Abstract
Francisella tularensis is a gram-negative, highly infectious, aerosolizable facultative intracellular pathogen that causes the potentially life-threatening disease tularemia. To date there is no approved vaccine available, and little is known about the molecular mechanisms important for infection, survival, and dissemination at different times of infection. We report the first whole-genome screen using an inhalation mouse model to monitor infection in the lung and dissemination to the liver and spleen. We queried a comprehensive library of 2,998 sequence-defined transposon insertion mutants in Francisella novicida strain U112 using a microarray-based negative-selection screen. We were able to track the behavior of 1,029 annotated genes, equivalent to a detection rate of 75% and corresponding to ∼57% of the entire F. novicida genome. As expected, most transposon mutants retained the ability to colonize, but 125 candidate virulence genes (12%) could not be detected in at least one of the three organs. They fell into a variety of functional categories, with one-third having no annotated function and a statistically significant enrichment of genes involved in transcription. Based on the observation that behavior during complex pool infections correlated with the degree of attenuation during single-strain infection we identified nine genes expected to strongly contribute to infection. These included two genes, those for ATP synthase C (FTN_1645) and thioredoxin (FTN_1415), that when mutated allowed increased host survival and conferred protection in vaccination experiments.
The three subspecies of Francisella tularensis and the highly related Francisella novicida are gram-negative facultative intracellular bacteria that cause the potentially life-threatening disease tularemia in a large number of mammals (9, 25, 27, 33). Infections can occur by inhalation, ingestion, exposure to infected animals, or transmission from an arthropod vector. Disease manifestation varies depending on the route of inoculation and ranges from respiratory oropharyngeal to ulceroglandular tularemia. These four bacteria show different virulence profiles in humans. F. tularensis subsp. tularensis (type A) is the most virulent subspecies of Francisella and, along with F. tularensis subsp. holartica (type B) and F. tularensis subsp. mediasiatica, is a human pathogen. Francisella novicida causes disease only in immunocompromised humans and therefore is not considered a human pathogen. Because of its high infectivity, severe virulence, ease of aerosol dissemination, and broad host and geographic distribution, F. tularensis subsp. tularensis is listed as a category A potential agent of bioterrorism. However, all subspecies and F. novicida share 95% DNA sequence identity, suggesting that F. tularensis subsp. tularensis and F. novicida share similar virulence genes (23, 29). Because F. novicida is more genetically amenable and safer to manipulate (it is a biosafety level 2 pathogen) but causes a disease in mice similar to that caused by the most virulent F. tularensis subsp. tularensis strains, F. novicida serves as a good experimental model to study Francisella pathogenesis targeted toward the development of a tularemia vaccine.
Our understanding of Francisella pathogenesis is advancing rapidly. Sequencing of strains from several different species and the development of tools for genetic manipulation have enabled researchers to identify novel virulence factors that impair macrophage function and disrupt the host immune response(5, 6, 13, 42). Despite recent advances, however, many important questions remain. It is unknown whether certain genes or virulence factors predominate during different routes of infection or bacterial residence in different tissues, including lung, liver, spleen, and skin, and therefore play a role in Francisella tularensis' ability to cause a wide spectrum of disease. Acceptable vaccine candidates ideally infect transiently but disseminate to lung, liver, and/or spleen before they are subsequently cleared, allowing sufficient time and exposure for generation of a robust immune response. Because of Francisella tularensis' ability to cause severe disease with a low infectious dose by the aerosol route, we chose to use an inhalation model in mice for our experiments. In our genome-wide negative-selection screen in F. novicida, we identified genes required for systemic infection in mice by analyzing mutant clone behavior in lung and after dissemination to the liver and spleen. As expected most mutants persisted in lung and were able to colonize liver and spleen. We found clones mutant in 44 genes lost in lung and clones mutant in 81 genes lost in liver and/or spleen but present in lung. If loss of a clone in lung occurred, the attenuated mutants in general persisted for the first 24 h and disseminated to liver and/or spleen and then were lost in lung after 48 and 72 h of infection. These genes with a strong phenotype in all three organs might be attractive vaccine candidates.
MATERIALS AND METHODS
Bacterial growth and mutant pool assembly.
All bacteria were cultured in tryptic soy broth (TSB) medium or tryptic soy agar (TSA) plates supplemented with 0.1% l-cysteine, 0.2% glucose, and kanamycin (15 μg/ml) at 37°C. A total of 11 pools of mutants were prepared from a sequence-defined transposon mutant library of F. novicida. The mutant library (http://www.francisella.org/) was generated by random transposon mutagenesis of F. novicida strain U112 as part of a screen for essential genes (12). The transposon mutant library was plated from 96-well frozen stock plates onto TSA plates using a floating pin replicator (V&P Scientific, Inc.) and incubated for 48 h. A half patch of each mutant was combined into each of two freezer vials with 1 ml TSB and 20% glycerol. After freezing at −80°C, titers of viable counts (CFU) in the frozen cultures were determined and the second vial used directly for aerosol infection experiments. For the primary screen, three 96-well plates from the original library were combined for each pool. For the secondary screen, two smaller pools containing 84 mutants each were selected from the 96-well frozen stock plates of the original library and combined. These mutants were identified as being lost in lung, liver, and/or spleen in the primary screen. In addition, both pools were supplemented with four controls selected from the primary screen as clones present at all three time points in all organs.
Screening of transposon mutant pools in mice.
C57BL/6 mice were purchased from The Jackson Laboratory and housed under specific-pathogen-free conditions. All animal experiments were performed using biosafety level 2 precautions in the animal facility of Harborview Medical Center. This work was approved by the Institutional Animal Care and Use Committee of the University of Washington. For each pool, 16 mice were exposed to aerosolized bacteria using a nose-only inhalation system as previously described (16). Bacterial deposition in the lungs in each experiment was determined by quantitative culture of homogenized lung tissue harvested from four mice euthanized immediately after exposure (0 h, deposition). The remaining 12 mice were sacrificed at 24 h, 48 h, and 72 h (four mice each) after being observed daily for clinical manifestation of disease. The inocula across all 11 pools ranged from 2,500 to 10,000 CFU/lung. The average pool size was 282 mutants, and therefore each mutant was represented at least 10 times, maximizing the representation of each mutant in the pool. Organs (lung, liver, and spleen) from each mouse at each time point were separately homogenized, serial dilutions were plated on TSA-kanamycin plates, and bacteria were grown for 48 h at 37°C and harvested from plates containing at least 3,000 CFU. For the deposition (0-h) time point, the entire lung homogenate was plated and combined to guarantee that all the grown clones were represented in the input pool. Genomic DNA was isolated from the deposition and output cultures using a Wizard genomic DNA preparation kit (Promega). The genomic DNAs from the outputs of all four mice for each time point and organ were pooled for microarray experiments.
Microarray tracing of transposon mutant behavior within pools.
The Francisella microarray used was a 70-mer oligonucleotide array that includes duplicate spots representative of 1,681 genes, equivalent to 93% of the entire genome. Since the different Francisella subspecies and strains F. novicida, Schu4, LVS, and type B strains are closely related at the genomic level, the array was customized to include the entire F. novicida genome plus genes unique to the other strains. The complete list of sequences printed on the array is available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11297. The spotted arrays were printed by the Genomics Shared Resource of the Fred Hutchinson Cancer Research Center.
Sequences flanking one side of the transposons in each pool were amplified in a two-step PCR and labeled for microarray hybridization. Approximately 1 μg of genomic DNA was digested using HindIII in a 20-μl reaction mixture. Each digest was cleaned with DNA Clean and Concentrator-5 (Zymoresearch) and eluted with 20 μl water to give a final concentration of 50 ng/μl. Agarose gel electrophoresis was used to control for complete digestion. Ligation was performed using the T4 ligase kit (Invitrogen) on 200 ng of digested genomic DNA with a total of 74 pmol of the Y-linker, using primers TA1 and TA2 at 16°C for 12 h as described previously (39). The ligation reaction was stopped by incubation at 65°C for 10 min, cleaned, and verified by agarose gel electrophoresis. The ligated DNA was used as a template in a PCR with 2 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-Cl, 15 mM MgCl2), 2 μl of 10 mM deoxynucleoside triphosphate mix (dG, dA, dC, and dT), and 1 μl each of 40-μM transposon-specific primer TA3 (ACCTGATTGCCCGACATTATCGCGAGCC) and linker-specific primer TA4 (ACGCACGCGACGAGACGTAGC). The PCR was initiated by incubation at 95°C for 2 min and then cycled 22 times between 94.5°C for 30 s and 72°C for 90 s. A second, nested PCR was done to incorporate aminoallyl-dUTP for Cy dye coupling. In a 100-μl reaction mixture, 10 μl of the previous PCR product was added to 10 μl 10× PCR buffer, 4 μl 3 mM deoxynucleoside triphosphate-UTP mix (3 mM dG, dA, and dC; 1.2 mM dT; 1.8 mM aminoallyl-dU), and 1 μl of the TA4 linker-specific primer as well as a nested transposon-specific primer, TA6 (GCGGCCTCGAGCAAGACGTTTCCCGTTG). The PCR program was the same as in the first PCR step for 12 cycles. The efficiency of the PCR amplifications was evaluated using 10 μl from the second PCR for agarose gel electrophoresis. The remainder of the PCR product (90 μl) was cleaned as described above and eluted with a total of 15 μl 50 mM NaHCO3. Coupling to Cy dyes was performed using the postlabeling kit from Amersham. Each tube containing the Cy3 or Cy5 dye was diluted with 11 μl of dimethyl sulfoxide, and 5.5 μl of the reconstituted dye was added to the cleaned PCR product.
