Abstract
Corticotropin releasing factor receptor type 1 (CRF1), a coordinator of the body responses to stress, is also expressed in human skin, where it undergoes alternative splicing. Since the epidermis is continuously exposed to the environmental stress, human keratinocytes were chosen to study the biological role of CRF1 alternative splicing. The expression pattern of CRF1 isoforms depended on cell density, presence or absence of serum, and exposure to ultraviolet irradiation (UVR). Only two isoforms α and c were predominantly localized to the cell membrane, with only CRF1α being efficient in stimulating cAMP responding element (CRE). CRF1d, f and g had intracellular localization, showing no or very low (g) activation of CRE. The co-expression of CRF1α with d, f or g resulted in intracellular retention of both isoforms suggesting dimerization confirmed by detection of high molecular weight complexes. The soluble isoforms e and h were diffusely distributed in the cytoplasm or localized to the ER, respectively, and additionally found in culture medium. These findings suggest that alternatively spliced CRF1 isoforms can interact and modify CRF1α subcellular localization, thus affecting its activity. We suggest that alternative splicing of CRF1 may play an important role in the regulation of skin cell phenotype with potential implications in pathology.
At least 70% of human genes and the majority of G protein-coupled receptors (GPCR) undergo alternative splicing, increasing the functional capability of the genome (Johnson et al., [2003]; Wang and Cooper, [2007]; Einstein et al., [2008]). Nevertheless, the biological significance of this process remains unclear, although splicing has been implicated in many disorders (Wang and Cooper, [2007]).
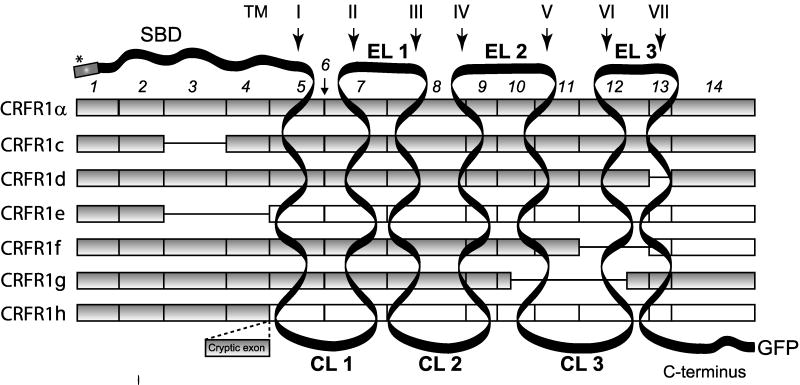
One of the most important elements of the body response to stress is the corticotropin releasing factor receptor type (CRF1) with its peptide ligands CRF and urocortin (Spiess et al., [1981]; Vale et al., [1981]; Perrin and Vale, [1999]; Hillhouse et al., [2002]; Hillhouse and Grammatopoulos, [2006]). Depending on its anatomical location, the CRF1 signaling system can regulate hypothalamic-pituitary-adrenal axis (HPA), as well as behavioral, autonomic, endocrine, reproductive, cardiovascular, gastro-intestinal, metabolic and immune systemic functions, or regulate homeostasis and viability of peripheral organs. The CRF1 gene belongs to family B1 of GPCRs (Hillhouse and Grammatopoulos, [2006]; Perrin et al., [2006]; Slominski et al., [2006b]) and codes several splicing variants, with at least eight found in humans, named α, β, c, d, e, f, g and h (Fig. 1). All of the defined CRF1 isoforms except β were detected in human skin (Pisarchik and Slominski, [2001], [2004]). The CRF1 isoforms could be divided into three groups: (1) full-length receptors - α and β (2) with deletion within the N-terminal extracellular domain (ECD) represented by isoforms c, e; (3) with partial or full deletion of seven transmembrane domain (7TM) - isoforms d, e, f, g, and h (Slominski et al., [2006b]). Although expression of different splicing variants of CRF1 receptor is well documented in several human organs (Pisarchik and Slominski, [2001], [2004]; Hillhouse and Grammatopoulos, [2006]; Slominski et al., [2006b]; Zmijewski et al., [2007]), their functions and significance are still unknown.
Figure 1.
Alternatively spliced isoforms of CRF 1. The CRF1 gene consists of 14 exons, which undergo alternative splicing, generating at least eight isoforms, seven of which are found in human skin (Pisarchik and Slominski, [2001]; Slominski et al., [2006a]). CRF1 splicing variants differ in the number of coding exons (filled squares). Excluded exons are shown as a line and exons with frame shift as white squares. ECD: extracellular substrate binding domain, EL: extracellular loops, CL: and three cyptoplasmatic loops. Signal peptide is marked by an asterisk. The constructs used in the present work also contain either V5, green fluorescent (EGFP) or red fluorescent (dsRED) proteins fused with C-terminus of each isoform.
Skin forms a self-regulating barrier between internal and external environment and is continuously subjected to different types of stressors (Slominski et al., [2006b], [2007a]). Therein, it is not than surprising that it expresses its own stress response coordinating system, including CRF1, to maintain or adjust local homeostasis disturbed by environmental factors or internal pathology (Slominski et al., [1995], [1998], [2004]). Human epidermis expresses CRF1 with a unique pattern of splicing variants characteristic for each skin cell type and regulated by external conditions (Pisarchik and Slominski, [2001], [2004]; Slominski et al., [2006b]). Previously, it was shown that CRF1α is a main isoform coupled to cAMP, IP3 and calcium signaling (Pisarchik and Slominski, [2004]), while CRF1 β was described as a pro-CRF1 form of receptor with its unique function in endometrium (Pisarchik and Slominski, [2004]; Hillhouse and Grammatopoulos, [2006]; Jin et al., [2007]; Teli et al., [2008]). Isoform c has impaired ligand binding (Karteris et al., [1998]; Hillhouse and Grammatopoulos, [2006]) and isoform d is poorly coupled to G proteins (Grammatopoulos et al., [1999]) due to deletion within the ECD or 7TM domains, respectively. Assigning function to the other isoforms is problematic, because their activity could only be detected in the presence of CRF1α (Pisarchik and Slominski, [2004]). We have speculated recently that alternative splicing would determine the intracellular localization of CRF1 isoforms and their biological activity (Slominski et al., [2006b]). It was also suggested that presumed “soluble isoforms” e and h (similar to sCRF2α (Chen et al., [2005])) could be secreted into extracellular environment (Jin et al., [2007]).
In order to define the localization and function of CRF1 isoforms we constructed a library of CRF1 isoforms, fused with fluorescent tags. The subcellular localization of CRF1 isoforms and their interactions were investigated using immortalized human epidermal keratinocytes (HaCaT). Additionally, the expression of CFR1 isoforms was also studied under normal, stress or pathological conditions in keratinocytes and in selected skin samples.
Materials and Methods
Cell culture, transfection and luciferase assay
HaCaT keratinocytes were grown in DMEM plus 10% fetal bovine serum (FBS) as described (Pisarchik and Slominski, [2004]) unless otherwise specified in the figure legend. Prior to transfection, cells were grown until they reached 70–80% of confluency and were transfected with the plasmids (Table S1) by using Lipofectamine and Plus reagent (Invitrogen, Carlsbad, CA). The dual luciferase reporter gene assays (Promega, Madison, WI) were conducted by using pCRE-luc vector containing luciferase gene under control of CRE transcription element and phRL-TK plasmid coding Renilla luciferase (used as normalization control; Promega) (Pisarchik and Slominski, [2004]; Zmijewski et al., [2007]).
