Abstract
Many proteins that function in the transcription, maturation, and export of metazoan mRNAs are concentrated in nuclear speckle domains, indicating that the compartment is important for gene expression. Here, we show that the NS1 protein of influenza B virus (B/NS1) accumulates in nuclear speckles and causes rounding and morphological changes of the domains, indicating a disturbance in their normal functions. This property was located within the N-terminal 90 amino acids of the B/NS1 protein and was shown to be independent of any other viral gene product. Within this protein domain, we identified a monopartite importin α binding nuclear localization signal. Reverse-genetic analysis of this motif indicated that nuclear import and speckle association of the B/NS1 protein are required for the full replication capacity of the virus. In the late phase of virus infection, the B/NS1 protein relocated to the cytoplasm, which occurred in a CRM1-independent manner. The interaction of the B/NS1 protein with nuclear speckles may reflect a recruitment function to promote viral-gene expression. To our knowledge, this is the first functional description of a speckle-associated protein that is encoded by a negative-strand RNA virus.
The nucleus of a vertebrate cell is highly organized in nonmembranous domains that exert distinct biochemical activities involved in gene expression (39). This partition gives rise to discrete structures, such as nuclear speckles, nucleoli, Cajal bodies, and promyelocytic leukemia protein (PML) bodies, which can be visualized by staining for antigens accumulating in these nuclear domains (4, 30). The concentration of proteins with functions in the same process in one nuclear compartment supports the spatial and temporal integration of tightly coupled nuclear processes, such as the transcription, splicing, and export of mRNA (46, 47).
Recent studies have shed light on the components and functions of several nuclear domains. Cajal bodies and nuclear speckles are enriched in spliceosomal small nuclear ribonucleoproteins (snRNPs) and have a specific role(s) in the biogenesis of cellular RNAs (9, 66). Nuclear speckles are defined by the irregular and punctate immunofluorescent staining patterns of RNA-processing factors, such as the serine/arginine-rich (SR) splicing factor SC35, and correspond largely to the interchromatin granule clusters (66). The current concept is that the enrichment of a given protein in speckles is mediated by its function and interactions with other factors residing in interchromatin granule clusters, although the existence of specific targeting or retention signals cannot be ruled out (61). Originally, it was proposed that nuclear speckles are mainly storage sites for RNA-processing factors from which they were recruited to sites of active transcription (36, 54, 77). However, more recent findings also suggest an active role of the compartment in mRNA biogenesis (7, 51, 59). The structural organization of nuclear speckles and their morphological appearance are tightly associated with the metabolism of the cell and seem to be regulated by phosphorylation and dephosphorylation events of SR proteins (13, 60, 75). Consequently, inhibition of RNA polymerase II transcription or heat shock leads to an enlarged and rounded appearance of the otherwise rather irregularly shaped speckles (39).
Influenza A and B viruses are major respiratory pathogens that replicate and transcribe their RNA genomes in the nucleus of the infected cell through a virus-encoded RNA-dependent RNA polymerase (56). The nuclear replication requires the virus to recruit cellular posttranscriptional activities to support its propagation. Hence, export of the viral genomic RNA late in infection is facilitated by the CRM1-dependent export pathway that is accessed by the viral nuclear export protein (21). However, other events of viral-gene expression are less well understood. For instance, efficient export of metazoan mRNA transcripts in vivo is tightly linked to their synthesis by the cellular RNA polymerase II, which involves a rapid interaction of maturation factors with the nascent transcript via its C-terminal domain (2). In this respect, influenza virus mRNAs are disadvantaged, as they are produced by the viral RNA polymerase, leaving open the question of how they are integrated into cellular transport pathways.
The focus of the present study was on the 281-amino-acid NS1 protein expressed by influenza B virus, which forms homodimers and binds to single- and double-stranded RNAs in vitro (70). This protein localizes to the nucleus during infection (53), but we do not know about its nuclear function(s), nor have the signals that mediate its trafficking been defined. The B/NS1 protein was previously shown to inhibit antiviral responses by blocking the induction of type I interferons (IFN) and the kinase PKR, which are most likely cytosolic activities (15, 16, 18). These functions are conserved in the influenza A virus NS1 protein (A/NS1), although the two proteins have less than 25% sequence identity (3, 49, 50, 55). Interestingly, the B/NS1 protein does not share the inhibitory activities of the A/NS1 protein in multiple steps of cellular-RNA maturation, including pre-mRNA splicing, polyadenylation, and export of cellular RNAs (12, 24, 27, 43, 52, 63, 70, 74). Those activities are believed to weaken host cell gene expression and have been suggested to depend on interactions with a number of cellular partners, including the cleavage and polyadenylation specificity factor 30-kDa, poly(A) binding protein 2 (PABP2), NS1-BP, RaeI, and NXF1/TAP, the major export receptor of cellular mRNA (12, 52, 63, 74).
Here, we demonstrate that the B/NS1 protein enters the nucleus and accumulates in SC35-containing speckles, leading to a coalesced appearance of these domains. Mutational analyses identified a nuclear localization signal (NLS) at NS1 amino acids 46 to 57 and determined the N-terminal 90 amino acids to be the shortest element that confers speckle association. Intriguingly, the B/NS1 protein relocalized to the cytoplasm late in infection, which is consistent with its known role in antagonizing cytoplasmic sensor proteins for viral RNA. The attenuation of recombinant viruses expressing NS1 mutant proteins with impaired speckle association suggested that this activity contributes to viral propagation. This report adds influenza B virus to the group of viral pathogens known to target nuclear speckles and provides a framework for more detailed investigations of this specific virus-host interaction.
