Abstract
Background
Prostate cancer is thought to arise as a result of oxidative stresses and induction of antioxidant electrophile defense (phase 2) enzymes has been proposed as a prostate cancer prevention strategy. The isothiocyanate sulforaphane, derived from cruciferous vegetables like broccoli, potently induces surrogate markers of phase 2 enzyme activity in prostate cells in vitro and in vivo. To better understand the temporal effects of sulforaphane and broccoli sprouts on gene expression in prostate cells, we carried out comprehensive transcriptome analysis using cDNA microarrays.
Methods
Transcripts significantly modulated by sulforaphane over time were identified using StepMiner analysis. Ingenuity Pathway Analysis (IPA) analysis was used to identify biological pathways, networks and functions significantly altered by sulforaphane treatment.
Results
StepMiner and IPA revealed significant changes in many transcripts associated with cell growth and the cell cycle, as well as a significant number associated with cellular response to oxidative damage and stress. Comparison to an existing dataset suggested that sulforaphane blocked cell growth by inducing G2/M arrest. Cell growth assays and flow cytometry analysis confirmed that sulforaphane inhibited cell growth and induced cell cycle arrest.
Conclusions
Our data suggests that in prostate cells sulforaphane primarily induces cellular defenses and inhibits cell growth by causing G2/M phase arrest. Furthermore, based on the striking similarities in the gene expression patterns induced across experiments in these cells, sulforaphane appears to be the primary bioactive compound present in broccoli sprouts, suggesting that broccoli sprouts can serve as a suitable source for sulforaphane in intervention trials.
Keywords: Sulforaphane, microarray, gene expression, prostate cancer, cancer prevention
Introduction
Consumption of cruciferous vegetables has been associated with a significantly decreased risk of several malignancies, including prostate cancer, and this reduction has been attributed to the isothiocyanate sulforaphane (1-12). Sulforaphane will reduce the incidence, multiplicity, and rate of development of mammary tumors in dimethylbenz(a)anthracene-treated rats (13). Its most striking feature, the ability to induce cellular carcinogen defenses, was the basis for its isolation from broccoli and has been the focus of investigations into its anticancer activity (14,15). However, sulforaphane can also block cell growth, largely through effects on cell cycle regulatory proteins, suggesting that it can affect carcinogenesis through other mechanisms (16).
Growing evidence suggests that prostate cancer, like several other malignancies, arises as a result of chronic oxidative stress (17,18). Based on these observations, strategies to induce cellular defenses against oxidative genotoxic stresses, including induction of anti-carcinogen “phase 2” enzymes, have been proposed as potential means of cancer prevention (19). In a screen of candidate phase 2 enzyme inducing compounds, we have demonstrated robust induction of phase 2 enzymes in prostate epithelial cells after treatment with candidate chemopreventive compounds of diverse chemical classes (20). Of the compounds tested, sulforaphane emerged as one of the most potent phase 2 enzyme inducing agents in prostate cells in vitro (21). In addition, oral feeding of sulforaphane to F-344 rats increases nicotinamide quinone oxidoreductase (NQO1), total glutathione transferase and mu-class glutathione transferase enzyme activities in prostate tissues in vivo (22).
We have used comprehensive gene expression profiling to provide insights into the mechanisms of action of several candidate prostate cancer preventive agents (23-25). Detection of transcripts whose levels changed by 2-fold or more over a time course after treatment and comparison of transcript profiles to existing gene expression datasets uncovered unsuspected transcriptional features underlying alterations in cell growth, apoptosis and androgen signaling. Recently, statistical tools have been developed to analyze microarray datasets to identify transcripts significantly modulated over a time course. One unique tool, StepMiner, identifies genes that undergo a significant binary shift in expression levels over time and identifies the time at which these transitions occur (26). Here we use StepMiner to identify comprehensive gene expression changes induced in the prostate cancer cell line LNCaP by two different concentrations of sulforaphane and an aqueous extract of broccoli sprouts. While some expression changes differ between the experiments, the core set of transcripts modulated suggests that sulforaphane works largely through induction of cellular defenses and modulation of cell cycle regulatory genes.
Materials and Methods
Cell Culture and Treatment
LNCaP cells were obtained from the ATCC (Manassas, VA) and grown in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml) in a humidified atmosphere at 37°C and 5% CO2. When cells reached 75% confluence, they were treated with either 10 μM or 25 μM L-sulforaphane (LKT Laboratories, St Paul, MN) dissolved in DMSO, or DMSO only (controls). Lyophilized three-day-old broccoli sprouts were obtained as a powder (4.35 mg sulforaphane/gm) (Natural Sprout Company, Springfield, MO) and dissolved in DMSO to a final sulforaphane concentration of 10 μM that was used to treat the cells. The concentration of DMSO in media did not exceed 0.01%. Cells treated with sulforaphane, sprout extract, and DMSO alone were harvested at various times after treatment by scraping in TRIzol solution (InVitrogen, Carlsbad, CA) and total RNA was isolated according to manufacturer’s instructions.