Construction of CRF1 isoforms fused with EGFP and V5 tag
The construction of plasmids overexpressing CRF1 isoforms fused with EGFP or V5-tag followed a general strategy, as reported previously (Pisarchik and Slominski, [2004]) and also described in detail in the Supplementary data section. The primers used for cloning are summarized in Table S1.
CRF1-EGFP constructs
Full-length CRF1α, f, g and h DNA were amplified from corresponding construct reported previously (Pisarchik and Slominski, [2004]) and primers: forward MZ006
(5′-AAAAGCTTAGGACCCGGGCATTCAGGA-3′) and reverse MZ007
(5′-AAGAATTCTGAGACTGCTGTGGACTGCT-3′) or MZ005
(5′-AAGAATTCTGATCCCCCCAGCCACAG-3′, only f) or MZ008
(5′-AAGAATTCTTCTGTCCCACCACGGTGTGCTC-3′ only for h).
Cloning of CRF1c, d, and e splicing variants required two steps PCR amplification. In the first step, fragments spanning exon: 1–2 (forward MZ006:
5′-AAAAGCTTAGGACCCGGGCATTCAGGA-3′ and reverse MZ010:
5′-CCAGCAACATCTCAG ACAATGGCTAC-3′) and 4–14 (forward MZ009:
5′-CCAGCAACATCTCAG ACAATGGCTAC-3′ and reverse MZ007:
5′-AAGAATTCTGAGACTGCTGTGGACTGCT-3′) for isoform c; 1–12 (forward MZ006:
5′-AAAAGCTTAGGACCCGGGCATTCAGGA-3′ and reverse MZ012:
5′-GATGGCAGAACGGAC CTGGAAGGATTC-3′) and 14 (forward MZ011:
5′-CCTGGAATCCTTCCAG GTCCGTTCTG-3′ and reverse MZ007:
5′-AAGAATTCTGAGACTGCTGTGGACTGCT-3′) for isoform d; 1–2 (forward MZ006:
5′-AAAAGCTTAGGACCCGGGCATTCAGGA-3′ and reverse MZ002
5′-TGGTAGTGCACCTTGCTTTTTTTATGAGATGTTGC-3′) and 5–7(forward E25
5′-AAAAAAAGCAAGGTGCACTACCA-3′ and reverse MZ003:
5′-AAGAATTCTTCGTGGAGTAGGTGAGCACGATG-3′, covering the predicted stop codon)
for isoform e; were amplified separately, using the PCR method and pCRF1α as template. In the second step, the full-length sequence was assembled with flanking primers: MZ006 and MZ007forCRF1c and d; MZ006 and MZ003 for CRF1e. Assembly was possible because primers: MZ010 and MZ009; MZ012 and MZ011; MZ002 and E25, respectively, possessed, in part, complimentary fragments.
Final PCR fragments were purified using a GFX gel band purification kit (Amersham Biosciences, Piscataway, NJ), digested by HindIII and EcoRI enzymes and cloned in the expression vector pEGFP-N2 (Clontech Laboratories, Inc., Mountain View, CA).
CRF1α-dsRED
The coding sequence of CRF1 was amplified using PCR with CRF1α V5 template (Pisarchik and Slominski, [2004]) and primers: forward MZ006
(5′-AAAAGCTTAGGACCCGGGCATTCAGGA-3′ and reverse MZ048
(5′-GCAGAATTCTGACTGCTGTGGAC-3′). The PCR fragment was purified using a GFX gel band purification kit (Amersham Biosciences), digested by HindIII and EcoRI enzymes and cloned in expression vector pDsRed-Express-N1 (Clontech Laboratories, Inc.).
CRF1 isoforms fussed with V5 epitope
Construction of CRF1α f, g, and h was described previously (Pisarchik and Slominski, [2004]). To attach V5 epitope to the CRF1c d, e isoforms we amplified the corresponding DNA fragments with primers: forward MZ006 (E3 (Pisarchik and Slominski, [2004]) 5′-AAAAGCTTAGGACCCGGGCATTCAGGA-3′) and reverse E29 (Pisarchik and Slominski, [2004]) (5′-AAGAATTCTTGACTGCTGTGGACTGCT-3′) or reverse MZ004 (5′-AAGAATTCTTGTGGAGTAGGTGAGCACGATG-3′ only for CRF1e). DNA of plasmids: CRF1c-EGFP, CRF1d-EGFP, CRF1e-EGFP were used as templates. Final PCR fragments were purified using a GFX gel band purification kit (Amersham-Pharmacia-Biotech), digested by HindIII and EcoRI enzymes and cloned in expression vector pcDNA6/V5-His version B (Invitrogen).
All fragments were amplified using PCR Master Mix (Promega), 0.4 mM of each primers in unified conditions (2.5 min initial denaturation 95°C and 25 cycles: 94°C for 30 sec (denaturation), 56°C for 40 sec (annealing) and 72°C for 1.5 min (elongation); followed by elongation for 10 min at 72°C).
cDNA preparation and RT-PCRs
RNA preparation, quantification and quality control was performed as described previously (Slominski et al., [2007b]). RT-PCR, PCR reactions and primers sequences were as described previously (Slominski et al., [2007b]; Zmijewski et al., [2007]).
Cell fractioning and Western blotting
Protein samples preparation, membrane fraction isolation and PNGase F treatment were described previously (Slominski et al., [2007b]).
Detection of CRF1, EGFP or V5 tagged CRF1 was performed essentially as described (Slominski et al., [2006b]; Zmijewski et al., [2007]), except that anti-GFP antibodies were also used. Briefly, primary antibodies were used at following dilutions: anti-CRH-R1 1:100, anti-GFP 1:500 or anti-V5 1:500. Next, the HRP conjugated anti-rabbit secondary antibody (anti-goat 1:2,000 or anti-rabbit 1:4,000) were used and final detection was performed by using SuperSignal West Pico Substrate from Pierce (Rockford, IL). All primary and secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) except anti-V5 from Invitrogen.
Visualization of CRF1 immonoreactivity and CRF1 isoforms in cells and tissues, and co-localization with organellar markers
Cells were seeded in 8 well Lab-Tek II chamber slides (Nalge Nunc, Inc., Naperville, IL). Subconfluent cultures were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 (in PBS), blocked with 1% PBS and immunostained with goat anti-CRF1 antibody at a 1:100 dilution (Santa Cruz Biotechnology, Inc.) as described previously (Pisarchik and Slominski, [2004]). After washing, slides were incubated with FITC-conjugated anti-goat antibody (1:500 in 1% BSA in PBS), washed again, and mounted using VECTASHIELD Mounting Medium with Propidium Iodide (Vector Laboratories, Burlingame, CA). Similarly, formalin fixed de-paraffinized sections were incubated with antibodies against CRF1 (1:100) as described above. For the in vivo EGFP or RFP fluorescence experiments cells transfected with DNA of CRF1 isoforms fused with EGFP and/or dsRED (only CRF1 α) were observed at least 24 h after transfection. Additionally, the following organelle markers were used for in vivo staining: wheat germ agglutinin (WGA conjugated with Texas Red (5 μg/ml)) for Golgi apparatus (GA) (Molecular Probes, Portland, OR), Lysotracker DND99for lysosomes (Molecular Probes) and Mitotracker Deep Red 633 for mitochondria (Molecular Probes). For in vitro co-localization studies cells were processed as described above and stained with: anti-Golgi97 antibody (Molecular Probes) for Golgi apparatus, anti-DPI antibody (Santa Cruz Biotechnology) to detect endoplasmic reticulum and anti-keratinin 14 antibody for cytoskeleton (Santa Cruz Biotechnology). The Alexa Fluor 647 conjugated secondary antibody (1:1,000) was used for detection.
