the ability of both DnaC and DnaA to form right-handed spiral oligomers, as opposed to closed rings, led us to speculate that the initiator and helicase loader might engage each other directly. A corollary of this assumption was that nucleotide might control these heteromeric interactions as it does DnaC and DnaA self-assembly.
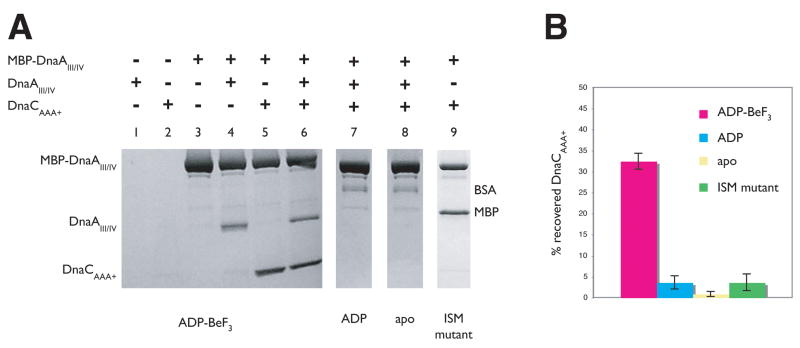
To test this model, we performed amylose resin pull-down assays using A. aeolicus DnaA tagged with MBP as bait and untagged A. aeolicus DnaCAAA+ as prey. Only the AAA+ and DNA-binding domains of DnaA (domains III and IV) were used in these assays, both because prior studies had shown this construct to be well behaved, and to rule out any possibility that DnaC might inadvertently associate with an unsatisfied contact surface on the N-terminal, DnaB-binding region of the initiator (domains I and II). The isolated ATPase domain of DnaC was employed for similar considerations. As a positive control, we first examined ability of tagged MBP-DnaAIII–IV to capture untagged DnaAIII–IV. Assays were conducted in the presence of ADP-BeF3 to generate permanently “activated” initiator and helicase loader states, and to simplify nucleotide addition experiments.
As expected due to its known self-association properties, untagged DnaAIII–IV associated with MBP-DnaAIII–IV, but not with free amylose resin (Figure 5A, lanes 1 and 4). Significantly, when the experiment was repeated using DnaCAAA+ as the prey, we found that a fraction of the protein associated with the resin only when MBP-DnaAIII–IV was present (Figure 5A, lanes 2 and 5, and Figure 5B). This interaction occurred regardless of whether untagged DnaAIII–IV was included with DnaCAAA+ in the binding reaction (Figure 5A, lanes 5–6), or if reciprocal pull-downs were preformed using MBP-tagged DnaCAAA+ and free DnaAIII–IV (not shown). Moreover, incubating MBP-DnaAIII–IV with the archaeal Orc1-1 initiator from S. solfataricus under identical reaction conditions failed to show any stable association between the two, indicating that the interaction between DnaA and DnaC does not result from the non-specific association of unsatisfied AAA+ domain surfaces (Sup. Figure 3). This finding demonstrates that the helicase loader can indeed interact with MBP-DnaAIII–IV.
Figure 5. The AAA+ domains of DnaC and DnaA interact in a nucleotide-dependent manner.
(A) Co-precipitation of DnaCAAA+ by domains III and IV of Aquifex aeolicus DnaA fused to maltose binding protein (MBP-DnaAIII–IV). The positions for MBP-DnaAIII–IV, untagged DnaAIII–IV, and DnaCAAA+ on the SDS PAGE gel are indicated. Apo - no nucleotide was added to reaction conditions; “ISM mutant” - double mutant F121D/Y165D. Free MBP present in lane 9 resulted from incomplete separation of the protein from the DnaCAAA+ ISM mutant during purification (see Sup. Material). (B) Quantitation of the results presented in (A). Experiments for each condition were repeated three times and averaged. Error bars represent the variance between experiments.
We next tested whether the interaction between DnaCAAA+ and MBP-DnaAIII–IV was dependent on the identity of the nucleotide present in the binding reaction. When incubated without nucleotide, or with ADP alone, untagged DnaA III–IV failed to associate with MBP-tagged DnaA III–IV (Figure 5A, lanes 7–8), consistent with prior studies suggesting that homomeric DnaA interactions are dependent on ATP or ATP mimetics (Speck and Messer, 2001; Speck et al., 1999). Strikingly, when DnaCAAA+ was present in the same mixture, it also failed to associate with either ADP-bound or apo MBP-DnaA III–IV (Figure 5A, lanes 7–8, and Figure 5B). Together, these findings demonstrate that the ATPase domain of DnaC is not only capable of binding to the ATPase and DNA binding regions of DnaA, but that this interaction is dependent upon the two proteins being in an ATP-bound state.