MATERIALS AND METHODS
Cells and viruses.
A549 cells, HeLa cells, and 293T cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and penicillin/streptomycin. Madin-Darby canine kidney type II (MDCKII) cells were grown in minimum essential medium supplemented as described above. All cells were maintained at 37°C and 5% CO2. Spodoptera frugiperda (Sf9) cells were used for baculovirus expression and maintained in TNM-FH medium as described previously (67). Influenza B/Yamagata/1/73 virus, recombinant influenza B/Lee wild-type (WT) virus, and influenza A/PR/8/34 virus were grown in the allantoic cavities of 11-day-old embryonated chicken eggs for 3 days at 33°C (type B) and for 2 days at 37°C (type A), respectively. The influenza B mutant viruses ΔNS1 and NS#1 (33 and 38), NS#2 (47 and 50), NS#3 (52, 53, and 54), NS#4 (58, 60, and 64), and NS#6 (77 and 78) harboring alanine replacements at the indicated NS1 amino acid positions have been described previously (16). All recombinant mutant viruses were grown in the allantoic cavities of 6-day-old embryonated chicken eggs for 3 days at 33°C. The clinical influenza B virus isolates B/Thuringen/2/06 and B/Berlin/37/06 were obtained from the German National Reference Centre for Influenza (Robert Koch-Institute, Berlin, Germany). Virus titers were determined on MDCK cells by plaque assay (type A virus) or by indirect immunofluorescence staining with a nucleoprotein (NP)-specific antibody (type B viruses) as described previously (15). Accordingly, viral titers are expressed as PFU or fluorescence-forming units (FFU)/ml, respectively.
Construction of plasmids.
The expression plasmids pcDNA3-B/NS1 and pHW-Lee-NS have been described previously (15). Also, the Escherichia coli glutathione S-transferase (GST)-importin α3 expression construct and the baculovirus vectors expressing fusion genes of GST and importins α1, α3, α4, α5, and α7 have been described elsewhere (48). The plasmids pLP-EGFP-C1-UAP56 (42) and pcDNA3-Myc/His-REF (73) were kindly provided by D. Goodrich (Roswell Park Cancer Institute, Buffalo, NY) and S. A. Wilson (Department of Molecular Biology and Biotechnology, Sheffield, United Kingdom), respectively. The plasmids pHM829 and pHM839 were kindly provided by T. Stamminger (Institute of Virology, Erlangen, Germany) (65). pEGFP-C1-B/NS1 was constructed by subcloning PCR-amplified B/NS1 cDNA between the HindIII and SalI restriction sites of pEGFP-C1 (Clontech, Heidelberg, Germany). The pGex-UAP56 expression plasmid was constructed by subcloning PCR-amplified UAP56 cDNA between the BamHI and XhoI restriction sites of pGex-5X-1. pHW-Lee-NS#2/3 expressing an NS1 protein with alanine replacements at positions 47, 50, 52, 53, and 54 was generated by introducing a corresponding mutant cDNA into pHW-Lee-NS#3. The expression plasmids pHM-B/NS1_N-WT, pHM-B/NS1_N-NLSmut, pHM-B/NS1_C, pHM-B/NS1aa46-56, pHM-B/NS1aa46-56#2/3, and pHM-B/NS1aa1-90 were generated with the In-Fusion Dry-Down PCR Cloning Kit (Clontech, Heidelberg, Germany). Primers including pHM-829 vector sequences encompassing the XbaI restriction site were used to amplify B/NS1 fragments of interest and to insert the resulting PCR fragments into the vector pHM829 according to the manufacturer's instructions. The expression plasmids pHM-B/NS1aa12-90, pHM-B/NS1aa12-104, pHM-B/NS1aa1-65, pHM-B/NS1aa1-73, pHM-B/NS1aa1-83, pHM839-B/NS1_N-WT, and pHM839-B/NS1_N-NLSmut were generated by inserting corresponding PCR products into the XbaI restriction site of pHM829 or pHM839, respectively.
Recovery of recombinant influenza B virus.
The influenza B virus mutant NS#2/3 (47, 50, 52, 53, and 54) was generated by transfecting 293T cells with the plasmids pHW-Lee-PB1, pHW-Lee-PB2, pHW-Lee-PA, pHW-Lee-HA, pHW-Lee-NA, pHW-Lee-NP, and pHW-Lee-M and a pHW-Lee-NS#2/3 construct using Lipofectamine 2000 (Invitrogen, Heidelberg, Germany) as described previously (15). At 72 h after transfection, the cell supernatant was inoculated into the allantoic cavities of 6-day-old embryonated chicken eggs, and stocks of recombinant virus were grown for 3 days at 33°C. The presence of the introduced mutations and the absence of unwanted mutations within the NS sequence were confirmed by restriction analysis and cycle sequencing of the amplified reverse transcription-PCR product.
Importin binding assay.