Images of cells were collected with Zeiss LSM 510 laser scanning microscope (Zeiss, Germany). For documentation of CRF1 immunoreactivity in skin sections Leica Digital Microscope DM4000B (Leica Microsystems Inc., Bannockburn, IL) was used. Cells incubated with corresponding non-immune serum were used as nonspecific immunostaining controls.
Statistical analyses
Data is presented as mean ± SEM (n = 3–4), and is analyzed with a Student’s t-test (for two groups) or one-way analysis of variance with appropriate post-hoc tests (for more than two groups) using Prism 4.00 (GraphPad Software, San Diego, CA). Statistically significant differences are denoted with asterisks: *P < 0.05, **P < 0.01, ***P < 0.005.
Results
Cell culture conditions and UVR regulate the expression of CRF1
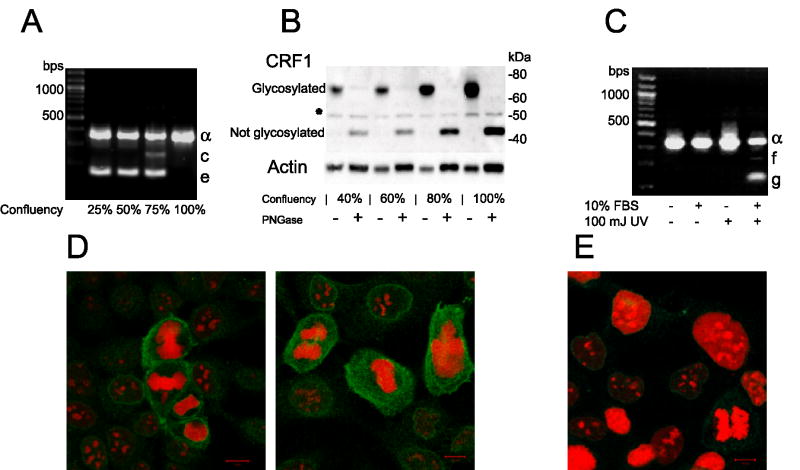
CRF1 gene expression and processing changed in keratinocytes during cell culture, being dependent on the cell density (Fig. 2A,B), for example, cultures confluent in 25–50% expressed only isoforms α and c, at 75% confluence they co-expressed CRF1e, whereas upon reaching 100% confluence, isoforms c and e disappeared and expression of CRF1 α increased. This process was accompanied by a gradual increase in the concentration of the mature (fully glycosylated) CRF1 α protein of approximately 60–70 kDa (Fig. 2B). The PNG-ase treatment removed the glycans to produce a protein of 45 kDa, which indicates N-glycosylation as a main posttranslational modification. Immunocytochemistry demonstrated CRF1 expression in both dividing and non-dividing cells with additional increased cytoplasmic CRF1 immunoreactivity in mitotic cells (Fig. 2D). An environmental stressor, UVB, either induced expression of additional isoforms f and g or increased expression of CRF1 α depending on the absence or presence of serum in the culture medium (Fig. 2C).
Figure 2.
Environmentally regulated expression of CRF1 in human keratinocytes. A: Alternative splicing of CRF1 in HaCaT keratinocytes is dependent on the cell density expressed as a percent of confluency. B: The expression of the mature (glycosylated) CRF1 α protein increases with increased cell culture density. Treatment with PNGase (+) generated a protein corresponding to the processed CRF1 α (deglycosylated) with approximate MW of 45 kDa (see Materials and Methods Section). * unknown protein, which may represent non-specific immunoprecipitation or partially glycosylated c or e isoforms. C: UVR effect on CRF1 mRNA expression in HaCaT keratinocytes is dependent on the presence or absence of serum in the culture medium. D: CRF1 is expressed in dividing and non-dividing cells. CRF1 immunoreactivity is shown in green and nuclear staining with propidium iodide in red. E. Negative control: cells incubated only with secondary antibody and stained with propidium iodide (red). The culture growth phase is expressed as a percent of confluence (A,B).
Subcellular localization of CRF1 isoforms
When control (non-transfected) keratinocytes were stained with anti-CRF1 antibodies, the receptor immunoreactivity was detected both on the cell membrane and intracellularly (Fig. S1). Similarly, studies of in situ immunofluorescence in skin biopsies have shown heterogeneous localization of the CRF1 immunoreactivity in both membrane and cytoplasmatic locations (Fig. 3).
Figure 3.
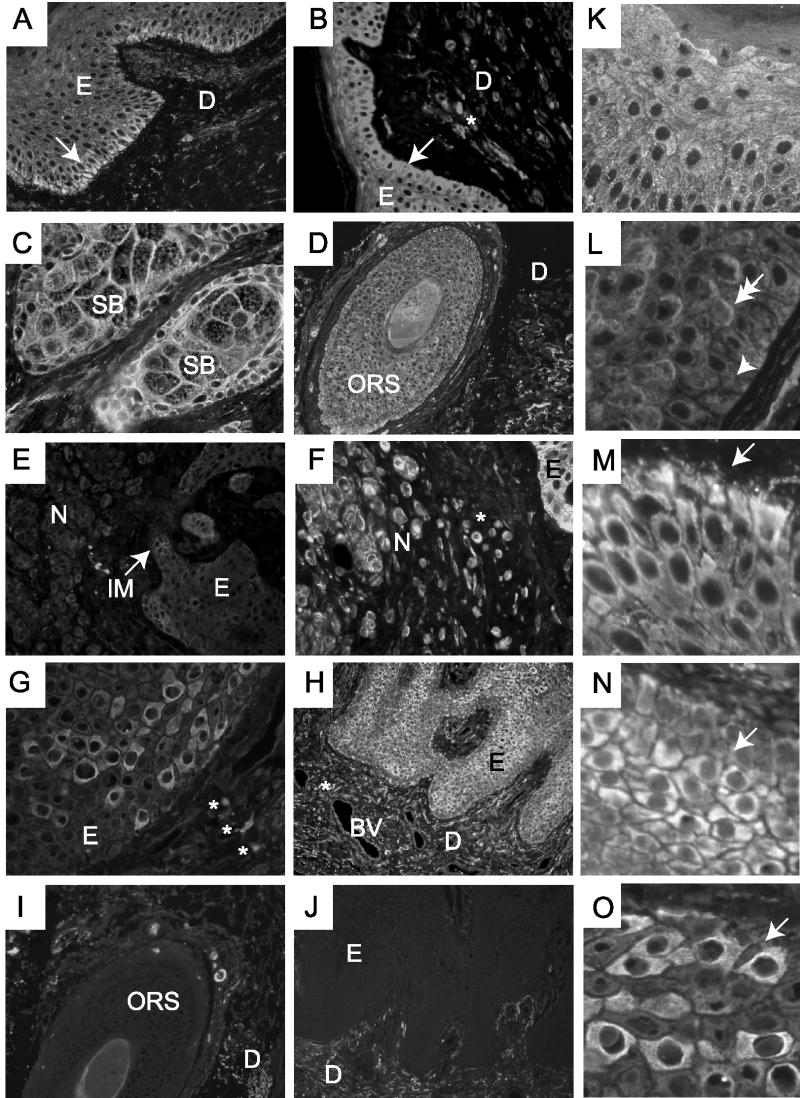
Differential expression of CRF1 in human skin. A, K, M: Normal skin, (B) intradermal nevus, (C) sebaceous glands, D, L: hair follicle, E, F: compound melanocytic nevus, G, H, N, O: psoriasis. Negative controls: (I) hair follicle, (J) epidermis. Panels K–N show magnified fragments of skin cross-sections. The skin layers and appendages are depicted as: EP, epidermis; DR or D, dermis; SB, sebaceous gland; ORS, outer root sheath; JM, junctional melanocytes; NE, nevocytes; SB, stratum basale; SS, stratum spinosum; BV, blood vessel. The position of the basal layer of keratinocytes is shown with an arrow (Panels A and B). Potential membrane (double-headed arrow) or cytosolic (arrow-head) localization of CRF1 immunoreactivity is shown in Panel L. *: CRF1 posivitive nevocytes in B and F, and CRF1 positive immune cells in G.