In each case, the input (0 h, deposition) pool DNA was labeled with Cy3 (green) and the output pool (24 h, 48 h, and 72 h) was labeled with Cy5 (red). Therefore, the red/green (R/G) ratio for each gene provides a measure of fitness for any given gene mutated in the input pool: a yellow spot (R/G, 1) present in the input and output represents no phenotype, and a green spot (R/G, <1) present in input but absent in output pool represents a candidate mutant. After Cy dye labeling, the input and output labeled DNAs for each experiment were combined, cleaned as described above, eluted with 13 μl of Tris-EDTA, and prepared for hybridization by adding 1 μl each of salmon sperm DNA and yeast tRNA, 5 μl 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 2 μl 1% SSC. The prepared probes were then denatured at 99°C for 2 min and centrifuged at 13,000 rpm for 2 min, and the supernatant was added to the array for overnight hybridization at 65°C.
Microarray scanning and analysis were performed on a GenePix 4000B scanner (Axon) using GenePix Pro 6.0 software (Axon). Spots were filtered for slide abnormalities, and data extracted for duplicate spots per gene were averaged. Because each pool contained on average 282 insertions, most of the 1,681 gene spots on the array were expected to give a background signal. Thus, we expected roughly 600 spots in each experiment array to give a strong hybridization signal, but the actual number would vary from experiment to experiment. To select spots for analysis, we first computed the average background-subtracted signal intensity in the Cy3 channel (input pool) of all the spots except the brightest 600 spots. We then chose for further analysis spots whose intensity was greater than one standard deviation from this mean value. The log2 R/G was collected for each of these gene spots. Two data sets were collected for each experiment (pool): one where amplification of flanking DNA was performed from the left (reverse) side of the transposon and a second where amplification of flanking DNA was performed from the right (forward) arm of the transposon.
Single-strain and protection studies.
Mice were exposed to aerosolized mutant bacteria using the same nose-only inhalation system as described for the mutant pool screen. Bacterial deposition in the lungs was determined by quantitative culture of homogenized lung tissue harvested from mice killed immediately after exposure. Mice were observed daily for clinical manifestation of disease by macroscopic appearance and by measurement of ventral surface body temperature, using an infrared thermometer (Raytek). Mice were euthanized if they manifested signs of severe illness (hunched posture, piloerection, crusted eyes, labored breathing, loss of resistance to handling, and surface temperature of <25°C). For protective immunity studies, four mice that had been infected with 149 CFU of the atpC mutant were challenged with 25 CFU of wild-type bacteria at 30 days postinfection, and survival of the mice was recorded.
Bacterial burden in mouse organs.
Mice were exposed to aerosolized wild-type or single-mutant bacteria as described above. Immediately after infection and at 1, 3, 7, and 14 days postinfection, mice were sacrificed. Initial bacterial deposition was determined by quantitative culture of homogenized lung tissue harvested from four mice immediately after infection. At the indicated time points, left lungs, livers, and spleens from four mice were homogenized in 1 ml phosphate-buffered saline for quantitative culture.
In vitro replication of attenuated mutants in a mouse alveolar macrophage cell line.
MH-S mouse alveolar macrophage cells (ATCC) were grown in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-Glutamine, 10 mM HEPES, and 1% penicillin-streptomycin (10,000 IU/ml; GIBCO) (supplemented RPMI) and incubated at 37°C with 5% CO2. The MH-S cells were washed four times with supplemented RPMI without antibiotics, after which they were seeded in 24-well plates (coated with 0.01% poly-l-lysine) at 105 cells/well. The plates were then incubated overnight for cell adhesion to occur. Wild-type F. novicida and transposon mutant strains FTN_1415 and FTN_1645 were grown over night in TSB at 37°C with shaking at 200 rpm. The bacterial strains were transferred to a 50-ml conical tube and harvested by centrifugation at 4°C at 3,000 rpm for 20 min. The bacterial pellet was washed twice with chilled Dulbecco phosphate-buffered saline and suspended in 5 ml of supplemented RPMI (without antibiotics). Each well containing MH-S cells were then infected with 1 × 107 CFU/500 μl for a multiplicity of infection of 100. Two hours after infection, each well was washed with supplemented RPMI without antibiotics to remove nonadherent bacteria, and 500 μl of fresh medium was added. Immediately thereafter, monolayers were lysed to harvest intracellular bacteria by adding 500 μl of 0.2% saponin to give a final concentration of 0.1% saponin in each well. Serial dilutions were plated on TSA supplemented with 0.1% cysteine and incubated for 48 h at 37°C to determine CFU. Bacteria were similarly harvested from lysed monolayers at 24 h and 48 h postinfection. Parallel wells of both uninfected and infected MS-S cells were stained with Trypan blue at the end of the experiment to monitor cell viability. In both cases, viability was estimated to be >90%.
Microarray data accession number.
The microarray data from these experiments have been submitted to Gene Expression Omnibus (www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE11297.
RESULTS
Screening of a comprehensive Francisella novicida transposon library for colonization and dissemination in a mouse inhalation infection model.
We used a comprehensive library of sequence-defined transposon insertion mutants of F. novicida strain U112 (12) to screen for genes required to establish lethal infection after aerosol exposure. This library contains 3,072 transposon mutant clones with two insertion alleles for the majority of nonessential genes. In order to screen as many mutants as possible using the lowest number of mice, we infected mice with pools of mutants and monitored the fate of these mutant clones using an adaptation of microarray tracking of transposons mutants (1). The pools were assembled by combining three 96-well plates of the parent library into one pool. The parent library was not arrayed in genome order. Therefore, the genomic distribution of individual mutants comprising each pool was essentially random.
There are roughly 1,800 genes in the F. novicida genome, of which approximately 312 are essential, leaving approximately 1,488 genes that can tolerate mutations by transposon insertions (12). We were able to grow 98% (2,998) of the 3,072 transposon mutants in the library on TSA plates and assembled a total of 11 pools with an average pool size of 282 mutants. Of these strains, 2,886 had transposon insertions annotated as genes in F. novicida and were used for further analysis. Results of transposon insertions in noncoding regions (intergenic) or insertion sequence elements are not included in the results. About 70% of the genes have transposon insertions in two different location of the same gene, and thus the library represents 1,414 unique genes with transposon insertions.
The microarray used to monitor the behavior of transposon clones contained probes representing 1,681 Francisella genes on our array and allowed us to collect data for 1,380 of the 1,414 unique genes in the library, corresponding to ∼77% of the F. novicida genome. We infected 16 C57BL/6 mice per pool by aerosol using a nose-only inhalation system (16) and harvested bacteria from lung, liver, and spleen for four mice at 24 h, 48 h, and 72 h after infection for generation of output pools. To obtain an accurate measure of the input pool, we also collected the bacterial clones successfully delivered to the lungs immediately after nebulization from four mice (deposition, 0 h). Mice were infected with approximately 1 × 104 CFU/lung (range, 1.6 × 103 to 2 × 104). As observed for single-strain wild-type infection at this dose, the bacteria initially grew rapidly, increasing by 3 to 4 log units in the first 24 h, reaching 1 × 108 to 109 CFU per lung over the next 48 h (72-h time point), and dying within the next 24 h, by day 4 (Fig. 1). By 24 h bacteria had disseminated to additional tissues and were recovered from both liver and spleen, where they appeared to further replicate.
FIG. 1.
Wild-type or mutant pool total bacterial load for lung, liver, and spleen. Lung, liver, and spleen data were averaged across all four mice per time point and across all 11 pools per organ. Mice were infected with pools of ∼282 mutants per pool, and depositions ranging from 1.6 × 103 to 2 × 104 CFU/lung across all 11 pools show similar overall growth and dissemination rates as high-dose (1.7 × 104 CFU) single wild-type strain infections.