Microarray Hybridizations and Data Analysis
Gene expression analysis was performed using spotted cDNA microarrays manufactured at Stanford University containing 42,000 elements representing 24,164 genes. Total RNA (80 μg) isolated from sulforaphane treated and control cells was reverse transcribed and the resulting cDNAs were fluorescently labeled by incorporation of Cy-5 (SFN treated) or Cy-3 (control) labeled dUTP during the reaction. Labeled cDNAs from treated and control cells matched by time point were mixed and hybridized to the microarrays according to previously described methods (25). After 14 h the microarrays were washed in SSC, dried and scanned with a GenePix microarray scanner.
Fluorescence intensities for each channel of the scanned microarrays in both the spots and background were determined using GenePix software. Artifacts and spots of insufficient quality on visual inspection were excluded from analysis. Data files containing fluorescence ratios for each spot were entered into the Stanford Microarray Database, and compiled experiments were further analyzed with hierarchical clustering software and visualized with Treeview software (27,28). The raw data from all experiments is available for downloading and has been deposited in GEO.
StepMiner Analysis
The microarray datasets contain seven replications for the zero hour (no treatment) and eight time points (2, 4, 6, 8, 12, 18, 24 and 36 hours) for each of the following treatments: broccoli sprouts, 10uM sulforaphane and 25uM sulforaphane. Three time course microarray datasets were constructed by appending the seven zero hour microarrays to the beginning of the time courses for each treatment. These time course datasets were analyzed using StepMiner (26). The StepMiner algorithm analyzes microarray time courses by identifying genes that undergo abrupt transitions in expression level, and the time at which the transitions occur. Genes that are significantly (p-value < 0.05) up-regulated and down-regulated were retrieved using StepMiner. The common significantly regulated genes in all three time courses were found by intersecting the significant genes from each time course experiment.
Advanced gene set analysis
The Unigene clusters ID for the 575 significant genes that were common to all three time course experiments were retrieved. These genes were then analyzed using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) (29). Significant biological functions, networks and pathways from this analysis were reported along with p-values for the probability of enrichment of these pathways.
Comparison with cell cycle genes
The significant genes in each experiment were analyzed for enrichment of cell cycle related genes. The list of cell cycle genes with their annotations was downloaded from http://genome-www.stanford.edu/Human-CellCycle/Hela/data.shtml (30). A simple hypergeometric test was performed to test the enrichment of genes related to different cell cycle phases in each time course experiment.
Cell growth Assays
LNCaP cells were cultured with or without sulforaphane in a range of concentrations for 3 days, during which time the cells did not achieve confluency. Cells were harvested by trypsinization and counted on a hemacytometer using a light microscope. Cell viability was determined by the trypan blue (0.1% w/v) exclusion assay.
Cell Cycle Analysis
Percentage of actively growing cells was determined using a propidium iodide (PI) based fluorescence assay. Briefly, 6×105 cells were plated in 6 cm culture dishes under the conditions mentioned above. Upon reaching 75% confluence cells were treated with DMSO alone, 10μM sulforaphane in DMSO, or 25μM sulforaphane in DMSO. After 8 hrs, 18 hrs and 24 hrs the media were aspirated and the adherent cells were trypsinized. Adherent and floating cells were collected and suspended in PBS containing 5 mM of EDTA. Approximately 1-2 × 106 cells were fixed in 70% ice-cold ethanol and left overnight at 4°C. Cells were collected by centrifugation at 1000 - 1200 rpm and gently suspended in 0.5 ml PBS plus 0.5 mM EDTA. The RNA was digested with Rnase A (10 μg/ml) for 30 min at 37°C. Before analysis the total volume was brought up to 1 ml and propidium iodide was added to a final concentration of 40 μg/ml. DNA content per cell was determined using a Beckman Coulter FACS machine and the data were analyzed with XL System II version 3.0 software (Beckman Coulter, Inc., Fullerton, CA). All experiments were performed in triplicate with identical results.
Results
Sulforaphane is thought to be primarily responsible for the cancer chemopreventive activities of cruciferous vegetables, and broccoli sprouts are a rich source of sulforaphane and precursor glucosinolates. To evaluate the effects of sulforaphane and broccoli sprouts on gene expression in prostate cells, we used cDNA microarrays to assess global changes in transcript levels over time for sulforaphane 10 μM, sulforaphane 25 μM and lyophilized broccoli sprouts reconstituted in DMSO to a sulforaphane concentration of 10 μM. Transcripts were selected that had fluorescent intensities 1.5 times over the background and 2-fold or greater change in expression level over control in at least 2 experiments at any time point with 80% good data (not more than 20% of measurements discarded due to poor data quality for each entry). When genes were grouped by unsupervised hierarchical cluster analysis, highly similar patterns of expression were seen across each of the 3 experiments (not shown). Variation of the data selection and filtering criteria yielded highly similar gene expression patterns across experiments, demonstrating the robustness of the datasets.