GST-importin α1, α3, α4, α5, and α7 fusion proteins were expressed by recombinant baculovirus in Sf9 cells, and GST-importin α3 was also produced in E. coli BL21 cells. GST fusion proteins were purified as described elsewhere (22). B/NS1 and β-galactosidase (β-Gal)-green fluorescent protein (GFP) fusion proteins were translated and radioactively labeled with [35S]methionine/cysteine in vitro by using the TnT Coupled Reticulocyte Lysate Systems (Promega, Madison, WI), together with Pro-Mix (Amersham Biosciences). The polypeptides produced were reacted with glutathione-Sepharose-immobilized GST or GST-importin fusion proteins in binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 0.5% Triton X-100) on ice for 60 min, followed by washing with the same buffer as described previously (22). GST-importin-bound 35S-labeled proteins were separated on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The gels were fixed and treated with Amplify reagent (Amersham Biosciences) as specified by the manufacturer, and the labeled proteins were visualized by autoradiography.
GST pull-down assay.
Glutathione-Sepharose beads were loaded with GST or GST-UAP56 protein expressed in bacterial lysates as described above. The B/NS1 protein was in vitro translated and radioactively labeled by using the TnT Coupled Reticulocyte Lysate System (Promega, Madison, WI), together with EasyTagMethionine L[35S] (Perkin-Elmer). Labeled B/NS1 protein was reacted with the coated glutathione-Sepharose beads in HEPES binding buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 0.05% Igepal CA630 detergent, and 0.01% bovine serum albumin) at 4°C for 2 h, followed by washings with the same buffer. The precipitated proteins were separated by SDS-gel electrophoresis and stained with Coomassie blue, and the labeled B/NS1 protein was visualized by autoradiography.
Fluorescence microscopy and image analysis.
For infection experiments, MDCK or A549 cells were grown on glass coverslips and infected with influenza virus at the indicated multiplicity of infection (MOI). To analyze CRM1-dependent transport, MDCK cells were infected with influenza B virus, and starting at 3.5 h postinfection (p.i.), the cells were left untreated or treated with 10 ng/ml leptomycin B (LMB) for 16.5 h. All transfection experiments were performed in cell suspension using Lipofectamine 2000 (Invitrogen, Heidelberg, Germany). For immunofluorescence staining, cells were seeded on coverslips. At the indicated time points, the cells were fixed with 2.5% paraformaldehyde for 15 min, followed by a permeabilization step with 0.2% Triton X-100 for 10 min. The cells were incubated with primary antibodies diluted in phosphate-buffered saline (PBS)/0.2% bovine albumin. After being washed, the cells were incubated with secondary antibodies in PBS for 1 h. Where indicated, cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml in PBS) for 10 min. Influenza virus NS1 proteins were detected with monospecific A/NS1- and B/NS1-specific rabbit antisera (16). Influenza B virus NP was detected with a primary monoclonal anti-NP antibody (AbD Serotec, Oxford, United Kingdom). For detection of nuclear subdomain marker proteins, the primary monoclonal antibodies anti-PML (PG-M3) (Santa Cruz Biotechnology, Inc., Heidelberg, Germany), anti-Sm Ab-1 (Y12) (Neomarkers, Freemont, CA), and anti-SC35 (BD Pharmingen, Heidelberg, Germany) were used. The primary antibodies mouse anti-myc (9E10) (Santa Cruz Biotechnology, Inc.) and rabbit anti-PABP2 serum (38) were used for colocalization analyses of speckle-associated nuclear proteins in transfected cells. Species-specific secondary immunoglobulin G (IgG) conjugated with Alexa Fluor dyes (anti-rabbit-488, anti-rabbit-594, anti-mouse-488, and anti-mouse-594; Molecular Probes, Leiden, The Netherlands) were used as secondary antibodies. Imaging of cells was performed using an LSM510 Meta confocal laser scanning microscope and a C-Apochromat 63/1.2 water objective lens (Zeiss, Jena, Germany) with a pinhole setting of 1. Data were analyzed and processed with the Zeiss LSM Image Browser 3.5 and Adobe Photoshop 4.0 software packages.
RESULTS
The B/NS1 protein accumulates in the nuclei of infected cells in a dot-shaped pattern.
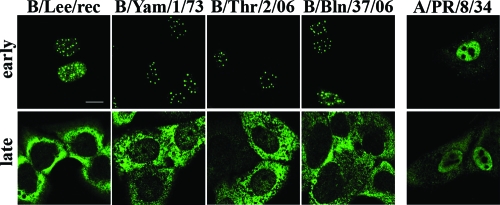
We stained human A549 lung epithelial cells after infection with the natural influenza B virus strains B/Yam/1/73, B/Thr/2/06, and B/Bln/37/06 and recombinant influenza B/Lee virus (15) for the localization of NS1 proteins and analyzed them by confocal laser scanning microscopy (CLSM) (Fig. 1). As a comparison, we localized in parallel the NS1 protein of the influenza A/PR/8/34 virus (subtype H1N1), which contains an N-terminal NLS but lacks a C-terminal nucleolar targeting signal found in most H3N2 subtype viruses (48). Interestingly, the cellular distributions of the A/NS1 and B/NS1 proteins during infection were very distinct. As shown previously, the A/NS1 protein localized to the nucleus and the cytoplasm early (4 h) and late (16 h) in infection (74). In contrast, the B/NS1 protein was detected exclusively in the nucleus early in infection, where it was observed in a bright dot-shaped pattern in addition to some diffuse nucleoplasmic staining. Late in infection, the protein was mostly relocalized to the cytoplasm of the cell. The patterns of B/NS1 distribution throughout infection were essentially the same for all four influenza B virus strains, although the homologous proteins differ by up to 10% in their primary sequences. This analysis showed a strikingly dynamic trafficking of the B/NS1 protein during virus infection and raised our interest in the targeted nuclear domain and the signals that control its specific intranuclear localization.