As a final test, we examined the ability of a mutant form of the DnaC AAA+ domain to bind MBP-DnaA III–IV. Incubation of a double ISM mutant of DnaCAAA+ (F121D, Y165D) with MBP-DnaA III–IV failed to reveal a stable interaction between the two, even though ADP·BeF3 was present, and the double mutant is competent to bind nucleotide (Figure 5A, lane 9, Figure 5B and Sup. Figure 1B). This result indicates that the association between DnaC and DnaA is likely mediated by conserved AAA+ interactions that are promoted by ATP.
DISCUSSION
DnaC is a DnaA paralog capable of forming helical oligomers
We have determined the structure of the ATPase region of the bacterial helicase loader, DnaC, in two different nucleotide states. The structures confirm that the AAA+ domain of DnaC is a close structural paralog of the bacterial initiator DnaA, supporting prior suggestions that these proteins arose from a common ancestral replication initiation factor (Iyer et al., 2004; Koonin, 1992). The structure also identifies several unanticipated, but conserved, amino acids as being the counterparts of well-established AAA+ motifs involved in ATP coordination. Biochemical and complementation assays confirm that these residues are important for binding ssDNA in vitro and for general DnaC function in vivo (Figures 2F and 4B). Unexpectedly, the structural congruence between DnaC and DnaA extends to the ability of the helicase loader to use an α-helical element (the ISM) for directing the formation of a right-handed helical oligomer, a structural state observed for ATP-bound DnaA (Erzberger et al., 2006) (Figure 3A). The resemblance is sufficiently close that the bipartite ATPase active site of DnaC is a near perfect duplicate of that seen in DnaA (Figures 2B, D and E).
The finding that DnaC can form right-handed spiral oligomers is surprising and unusual. Though highly diverse in function, AAA+ proteins commonly assemble into closed-ring hexamers or heptamers (Erzberger and Berger, 2006; Neuwald et al., 1999; Ogura and Wilkinson, 2001), or in the case of processivity clamp loader complexes, notched-ring pentameric particles (Bowman et al., 2004; Jeruzalmi et al., 2001). While AAA+ proteins occasionally have been observed to form helical filaments crystallographically (Guo et al., 2002), the coordination of nucleotide in these structures is invariably incomplete, typically due to the displacement of the Box VII arginine-finger from the catalytic center. By contrast, the active site seen in our DnaCAAA+ crystals appears properly formed, with nucleotide liganded by the appropriate retinue of conserved amino acids required for function. While the association of DnaC with DnaB probably precludes formation of a true filament in vivo, the spiral configuration observed here likely reflects a relevant mode of assembly accessed by the loader during its catalytic cycle.
Bacterial initiators and loaders use their AAA+ domains to facilitate crosstalk
When the structure of an ATP-bound form of bacterial DnaA was first determined, one explanation for the utility of an open-ring structure, as opposed to a closed-ring state, was that this configuration might allow the nucleotide-binding site of a terminal initiator subunit to interact with auxiliary replication factors. In this regard, the AAA+ module of DnaA was predicted to act not just as an ATP-regulated assembly platform, but also as a possible protein-protein docking station. Due to their similarity to DnaA, both DnaC and Hda, an AAA+ protein that stimulates the ATPase activity of DnaA (Su’etsugu et al., 2005), have been proposed as candidate binding partners whose AAA+ domains might engage the open ends of an assembled initiator complex (Erzberger et al., 2006; Mott and Berger, 2007).