The purpose of our screen was to identify genes that when inactivated by transposon insertion reduced the ability of the bacterium to establish a productive infection by the aerosol route of inoculation. Using this model, as few as 5 wild-type bacteria establish a productive infection resulting in death at 4 to 5 days and mice infected with as many as 104 bacteria still survive for 4 days, indicating little dose dependence on infection outcome in this dose range (44; and S. J. Skerett, unpublished data). With a pool size of approximately 300 clones and inocula of 104 CFU, we expected that each mutant should be represented by 30 CFU per animal, a dose sufficient to establish infection during single-strain infections. To further control for stochastic elimination of clones from the output pool, we pooled the genomic DNAs prepared from the output bacterial pools from each of the four mice for each time point for the analysis of each organ site. The behavior of clones in the input (deposition) and output pools were monitored using a two-step PCR to amplify the DNA next to the transposon insertion for hybridization to microarrays as outlined in Fig. 2. By labeling both sides of the transposon, we were able to track the behavior of 1,029 of the potentially identifiable 1,380 annotated genes represented in the 11 pools and on the array. This is equivalent to a detection rate of 75%. Of the 1,029 genes identified in our screen, 532 genes (52%) yielded results from both sides of the transposon, whereas 255 (25%) and 242 (24%) were uniquely detected by labeling the reverse and forward sides of the transposon, respectively.
FIG. 2.
Schematic of negative-selection screen for identification of genes essential for colonization and dissemination in mouse inhalation infection model. Separate infections by inhalation were performed for each of the 11 mutant pools. Four mice per time point were sacrificed at 0 h (deposition) and at 24 h, 48 h, and 72 h (different outputs) and genomic DNA isolated from lung, liver, and spleen for each mouse separately at all time points. After combining DNA from all four mice per time point and organ, DNAs next to the transposon insertions were amplified in a two-step PCR to label deposition DNAs with Cy3 and output DNAs with Cy5. The resulting log2 (Cy5/Cy3) provided an indication of the representation of individual mutants in the output pool compared to the input pool.
In our primary screen, we collected data for three time points (24 h, 48 h, and 72 h) for all three organs. To assess the behavior of a given mutant clone, we required microarray data for two out of three time points per organ. A gene was called absent (clone lost during infection) in the output pool, implying a role in virulence, if the log2 ratio of deposition to output pool was ≤−1 at at least two out of three time points in lung, liver, or spleen. In parallel, a gene was called present (clone survived during infection) in the output pool if the log2 ratio of deposition to output pool was >−1 at at least two out of the three time points in lung, liver, or spleen. In the primary screen we received information for two different transposon insertions in 598 genes, and the results for these independent mutations showed agreement in 463 cases. In addition to labeling both sides of the transposon for all 11 pools (primary screen), labeling of only one side was repeated with two smaller pools containing 80 mutants and four controls (secondary screen). These mutants were identified as being lost in lung, liver, and/or spleen in the primary screen. The controls for the secondary screen were selected from the primary screen as clones present at all three time points in all three organs. Discordant results for one gene between labeling the two different sides of the transposon in the primary screen as well as the same side of the transposon between primary and secondary screens were not included in the candidate list of virulence genes.
As expected, most transposon mutants were present in both the deposition and output pools, but 125 genes (12%) were no longer detected in at least one of the three organs (Table 1). We observed transposon insertions in 44 genes (35%) lost in lung and in 81 (65%) genes lost in liver and/or spleen but present in lung (Table 1). When loss occurred in the in lung, most of the attenuated clones still could be detected at the 24-h time point and were able to disseminate to liver and/or spleen but then appeared to be lost in lung after 48 h and 72 h of infection. Even when the attenuated clones showed loss in lung at the 24-h time point, many still disseminated to liver and/or spleen, indicating that under the condition of high inocula used in our experiment, dissemination from the lung to other organs is quite rapid. Nine out of the 125 genes with a phenotype were lost in all three organs. These were tyrA (FTN_0055) and folD (FTN_0417), both involved in amino acid metabolism; the intracellular growth locus protein D gene (iglD); atpC (regulatory subunit of ATP synthase); pip (proline iminopeptidase); the thioredoxin gene (FTN_1415); two genes of unknown function (FTN_0400 and FTN_0430); and a gene for a hypothetical protein (FTN_1351).
TABLE 1.
List of genes showing a phenotype in lung, liver, and/or spleena
| FTN | Gene name | Protein descriptionb | Function class | Organ(s) showing loss |
|---|---|---|---|---|
| FTN_0008 | 10 TMS drug/metabolite exporter protein | Defense mechanisms | Spleen | |
| FTN_0012 | Hypothetical protein | Hypothetical, novel | Lung | |
| FTN_0023 | tmpT | Thiopurine S-methyltransferase | General function prediction only | Liver |
| FTN_0028 | Conserved hypothetical membrane protein | Function unknown | Spleen | |
| FTN_0031 | Transcriptional regulator, LysR family | Transcription | Spleen | |
| FTN_0045 | Protein of unknown function | Function unknown, novel | Liver | |
| FTN_0055 | tyrA | Prephenate dehydrogenase | Amino acid transport and metabolism | Lung, liver, spleen |
| FTN_0111 | ribH | Riboflavin synthase beta-chain | Coenzyme transport and metabolism | Lung |
| FTN_0132 | Protein of unknown function | Function unknown, novel | Spleen | |
| FTN_0133 | RNase II family protein | Transcription | Spleen | |
| FTN_0143 | Monovalent cation:proton antiporter | Inorganic ion transport and metabolism | Lung, spleen | |
| FTN_0169 | Conserved hypothetical membrane protein | Function unknown | Spleen | |
| FTN_0199 | cyoE | Heme O synthase | Posttranslational modification, protein turnover, chaperones | Liver |
| FTN_0205 | Conserved hypothetical protein | General function prediction only | Lung, liver | |
| FTN_0208 | Hypothetical membrane protein | Hypothetical, novel | Spleen | |
| FTN_0217 | l-Lactate dehydrogenase | Energy production and conversion | Spleen | |
| FTN_0280 | Hypothetical protein | Hypothetical, novel | Lung, spleen | |
| FTN_0281 | Protein of unknown function | Function unknown, novel | Spleen | |
| FTN_0292 | Protein of unknown function | Function unknown, novel | Spleen | |
| FTN_0297 | Conserved protein of unknown function | General function prediction only | Spleen | |
| FTN_0298 | gplX | Fructose 1,6-bisphosphatase II | Carbohydrate transport and metabolism | Lung, spleen |
| FTN_0299 | putP | Proline:Na+ symporter | Amino acid transport and metabolism | Liver |
| FTN_0302 | Hypothetical protein | Hypothetical, novel | Spleen | |
| FTN_0326 | Conserved hypothetical protein | Hypothetical, conserved | Lung | |
| FTN_0340 | Protein of unknown function | Function unknown, novel | Lung, spleen | |
| FTN_0344 | Aspartate:alanine exchanger family protein | General function prediction only | Spleen | |
| FTN_0347 | fkpB | FKBP-type peptidyl-prolyl cis-trans isomerase | Posttranslational modification, protein turnover, chaperones | Liver |
| FTN_0351 | Hypothetical protein | Hypothetical, novel | Liver | |
| FTN_0365 | Conserved hypothetical membrane protein | General function prediction only | Spleen | |
| FTN_0393 | Conserved protein of unknown function | Function unknown, conserved | Lung, spleen | |
| FTN_0395 | Transcriptional regulator, ArsR family | Transcription | Spleen | |
| FTN_0397 | gpsA | Glycerol-3-phosphate-dehydrogenase [NAD+] | Energy production and conversion | Spleen |
| FTN_0400 | Protein of unknown function | General function prediction only | Lung, liver, spleen | |
| FTN_0402 | aroC | Chorismate synthase | Amino acid transport and metabolism | Spleen |
| FTN_0417 | folD | Methylene tetrahydrofolate enzyme/cyclohydrolase/dehydrogenase | Coenzyme transport and metabolism | Lung, liver, spleen |
| FTN_0426 | Conserved protein of unknown function | Function unknown | Lung, liver | |
| FTN_0430 | Protein of unknown function | Function unknown, novel | Lung, liver, spleen | |
| FTN_0437 | Hydrolase, HD superfamily | General function prediction only | Lung, spleen | |
| FTN_0438 | rrmJ | 23S rRNA methylase | Translation, ribosomal structure, and biogenesis | Lung |