FIG. 1.
The B/NS1 proteins accumulate in a dot-shaped pattern in the nucleus early in infection. Human lung epithelial A549 cells were infected with the influenza B virus strains B/Lee, B/Yamagata/1/73 (B/Yam/1/73), B/Thuringen/2/06 (B/Thr/2/06), and B/Berlin/37/06 (B/Bln/37/06) and influenza A virus A/PR/8/34 at an MOI of 2. Infected cells were processed for microscopic analysis at early (4 to 6 h p.i.) or late (16 h p.i.) times of infection. The cells were stained with primary rabbit antiserum specific for the A/NS1 or the B/NS1 protein, which was followed by detection with secondary α-rabbit Alexa 488 antibodies. The cells were analyzed by CLSM using the 488-nm laser setting. Scale bar = 10 μm.
The B/NS1 protein colocalizes with the splicing factor SC35 in nuclear speckles.
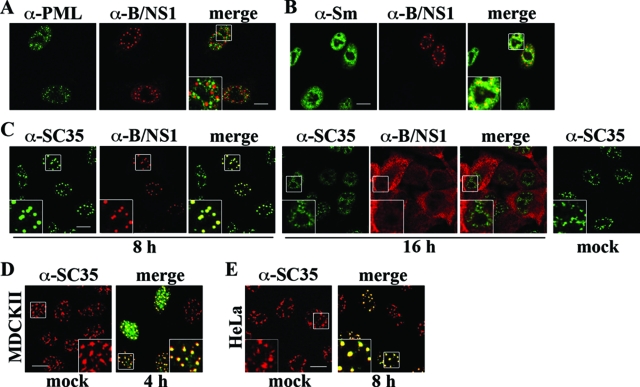
A549 cells were infected with influenza B virus and double stained for the B/NS1 protein and marker antigens of certain nuclear domains, including PML, uridine-rich snRNPs, and SC35 (Fig. 2A to C). Our analysis showed that the B/NS1 protein did not accumulate in PML bodies (Fig. 2A). The snRNPs U1, U2, U4/U6, and U5 have essential roles in the processing of cellular pre-mRNAs and can be detected by the conserved Sm autoantigen in Cajal bodies and nuclear speckles (8, 9). In virus-infected cells, we observed partial colocalization of the B/NS1 and snRNP signals early in infection, which did not include Cajal bodies (Fig. 2B). This staining pattern indicated an association of the viral protein with nuclear speckles, for which the non-snRNP splicing factor SC35 serves as a marker antigen (26). In fact, intense colocalization of the B/NS1 protein and SC35 was observed in a bright dot-like pattern early in infection (Fig. 2C). Interestingly, the SC35 domains had a rounded and coalesced shape after virus infection, which clearly differed from the irregular and cloudy forms observed in mock-infected cells (Fig. 2C, mock). Late in infection, the colocalization of B/NS1 and SC35 was no longer seen. SC35 was distributed throughout the nucleoplasm, whereas the viral B/NS1 protein was mainly cytoplasmic (Fig. 2C). At early times of infection, the B/NS1 protein also colocalized with SC35 in infected MDCK cells (Fig. 2D) and human HeLa cells (Fig. 2E). In addition, B/NS1 was detected in the same typical punctate pattern in mouse embryo fibroblasts (data not shown). Thus, we concluded that the B/NS1 protein accumulates in nuclear speckles, possibly in all types of mammalian cells.
FIG. 2.
The B/NS1 protein colocalizes with the splicing factor SC35 in nuclear speckles, leading to a coalesced appearance of these domains early in infection. A549 cells were mock infected or infected with influenza B/Yam/1/73 virus for 8 (A, B, and C) or 16 (C) h. Subsequently, the cells were fixed, permeabilized, and double stained with B/NS1-specific rabbit serum (α-B/NS1), together with monoclonal antibodies recognizing either PML bodies (A, α-PML), snRNPs (B, α-Sm), or the speckle marker antigen SC35 (C, α-SC35), followed by detection with secondary α-rabbit IgG-Alexa 594 and α-mouse IgG-Alexa 488 antibodies, respectively. MDCKII cells (D) and HeLa cells (E) were mock infected (left) or infected with influenza B/Lee/rec virus (right). Cells were stained for SC35 only (D and E, mock) or the SC35 (red signal) and B/NS1 (green signal) proteins together (D and E, virus) as described for panel B. The stained cells were analyzed by CLSM. Scale bar = 10 μm. The insets show enlargements of the indicated areas (white squares).
Accumulation in nuclear speckles is an inherent function of the B/NS1 protein.