Our discovery that the DnaC AAA+ domain can adopt an oligomerization state similar to that of DnaA is consistent with this hypothesis, and prompted us to test whether the two proteins could interact directly. Using pull-down assays, we found that this association can indeed occur: the isolated ATPase domain of DnaC can bind to a DnaA construct lacking its N-terminal DnaB-binding domain, but retaining its ATPase and duplex DNA binding regions (Figure 5). Moreover, this interaction is specific for an ATP-like state, and relies on the integrity of the DnaC AAA+ oligomerization interface, as pull-downs performed in either ADP or nucleotide-free conditions, or with a DnaC ISM mutant, fail to show any stable DnaCAAA+ association. These findings suggest that DnaC serves in functions beyond the loading of the DnaB helicase onto ssDNA. In particular, our data support an unanticipated role for DnaC as a molecular adaptor that can specifically recognize an ATP-activated DnaA initiator, most likely through direct AAA+/AAA+ interactions (Figure 6A).
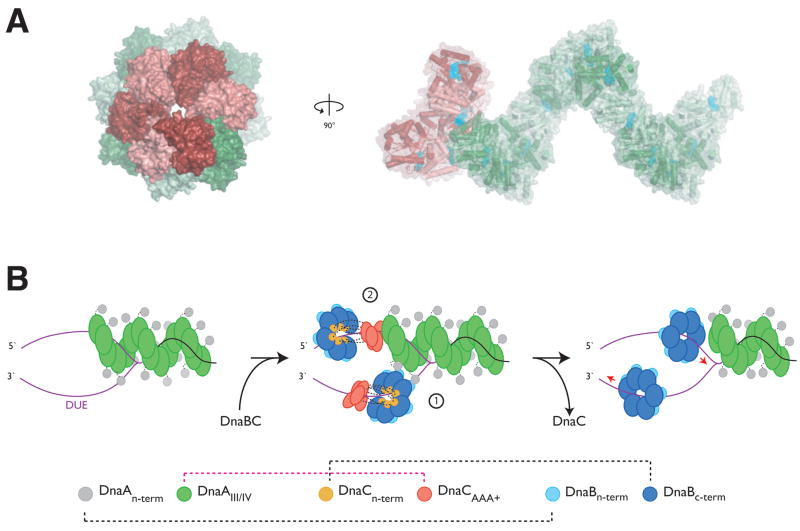
Figure 6. Model for DnaC/DnaA crosstalk and helicase deposition.
(A) Structural model for oligomeric DnaC:DnaA interactions. The figure was generated by superimposing the last subunit of a six-subunit DnaCAAA+ oligomer onto the end of a twelve-subunit DnaA filament assembly. Axial and side views are shown. Cyan spheres represent bound nucleotide. (B) Model for the symmetric loading of two replicative helicases at oriC. Left – DnaA assembles at oriC and melts the DUE (purple strands). Middle, (1) – helicase loading on the bottom DUE strand is facilitated through direct DnaA:DnaB interaction. Middle, (2) - DnaC, through a specific interaction with ATP-charged DnaA, recruits the helicase destined for the top strand to oriC. Right – ATP hydrolysis and loss of DnaC frees both DnaB hexamers to migrate to their proper fork positions.
Implications for the construction of a bidirectional replication fork
Why might DnaC and DnaA interact? During helicase loading in E. coli, two DnaB hexamers must ultimately positioned in opposite directions on the two strands of the unwound DUE to create a bidirectional replication fork. Biochemical and electron microscopy studies have indicated that the DnaA nucleoprotein complex forms off to one side of the DUE (Funnell et al., 1987). This organization raises a paradox as to how an assymetric initiation assembly facilitates the symmetric loading of two helicases onto this region. We propose that the ability of DnaC to directly engage an available end of the DnaA oligomer provides a missing piece toward solving this long-standing puzzle.
Following binding to oriC, DnaA promotes the melting of three AT-rich regions (L, M, and R) in the DUE in preparation for the loading of DnaB (Bramhill and Kornberg, 1988). During this process, DnaA is thought to associate predominantly with the “upper” strand of the melted origin region (Figure 6B, left) (Speck and Messer, 2001). It is known that different faces of DnaB are recognized by the initiator and helicase loader: the N-terminal domain of DnaA binds to the N-terminal side of the helicase, while the N-terminal domain of DnaC binds to the C-terminal face (Barcena et al., 2001; Galletto et al., 2000; Ludlam et al., 2001; Saluja and Godson, 1995; Seitz et al., 2000; Sutton et al., 1998). On artificial forked DNA substrates that mimic a melted oriC, DnaA can recruit a single DnaBC complex to the lower strand of the unwound origin, suggesting that direct initiator:helicase interactions may be responsible for facilitating the deposition of this complex (Figure 6B, middle, stage 1) (Weigel and Seitz, 2002). How the second helicase is loaded has remained unclear: since DnaA is offset to the right side of the DUE, an identical interaction between the initiator and DnaB would result in a mismatch between the orientation of the helicase and the polarity of the upper DUE strand. A direct contact between DnaC and an ATP-activated DnaA oligomer resolves this problem, allowing the second helicase to be placed with the proper geometry on this DNA segment through its interaction with the helicase loader (Figure 6B, middle, stage 2).