| FTN_0488 | cspC | Cold shock protein, DNA binding | Transcription | Spleen |
| FTN_0537 | Proton-dependent oligopeptide transporter family protein | Amino acid transport and metabolism | Liver, spleen | |
| FTN_0554 | RNA methyltransferase, SpoU family | Translation, ribosomal structure, and biogenesis | Liver | |
| FTN_0571 | Amino acid-polyamine-organocation superfamily protein | Amino acid transport and metabolism | Lung | |
| FTN_0578 | Major facilitator superfamily transport protein | Carbohydrate transport and metabolism | Liver | |
| FTN_0582 | gph | Phosphoglycolate phosphatase | General function prediction only | Liver |
| FTN_0609 | pnp | Polyribonucleotide nucleotidyltransferase | Translation, ribosomal structure, and biogenesis | Lung |
| FTN_0646 | cscK | ROK family protein | Transcription | Spleen |
| FTN_0678 | Drug:H+ antiporter-1 (DHA1) family protein | Carbohydrate transport and metabolism | Lung | |
| FTN_0689 | ppiC | Parvulin-like peptidyl-prolyl isomerase domain | Posttranslational modification, protein turnover, chaperones | Liver, spleen |
| FTN_0690 | deaD | DEAD-box subfamily ATP-dependent helicase | DNA replication, recombination, modification, and repair | Spleen |
| FTN_0710 | Type I restriction-modification system, subunit R (restriction) | DNA replication, recombination, modification, and repair | Liver | |
| FTN_0713 | ostA2 | Organic solvent tolerance protein OstA | Cell wall/membrane/envelope biogenesis | Lung, spleen |
| FTN_0720 | Transcriptional regulator, LclR family | Transcription | Spleen | |
| FTN_0737 | potI | ATP-binding cassette putrescine uptake system | Amino acid transport and metabolism | Liver, spleen |
| FTN_0740 | Protein of unknown function | Function unknown, novel | Spleen | |
| FTN_0768 | tspO | Tryptophan-rich sensory protein | Signal transduction and regulation | Spleen |
| FTN_0787 | rep | UvrD/REP superfamily I DNA and RNA helicases | DNA replication, recombination, modification, and repair | Lung, spleen |
| FTN_0810 | ROK family protein | Transcription | Spleen | |
| FTN_0824 | Major facilitator superfamily transport protein | Carbohydrate transport and metabolism | Liver, spleen | |
| FTN_0825 | Aldo/keto reductase family protein | Energy production and conversion | Liver, spleen | |
| FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) | General function prediction only | Lung, liver |
| FTN_0853 | sufD | SufS activator complex, SufD subunit | Posttranslational modification, protein turnover, chaperones | Spleen |
| FTN_0857 | Conserved hypothetical protein | Hypothetical, conserved | Spleen | |
| FTN_0873 | dcd | dCTP deaminase | Nucleotide transport and metabolism | Liver |
| FTN_0877 | cls | Cardiolipin synthetase | Lipid transport and metabolism | Spleen |
| FTN_0881 | Fe2+/Zn2+ uptake regulator protein | Transcription | Lung | |
| FTN_0918 | Conserved protein of unknown function | Function unknown, conserved | Spleen | |
| FTN_0933 | Protein of unknown function | Function unknown, novel | Lung | |
| FTN_0938 | Hypothetical protein | Hypothetical, novel | Spleen | |
| FTN_1006 | Transporter-associated protein, HlyC/CorC family | Inorganic ion transport and metabolism | Spleen | |
| FTN_1015 | Isochorismatase family protein | Secondary metabolite biosynthesis, transport, and catabolism | Spleen | |
| FTN_1016 | Conserved protein of unknown function | Function unknown, conserved | Spleen | |
| FTN_1029 | Conserved protein of unknown function | Function unknown, conserved | Lung, liver | |
| FTN_1030 | lipA | Lipoic acid synthetase | Coenzyme transport and metabolism | Spleen |
| FTN_1055 | lon | DNA-binding, ATP-dependent protease La | Posttranslational modification, protein turnover, chaperones | Liver |
| FTN_1063 | tRNA-methylthiotransferase MiaB protein | Translation, ribosomal structure and biogenesis | Lung | |
| FTN_1098 | Conserved hypothetical membrane protein | Hypothetical, conserved | Lung | |
| FTN_1107 | metIQ | Methionine uptake transporter family protein | Inorganic ion transport and metabolism | Lung, spleen |
| FTN_1133 | Protein of unknown function | Function unknown, novel | Lung, spleen | |
| FTN_1148 | Glycoprotease family protein | Posttranslational modification, protein turnover, chaperones | Spleen | |
| FTN_1157 | GTP-binding translational elongation factor Tu and G family protein | Signal transduction mechanisms | Liver | |
| FTN_1165 | Predicted ATPase of the PP-loop superfamily | Cell cycle control cell division | Spleen | |
| FTN_1177 | sbcB | Exodeoxyribonuclease I | DNA replication, recombination, modification, and repair | Lung, spleen |
| FTN_1196 | Conserved protein of unknown function | Function unknown, conserved | Liver, spleen | |
| FTN_1199 | Conserved protein of unknown function | Function unknown, conserved | Spleen | |
| FTN_1200 | capC | Capsule biosynthesis protein CapC | Cell wall/membrane/envelope biogenesis | Spleen |
| FTN_1209 | cphB | Cyanophycinase | Inorganic ion transport and metabolism | Lung, spleen |
| FTN_1218 | Glycosyltransferase, group 1 | Cell wall/membrane/envelope biogenesis | Liver, spleen | |
| FTN_1232 | Conserved hypothetical membrane protein | Function unknown | Spleen | |
| FTN_1243 | recO | RecFOR complex, RecO component | DNA replication, recombination, modification, and repair | Lung, spleen |
| FTN_1272 | Proton-dependent oligopeptide transporter family protein | Amino acid transport and metabolism | Spleen | |
| FTN_1292 | Solute:sodium symporter | Amino acid transport and metabolism | Spleen | |
| FTN_1298 | GTPase of unknown function | General function prediction only | Liver | |
| FTN_1312 | Conserved hypothetical protein | Hypothetical, conserved | Lung, liver | |
| FTN_1321 | iglD | Intracellular growth locus protein D | Function unknown, novel | Lung, liver, spleen |
| FTN_1349 | Hypothetical protein | Function unknown | Liver | |
| FTN_1351 | Conserved hypothetical protein | Function unknown | Lung, liver, spleen | |
| FTN_1355 | Regulatory factor, Bvg accessory factor family | Transcription | Liver | |
| FTN_1362 | Hypothetical protein | Hypothetical, novel | Lung | |
| FTN_1400 | S-Adenosylmethionin-dependent methyltransferase | General function prediction only | Spleen | |
| FTN_1415 | Thioredoxin | Posttranslational modification, protein turnover, chaperones | Lung, liver, spleen | |
| FTN_1439 | fadA | Acetyl coenzyme A acetyltransferase | Lipid transport and metabolism | Lung |
| FTN_1440 | Conserved hypothetical protein, thioesterase superfamily | Hypothetical, conserved | Spleen | |
| FTN_1455 | Conserved hypothetical protein | Function unknown | Liver | |
| FTN_1459 | Short chain dehydrogenase | General function prediction only | Lung | |
| FTN_1464 | lepB | Signal peptidase I | Intracellular trafficking and secretion | Spleen |
| FTN_1467 | proC | Pyrroline-5-carboxylate reductase | Amino acid transport and metabolism | Liver |
| FTN_1471 | pcs | (CDP-alcohol) phosphatidyltransferase | Lipid transport and metabolism | Spleen |
| FTN_1507 | Pilus assembly protein | Intracellular trafficking and secretion | Lung | |
| FTN_1534 | Conserved protein of unknown function | Function unknown, conserved | Liver | |
| FTN_1536 | Amino acid-polyamine-organocation superfamily protein | Amino acid transport and metabolism | Spleen | |
| FTN_1587 | Protein of unknown function | Function unknown, novel | Spleen | |
| FTN_1588 | Major facilitator superfamily transport protein | Carbohydrate transport and metabolism | Spleen | |
| FTN_1611 | Major facilitator superfamily transport protein | Carbohydrate transport and metabolism | Spleen | |
| FTN_1645 | atpC | ATP synthase, F1 sector, subunit epsilon | Energy production and metabolism | Lung, liver, spleen |
| FTN_1665 | Magnesium chelatase | Posttranslational modification, protein turnover, chaperones | Spleen | |
| FTN_1673 | nuoH | NADPH dehydrogenase I, H subunit | Energy production and conversion | Liver |
| FTN_1726 | Pyridoxal-dependent decarboxylase | Amino acid transport and metabolism | Liver | |
| FTN_1731 | pip | Proline iminopeptidase | General function prediction only | Lung, liver, spleen |
| FTN_1753 | Rieske (2Fe-2S) domain protein | Inorganic ion transport and metabolism | Spleen | |
| FTN_1756 | bcp | Bacterioferritin comigratory protein | Posttranslational modification, protein turnover, chaperones | Liver |
| FTN_1760 | Zinc-binding alcohol dehydrogenase | Energy production and conversion | Liver, spleen | |
| FTN_1778 | trpE | Anthranilate synthase component I | Amino acid transport and metabolism | Lung, liver |
Boldface indicates genes that were lost in all three organs.
TMS, transmembrane alpha helical spanner; FKBP, FK506 binding protein; ROK, repressors, open reading frames, and kinases.
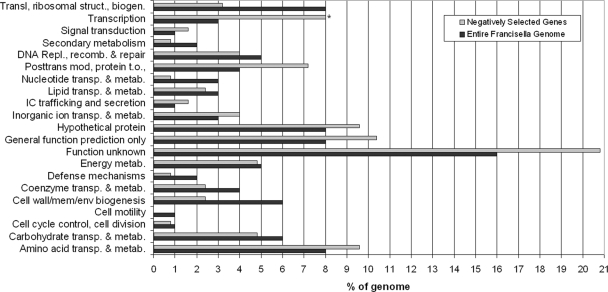
Candidate virulence genes are enriched for unknown/hypothetical genes and genes involved in transcription.