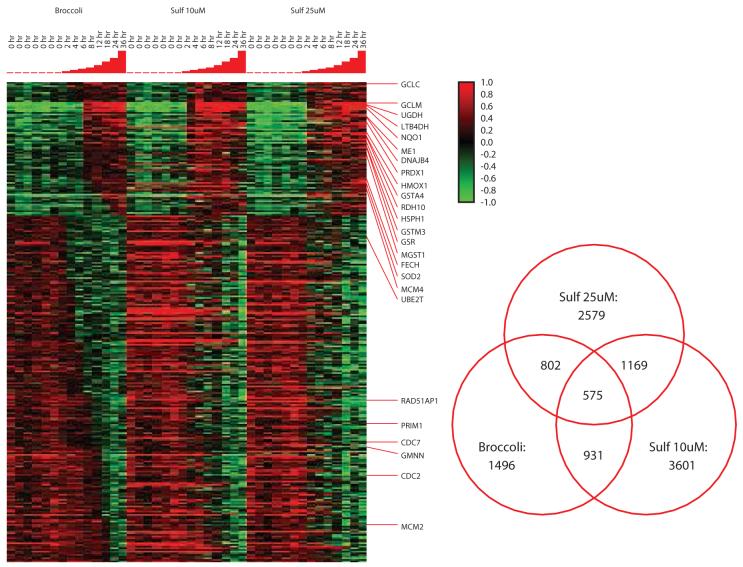
Selection of transcripts using arbitrary criteria (e.g. 2-fold over control) can lead to false positive calls because of noise in one or more samples or because of false negative calls for transcripts that significantly change but fail to reach the arbitrarily selected threshold. StepMiner iteratively evaluates time course data for a significant change (or “step”) in transcript abundance by comparing expression levels of early and late time points. StepMiner allows automated identification of transcripts with statistically significant changes and the point at which the change occurs. StepMiner identified 2579 transcripts altered by sulforaphane 25 μM, 3601 by sulforaphane 10 μM and 1496 altered by broccoli sprouts. As shown in figure 1, for the 575 transcripts common to the 3 datasets, expression patterns across the time points were highly similar. The degree of overlap between the datasets was highest between sulforaphane 10 μM and broccoli sprouts which was not surprising since the sprout extract was reconstituted to a sulforaphane concentration of 10 μM.
Figure 1.
StepMiner analysis of 575 transcripts modulated in common by broccoli sprouts, sulforaphane 10 μM and sulforaphane 25 μM. Individual transcripts are displayed in rows while experiments are represented in columns. Red indicates relative induction of transcripts while green represents relative decrease in expression levels, while the degree of color saturation corresponds to the degree of change (key to the right of the heat map). At the top of the cluster, genes induced include many phase 2 enzymes. To the right, a Venn diagram illustrates the total number of transcripts identified as significantly changes over time by StepMiner analysis with the corresponding overlaps between any two and all three of the datasets.
Sulforaphane produced a distinct, robust change (≥ 2-fold) in transcript levels for many genes that began within 4 hours of treatment and persisted for at least 24 hours. The most striking increases in transcript levels were observed in genes associated with electrophile defense (phase 2 enzymes) and included NQO1, leukotriene B4 dehydrogenase (LTB4DH), malic enzyme (ME1), thioredoxin reductase (TXNRD1), glutathione s-transferase mu (GSTM1), microsomal glutathione S-transferase (MGST1) superoxide dismutase (SOD1) and peroxiredoxin (PRDX1). Changes in transcript levels for NQO-1, γ-glutamylcysteine synthase (GCLM) and microsomal glutathione S-transferase seen in the microarray experiments were consistent with our previous results showing sulforaphane-induced transcript levels by northern blot analysis and parallel increases in NQO1 enzymatic activity and cellular glutathione levels (21). In addition, we confirmed increased expression of LTB4DH and GCLM and decreased expression of JUN in sulforaphane treated cells by quantitative RT-PCR and noted similar degrees of change in expression compared to the microarray experiments (not shown).