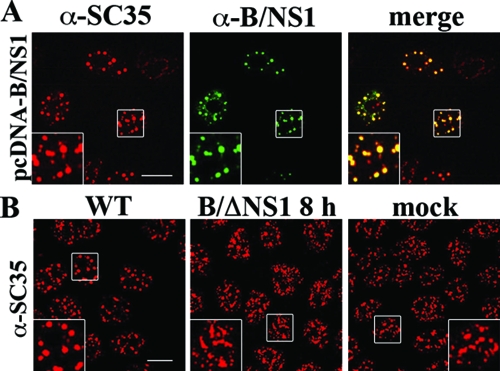
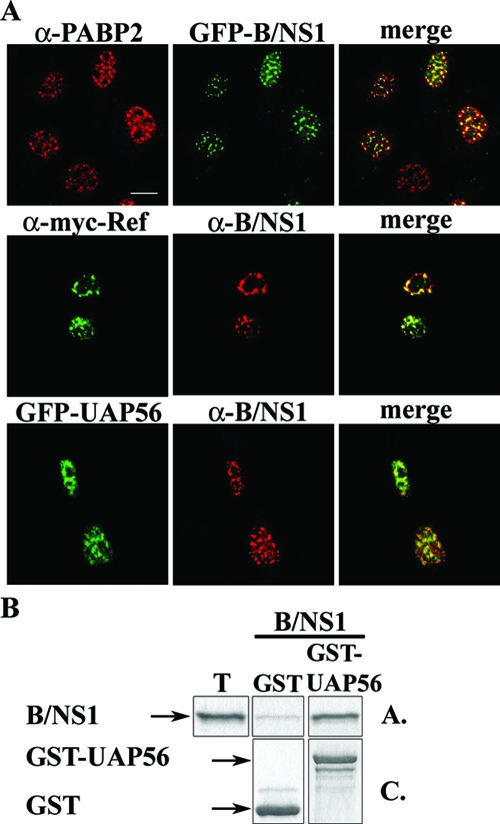
Next, we investigated whether other viral proteins are required for the association of the B/NS1 protein with nuclear speckles. Hence, we expressed B/NS1 from a transfected plasmid and found that it colocalized with SC35 and also induced rounding of nuclear speckles in this experimental setting (Fig. 3A). In contrast to infected cells, the B/NS1 protein remained stably associated with nuclear speckles upon transient expression for more than 36 h posttransfection and did not relocalize to the cytoplasm. Next, we infected cells with a mutant influenza B virus lacking the entire NS1 gene (ΔNS1) (15) (Fig. 3B). Successful infection was confirmed in parallel cultures by positive NP staining of all cells (data not shown). Significantly, there was no morphological change of nuclear speckles in comparison to mock infection, whereas the SC35 domains were coalesced in WT-virus-infected cells (Fig. 3B). These findings indicated that the interaction with nuclear speckles and their change in morphology are inherent activities of the B/NS1 protein, which does not require any other viral gene product. To confirm that the B/NS1 protein accumulates in SC35-positive domains, we localized the viral protein, together with other cellular speckle-associated factors (Fig. 4A). In fact, B/NS1 and the GFP-B/NS1 fusion protein partially colocalized with PABP2, UAP56, and Aly/REF, which are involved in the maturation and/or transport of cellular RNA transcripts (28, 38, 78). We note that the rounding of the speckled pattern of those cellular factors was less prominent than the SC35 signals in virus-infected cells. In GST pull-down analysis, we observed binding of the B/NS1 protein to UAP56, indicating that the colocalization in cells reflects a physical interaction, at least for this factor (Fig. 4B).
FIG. 3.
The B/NS1 protein accumulates in nuclear speckles in the absence of other influenza B virus proteins. (A) MDCK cells were transfected with the expression plasmid pcDNA3-B/NS1; 36 h posttransfection, the cells were fixed, permeabilized, and stained for the B/NS1 and SC35 proteins. (B) MDCK cells were either mock infected (mock) or infected with WT influenza B virus (WT) or ΔNS1 virus (B/ΔNS1) at an MOI of 5 to infect all cells. The cells were fixed at 8 h p.i. and stained for the SC35 protein, and micrographs were captured by CLSM using the 488-nm and 594-nm laser settings. Scale bar = 10 μm. The insets show enlargements of the indicated areas (white squares).
FIG. 4.
The B/NS1 protein colocalizes with the speckle-associated proteins PAPB2, REF, and UAP56. (A) MDCKII cells were transfected with a plasmid expressing autofluorescent GFP-B/NS1 fusion protein (top row) or with a combination of pEGFP-UAP56 and pcDNA3-B/NS1 (bottom row). (Middle row) HeLa cells were transfected with the expression plasmids pcDNA-myc-His-REF and pcDNA3-B/NS1. Twenty-four hours after transfection, the cells were fixed, permeabilized, and stained as indicated for endogenous PABP2 (top row) and for myc-tagged REF and expressed B/NS1 proteins (middle and bottom rows) using primary myc antibodies and B/NS1-specific rabbit antiserum, together with species-specific Alexa 488- or Alexa 549-labeled secondary antibodies. The cells were analyzed by CLSM, and micrographs were captured using the 488-nm and 594-nm laser settings. Scale bar = 10 μm. (B) In vitro-translated and 35S-labeled B/NS1 protein was reacted with glutathione-Sepharose-immobilized GST or GST-UAP56 fusion protein. After being washed, the precipitated proteins were separated by SDS-gel electrophoresis. The proteins were visualized by Coomassie blue staining (C.) and autoradiography (A.). T, input control corresponding to 3% of the amount used in each binding reaction.
The N-terminal region of B/NS1 mediates nuclear targeting and speckle association.