The genes we identified as candidate virulence factors in our screens fell into a variety of functional categories, with about one-third annotated as having unknown function or hypothetical. Genes in this category often (50%) were conserved in other sequenced organisms. Several categories, such as signal transduction mechanisms, posttranslational modification/protein turnover/chaperones, genes involved in amino acid metabolism/transport, and genes of unknown function, appeared to be enriched in our group of 125 candidate virulence genes compared to their representation in the entire Francisella genome (Fig. 3). Only genes involved in transcription showed statistically significant enrichment (P = 0.0017) as calculated by the chi-square test after Bonferroni correction for multiple comparisons. Our screen queried transposon mutants in 44 of these genes, and we obtained usable data for 32 transcription genes. One-third of the 32 identified transcription genes (10 genes) are included in our group of 125 candidate virulence genes. These include putative transcription factor genes FTN_0031 (LysR family), FTN_0395 (ArsR family), FTN_0720 (IclR family), and FTN_0646 (ROK family); a cold shock regulator gene (cspA); and FTN_1355, which shows homology to the Bvg accessory factor family that plays an important role in virulence of Bordetella (11, 41, 46). Additional mutants in this category were rpoC, FTN_0810, a RNase II family protein gene (FTN_0133), and a putative Fe2+/Zn2+ uptake regulator protein gene (FTN_0881).
FIG. 3.
Enrichment of functional categories in aerosol infection mutants. The percentage of genes in each category relative to genes with an infection phenotype (gray) or in the Francisella genome (black) is shown. *, significant enrichment (P = 0.0017 by chi-square test with Bonferroni correction for multiple comparisons). Abbreviations: IC, intracellular; trans, translation; mod, modification; t.o., turnover; sec, secondary; metab, metabolism; biosynth, biosynthesis; catab, catabolism.
Genes lost in output pools of all three organs show pronounced attenuation during single-strain infection.
We further investigated eight candidate virulence genes with a range of behaviors in our pooled mutant screens, as shown in Fig. 4. Some showed strong, consistent loss in lung and/or organs to which bacteria disseminated (liver/spleen), while others showed variable phenotypes in the two screens. These eight mutants (Table 2) were singly infected into mice by the aerosol route with a targeted deposition of 100 CFU/lung. The actual depositions ranged from 21 to 19,489 CFU/lung after aerosol inoculation. Previous work showed that mice infected with 5 to 10,000 CFU/lung via exposure to aerosolized wild-type Francisella novicida died within 4 to 5 days postinfection (S. J. Skerrett, unpublished data). Therefore, we expected mice infected with mutants with no attenuation to have a mean survival time of approximately 4 days that would not depend greatly on the actual inoculating dose.
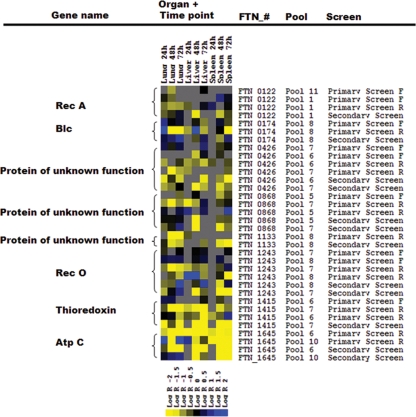
FIG. 4.
Heat map of genes validated by single-strain infections in vivo. Results [log2 (output/input)] from microarray experiments for eight mutants tested in vivo in separate infections for the primary screen (labeling both the forward and reverse sides of the transposon) and the secondary screen (labeling only the reverse side of the transposon) are shown. Results from transposon mutants included in different pools are shown separately. Color code for heat map: gray, no data obtained; blue, overrepresentation of output compared to input signal, log R > 1; black, same signal for output and input, log R = 0 (mutant present in output and input pools); yellow, overrepresentation of input compared to output signal, log R < −1 (negatively selected, mutant lost in output pool).
TABLE 2.
Survival of mice infected with candidate mutants in single-strain infections
| Locus mutated | Dose (CFU)a | Mean survival (days)b |
|---|---|---|
| None (wild-type strain U112) | 142 | 3.9 |
| FTN_0122 (recA) | 21 | 5.0 |
| FTN_0174 (blc) | 187 | 4.6 |
| FTN_0426 | 19,489 | 4.0 |
| FTN_0868 | 127 | 4.7 |
| FTN_1133 | 12,378 | 4.3 |
| FTN_1243 (recO) | 239 | 5.0 |
| FTN_1415 (thioredoxin, low) | 23 | 8.3 |
| FTN_1415 (thioredoxin, high) | 91 | 5.0 |
| FTN_1645 (atpC) | 149 | 30c |
Dose measured as deposition obtained from homogenized lungs of two mice immediately after exposure.
Survival experiments utilized four mice per strain, except for FTN_0174 and FTN_0868, for which eight mice were used.
The experiment ended at 30 days.
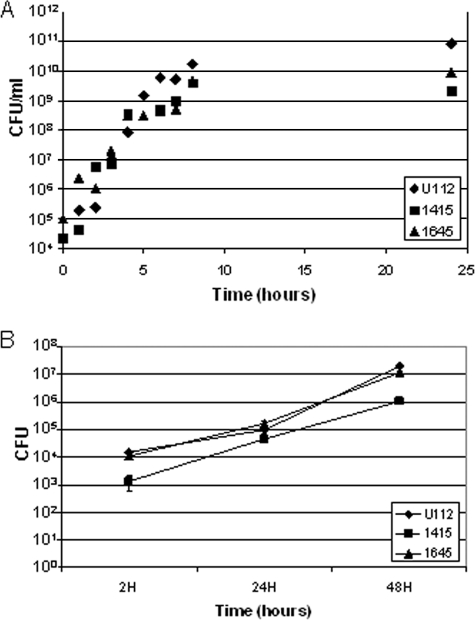
A putative ATP synthase epsilon subunit homolog (FTN_1645, atpC) mutant strain showed the most severe attenuation. Mice infected with this mutant showed no signs of illness and uniformly survived the infection (Table 2). The array results showed clear loss in the output pools for all three time points in all three organs in both the primary and secondary screens. Mice infected with a mutant with a thioredoxin domain-containing protein mutation (FTN_1415) showed no signs of sickness until day 7 but had to be sacrificed on each of the consecutive days, with a mean survival of 8.25 days across four mice. We repeated that infection, since the deposition of 23 CFU/lung was at the lower end of the dose range. The replicate infection with FTN_1415 resulted in a mean survival of 5 days, and the deposition of 91 CFU/lung was closer to the target of 100 CFU/lung. The dose dependence on the mean survival time observed for the thioredoxin mutant has not been observed for similar wild-type infections. The array profile of FTN_1415 showed loss in the output pool in lung and liver at at least two time points for the primary screen. The secondary screen confirmed these results but showed additional loss in spleen at the two tested time points. The avirulence phenotype of FTN_1645 and the attenuation of FTN_1415 were not due to in vitro growth defects in TSB compared with the wild-type strain (Fig. 5A). In addition, similar numbers of bacteria were recovered from cultured alveolar macrophages (MH-S cell line) after infection with both the attenuated mutants and the wild-type Francisella strain (Fig. 5B).
FIG. 5.
Severely attenuated mutants show normal growth in broth (TSB-C) (A) and a macrophage cell line (MH-S) (B). (A) Growth in broth of FTN_1415 (▪) and FTN_1645 (▴) in comparison to wild type (U112, ♦). (B) Growth in MH-S mouse alveolar macrophages of FTN_1415 and FTN_1645 compared to wild type. Results are representative of three independent experiments; error bars are standard errors of the means.
We examined the behavior of three candidate virulence genes from our screens with unknown function based on sequence analysis: FTN_0426, FTN_0868, and FTN_1133. The depositions for the FTN_0426 and FTN_1133 mutants were 19,489 and 12,378 CFU/lung, at the high end of the dose range, whereas the FTN_0868 mutant deposition of 127 CFU/lung was closer to the target dose of 100 CFU. FTN_0426 was assembled in two different pools and showed loss in the output pool for only one organ, either lung or liver, in the primary and secondary screens, respectively. Upon single-strain infection, the FTN_0426 mutant showed no attenuation, with the same survival times for infected mice as those with wild-type infection. In contrast, the FTN_1133 and FTN_0868 mutants were both lost in two organs (lung and liver or spleen) in either the primary or secondary screen but showed survival in the other screen. Both mutants were classified as very slightly attenuated, giving mean survival times of 4.3 and 4.7 days, respectively. The same survival times (mean of 4.6 days) and slightly attenuated phenotype was also found for FTN_0174 (blc), encoding a putative outer membrane protein of unknown function which, showed loss in liver during the primary screen only.
We also investigated the contribution to infection of genes involved in DNA repair and homologous recombination during infection. This functional category was enriched in the virulence gene candidate list compared to the whole genome in a preliminary analysis, and DNA repair proteins seem to promote infection and disease caused by Salmonella enterica serovar Typhimurium, another bacterial pathogen that resides within macrophages (4, 36). The mean survival times of 5 days for infection with recA (FTN_0122) and recO (FTN_1243) mutants were slightly higher than those for the previously described mutants and 1 day longer than those for infections with the wild type. The array data for recA showed loss in lung for the primary but not the secondary screen, whereas recO was lost in lung only for the primary screen and in spleen only for the secondary screen.