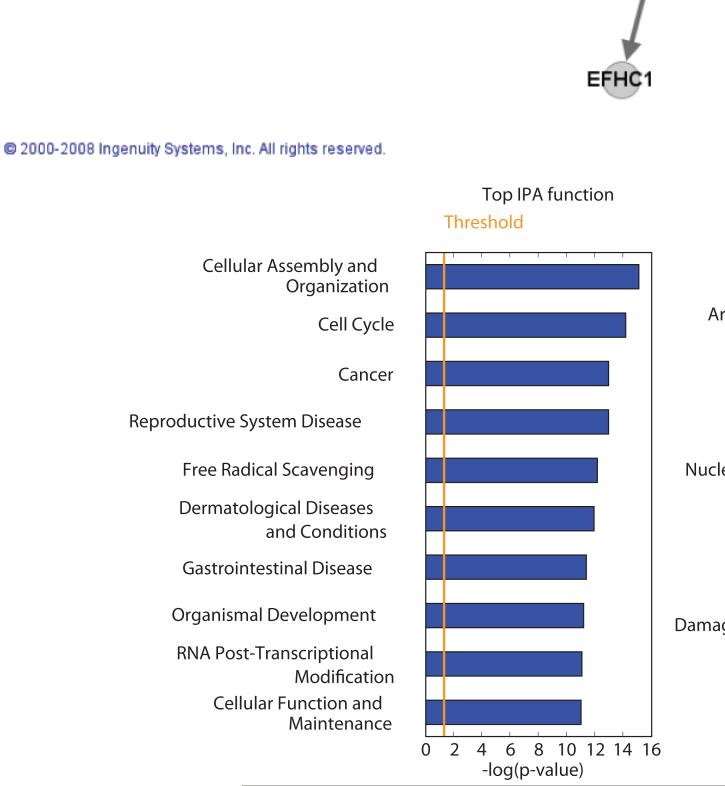
To better understand the effects of sulforaphane on prostate cancer cells we used Ingenuity Pathway Analysis (IPA) to test for enrichment of transcriptional networks, pathways and functions using the 525 genes modulated in common between the 3 experiment sets. Cell cycle regulation was the most significantly enriched network showing alterations in gene expression (Figure 2 at the top), and the second most altered function identified. Pathway analysis also confirmed that the cell cycle, particularly G2/M damage checkpoint regulation, showed significant alteration in the genes common to the 3 experiments. Not surprisingly, the most highly significantly enriched pathway involved Nrf2 signaling, known to be the principle regulatory element involved in induction of phase 2 enzyme gene expression. Consistent with this observation was significant enrichment of free radical scavenging under function and pathway enrichment for glutathione and glutamate metabolism, all likely reflecting induction of the phase 2 enzyme response. Aryl hydrocarbon metabolism was also significantly affected by sulforaphane, although many of these genes were down-regulated. IPA analysis did not reveal other major networks, pathways or functions that provided additional insights into the mechanisms of action of sulforaphane.
Figure 2.
At top, the cell cycle regulatory network identified by Ingenuity Pathway Analysis as the most significantly altered network by sulforaphane in the 575 genes from the StepMiner analysis. Gene names in bold correspond to transcripts significantly modulated by sulforaphane. Below are cellular functions most highly enriched in the StepMiner gene set (left) and cell pathways significantly modulated (right) in sulforaphane-treated LNCaP cells.
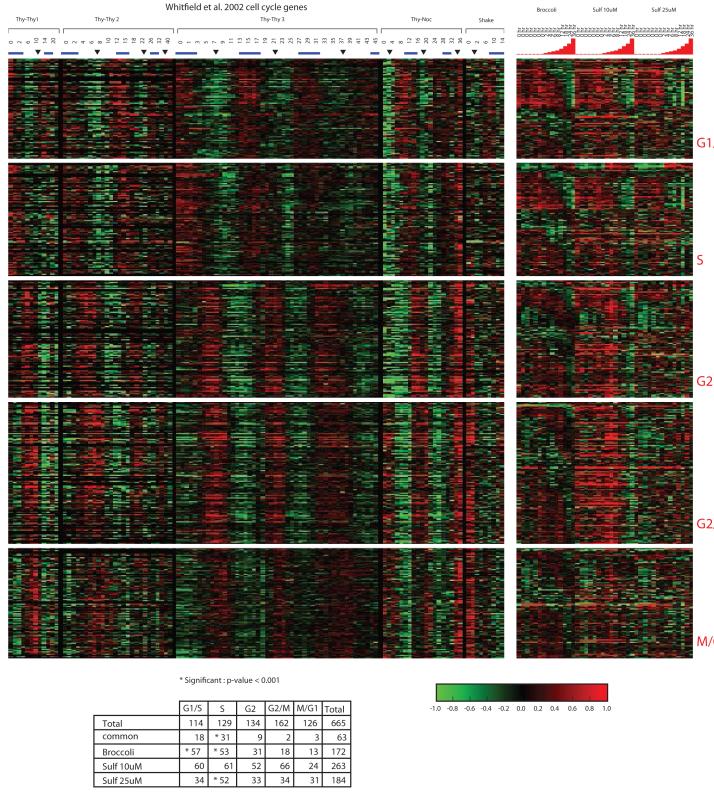
Based on the significant enrichment of transcripts involved in cell growth and cell cycle regulation, we investigated the effects of sulforaphane on cell cycle regulatory genes by comparing the genes modulated by sulforaphane to a set of genes found to vary cyclically as synchronized HeLa cells pass through the cell cycle (Figure 3). Sulforaphane showed significant enrichment for transcripts in G1/S and S phase and, in general, levels of these transcripts showed decreased expression over time after treatment with sulforaphane. In transcripts associated with G2 and G2/M phases, many transcripts showed increased expression over time after sulforaphane treatment, particularly in broccoli sprout extract and sulforaphane 25 μM. In the Whitfield dataset, increased expression of transcripts associated with particular phases of the cell cycle was observed in cells arrested at that phase in the cell cycle (30). Since sulforaphane showed increased expression for many transcripts associated with G2/M we hypothesized that it inhibits cell growth and acts by inducing cell cycle arrest in G2/M.