Such heterogeneous CRF1 staining was found in keratinocytes of the epidermis (Fig. 3A,B,F,G,K,M), hair follicle (Fig. 3D,L), in sebocytes (Fig. 3C), in melanocytes of the intradermal or compound nevus (Fig. 3E,F) and in blood vessels and immune cells of the dermis (Fig. 3H). The strongest CRF1 immunoreactivity is detected in keratinocytes. In normal epidermis a gradient of expression is seen with the highest immunoreactivity in the basal layers (Fig. 3A, B,K,M). This pattern was reversed in psoriasis, where the epidermis showed weaker staining of stratum basale with an increased stain in the stratum spinosum with focal or random patterns of immunoreactivity (Fig. 3G,H,N). A random pattern of expression was also seen in inflammatory dermal cells of psoriatic skin (Fig. 3H), and is comparatively weaker in nevocytes (Fig. 3E,F).
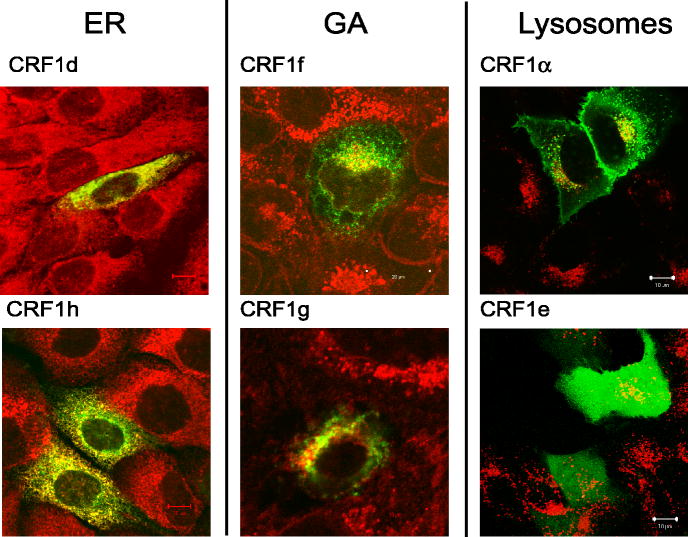
To find a mechanistic explanation for the above phenomena we used the CRF1 α, c, d, e, f, g, and h with EGFP, dsRED and V5 on C-terminus of the receptor to investigate their intracellular localization in keratinocytes (see Supplementary data (SD) for additional information and details on cloning procedures). The distinct cellular compartments including endoplasmic reticulum (ER), Golgi apparatus (GA), lysosomes, mitochondria and cytoskeleton have also been stained with selective markers to define precise receptor protein localization. The detailed co-localization study was presented in SD (Fig. S1–8). As predicted (Slominski et al., [2006b]), only isoforms α and c showed predominant expression in the cell membrane of transfected cells with the remaining isoforms having intracellular localization (Fig. 4 and Fig. S1–8). Isoforms with partial (d, f, and g) or entire deletion (h) of 7-TM domain were found mostly in ER (d and h) or GA (f and g). The CRF1e was expressed ubiquitously inside the cytoplasm and was the only isoform found in the nucleus (Fig. 4 and Fig. S5). Additionally, CRF1 α-EGFP fluorescence was found in lysosomes, but none of the isoforms tested, co-localized with mitochondria or cytoskeleton markers (Fig. 4 and Fig. S1).
Figure 4.
Intracellular localization of CRF1-EGFP isoforms in HaCaT keratinocytes. Cell staining for GA was performed with anti-Golgin97 (1:100) and for ER with anti-PDI (1:100). The Alexa Fluor 647 conjugated secondary antibody (1:1,000) was used for detection. EGFP fused isoforms are shown in green, organelles in red and co-localization in yellow.
To test whether fusion with EGFP could affect localization of CRF1 isoforms, keratinocytes were transfected with constructs carrying CRF1 fused with V5 tag and the V5 tag was subsequently detected by immunocytochemistry. Similarly to EGFP-fused receptors, isoform α was found on the cell membrane, while isoforms f and h showed intracellular localization (Fig. S9). Furthermore, the ligand binding capability of EGFP fused isoform α was demonstrated by co-localization of CRF1α-EGFP with CRF tagged with rhodamine (Fig. S10A,B). The treatment of keratinocytes expressing CRF1 α,-EGFP with CRF (100 nM) resulted in internalization of the receptor (Fig. S10C,D). These findings indicate that the presence of EGFP tag neither affected the localization nor CRF1 activity.
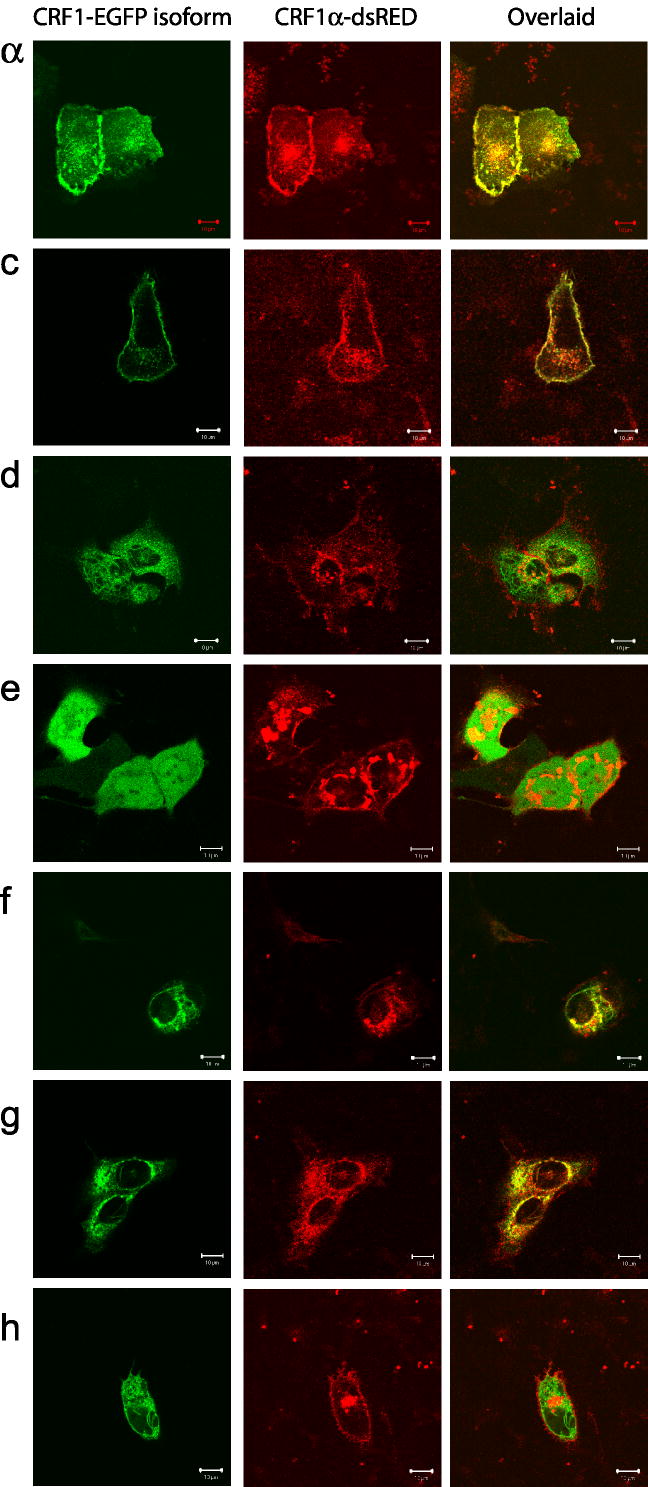
Co-localization of CRF1α-dsRED with membrane bound CRF1-EGFP isoforms
CRF1α-EGFP and CRF1c-EGFP co-localized with CRF1α-dsRED predominantly on the cell surface (Fig. 5). Surprisingly, co-expression of isoform α, with isoforms d, f, and g resulted in the intracellular retention of the CRF1α with a partial subcellular co-localization with co-transfected isoforms suggesting that those isoforms formed complexes (presumably heterodimers). A process of homo or heterodimerization for CRF1 has been described previously (Kraetke et al., [2005]; Mikhailova et al., [2007]; Young et al., [2007]).