Comparison of the mean survival times for the mutant clones during single-strain infection (Table 2) with the extent of clone loss observed by microarray during the mutant pool infections (Fig. 4) suggests a relationship between the extent of loss and virulence of the mutant strain. Loss in both the primary and secondary screens in lung and at least one dissemination organ (liver or spleen) for two or three time points was predictive of virulence attenuation as measured by increased mean survival of mice during single-strain aerosol infection. Mutant clones with variable results in different screens or among different organs showed detectable but much milder attenuation.
Bacterial replication of attenuated mutants in mice.
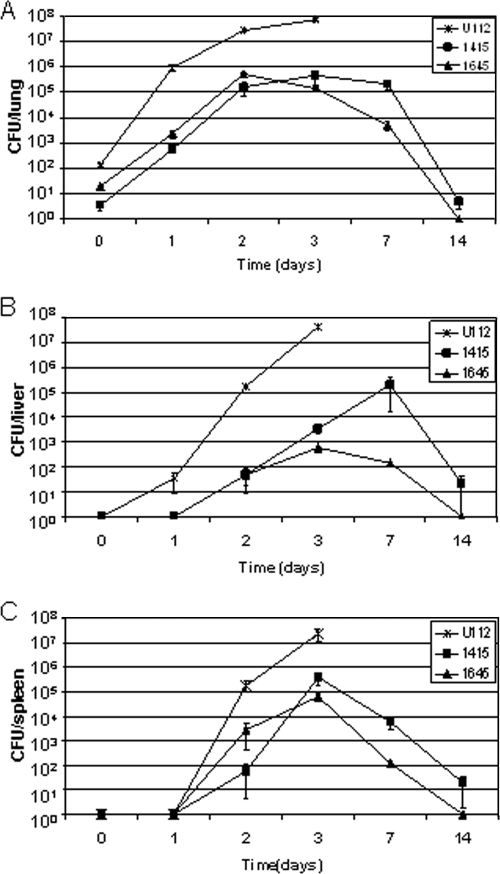
To determine the kinetics of bacterial replication and spread following infection, C57BL/6 mice were infected by aerosolization with the wild type or the two most attenuated mutants (atpC [FTN_1645] and thioredoxin domain-containing protein [FTN_1415]). Lungs, livers, and spleens were harvested at 1, 2, 4, 7, and 14 days postinfection and plated to determine individual organ burdens (Fig. 6). Mutant bacteria were able to efficiently replicate in the lung in vivo despite a lower-than-targeted deposition (atpC, 19 CFU/lung; FTN_1415, 4 CFU/lung; wild type, 142 CFU/lung). The mutants achieved bacterial loads of 103 and 105 CFU in the lungs at the 24-h and 48-h time points, while wild-type bacterial loads were 2 log units higher (105 and107) for the same time points and mice maintained the same organ burden for 1 more day before they died. After the 48-h time point, the atpC mutant began to be slowly cleared from the lungs, and it could not be detected at 14 days. In contrast, the thioredoxin mutant maintained the lower load in the lungs at 72 h and 7 days, while at 14 days the load had decreased, indicating that it was now being cleared. Clinically, three out of the four mice showed no signs of infection until they were sacrificed at day 14, while one FTN_1415-infected mouse was moribund at day 11 and had to be sacrificed. In comparison, two previous infections examining survival over a 30-day time period with FTN_1415 and higher depositions of 23 and 91 CFU/lung showed mean survival times of 8.25 and 5 days, indicating a dose-dependent effect on survival for FTN_1415.
FIG. 6.
Bacterial burdens for most strongly attenuated mutants and the wild type in lung (A), liver (B), and spleen (C). C57BL/6 mice were infected by aerosol with the wild type (142 CFU), FTN_1415 (4 CFU), and FTN_1645 (9 CFU) in individual infections, and four mice were sacrificed at each time point. The data presented are the mean bacterial loads, and error bars show standard errors of the means. All mice infected with the wild type died at day 3. One mouse infected with FTN_1415 mutant died at day 11.
Despite lower-than-wild-type bacterial loads in the lungs, both mutants were able to disseminate to the spleen and liver. In spleen, both mutants replicated for the first 72 h postinfection to approximately 105 CFU/spleen, which is about 2 log units less than wild-type levels for that time point. After 72 h the atpC mutant began to be cleared, and it was not detected at 14 days. The thioredoxin mutant behaved similarly, except at 14 days bacteria could still be detected (19 CFU/lung). In liver the atpC mutant showed a profile similar to that in spleen, with a peak bacterial load 4 log units lower than that of the wild type at 72 h, fewer but detectable bacteria at 7 days, and no bacteria detectable at 14 days. The FTN_1415 mutant showed better replication in the liver; it increased to a peak of 105 CFU/liver at 7 days and then decreased to 22 CFU/liver at 14 days. These data indicate that mice infected with the atpC mutant reach, except for in liver, the same peak organ burden as mice infected with the less attenuated FTN_1415-mutant but are able to clear the infection faster and entirely. Compared to the wild type, the peak organ burden for FTN_1645 was 2 log units lower in lung and spleen, whereas liver counts were 4 log units lower. Thus, mice infected with the attenuated mutant clones showed dissemination to spleen and liver, similar to that for infection with wild-type bacteria, but controlled mutant bacterial replication and were able to clear the mutant clones from all three organs.
Protection against lethal challenge with wild-type F. novicida.
To determine if mice that survived initial aerosol infection were protected from subsequent challenge with wild-type Francisella, the surviving atpC mutant-infected mice were challenged with a lethal dose of wild-type Francisella (25 CFU/lung) at 30 days after the initial infection. Four mice that were initially infected with the atpC mutant acquired complete protective immunity and survived without signs of illness for a 30-day period. As a control, naïve C57BL/6 mice were infected with wild-type bacteria at the same dose, and all died at day 4 postinfection. The bacterial burden in the surviving wild-type-challenged and initially atpC mutant-infected mice was investigated after 30 days, and this showed no detectable bacteria in all three organs.
DISCUSSION
This is the first whole-genome screen in Francisella that has used an inhalation model and, in addition to looking at colonization in the organ of infection, also looked at dissemination to liver and spleen. This is particularly important since inhalation and skin contact are the two most common modes of human F. tularensis infection, and aerosolized F. tularensis is considered a prime candidate for use as a biological warfare agent (8, 25). Other studies have shown that mice challenged by aerosol or intradermally with low doses of virulent type A and type B strains had extensive and severe histological changes in liver and spleen but much more limited changes in lungs, even in mice challenged by aerosol (7). Thus, it appears that regardless of the route of infection, systemic rather than pulmonary infection was the likely cause of death following low-dose challenge with virulent F. tularensis (7). Previous screens either have used intranasal infection and measured colonization in lung with Francisella tularensis subsp. holartica LVS strain (40) or have used intraperitoneal (i.p.) infection and evaluated colonization in spleen with F. novicida (43). In contrast, our study used aerosol infection and evaluated colonization in the lung and systemic dissemination to liver and spleen with F. novicida.
As expected, our screen identified a number of genes annotated to function in basic metabolism and physiology (amino acid, carbohydrate, lipid, and nucleotide transport and metabolism) to promote colonization. Another expected class of genes was those associated with bacterial cell surface structures (cell wall/lipopolysaccharide/capsule). FTN_1218, a glycosyltransferase gene, showed a phenotype in spleen and liver and is involved in various biosynthetic processes, including lipopolysaccharide core biosynthesis. A previous study has shown that modifications of lipid A, the biologically active component of gram-negative bacterial lipopolysaccharide, can alter recognition by the host innate immune system, leading to attenuation in virulence (18). We also identified two genes of the capBAC locus, capA (FTN_1199) and capC (FTN_1200), as being lost in spleen. This locus was characterized in the LVS screen (40), where capBCA were shown to be essential for in vivo survival during intranasal infection of mice, and it also showed a phenotype in the F. novicida i.p. infection screen (43).
The only functional category significantly enriched in the candidate virulence genes identified in our screen was genes involved in transcription (Fig. 3). Genes in this category likely regulate functions important for intracellular growth and survival of the bacterium. For example, the Escherichia coli homologue of CspC upregulates the expression of rpoS, a gene that encodes a global stress response regulator. Recent work found CspC among a number of stress response proteins regulated by MglA, a previously characterized transcriptional regulator of Francisella tularensis virulence (15).