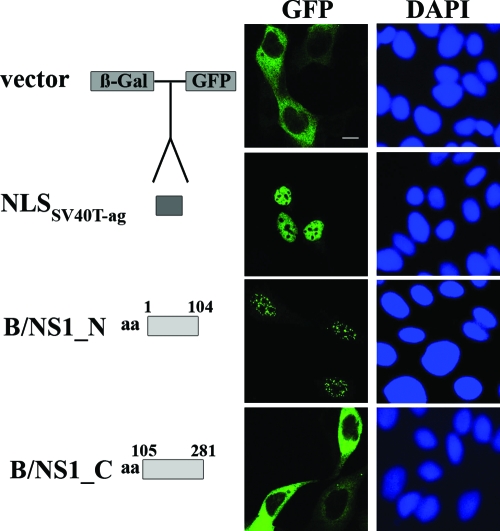
After having identified SC35-positive speckles as nuclear accumulation sites of the B/NS1 protein, we determined the signals that mediated this specific localization. We used an established assay that employs plasmid-based expression and intracellular detection of autofluorescent β-Gal-GFP fusion proteins (65). First, β-Gal-GFP fusion genes with the coding sequences of the N-terminal (amino acids 1 to 104) and C-terminal (amino acids 105 to 281) parts of the B/NS1 protein were generated. Transient expression revealed that the N-terminal region of B/NS1 mediates translocation of β-Gal-GFP fusion protein into the nucleus, where it was detected in a punctate pattern (Fig. 5). In contrast, the β-Gal-GFP protein alone or when fused to the C-terminal B/NS1 fragment remained in the cell cytoplasm (Fig. 5). A β-Gal-GFP fusion protein containing the NLS of the large T antigen (T-Ag) of simian virus 40 (SV40) localized to the nucleoplasm of the cell, ruling out the possibility that the core fusion protein alone accumulated in a punctate pattern when translocated to the nuclear compartment (Fig. 5). Hence, the N-terminal 104 amino acids contain a signal(s) that is necessary and sufficient for nuclear import and for speckle association of the B/NS1 protein.
FIG. 5.
The signals that mediate nuclear localization and speckle association of the B/NS1 protein reside in the N-terminal region. The coding sequence for the N-terminal part (B/NS1_N; amino acids [aa] 1 to 104) and the C-terminal part (B/NS1_C; aa 105 to 281) of influenza B virus NS1 protein, respectively, was inserted in frame between the β-Gal and GFP sequences in the vector pHM829. The vector pHM839 contains an additional insertion of the coding sequence for the SV40 T-Ag NLS between the β-Gal and GFP genes. MDCK cells were transfected with the indicated expression plasmids. Twenty-four hours posttransfection, the cells were fixed and permeabilized. DAPI was used to stain the nuclei of the cells. GFP signals were recorded by CLSM. DAPI signals were analyzed by fluorescence microscopy. The expression constructs are shown schematically on the left. Scale bar = 10 μm.
Characterization of an NLS in the B/NS1 protein at amino acids 46 to 56 and its role in virus replication.
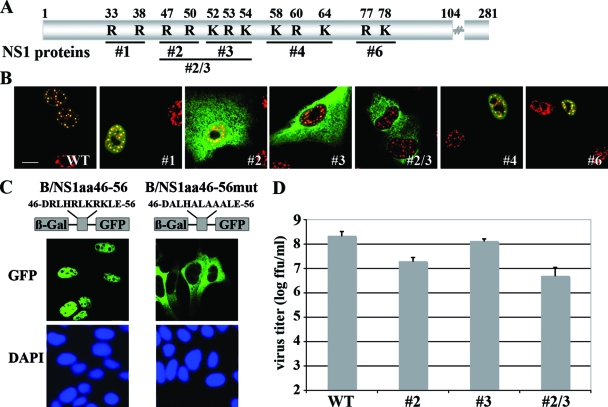
To narrow down the signal(s) mediating nuclear trafficking of the B/NS1 protein, we conducted a mutational analysis. Classical monopartite NLSs, such as the signal of the SV40 T-Ag, consist of clusters of three to six basic amino acids that are recognized by members of the importin α receptor family (40). The B/NS1 protein carries seven highly conserved clusters of two to three basic amino acids in the N-terminal RNA-binding domain (18), but their roles in nuclear localization have not been examined. To assess the contributions of the N-terminal clusters, we analyzed a set of recombinant influenza B viruses expressing NS1 mutant proteins with alanine replacements of basic amino acids within one or two of the clusters (16) (Fig. 6A). The localization of the B/NS1 mutant proteins and the SC35 antigen by confocal microscopy revealed that the basic-to-alanine exchanges within the B/NS1 sequence 47 to 54 strongly impaired nuclear accumulation, indicating that this stretch is important for nuclear transport (Fig. 6B). Interestingly, a complete loss of nuclear localization was observed only for the combined NS1 mutant no. 2/3 (47, 50, 52, 53, and 54), whereas the separate replacements at positions 47 and 50 or at 52, 53, and 54 still allowed a fraction of the proteins to enter the nucleus and associate with SC35-reactive domains (Fig. 6B). In contrast, none of the other mutant NS1 proteins was affected in nuclear migration and speckle association (Fig. 6B). A hallmark of NLS sequences is that they can target an unrelated protein to the nucleus (40). When B/NS1 amino acids 46 to 56 were fused to the β-Gal-GFP protein, those 11 amino acids were sufficient to translocate the β-Gal-GFP fusion protein into the nucleus, where it was diffusely distributed (Fig. 6C). Replacing the five basic amino acids with alanines at NS1 positions 47, 50, 52, 53, and 54 within the β-Gal-GFP fusion construct completely abrogated nuclear localization, suggesting that these amino acids form the nuclear targeting signal of the B/NS1 protein (Fig. 6C).