Figure 5.
Co-localization of CRF1α with isoforms c–h. HaCaT keratinocytes were co-transfected with CRF1α fused with dsRED and CRF1 isoforms tagged with EGFP. Slides were visualized with a laser scanning confocal fluorescent microscope - LSM 510 equipped with Plan-Neofluor oil immersion 40x objective with suitable filter setup. CRF1α-dsRED is shown in red, EGFP fused isoforms in green, and co-localization in yellow.
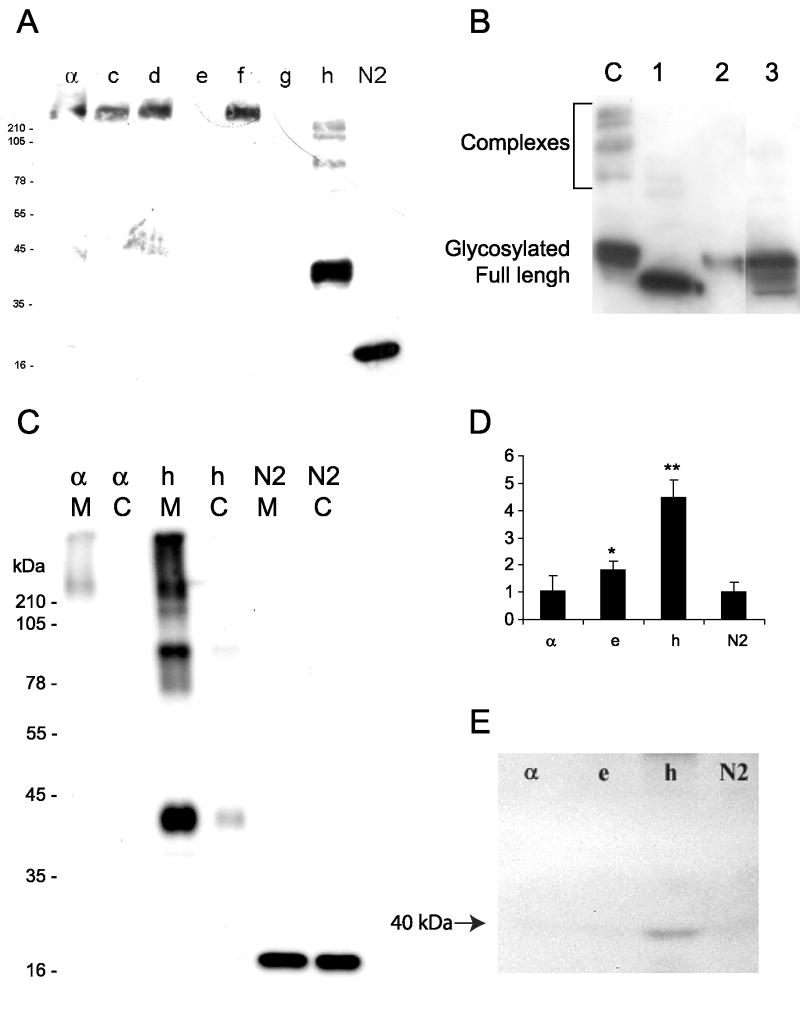
To study further this phenomenon, we performed western blotting analyses (Fig. 6A). The presence of high molecular weight complexes (HMWS) of CRF1 receptors was detected after separation of proteins under denaturating conditions using standard SDS-PAGE electrophoresis (Fig. 6A). The HMWS were dissolved by treatment with PNGase (deglycosylation), 6 M guanidinium chloride or DTT, as is shown for CRF1h as an example (Fig. 6B). These findings are consistent with previous detection of HMWS in COS cells overexpressing CRF1 isoforms (Pisarchik and Slominski, [2004]) or of native receptors in different tissues (Sydow et al., [1997]; Cao et al., [2005]; Slominski et al., [2006a], [2007b]).
Figure 6.
Oligomerization, glycosylation and different localization of soluble CRF1 isoforms in human HaCaT keratinocytes. Cells were transfected with plasmid DNA coding fusion of CRF1 isoforms with EGFP (a, c–h) or plasmids coding EGFP alone (N2). The EGFP tagged receptors was detected in cell lysates by Western Blotting (WB) with mouse anti-GFP antibody (1:500) and secondary anti-mouse antibody (1:4,000). A: High molecular weight complexes (HMCs) of isoforms: α c, d, f, and h. B: Dissociation of HMCs of CRF1h isoform; C, untreated control; 1, lysate treated with PNG-ase F, 2–6 M Guanidinium hydrochloride, 3–50 mM DTT. C: Detection of CRF1-EGFP fusion protein in membrane (M) or cytoplasm (C) fractions isolated from cells (C) or conditioned media (D, E). Extracellular localization of CRF1-EGFP fusion proteins detected by direct fluorescence of EGFP (D) or WB of proteins precipitated from media with TCA followed by detection with anti-GFP antibodies (mouse anti-GFP 1:500, anti-mouse 1:4,000) (E).
Soluble isoforms
The coding sequences of CRF1 e and h lack the entire 7-TM domain (Pisarchik and Slominski, [2001]; Slominski et al., [2006b]), but contain signaling peptides indicating that these isoforms can be secreted from the cells (Slominski et al., [2006b]). Indeed, the presence of green fluorescence of CRF1e-EGFP and CRF1h-EGFP fusion proteins was detected in the media collected from HaCaT keratinocytes cultures transfected with appropriate plasmids (Fig. 6D). The fluorescence of media from cells expressing CRF1h-EGFP or CRF1e-EGFP were at least 4 and 2 times higher, respectively, than media collected from keratinocytes transfected with CRF1α, or a vector containing EGFP only (Fig. 6D). The secretion of CRF1h-EGFP protein into the media was further confirmed by WB detection of protein with Mr of approximately 40 kDa, which after subtraction of EGFP (27 kDa) gave Mr of 13 kDa that was predicted for CFR1h (Fig. 6E).