Several more of our identified candidate virulence genes overlapped with those described in previous screens (40, 43). Weiss et al. identified mutants in 164 F. novicida genes (∼9% of the genome, versus ∼7% in our screen) that were absent in the spleen after i.p. infection (43). The overlap with our screen included 13 genes, of which 5 have no functional annotation. For 11 of the 13 overlapping genes, including the 5 of unknown function, we confirmed loss in spleen, though we used an aerosol rather than i.p. route of infection. In fact, three genes (FTN_0430, FTN_0417, and iglD [FTN_1321]) were lost for all three organs in the primary screen, and loss could be confirmed for two genes in a subset of the organs in the secondary screen. IglD is part of the pathogenicity island, and its role in virulence has been described previously (3, 14, 24, 26, 32). Another gene identified in both screens was FTN_0720, a homologue of the IclR family of transcriptional regulators. We observed loss of this mutant in spleen after aerosol infection, while Weiss et al. observed attenuation during subcutaneous. and i.p. infections (43). Thus, FTN_0720 appears to be required for full virulence regardless of route of inoculation. Similar results were observed for FTN_1471 (pcs), a gene involved in fatty acid and lipid metabolism. For 2 out of the 13 genes overlapping between our screen and that of Weiss et al., a conserved hypothetical gene (FTN_1312) and xasA, an acid/polyamine/choline superfamily protein involved in amino acid transport (FTN_0571), we did not confirm loss in the output pool for spleen but did see loss in lung and/or liver. As the role in virulence was not subsequently validated for either of these genes in the previous studies, they may represent false positives. Alternatively, mutation of these genes may result in a partial virulence phenotype that depends on inoculation size or site, which differed from those in our study.
The Francisella tularensis subsp. holartica LVS intranasal infection screen identified 95 mutants attenuated for colonization of the lung (40). Three of the six genes overlapping with our screen (lon, encoding the ATP-dependent protease La that also binds strongly to DNA [FTN_1055], capC [described above], and a gene for a conserved protein of unknown function [FTN_1199]) showed a phenotype in our screen. We did not receive data for the lon gene in lung, but we found capC and FTN_1199 mutants present in the output of lung at all time points. In contrast, all three mutants were lost in liver or spleen in our screen, suggesting that these genes are involved in dissemination or survival in distal organ sites if not absolutely required in the lung under the high-dose conditions used in our screen. Our results for the remaining three genes showed inconsistent loss in lung and spleen or in spleen only.
All three screens investigated the genes within the Francisella pathogenicity island. Weiss et al. found loss for all mutants with mutations mapping to genes within the island (43), whereas Su et al. (40) observed loss only for mutants in the intracellular growth locus (iglA, iglB, and iglC). All genes of the Francisella pathogenicity island were represented in the 11 deposition pools of our screen, but we received results in the output pool for only a subset of genes. FTN_1321 (iglD) and FTN_1312 showed loss in either lung and liver or all three organs. All the other genes with data (FTN_1311, FTN_1314, FTN_1320, FTN_1324 [iglA], FTN_1325 [pdpD], and FTN_1326) appeared to be present, including in spleen.
The small overlap of only 13 genes with the results from Weiss et al. and 6 genes with those from Su et al. indicate that route of exposure or differences in the complexity of the pools used for infection may influence outcomes in the primary and disseminating organs. Alternatively, this may reflect the presence of redundant pathways contributing to survival in the host such that inactivation of any one pathway gives only partial or variable attenuation of infection. A closer examination of our results suggests that the lack of a phenotype for these and other genes in our screen results from the stringent cutoff that we used for inclusion as candidate virulence genes. A less stringent cutoff that does not exclude inconsistencies between the primary and secondary screens would have doubled the number of genes overlapping with the results from Weiss et al. to 26 genes. Since we pooled outputs from four animals in our experiment, a clone lost in a subset of animals could still give a robust signal in the output pool. While we sought to identify the most severely attenuated mutants, study of genes with partial phenotypes could yield important information regarding mechanisms of pathogenesis.
We chose eight genes with a range of behaviors in the infection pools and from several different functional categories to be validated by single-strain infections. In general, mutants showing inconsistent phenotypes across organs or screens (Fig. 4) showed little or no phenotype when tested individually for the ability to induce disease and death (Table 2). In contrast, two genes, those for a putative ATP synthase epsilon subunit homolog (atpC, FTN_1645) and a thioredoxin (FTN_1415), showed consistent loss in lung and/or dissemination organs (liver/spleen) in multiple screens (Fig. 4) and either complete or much longer mean survival of infected mice during single-strain infections (Table 2). Thus, the degree of virulence attenuation during single-strain infection correlated with observed behavior during complex pool infections measured by microarrays.
The mutation in the most severely attenuated mutant confirmed in our screen mapped to the epsilon subunit of FoF1-ATP synthase. FoF1-ATP synthases are membrane-bound enzymes catalyzing synthesis of ATP from ADP and inorganic phosphate using the energy of transmembrane electrochemical potential difference. The enzyme can also work as an ATPase, hydrolyzing ATP to generate an electrochemical gradient of protons (2, 37, 48). The enzyme consist of two main subcomplexes: F1 contains five subunits (α, β, γ, δ, and ɛ), has the catalytic sites of the enzyme, and is cytosolic, whereas Fo consists of three subunits (a, b, and c), is embedded in the membrane, and translocates protons across the membrane. In F. novicida, all the ATPase genes are arranged in a single operon (31). This enzyme is generally essential, and transposon insertions were recovered only in the epsilon subunit during construction of the transposon mutant library (12). The epsilon subunit is thought to be a regulatory subunit (10) which lies at the interface of F1 and Fo (21) and acts as an inhibitor of ATP hydrolysis activity in F1 alone, as well as in the entire F1Fo enzyme (19, 38). The mutant that we evaluated had a transposon insertion in the C-terminal domain of the epsilon subunit. Several studies have shown that the C-terminal domain in not absolutely required for oxidative phosphorylation but is necessary for the inhibition of ATPase activity (20, 47). This transposon mutant was able to grow on succinate, suggesting that oxidative phosphorylation is indeed intact (M. Enstrom and C. Manoil, unpublished observations). Thus, ATPase inhibition activity appears to be required for Francisella virulence.
The protection afforded by primary infection with the atpC mutant against secondary airborne challenge with wild-type Francisella is not a universal result with attenuated mutants: previous reports have shown that infection with an iglC mutant is protective against nasal challenge (28), whereas infection with an mglA mutant was not protective (44). In contrast, lipid A mutant strains provided protection against lethal wild-type Francisella infection via both pulmonary and subcutaneous routes of infection (18), and some purine mutants elicited protective immunity in a systemic challenge model (30). These observations suggest that the capacity of the atpC mutant to cause systemic infection was important to the development of protective immunity.
In addition to the avirulent phenotype of an atpC gene (FTN_1645) transposon mutant, a transposon insertion in FTN_1415, encoding a thioredoxin, showed a dose-dependent survival that was significantly higher than that for all the other mutants tested in single-strain infections. Thioredoxins are often regulated by external factors and fulfill a number of different important cellular functions in all living organisms, including DNA synthesis and protein repair by acting as hydrogen donor for peroxidases (17). In E. coli, proteomic analysis revealed many thioredoxin-targeted proteins involved in at least 26 distinct cellular processes that include cell division, transcriptional regulation, energy transduction, protein folding and degradation, and biosynthetic pathways (22). In addition, thioredoxins play an important role in the oxidative stress response by functioning as a scavenger for reactive oxygen species. Since Francisella tularensis resides in mononuclear phagocytes, thioredoxin's role in scavenging free radicals produced by mononuclear phagocytes upon infection could be a mechanism for resistance to intracellular killing. Interestingly, this thioredoxin mutant was able to grow in cultured macrophages, though the macrophages were not activated. A recently published proteomics study (15) found that MglA, a transcriptional regulator of Francisella tularensis' virulence, regulates many genes involved in responses to starvation and oxidative stress, including those for thioredoxin (FTN_1415) glutathione synthetase (FTN_0804), peroxiredoxins (FTN_0958 and FTN_0973), peroxidase (FTN0633), NADPH-quinone reductase (FTN_0840), and glutaredoxins (FTN_1033 and FTN_0982). All of these proteins have redundant functions that participate in resistance to reactive oxygen species. For a subset of these genes we received results in our screen. In addition to thioredoxin mutants, we also found NADPH-quinone reductase mutants lost in lung and liver, whereas the peroxiredoxin (FTN_0973) mutants and the peroxidase mutants did not show a phenotype in the screen. Thus, thioredoxin may be a dominant defense against oxidative damage during infection.
One of the targets of reactive oxygen species in the bacterial cell is DNA, and in another intracellular pathogen of macrophages, Salmonella enterica serovar Typhimurium, recombination-based DNA repair proteins promote survival in the host (4, 36). Among the mutants we tested that showed a slight increase in mean survival during single-strain infections were recA and recO mutants, which showed up to 1 day longer survival than the wild type. This nearly wild-type behavior of the recombination-base DNA repair homologue mutants could suggest that Francisella does not experience a significant amount of DNA damage during infection. This prediction is consistent with the relative absence of inflammation even in the context of high bacterial loads in the lung (34, 35, 45). Alternatively, Francisella has mechanisms, which may include upregulation of thioredoxin, to neutralize reactive oxygen and nitrogen species that it does encounter.