FIG. 6.
The basic amino acids at positions 46 to 56 mediate nuclear localization of the B/NS1 protein and contribute to efficient viral replication. (A) Schematic representation of the primary structure of the B/NS1 protein with indicated positions of basic amino acid clusters that are replaced by alanine residues in the mutant proteins no. 1, no. 2, no. 3, no. 2/3, no. 4, and no. 6 expressed by recombinant influenza B viruses. (B) Intracellular localization of WT and mutant B/NS1 proteins expressed by recombinant influenza B viruses. A549 cells were infected with either WT or the indicated mutant influenza B viruses at an MOI of 1. The cells were fixed at 8 h p.i., followed by permeabilization and staining for the B/NS1 and SC35 proteins. (C) The B/NS1 amino acids 46 to 56 mediate nuclear localization of a β-Gal-GFP fusion protein. A schematic representation of the β-Gal-GFP fusion protein containing the suspected NLS at positions 46 to 56 and the mutant sequence with alanine replacements of the five basic amino acids is shown on the right. The sequences encoding amino acids 46 to 56 (B/NS1_aa46-56) or a mutant derivative thereof (B/NS1_aa46-56mut) were fused to the coding sequences for β-Gal and GFP. MDCK cells were transfected with the indicated expression plasmids, and the cells were fixed at 24 h posttransfection and processed for microscopic analysis. The cells were analyzed by CLSM. DAPI signals were recorded by fluorescence microscopy. Scale bar = 10 μm. (D) Six-day-old embryonated chicken eggs were inoculated with 1,000 FFU of recombinant B/Lee WT or mutant viruses no. 2, no. 2/3, and no. 3 and were incubated for 72 h at 33°C. Virus titers were determined as described in Materials and Methods. The indicated values represent the averages of at least three independent experiments. The error bars indicate the standard deviations of the means.
Next, we assessed the contribution of B/NS1 amino acids 47 to 54 to virus replication by comparing the growth of WT and mutant viruses in 6-day-old embryonated chicken eggs. These hosts provide an IFN-free environment and were chosen to avoid possible effects of this antiviral cytokine on virus growth (16). It was found that the mutant viruses replicated to titers up to 2 orders of magnitude lower (no. 2, 2.0 × 10e7; no. 3, 1.3 × 10e8; and no. 2/3, 5.0 × 10e6 FFU/ml) than that of recombinant WT virus (2.2 × 10e8 FFU/ml) (Fig. 6D). These results indicate a necessity for nuclear accumulation and possibly speckle association of the NS1 protein for full virus replication.
We also analyzed the abilities of the B/NS1 protein and its putative NLS to mediate interaction with the mammalian nuclear import factor importin α. Five importin α isoforms were expressed as GST fusion proteins in Sf9 insect cells, and pull-down analysis was carried out with 35S-labeled B/NS1 protein generated by in vitro translation. All the Sf9-expressed GST-importin α isoforms (α1, α3, α4, α5, and α7) bound the NS1 protein (Fig. 7A). The viral protein also interacted with the E. coli-expressed GST-importin α6 (a testis-specific isotype), whereas no binding to GST-importin β was found (data not shown). We then analyzed the binding of β-Gal-GFP fusion protein containing B/NS1 amino acids 46 to 56 to GST-importin α3. The peptide sequence mediated tight binding to importin α3, which was more efficient than that of the T-Ag NLS of SV40 that served as a positive control (Fig. 7B). The replacement of the five basic amino acids within this 11-amino-acid stretch by alanines abolished all binding of the fusion protein to importin α3 (Fig. 7B). Thus, the results of the GST-importin α pull-down experiments confirmed the identification of an NLS at positions 46 to 56 that mediates nuclear localization and binding to importin α.
FIG. 7.
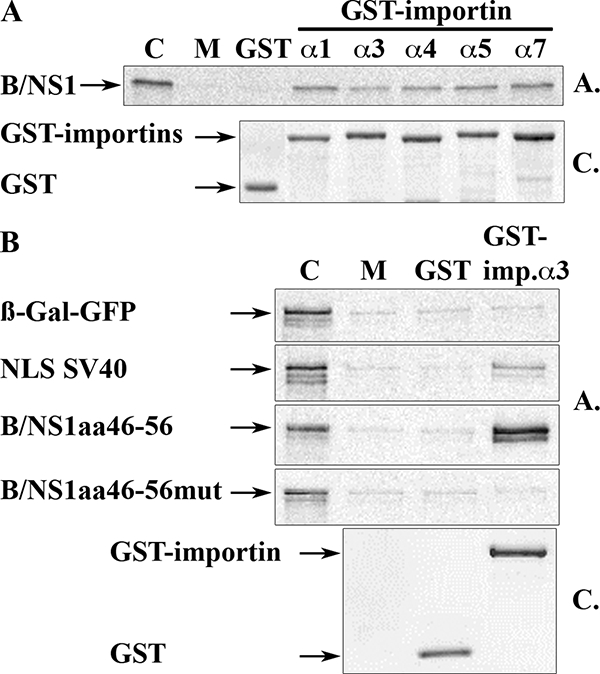
The B/NS1 protein binds to importin α isoforms via amino acids 46 to 56. (A) 35S-labeled and in vitro-translated B/NS1 protein was allowed to bind to Sf9-expressed and glutathione Sepharose-immobilized GST or GST-importin α1, α3, α4, α5, or α7. After being washed with binding buffer, importin-bound NS1 was dissolved in Laemmli sample buffer, separated by SDS-gel electrophoresis, and visualized by autoradiography (gel A.). Protein samples of the same GST pull downs were run on a similar gel, followed by staining with Coomassie brilliant blue (gel C.) to visualize the numbers of glutathione-Sepharose-immobilized GST- and GST-importin α isoforms. Lane C is the input control corresponding to 10% of the amount used in each binding reaction. Lane M shows the amount of in vitro-translated NS1 protein that was retained by unloaded glutathione-Sepharose matrix. (B) 35S-labeled and in vitro-translated β-Gal-GFP and its fusion proteins containing the SV40 T-Ag NLS, B/NS1aa46-56, or B/NS1aa46-56mut were allowed to bind to E. coli-expressed and glutathione-Sepharose-immobilized GST and GST-importin α3 as indicated. The precipitated fusion proteins were dissolved in Laemmli sample buffer, separated by SDS-gel electrophoresis, and visualized by autoradiography (gel A.). Aliquots of the glutathione-Sepharose-immobilized GST and GST-importin α3 proteins were separated on a similar gel that was stained with Coomassie brilliant blue (gel C.). Lane M shows the amount of in vitro-translated fusion protein that was retained by unloaded glutathione-Sepharose matrix.