Following intracellular localization of soluble isoforms by direct immunofluorescence (see Fig. 4 and Fig. S1), we analyzed their effects on CFR1α, expression pattern. The co-expression experiment of CFR1α, with CRF1e showed that CRF1α, aggregated near the nucleus, without a significant co-localization with isoform e (Fig. 5), suggesting that CRF1e can modulate its localization without a direct interaction. However, the intracellular fluorescence of CRF1 e-EGFP decayed within 48–72 h (not shown), suggesting a fast protein turnover or rapid attenuation of translation of CRF1e mRNA. This is in contrast to other isoforms including α. and h whose long fluorescence lifetime (2–3 weeks) has prompted us to study more stable isoform h.
The WB analyses were consistent with confocal microscopy and showed the CRF1 h immunoreactivity both in crude membrane and cytoplasmic fractions, while CRF1 α was found exclusively in the membranes (Fig. 6C). Further investigation showed that the isoform h formed stable high molecular weight complexes, similarly to CRF1 α, c, d, or f (Fig. 6A); these complexes were dissolved by treatment with PNGase, 6 M guanidinium chloride or DTT (Fig. 6C). This suggests that, at least for isoform h, the presence of 7TM domain is not a prerequisite for dimerization.
CRF1 signaling in keratinocytes
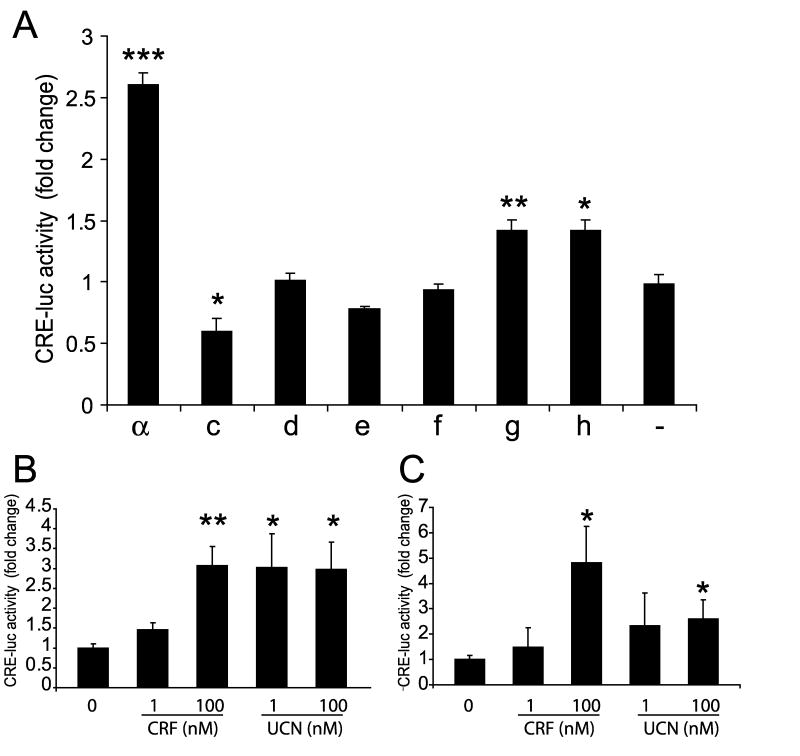
To study CRF1 signaling in keratinocytes we measured the transcriptional activity of the cAMP responsive element (CRE) using CRE-luc reporter gene (Pisarchik and Slominski, [2004]). Human HaCaT keratinocytes were transfected with plasmids carrying coding sequences of CRF isoforms α, c, d, e, f, g, or h. A 2.5-fold stimulation of CRE was observed in keratinocytes overexpressing CRF1 α, 24 h after CRF treatment. Lower (1.4-fold), but statistically significant, stimulation was observed only for two isoforms CRF1g and h, while overexpression of isoform c inhibited activity of CRE by 45% (Fig. 7). Isoforms d, e, and f showed no detectable activity.
Figure 7.
CRF stimulation of cAMP responsive element (CRE) in HaCaT keratinocytes overexpressing CRF1 isoforms. HaCaT cells were co-transfected with DNA constructs carrying different CRF1 isoforms, as indicated, and with CRE-luc and pRL-TK (Renilla luciferase) plasmids. 24 h after transfection cells were treated with CRF (100 nM). The activity of luciferase was measured using a dual luciferase assay. Results are shown as a fold change versus control. *P< 0.05, **P< 0.01, ***P< 0.005.
Discussion
At least 7 splicing variants of CRF1 receptor (α, c, d, e, f, g, and h) were detected in human skin (Pisarchik and Slominski, [2001], [2004]), representing a variety of N-terminal and C-terminal deletions and insertions, with or without frame-reading shifts and generation of presumed soluble isoforms (see Fig. 1) (Pisarchik and Slominski, [2001]; Hillhouse and Grammatopoulos, [2006]; Slominski et al., [2006b]). In the present study we define their subcellular localizations and demonstrate that their differential expression and/or co-expression can play an initial role in CRF signaling in human skin cells (Figs. 2–4).
CRF1 stimulates cAMP, IPS and Ca2+ in epidermal keratinocytes with downstream phenotypic effects such as stimulation of differentiation and secretory activities (Pisarchik and Slominski, [2001], [2004]; Hillhouse and Grammatopoulos, [2006]; Slominski et al., [2006a],[b]). High levels of CRF1 in lysates of confluent cells that express solely isoform α, under such conditions (Fig. 2A,B) suggests that CRF1 α. is also the most stable and necessary for cellular functions of normal keratinocytes. Accordingly, its expression was increased by UVB under starvation (Fig. 2C), and only isoform α represented the fully functioning receptor capable to activate CRE with other showing none (d, e, f) or very small effect (c, g, h) (Fig. 7). The latter is in agreement with our previous observations in COS cells showing that overexpression of isoforms e, f, g, and h was not sufficient to induce significant transcriptional activity and the modulatory role of some of them (h) could be detected when co-expressed with isoform α, (Pisarchik and Slominski, [2001], [2004]). The potential role for co-expressing isoforms in keratinocytes was suggested by regulation of their expression by cell density and UVB (Fig. 2). Furthermore, in myometrium, alternatively spliced CRF1 β isoform plays a modulatory role of CRF signaling (Markovic et al., [2006]). Therefore, extensive in situ localization studies were performed to better define the role of alternatively spliced isoforms in keratinocyte functions.