Figure 3.
Comparison of genes modulated as cells pass through the cell cycle from Whitfield et al to the transcript profiles significantly altered by broccoli sprouts, sulforaphane 10 μM and sulforaphane 25 μM. Over the time course, the majority of transcripts associated with G1/S and S phases show declining expression, while many associated with G2 and G2/M show increasing expression. Hypergeometric test confirms enrichment of transcripts in G1/S and S, with corresponding decreased gene expression levels.
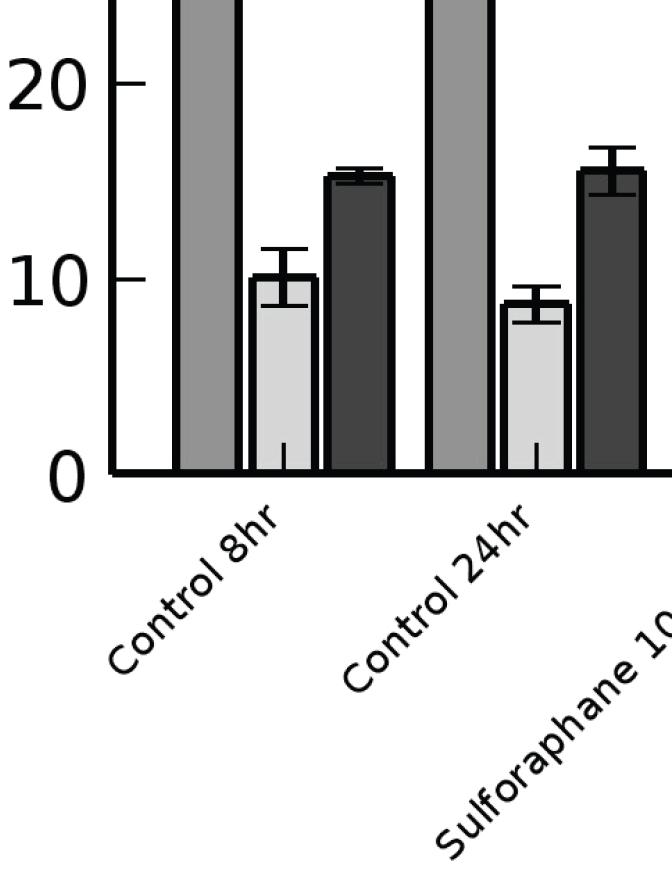
To test whether sulforaphane affects cell growth, we tested the effects of serial increases in sulforaphane concentrations in the media on cell number after 72 hours of growth. As seen in Figure 4A, increasing concentration of sulforaphane was associated with decreased cell number. We observed no increase in the number of apoptotic cells until sulforaphane concentrations in the media exceeded 25 μM, implying that the decrease in cell number was due to growth inhibition. To further evaluate the effects of sulforaphane on cell growth, we performed flow cytometry on LNCaP cells after treatment with sulforaphane (Figure 4B). By 8 hours after treatment sulforaphane and broccoli sprouts produced a significant decrease in the number of cells at G0/G1 or S phase and a significant increase in the number of cells in G2. A similar pattern was observed at 24 hours after treatment, implying that sulforaphane inhibits cell growth by inducing G2/M cell cycle arrest, leading to depletion of the number of cells in G0/G1 and S phases.
Figure 4.
Effects of sulforaphane on cell growth and cell cycle distribution of LNCaP cells.
A) Relative number of cells after treatment for 72 hours with sulforaphane. Loss of cell viability was observed at concentrations of 25 μM and above.
B) Flow cytometry analysis of the relative number of cells in each of the phases of the cell cycle after 8 and 24 hours of sulforaphane treatment at either 10 or 25 μM. Significant changes in the relative proportion of cells is indicated. Sulforaphane treatment resulted in increased numbers of cells in G2/M and a relative decrease in the number of cells in G1 and S phases.
Discussion
StepMiner and Ingenuity Pathway Analysis demonstrate that sulforaphane induces transcriptional changes primarily in cellular (carcinogen) defenses and cell cycle regulation in the prostate cancer cell line LNCaP. Particularly striking is the significant induction of phase 2 enzymes that encode for diverse enzymes involved in reduction of electrophiles. Genome-wide approaches have shown that sulforaphane produces similar broad induction of phase 2 enzymes in a variety of cell lines in vitro and tissues in rodents in vivo (31-35). The similarity in the precise transcripts induced and significant degree of induction suggests that regulation of these transcripts is highly conserved between species and tissues. In agreement with the IPA analysis, many of the genes induced are regulated by nrf-2, demonstrated by the attenuation of induction in mice engineered to lack nrf-2 expression (33,34,36). Therefore, sulforaphane is highly effective at inducing cellular defenses and is capable of inducing these changes in various tissues (including the prostate) in vitro and in vivo.