The two attenutated/avirulent mutants discussed above (thioredoxin and atpC) are part of a group of nine genes (Table 1) that were all lost in all three organs. In particular, the partial loss-of-function mutation in atpC was both avirulent and provided protection from subsequent challenge with wild-type bacteria. Study of additional genes identified by our screen, particularly those genes lost in the output pools of all three organs, may lead to further advances in understanding Francisella virulence mechanisms and identify additional vaccine candidates.
Acknowledgments
The project described here was supported by grant U54 AI057141 from the NIH.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
We thank the FHCRC Genomics Shared Resources for experimental support.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 27 October 2008.
REFERENCES
- 1.Baldwin, D. N., B. Shepherd, P. Kraemer, M. K. Hall, L. K. Sycuro, D. M. Pinto-Santini, and N. R. Salama. 2007. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect. Immun. 751005-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, P. D. 1997. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66717-749. [DOI] [PubMed] [Google Scholar]
- 3.Brotcke, A., D. S. Weiss, C. C. Kim, P. Chain, S. Malfatti, E. Garcia, and D. M. Monack. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 746642-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., C. J. Lipps, M. Y. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7933-936. [DOI] [PubMed] [Google Scholar]
- 5.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 735892-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlan, J. W., W. Chen, H. Shen, A. Webb, and R. KuoLee. 2003. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb. Pathog. 34239-248. [DOI] [PubMed] [Google Scholar]
- 8.Darling, R. G., C. L. Catlett, K. D. Huebner, and D. G. Jarrett. 2002. Threats in bioterrorism. I. CDC category A agents. Emerg. Med. Clin. N. Am. 20273-309. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feniouk, B. A., and M. Yoshida. 2008. Regulatory mechanisms of proton-translocating F(O)F(1)-ATP synthase. Results Probl. Cell Differ. 45279-308. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs, T. M., H. Deppisch, V. Scarlato, and R. Gross. 1996. A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J. Bacteriol. 1784445-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher, L. A., E. Ramage, M. A. Jacobs, R. Kaul, M. Brittnacher, and C. Manoil. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. USA 1041009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 715940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 21553-56. [DOI] [PubMed] [Google Scholar]
- 15.Guina, T., D. Radulovic, A. J. Bahrami, D. L. Bolton, L. Rohmer, K. A. Jones-Isaac, J. Chen, L. A. Gallagher, B. Gallis, S. Ryu, G. K. Taylor, M. J. Brittnacher, C. Manoil, and D. R. Goodlett. 2007. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. J. Bacteriol. 1896580-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajjar, A. M., M. D. Harvey, S. A. Shaffer, D. R. Goodlett, A. Sjostedt, H. Edebro, M. Forsman, M. Bystrom, M. Pelletier, C. B. Wilson, S. I. Miller, S. J. Skerrett, and R. K. Ernst. 2006. Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect. Immun. 746730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong, W., M. K. Cha, and I. H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 2752924-2930. [DOI] [PubMed] [Google Scholar]
- 18.Kanistanon, D., A. M. Hajjar, M. R. Pelletier, L. A. Gallagher, T. Kalhorn, S. A. Shaffer, D. R. Goodlett, L. Rohmer, M. J. Brittnacher, S. J. Skerrett, and R. K. Ernst. 2008. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 4e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, Y., T. Matsui, N. Tanaka, E. Muneyuki, T. Hisabori, and M. Yoshida. 1997. Thermophilic F1-ATPase is activated without dissociation of an endogenous inhibitor, epsilon subunit. J. Biol. Chem. 27224906-24912. [DOI] [PubMed] [Google Scholar]
- 20.Kato-Yamada, Y., D. Bald, M. Koike, K. Motohashi, T. Hisabori, and M. Yoshida. 1999. Epsilon subunit, an endogenous inhibitor of bacterial F(1)-ATPase, also inhibits F(0)F(1)-ATPase. J. Biol. Chem. 27433991-33994. [DOI] [PubMed] [Google Scholar]
- 21.Keis, S., A. Stocker, P. Dimroth, and G. M. Cook. 2006. Inhibition of ATP hydrolysis by thermoalkaliphilic F1Fo-ATP synthase is controlled by the C terminus of the epsilon subunit. J. Bacteriol. 1883796-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, J. K., S. Tabor, and C. C. Richardson. 2004. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 1013759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37153-159. [DOI] [PubMed] [Google Scholar]
- 24.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLendon, M. K., M. A. Apicella, and L. A. Allen. 2006. Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annual Rev. Microbiol. 60167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 1866430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. 2967-978. [DOI] [PubMed] [Google Scholar]
- 28.Pammit, M. A., E. K. Raulie, C. M. Lauriano, K. E. Klose, and B. P. Arulanandam. 2006. Intranasal vaccination with a defined attenuated Francisella novicida strain induces gamma interferon-dependent antibody-mediated protection against tularemia. Infect. Immun. 742063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrosino, J. F., Q. Xiang, S. E. Karpathy, H. Jiang, S. Yerrapragada, Y. Liu, J. Gioia, L. Hemphill, A. Gonzalez, T. M. Raghavan, A. Uzman, G. E. Fox, S. Highlander, M. Reichard, R. J. Morton, K. D. Clinkenbeard, and G. M. Weinstock. 2006. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J. Bacteriol. 1886977-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quarry, J. E., K. E. Isherwood, S. L. Michell, H. Diaper, R. W. Titball, and P. C. Oyston. 2007. A Francisella tularensis subspecies novicida purF mutant, but not a purA mutant, induces protective immunity to tularemia in mice. Vaccine 252011-2018. [DOI] [PubMed] [Google Scholar]
- 31.Santana, M., M. S. Ionescu, A. Vertes, R. Longin, F. Kunst, A. Danchin, and P. Glaser. 1994. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 1766802-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santic, M., M. Molmeret, J. R. Barker, K. E. Klose, A. Dekanic, M. Doric, and Y. Abu Kwaik. 2007. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell. Microbiol. 92391-2403. [DOI] [PubMed] [Google Scholar]
- 33.Santic, M., M. Molmeret, K. E. Klose, and Y. Abu Kwaik. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 1437-44. [DOI] [PubMed] [Google Scholar]
- 34.Saslaw, S., H. T. Eigelsbach, J. A. Prior, H. E. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107702-714. [DOI] [PubMed] [Google Scholar]
- 35.Saslaw, S., H. T. Eigelsbach, H. E. Wilson, J. A. Prior, and S. Carhart. 1961. Tularemia vaccine study. I. Intracutaneous challenge. Arch. Intern. Med. 107689-701. [DOI] [PubMed] [Google Scholar]
- 36.Schapiro, J. M., S. J. Libby, and F. C. Fang. 2003. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc. Natl. Acad. Sci. USA 1008496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senior, A. E. 1988. ATP synthesis by oxidative phosphorylation. Physiol. Rev. 68177-231. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. B., and P. C. Sternweis. 1977. Purification of membrane attachment and inhibitory subunits of the proton translocating adenosine triphosphatase from Escherichia coli. Biochemistry 16306-311. [DOI] [PubMed] [Google Scholar]
- 39.Stewart, G. R., J. Patel, B. D. Robertson, A. Rae, and D. B. Young. 2005. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 1269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su, J., J. Yang, D. Zhao, T. H. Kawula, J. A. Banas, and J. R. Zhang. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 753089-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sukumar, N., M. Mishra, G. P. Sloan, T. Ogi, and R. Deora. 2007. Differential Bvg phase-dependent regulation and combinatorial role in pathogenesis of two Bordetella paralogs, BipA and BcfA. J. Bacteriol. 1893695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telepnev, M., I. Golovliov, T. Grundstrom, A. Tarnvik, and A. Sjostedt. 2003. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 541-51. [DOI] [PubMed] [Google Scholar]
- 43.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA 1046037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, T. E., M. R. Pelletier, M. C. Majure, A. Lembo, A. M. Hajjar, and S. J. Skerrett. 2008. Inhalation of Francisella novicida ΔmglA causes replicative infection that elicits innate and adaptive responses but is not protective against invasive pneumonic tularemia. Microbes Infect. 10773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, J. D., J. R. Rooney, P. A. Prickett, E. B. Derrenbacher, C. W. Beard, and W. R. Griffith. 1964. Pathogenesis of experimental respiratory tularemia in monkeys. J. Infect. Dis. 114277-283. [DOI] [PubMed] [Google Scholar]
- 46.Wood, G. E., and R. L. Friedman. 2000. The Bvg accessory factor (Baf) enhances pertussis toxin expression in Escherichia coli and is essential for Bordetella pertussis viability. FEMS Microbiol. Lett. 19325-30. [DOI] [PubMed] [Google Scholar]
- 47.Xiong, H., D. Zhang, and S. B. Vik. 1998. Subunit epsilon of the Escherichia coli ATP synthase: novel insights into structure and function by analysis of thirteen mutant forms. Biochemistry 3716423-16429. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, M., E. Muneyuki, and T. Hisabori. 2001. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2669-677. [DOI] [PubMed] [Google Scholar]