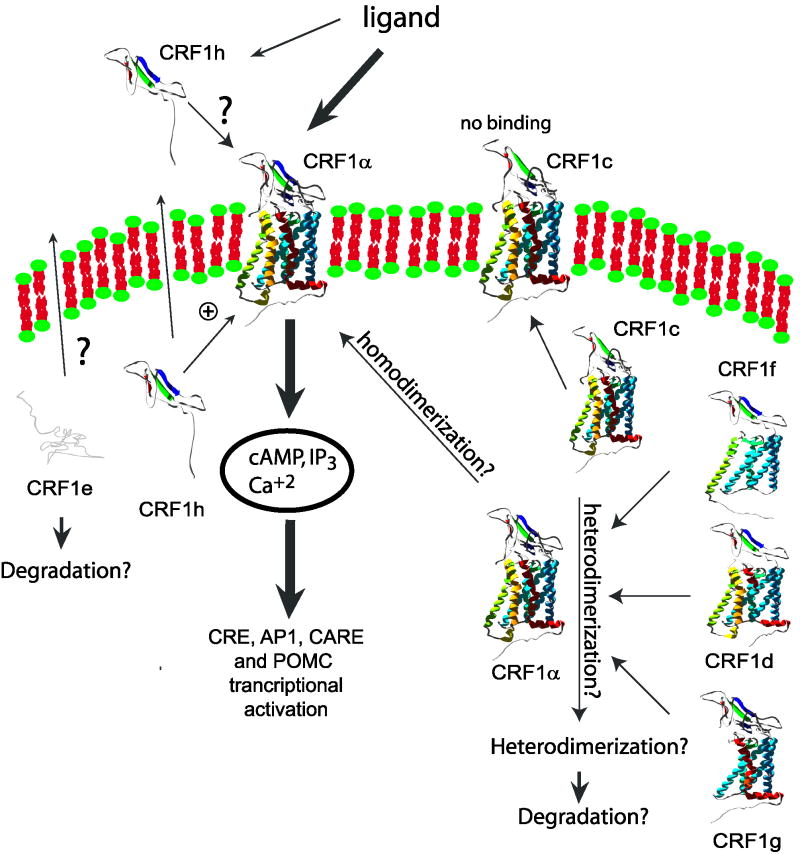
In epidermal keratinocytes, CRF1 isoforms d–h were found intracellularly (Fig. 4), which in part explains the absence of or very low ligand induced activity, because plasma membrane receptors were not available for binding. The extracellular detection of soluble isoform h, of which N-terminus contains the CRF binding sequence, suggests its role in the regulation of ligand availability for native CRF1 α by its retention or protection from degradation, which may explain stimulation of CRE in keratinocytes overexpressing CRF1h. Analogical soluble isoform-sCRF2α. (soluble isoform of CRF2) was found to bind several CRF-like ligands and inhibited CRF and Ucn I cellular responses when purified protein was added to media (Chen et al., [2005]). It is possible that soluble isoforms of CRF receptors might have different functions when expressed in cells or present in the media. Most of the isoforms co-localized with CRF1 α leading to its retention inside of the cell (Fig. 5). By analogy to a documented formation of heterodimers for other GPCR’s (Slominski et al., [2006b]; White et al., [2007]; Gurevich and Gurevich, [2008]), we suggest that similarly the retention of CRF1 α. is secondary to the formation of oligomers. This is further supported, by WB analyses showing that CRF1 isoforms can form HMWC, which are stable under mild denaturing conditions (Fig. 6, and previous findings in COS-7 cells; Slominski et al., [2006b], [2007b]), but dissolved by high salt, DTT or deglycosylation (Fig. 6B). This is also consistent with recent reports showing that CRF1 can form dimers (Kraetke et al., [2005]) and heterodimers with calcitonin or vasopressin V1b receptors (Mikhailova et al., [2007]; Young et al., [2007]). Theoretically, partial deletion of a transmembrane fragment (as for isoform d, f, and g) could cause lower stability of the receptor and its hetero-homodimers, resulting in premature degradation through quality control mechanism (Bulenger et al., [2005]; Conn et al., [2007]; Gurevich and Gurevich, [2008]). Taken together, alternative splicing and dimerization of CRF1 isoforms create additional level of regulation of CRF signaling, as it is proposed in Figure 8. The main implications of CRF1 splicing could be: (i) alternative splicing is lowering the pull of fully active isoform α, (all isoforms); (ii) isoform α, is sequestered from its proper membrane localization (isoforms d, f, g); (iii) partial inactivation of membrane bound receptor (isoform c also as possible heterodimerwith α); (iv) modulation of signaling (isoforms c, g, and h).
Figure 8.
Hypothetic model of regulation of CRF1 signaling by alternatively spliced CRF1 isoforms.
The complex, but very efficient regulation of CRF signaling by alternative splicing of CRF1 receptor in human skin may reflect and enable fast adaptation to the changing condition and counteract stressors from the surrounding environment. In this model, fast growing dividing cells, expressing multiple isoforms of receptor, would decrease their ability to produce a stress response (CRF signaling); this would ensure proper growth of the basal layer of epidermis even under stress stimuli. Deregulation of CRF signaling may result in pathological condition such as psoriasis (Fig. 3). Indeed, down regulation of CRF1 was observed in affected samples from a psoriasis patient (Tagen et al., [2007]). Thus, CRF1 receptor and its splicing variants may represent a possible target for the treatment of inflammatory or hyperproliferative skin diseases. In conclusion, our in situ studies explain why a majority of CRF1 spliced forms have minimal or no phenotypic effect, showing that they can interact and modify CRF1 α. subcellular localization and suggesting regulation of its activity through mechanisms outlined in Figure 8.
Supplementary Material
Acknowledgments
Funded by: NIH; Grant Number: AR047079, AR052190
The work was supported by a National Institutes of Health grant AR047079 (AS). Authors would like to thank Dr. Jacquline Granese for the immunofluorescence stains of skin biopsies, and Dr. Trevor Sweatman for critical reading of the manuscript and helpful suggestions. Confocal microscopy was performed on equipment obtained through Shared Instrumentation Grant from National Center for Research Purposes at the National Institutes of Health (S10 RR13725-01).
Footnotes
This article is a US Government work and, as such, is in the public domain in the United States of America.