Since prostate cancer loses expression of a critical phase 2 enzyme (GSTP1) early in its development, phase 2 enzyme induction represents an attractive approach to prostate cancer chemoprevention (37). The robustness and breadth of carcinogen defense induction by sulforaphane, and the demonstration that induction within prostate tissues follows oral administration of sulforaphane in rats identify it as an important candidate preventive agent (22). Broccoli sprouts contain high levels of sulforaphane and its precursor glucosinolates, and have been used as a source of sulforaphane in intervention trials (38,39). In our time course experiments, lyophilized broccoli sprouts produced gene expression changes that were largely similar to those produced by a comparable dose of pure sulforaphane. Indeed, the similarity of the expression changes induced by sulforaphane and broccoli sprouts suggests that sulforaphane is the primary active micronutrient in broccoli sprouts and that broccoli sprouts are an acceptable means of administering sulforaphane in clinical trials.
Gene expression profiling also implicates inhibition of cell growth through cell cycle arrest as a second important feature of sulforaphane’s activity in prostate cells. After exposure to sulforaphane, transcripts associated with G2/M phase of the cell cycle were up regulated while those associated with other phases of the cell cycle decreased. This likely reflects a relative increase in the proportion of cells in G2/M due to cell cycle arrest by cells in that phase of the cell cycle. Other groups have demonstrated cell cycle arrest by sulforaphane at G2/M in prostate cancer cells. Several potential mechanisms have been implicated in causing cell cycle arrest including generation of reactive oxygen species, activation of JNK signaling, Chk2-mediated phosphorylation of Cdc25C, inhibition of histone deacetylase and decreased expression of cyclin D1 (16,40-46). Administration of sulforaphane will inhibit the growth of the prostate cancer cell line PC-3 in xenografts (47). While it is possible that growth inhibition in vivo is due to induction of apoptosis, accumulating evidence suggests that inhibition of cell growth is an important feature of sulforaphane’s activity in vivo.
It is notable that sulforaphane does not appear to modulate transcript levels in other critical pathways, making its transcriptional effects distinct from other candidate preventive agents. For example, androgen signaling is central to prostate cancer biology and we have shown previously widespread changes in androgen modulated transcripts in LNCaP cells treated with methylselenic acid, selenomethionine and resveratrol (23-25). Sulforaphane did not appear to significantly affect androgen signaling pathways based on Ingenuity Pathway Analysis or manual inspection of target genes. While the 2 selenium compounds and resveratrol increased expression of a few canonical phase 2 enzyme transcripts, none produced the broad induction apparent after sulforaphane treatment. Finally, all of the compounds influenced expression of genes modulated during the cell cycle, although different sets of genes are modulated, corresponding to the distinct phase of the cell cycle in which each compound induces arrest. Viewed as a whole, these data suggest that each of the 4 compounds we have analyzed induces unique effects in LNCaP cells and suggests that combining agents with distinct and complimentary effects on carcinogenesis might be an effective approach to prostate cancer progression.
It is unclear whether the transcriptional programs induced by sulforaphane in LNCaP cells in vitro are similar to those induced in vivo. However, the observation that oral feeding of sulforaphane to mice can induce phase 2 enzyme activity and broadly affect phase 2 enzyme transcript levels in several tissues suggests that such effects will occur in vivo (31-34). Furthermore, we have demonstrated that oral feeding of sulforaphane can increase the specific activities of NQO-1, total GST and mu-class GST in the prostate tissues of F344 rats (22). While plasma levels of sulforaphane can reach as high as 1-5 μg, they are well below the doses used in this study. However, Zhang et al. have demonstrated that sulforaphane is concentrated intracellularly after conjugation with glutathione (48). Sulforaphane glutathione conjugates are still capable of inducing a phase 2 enzyme response. Whether sulforaphane is concentrated in the prostate is unknown, and it will be important to assess tissue levels of sulforaphane to better understand its pharmacokinetics and whether induction is possible in man (49).
In summary, StepMiner and Ingenuity Pathway Analysis allowed identification of genes, networks, functions and pathways significantly modulated over a time course after treatment of LNCaP cells with sulforaphane. Sulforaphane induced broad expression of transcripts involved in cellular defenses and modulated transcripts associated with progression through the cell cycle. Broccoli sprouts induce many of the same transcripts as pure sulforaphane implying that sulforaphane is the primary active compound in broccoli sprouts, and that they are appropriate for use in intervention trials. These data provide additional insights and rationale that suggest sulforaphane should be investigated as a candidate prostate cancer chemopreventive agent.
Acknowledgments
Supported in part by the United States Army MRMC Prostate Cancer Research Program (DAMD17-98-1-8555), the Doris Duke Foundation (T98064) and NIH CA111782.