An interesting prediction arising from this model is that the helicase destined to lead the left replication fork should be loaded initially on the right side of the lower DUE strand, while the helicase that will generate the right replication fork would be deposited on the left side of the upper strand. Remarkably, O’Donnell and co-workers have shown that this precise asymmetric loading arrangement is formed during initiation, with the two helicases moving 5′ to 3′ past each other following deposition to establish the final geometry of the bidirectional replication fork (Figure 6B, right) (Fang et al., 1999). Our model provides a physical explanation for this initial loading configuration, suggesting that the left-fork helicase is first localized close to the initiator complex due to its direct interaction with DnaA, whereas the right-fork helicase is placed at a distance from the initiator through a DnaC coupler. How the DnaB ring is cracked to permit entry of ssDNA is not known, although the formation of a spiral DnaC oligomer structure on the surface of the helicase may provide a means for facilitating this event. Future studies will help test various predictions of this model and establish the functional significance of the DnaC:DnaA interaction further.
Conclusion
In summary, our analyses of DnaC have revealed a series of structural and biochemical properties that closely mirror those of the bacterial initiator, DnaA. These characteristics include a propensity to form right-handed helices when bound to appropriate nucleotide substrates and an ability to form hetero-oligomeric interactions when in an ATP-like state. The similarities between the two proteins are particularly interesting in that only a subset of bacteria actually contain a recognizable DnaC ortholog; in such species, it is tempting to speculate that DnaA may fulfill the role of both initiator and helicase loader, a unification of labor that may be paralleled in some instances by the archaeal AAA+ initiator, Cdc6/Orc1, in the recruitment and loading of the MCM helicase (Shin et al., 2008). Along the same lines, it is noteworthy that the eukaryotic Cdc6 protein acts in concert with the origin recognition complex (ORC) to help load the MCM2–7 helicase onto replication origins (Bell and Dutta, 2002; Donovan et al., 1997; Mendez and Stillman, 2003). In yeast, it has been shown the binding of ATP by the Orc1 subunit of ORC specifically recruits Cdc6 (Klemm and Bell, 2001; Randell et al., 2006; Speck et al., 2005); both proteins are also AAA+ ATPases (Neuwald et al., 1999; Speck et al., 2005). Thus, the coordination between DnaC and DnaA AAA+ domains may reflect part of a universal ATP-dependent strategy to help ensure that replicative helicases, which ultimately serve as the leading edge of the replication fork, are properly recruited and deposited at activated origins.
METHODS
Protein expression and purification/Crystallization/Data Collection and Structure Determination
Detailed procedures are provided in the Supplementary Material.
Genetic assays
Complementation assays were carried out in the host strain MG1655 dnaC2 thr+ using pINCnat (Allen and Kornberg, 1991), a plasmid containing the wild-type E. coli dnaC+ gene under arabinose promoter control. Following growth to mid-log in LB/Kan media at 30°C, cells were serially diluted and plated at 30°C and 42°C.
ssDNA and nucleotide binding assays
The binding of 5′ fluorescein-tagged dT25 oligonucleotide (Operon Technologies, Inc.) was monitored by fluorescence polarization using a Victor3 V (Perkin Elmer) multi-label plate reader. Samples were mixed with DNA in binding buffer for 10 minutes prior to readout.
Pull-downs
Co-precipitation studies were performed by incubation of amylose beads (New England Biolabs) with bait and prey proteins in binding buffer. Following washing, the samples were analyzed by SDS-PAGE. Additional details are provided in Supplementary Experimental Procedures.
Supplementary Material
Acknowledgments
We thank the Advanced Light Source beamline 8.3.1 staff for support and N. Crisona and K. Jude for providing critical reagents. N. Crisona, J. Corn, E. Dueber, and members of the Berger laboratory provided helpful comments and suggestions. This work was supported by a National Science Foundation Pre-doctoral Fellowship (to M.L.M), and by NIGMS (GM071747).