References
- Bulenger S, Marullo S, Bouvier M. Emerging role of homo- and heterodimerization in G-protein-coupled receptor biosynthesis and maturation. Trends Pharmacol Sci. 2005;26:131–137. doi: 10.1016/j.tips.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Cao J, Papadopoulou N, Kempuraj D, Boucher WS, Sugimoto K, Cetrulo CL, Theoharides TC. Human mast cells express corticotropin-releasing hormone (CRH) receptors and CRH leads to selective secretion of vascular endothelial growth factor. J immunol. 2005;174:7665–7675. doi: 10.4049/jimmunol.174.12.7665. [DOI] [PubMed] [Google Scholar]
- Chen AM, Perrin MH, Digruccio MR, Vaughan JM, Brar BK, Arias CM, Lewis KA, Rivier JE, Sawchenko PE, Vale WW. A soluble mouse brain splice variant of type 2alpha corticotropin-releasing factor (CRF) receptor binds ligands and modulates their activity. Proc Natl Acad Sci USA. 2005;102:2620–2625. doi: 10.1073/pnas.0409583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PM, Ulloa-Aguirre A, Ito J, Janovick JA. G protein-coupled receptor trafficking in health and disease: Lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacol Rev. 2007;59:225–250. doi: 10.1124/pr.59.3.2. [DOI] [PubMed] [Google Scholar]
- Einstein R, Jordan H, Zhou W, Brenner M, Moses EG, Liggett SB. Alternative splicing of the G protein-coupled receptor superfamily in human airway smooth muscle diversifies the complement of receptors. Proc Natl Acad Sci USA. 2008;105:5230–5235. doi: 10.1073/pnas.0801319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatopoulos DK, Dai Y, Randeva HS, Levine MA, Karteris E, Easton AJ, Hillhouse EW. A novel spliced variant of the type 1 corticotropin-releasing hormone receptor with a deletion in the seventh transmembrane domain present in the human pregnant term myometrium and fetal membranes. Mol Endocrinol. 1999;13:2189–2202. doi: 10.1210/mend.13.12.0391. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR monomers and oligomers: It takes all kinds. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillhouse EW, Grammatopoulos DK. The molecular mechanisms underlying the regulation of the biological activity of corticotropin-releasing hormone receptors: Implications for physiology and pathophysiology. Endocrine Rev. 2006;27:260–286. doi: 10.1210/er.2005-0034. [DOI] [PubMed] [Google Scholar]
- Hillhouse EW, Randeva H, Ladds G, Grammatopoulos D. Corticotropin-releasing hormone receptors. Biochem Soc Trans. 2002;30:428–432. doi: 10.1042/bst0300428. [DOI] [PubMed] [Google Scholar]
- Jin D, He P, You X, Zhu X, Dai L, He Q, Liu C, Hui N, Sha J, Ni X. Expression of corticotropin-releasing hormone receptor type 1 and type 2 in human pregnant myometrium. Reprod Sci. 2007;14:568–577. doi: 10.1177/1933719107307821. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative premRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Karteris E, Grammatopoulos D, Dai Y, Olah KB, Ghobara TB, Easton A, Hillhouse EW. The human placenta and fetal membranes express the corticotropin-releasing hormone receptor 1 alpha (CRH-1 alpha) and the CRH-C variant receptor. J Clin Endocrinol Metab. 1998;83:1376–1379. doi: 10.1210/jcem.83.4.4705. [DOI] [PubMed] [Google Scholar]
- Kraetke O, Wiesner B, Eichhorst J, Furkert J, Bienert M, Beyermann M. Dimerization of corticotropin-releasing factor receptor type 1 is not coupled to ligand binding. J Recept Signal Transduct Res. 2005;25:251–276. doi: 10.1080/10799890500468838. [DOI] [PubMed] [Google Scholar]
- Markovic D, Papadopoulou N, Teli T, Randeva H, Levine MA, Hillhouse EW, Grammatopoulos DK. Differential responses of corticotropin-releasing hormone receptor type 1 variants to protein kinase C phosphorylation. J Pharmacol Exp Therap. 2006;319:1032–1042. doi: 10.1124/jpet.106.107441. [DOI] [PubMed] [Google Scholar]
- Mikhailova MV, Mayeux PR, Jurkevich A, Kuenzel WJ, Madison F, Periasamy A, Chen Y, Cornett LE. Heterooligomerization between vasotocin and corticotropin-releasing hormone (CRH) receptors augments CRH-stimulated 3′, 5′-cyclic adenosine monophosphate production. Mol Endocrinol. 2007;21:2178–2188. doi: 10.1210/me.2007-0160. [DOI] [PubMed] [Google Scholar]
- Perrin MH, Vale WW. Corticotropin releasing factor receptors and their ligand family. Ann NYAcad Sci. 1999;885:312–328. doi: 10.1111/j.1749-6632.1999.tb08687.x. [DOI] [PubMed] [Google Scholar]
- Perrin MH, Grace CR, Riek R, Vale WW. The three-dimensional structure of the N-terminal domain of corticotropin-releasing factor receptors: Sushi domains and the B1 family of G protein-coupled receptors. Ann NY Acad Sci. 2006;1070:105–119. doi: 10.1196/annals.1317.065. [DOI] [PubMed] [Google Scholar]
- Pisarchik A, Slominski AT. Alternative splicing of CRH-R1 receptors in human and mouse skin: Identification of new variants and their differential expression. FASEB J. 2001;15:2754–2756. doi: 10.1096/fj.01-0487fje. [DOI] [PubMed] [Google Scholar]
- Pisarchik A, Slominski A. Molecular and functional characterization of novel CRFR1 isoforms from the skin. Eur J Biochem. 2004;271:2821–2830. doi: 10.1111/j.1432-1033.2004.04216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Ermak G, Hwang J, Chakraborty A, Mazurkiewicz JE, Mihm M. Proopiomelanocortin, corticotropin releasing hormone and corticotropin releasing hormone receptor genes are expressed in human skin. FEBS Lett. 1995;374:113–116. doi: 10.1016/0014-5793(95)01090-2. [DOI] [PubMed] [Google Scholar]
- Slominski A, Ermak G, Mazurkiewicz JE, Baker J, Wortsman J. Characterization of corticotropin-releasing hormone (CRH) in human skin. J Clin Endocrinol Metab. 1998;83:1020–1024. doi: 10.1210/jcem.83.3.4650. [DOI] [PubMed] [Google Scholar]
- Slominski A, Pisarchik A, Tobin DJ, Mazurkiewicz JE, Wortsman J. Differential expression of a cutaneous corticotropin-releasing hormone system. Endocrinology. 2004;145:941–950. doi: 10.1210/en.2003-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Zbytek B, Pisarchik A, Slominski RM, Zmijewski MA, Wortsman J. CRH functions as a growth factor/cytokine in the skin. J Cell Physiol. 2006a;206:780–791. doi: 10.1002/jcp.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Zbytek B, Zmijewski M, Slominski RM, Kauser S, Wortsman J, Tobin DJ. Corticotropin releasing hormone and the skin. Front Biosci. 2006b;11:2230–2248. doi: 10.2741/1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Wortsman J, Tuckey RC, Paus R. Differential expression of HPA axis homolog in the skin. Mol Cell Endocrinol. 2007a;265–266:143–149. doi: 10.1016/j.mce.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski AT, Zmijewski MA, Pisarchik A, Wortsman J. Molecular cloning and initial characterization of African green monkey (Cercopithecus aethiops) corticotropin releasing factor receptor type 1 (CRF1) from COS-7 cells. Gene. 2007b;389:154–162. doi: 10.1016/j.gene.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess J, Rivier J, Rivier C, Vale W. Primary structure of corticotropin-releasing factor from ovine hypothalamus. Proc Natl Acad Sci USA. 1981;78:6517–6521. doi: 10.1073/pnas.78.10.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow S, Radulovic J, Dautzenberg FM, Spiess J. Structure-function relationship of different domains of the rat corticotropin-releasing factor receptor. Brain Res. 1997;52:182–193. doi: 10.1016/s0169-328x(97)00256-8. [DOI] [PubMed] [Google Scholar]
- Tagen M, Stiles L, Kalogeromitros D, Gregoriou S, Kempuraj D, Makris M, Donelan J, Vasiadi M, Staurianeas NG, Theoharides TC. Skin corticotropin-releasing hormone receptor expression in psoriasis. J Invest Dermatol. 2007;127:1789–1791. doi: 10.1038/sj.jid.5700757. [DOI] [PubMed] [Google Scholar]
- Teli T, Markovic D, Hewitt ME, Levine MA, Hillhouse EW, Grammatopoulos DK. Structural domains determining signalling characteristics of the CRH-receptor type 1 variant R1beta and response to PKC phosphorylation. Cell Signal. 2008;20:40–49. doi: 10.1016/j.cellsig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat Rev. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- White JF, Grodnitzky J, Louis JM, Trinh LB, Shiloach J, Gutierrez J, Northup JK, Grisshammer R. Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc Natl Acad Sci USA. 2007;104:12199–12204. doi: 10.1073/pnas.0705312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SF, Griffante C, Aguilera G. Dimerization between vasopressin V1b and corticotropin releasing hormone type 1 receptors. Cell Mol Neurobiol. 2007;27:439–461. doi: 10.1007/s10571-006-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmijewski MA, Sharma RK, Slominski AT. Expression of molecular equivalent of hypothalamic-pituitary-adrenal axis in adult retinal pigment epithelium. J Endocrinol. 2007;193:157–169. doi: 10.1677/joe.1.06927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.