References
- 1.Cohen JH, Kristal AR, Stanford JL. Fruit and vegetable intakes and prostate cancer risk. J Natl Cancer Inst. 2000;92(1):61–68. doi: 10.1093/jnci/92.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Giovannucci E, Rimm EB, Liu Y, Stampfer MJ, Willett WC. A prospective study of cruciferous vegetables and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2003;12(12):1403–1409. [PubMed] [Google Scholar]
- 3.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55(3):224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain MG, Hislop GT, Howe GR, Ghadirian P. Plant foods, antioxidants, and prostate cancer risk: findings from case-control studies in Canada. Nutr Cancer. 1999;34(2):173–184. doi: 10.1207/S15327914NC3402_8. [DOI] [PubMed] [Google Scholar]
- 5.Kirsh VA, Peters U, Mayne ST, Subar AF, Chatterjee N, Johnson CC, Hayes RB. Prospective study of fruit and vegetable intake and risk of prostate cancer. J Natl Cancer Inst. 2007;99(15):1200–1209. doi: 10.1093/jnci/djm065. [DOI] [PubMed] [Google Scholar]
- 6.Kolonel LN, Hankin JH, Whittemore AS, Wu AH, Gallagher RP, Wilkens LR, John EM, Howe GR, Dreon DM, West DW, Paffenbarger RS., Jr. Vegetables, fruits, legumes and prostate cancer: a multiethnic case-control study. Cancer Epidemiol Biomarkers Prev. 2000;9(8):795–804. [PubMed] [Google Scholar]
- 7.Kune S, Kune GA, Watson LF. Case-control study of dietary etiological factors: the Melbourne Colorectal Cancer Study. Nutr Cancer. 1987;9(1):21–42. doi: 10.1080/01635588709513908. [DOI] [PubMed] [Google Scholar]
- 8.Le Marchand L, Hankin JH, Kolonel LN, Wilkens LR. Vegetable and fruit consumption in relation to prostate cancer risk in Hawaii: a reevaluation of the effect of dietary beta-carotene. Am J Epidemiol. 1991;133(3):215–219. doi: 10.1093/oxfordjournals.aje.a115865. [DOI] [PubMed] [Google Scholar]
- 9.Michaud DS, Pietinen P, Taylor PR, Virtanen M, Virtamo J, Albanes D. Intakes of fruits and vegetables, carotenoids and vitamins A, E, C in relation to the risk of bladder cancer in the ATBC cohort study. Br J Cancer. 2002;87(9):960–965. doi: 10.1038/sj.bjc.6600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghavan M, Knapp DW, Bonney PL, Dawson MH, Glickman LT. Evaluation of the effect of dietary vegetable consumption on reducing risk of transitional cell carcinoma of the urinary bladder in Scottish Terriers. J Am Vet Med Assoc. 2005;227(1):94–100. doi: 10.2460/javma.2005.227.94. [DOI] [PubMed] [Google Scholar]
- 11.Slattery ML, Kampman E, Samowitz W, Caan BJ, Potter JD. Interplay between dietary inducers of GST and the GSTM-1 genotype in colon cancer. Int J Cancer. 2000;87(5):728–733. [PubMed] [Google Scholar]
- 12.Tang L, Zirpoli GR, Guru K, Moysich KB, Zhang Y, Ambrosone CB, McCann SE. Consumption of raw cruciferous vegetables is inversely associated with bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(4):938–944. doi: 10.1158/1055-9965.EPI-07-2502. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci U S A. 1994;91(8):3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prochaska HJ, Santamaria AB, Talalay P. Rapid detection of inducers of enzymes that protect against carcinogens. Proc Natl Acad Sci U S A. 1992;89(6):2394–2398. doi: 10.1073/pnas.89.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci U S A. 1992;89(6):2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke JD, Dashwood RH, Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7(4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349(4):366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 19.Nelson WG. Prostate cancer prevention. Curr Opin Urol. 2007;17(3):157–167. doi: 10.1097/MOU.0b013e3280eb110f. [DOI] [PubMed] [Google Scholar]
- 20.Brooks JD, Goldberg MF, Nelson LA, Wu D, Nelson WG. Identification of potential prostate cancer preventive agents through induction of quinone reductase in vitro. Cancer Epidemiol Biomarkers Prev. 2002;11(9):868–875. [PubMed] [Google Scholar]
- 21.Brooks JD, Paton VG, Vidanes G. Potent induction of phase 2 enzymes in human prostate cells by sulforaphane. Cancer Epidemiol Biomarkers Prev. 2001;10(9):949–954. [PubMed] [Google Scholar]