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 3EC2 and 3ECC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen GC, Jr, Kornberg A. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J Biol Chem. 1991;266:22096–22101. [PubMed] [Google Scholar]
- Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- Barcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher M, Carazo JM. The DnaB.DnaC complex: a structure based on dimers assembled around an occluded channel. Embo J. 2001;20:1462–1468. doi: 10.1093/emboj/20.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Biswas SB, Flowers S, Biswas-Fiss EE. Quantitative analysis of nucleotide modulation of DNA binding by DnaC protein of Escherichia coli. Biochem J. 2004;379:553–562. doi: 10.1042/BJ20031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Carl PL. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109:107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- Davey MJ, Fang L, McInerney P, Georgescu RE, O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. Embo J. 2002a;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol. 2002b;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit Rev Biochem Mol Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- Fang L, Davey MJ, O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- Felczak MM, Kaguni JM. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J Biol Chem. 2004;279:51156–51162. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- Galletto R, Rajendran S, Bujalowski W. Interactions of nucleotide cofactors with the Escherichia coli replication factor DnaC protein. Biochemistry. 2000;39:12959–12969. doi: 10.1021/bi0012484. [DOI] [PubMed] [Google Scholar]
- Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J. Crystal structure of the delta’ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Ioannou C, Schaeffer PM, Dixon NE, Soultanas P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006;34:5247–5258. doi: 10.1093/nar/gkl690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- Klemm RD, Bell SP. ATP bound to the origin recognition complex is important for preRC formation. Proc Natl Acad Sci U S A. 2001;98:8361–8367. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB-dnaC protein complex. J Biol Chem. 1982;257:13770–13775. [PubMed] [Google Scholar]
- Koonin EV. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 1992;20:1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. Embo J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learn BA, Um SJ, Huang L, McMacken R. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc Natl Acad Sci U S A. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Bell SP. ATPase switches controlling DNA replication initiation. Curr Opin Cell Biol. 2000;12:280–285. doi: 10.1016/s0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol Microbiol. 2005;55:978–985. doi: 10.1111/j.1365-2958.2004.04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlam AV, McNatt MW, Carr KM, Kaguni JM. Essential amino acids of Escherichia coli DnaC protein in an N-terminal domain interact with DnaB helicase. J Biol Chem. 2001;276:27345–27353. doi: 10.1074/jbc.M101940200. [DOI] [PubMed] [Google Scholar]
- Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- Marszalek J, Zhang W, Hupp TR, Margulies C, Carr KM, Cherry S, Kaguni JM. Domains of DnaA protein involved in interaction with DnaB protein, and in unwinding the Escherichia coli chromosomal origin. J Biol Chem. 1996;271:18535–18542. doi: 10.1074/jbc.271.31.18535. [DOI] [PubMed] [Google Scholar]
- Matsui M, Oka A, Takanami M, Yasuda S, Hirota Y. Sites of dnaA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1985;184:529–533. doi: 10.1016/0022-2836(85)90299-2. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- Petsko GA. Chemistry and biology. Proc Natl Acad Sci U S A. 2000;97:538–540. doi: 10.1073/pnas.97.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Saluja D, Godson GN. Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43, and dnaB454. J Bacteriol. 1995;177:1104–1111. doi: 10.1128/jb.177.4.1104-1111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Weigel C, Messer W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol Microbiol. 2000;37:1270–1279. doi: 10.1046/j.1365-2958.2000.02096.x. [DOI] [PubMed] [Google Scholar]
- Shin JH, Heo GY, Kelman Z. The Methanothermobacter thermautotrophicus Cdc6-2 protein, the putative helicase loader, dissociates the minichromosome maintenance helicase. J Bacteriol. 2008;190:4091–4094. doi: 10.1128/JB.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. Embo J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. Embo J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Su’etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J Biol Chem. 2005;280:6528–6536. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- Sutton MD, Carr KM, Vicente M, Kaguni JM. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J Biol Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- Weigel C, Seitz H. Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol Microbiol. 2002;46:1149–1156. doi: 10.1046/j.1365-2958.2002.03232.x. [DOI] [PubMed] [Google Scholar]
- Wickner S, Hurwitz J. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc Natl Acad Sci U S A. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers HL, Bernander R. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J Bacteriol. 1998;180:1624–1631. doi: 10.1128/jb.180.7.1624-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.