- 22.Jones SB, Brooks JD. Modest induction of phase 2 enzyme activity in the F-344 rat prostate. BMC Cancer. 2006;6:62. doi: 10.1186/1471-2407-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones SB, DePrimo SE, Whitfield ML, Brooks JD. Resveratrol-induced gene expression profiles in human prostate cancer cells. Cancer Epidemiol Biomarkers Prev. 2005;14(3):596–604. doi: 10.1158/1055-9965.EPI-04-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Brooks JD. Selenomethionine induced transcriptional programs in human prostate cancer cells. J Urol. 2007;177(2):743–750. doi: 10.1016/j.juro.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Whitfield ML, Xu T, Botstein D, Brooks JD. Diverse effects of methylseleninic acid on the transcriptional program of human prostate cancer cells. Mol Biol Cell. 2004;15(2):506–519. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahoo D, Dill DL, Tibshirani R, Plevritis SK. Extracting binary signals from microarray time-course data. Nucleic Acids Res. 2007;35(11):3705–3712. doi: 10.1093/nar/gkm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, Eisen MB, Spellman PT, Brown PO, Botstein D, Cherry JM. The Stanford Microarray Database. Nucleic Acids Res. 2001;29(1):152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 30.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu R, Hebbar V, Kim BR, Chen C, Winnik B, Buckley B, Soteropoulos P, Tolias P, Hart RP, Kong AN. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther. 2004;310(1):263–271. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- 32.Hu R, Khor TO, Shen G, Jeong WS, Hebbar V, Chen C, Xu C, Reddy B, Chada K, Kong AN. Cancer chemoprevention of intestinal polyposis in ApcMin/+ mice by sulforaphane, a natural product derived from cruciferous vegetable. Carcinogenesis. 2006;27(10):2038–2046. doi: 10.1093/carcin/bgl049. [DOI] [PubMed] [Google Scholar]
- 33.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Cancer Lett. 2006;243(2):170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 34.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- 35.Traka M, Gasper AV, Smith JA, Hawkey CJ, Bao Y, Mithen RF. Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J Nutr. 2005;135(8):1865–1872. doi: 10.1093/jn/135.8.1865. [DOI] [PubMed] [Google Scholar]
- 36.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99(18):11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, Epstein JI, Isaacs WB, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998;7(6):531–536. [PubMed] [Google Scholar]
- 38.Fahey JW, Zhang Y, Talalay P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci U S A. 1997;94(19):10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shapiro TA, Fahey JW, Dinkova-Kostova AT, Holtzclaw WD, Stephenson KK, Wade KL, Ye L, Talalay P. Safety, tolerance, and metabolism of broccoli sprout glucosinolates and isothiocyanates: a clinical phase I study. Nutr Cancer. 2006;55(1):53–62. doi: 10.1207/s15327914nc5501_7. [DOI] [PubMed] [Google Scholar]
- 40.Chiao JW, Chung FL, Kancherla R, Ahmed T, Mittelman A, Conaway CC. Sulforaphane and its metabolite mediate growth arrest and apoptosis in human prostate cancer cells. Int J Oncol. 2002;20(3):631–636. doi: 10.3892/ijo.20.3.631. [DOI] [PubMed] [Google Scholar]
- 41.Cho SD, Li G, Hu H, Jiang C, Kang KS, Lee YS, Kim SH, Lu J. Involvement of c-Jun N-terminal kinase in G2/M arrest and caspase-mediated apoptosis induced by sulforaphane in DU145 prostate cancer cells. Nutr Cancer. 2005;52(2):213–224. doi: 10.1207/s15327914nc5202_11. [DOI] [PubMed] [Google Scholar]
- 42.Herman-Antosiewicz A, Xiao H, Lew KL, Singh SV. Induction of p21 protein protects against sulforaphane-induced mitotic arrest in LNCaP human prostate cancer cell line. Mol Cancer Ther. 2007;6(5):1673–1681. doi: 10.1158/1535-7163.MCT-06-0807. [DOI] [PubMed] [Google Scholar]
- 43.Singh SV, Herman-Antosiewicz A, Singh AV, Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L, Baskaran R. Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J Biol Chem. 2004;279(24):25813–25822. doi: 10.1074/jbc.M313538200. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Liu D, Ahmed T, Chung FL, Conaway C, Chiao JW. Targeting cell cycle machinery as a molecular mechanism of sulforaphane in prostate cancer prevention. Int J Oncol. 2004;24(1):187–192. [PubMed] [Google Scholar]
- 45.Xu C, Shen G, Chen C, Gelinas C, Kong AN. Suppression of NF-kappaB and NF-kappaB-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene. 2005;24(28):4486–4495. doi: 10.1038/sj.onc.1208656. [DOI] [PubMed] [Google Scholar]
- 46.Xu C, Shen G, Yuan X, Kim JH, Gopalkrishnan A, Keum YS, Nair S, Kong AN. ERK and JNK signaling pathways are involved in the regulation of activator protein 1 and cell death elicited by three isothiocyanates in human prostate cancer PC-3 cells. Carcinogenesis. 2006;27(3):437–445. doi: 10.1093/carcin/bgi251. [DOI] [PubMed] [Google Scholar]
- 47.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232(2):227–234. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Callaway EC. High cellular accumulation of sulphoraphane, a dietary anticarcinogen, is followed by rapid transporter-mediated export as a glutathione conjugate. Biochem J. 2002;364(Pt 1):301–307. doi: 10.1042/bj3640301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye L, Dinkova-Kostova AT, Wade KL, Zhang Y, Shapiro TA, Talalay P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin Chim Acta. 2002;316(1-2):43–53. doi: 10.1016/s0009-8981(01)00727-6. [DOI] [PubMed] [Google Scholar]