Speckle association of the B/NS1 protein requires the N-terminal region and basic amino acids within the NLS.
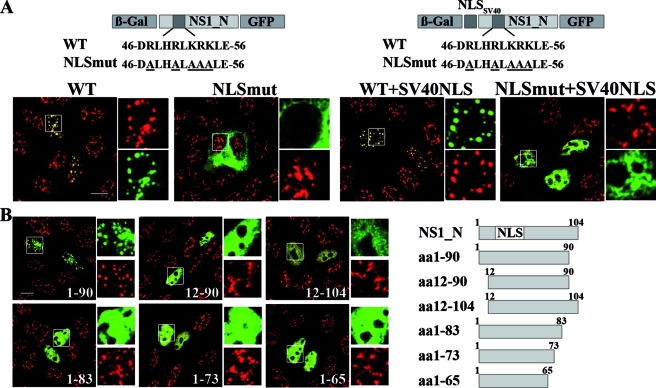
In further experiments, we identified the signals that are important for association of the B/NS1 protein with nuclear speckles. The data described above (Fig. 5) suggested that this information resides in the N-terminal 104 amino acids and that the basic clusters at positions 33 and 38; 58, 60, and 64; and 77 and 78 do not contribute to this specific localization. An expected prerequisite for speckle targeting of the viral protein is the ability to enter the nucleus. This prediction was evidenced by the nuclear exclusion of a β-Gal-GFP fusion protein carrying the N-terminal 104 amino acids of B/NS1 with a mutated NLS (Fig. 8A, compare WT and NLSmut on the left). The mutant B/NS1 fusion protein did not modify SC35 speckles, whereas those structures appeared coalesced in the WT situation (Fig. 8A, left). Remarkably, the arginine residues at positions 47 and 50 that were identified as part of the B/NS1 NLS (Fig. 6 and 7) were previously shown to also be essential for RNA binding (18). RNA recognition motifs have previously been implicated in the speckle targeting of some proteins, including the SR protein SF2/ASF (6). Thus, we hypothesized that the basic amino acids within the B/NS1 NLS are not only required for efficient nuclear import, but may in addition contribute to speckle association. To test this, it became necessary to relocate the cytoplasmic fusion protein into the nucleus. For this reason, we created two additional β-Gal-GFP expression constructs carrying the T-Ag NLS of SV40 fused to the N-terminal 104 amino acids of B/NS1 with intact (WT) or mutated (NLSmut) sequence at positions 47 to 54 (Fig. 8A, right). The additional NLS did not alter the accumulation of the WT fusion protein in nuclear speckles (Fig. 8A, left). As expected, the NLSmut fusion protein was relocated to the nucleus by the fused SV40 NLS (Fig. 8A, right). Importantly, the mutant fusion protein was distributed throughout the nucleoplasm, and there was no recognizable change of speckle morphology in these cells (Fig. 8A, right). These findings indicate that the basic amino acids within the NLS are important not only for nuclear localization of B/NS1, but also for its speckle association.
FIG. 8.
Speckle association of the B/NS1 protein requires an intact NLS and amino acids 1 to 90. (A) The schematic drawings at the top show the primary structure of the inserted B/NS1_N polypeptide within the constructed β-Gal-GFP fusion genes. The sequences of the B/NS1 WT and mutant NLS (NLSmut) and the position of the additional SV40 T-Ag NLS are also given. The coding sequences of B/NS1_N (amino acids 1 to 104) carrying a WT or mutant NLS were inserted into the vectors pHM829 and pHM839, respectively, thereby generating β-Gal-GFP fusion proteins of B/NS1_N without (left) or with (right) an additional SV40 T-Ag NLS. MDCK cells were transfected with plasmids expressing the indicated fusion proteins and were stained with SC35-specific antibody and secondary α-mouse IgG-Alexa 594 conjugate. The GFP and Alexa 594 signals were analyzed by CLSM, and fields with merged signals are shown. The white squares in the larger micrographs indicate areas that are expanded on the right, with separate recordings of both channels shown. (B) The scheme on the right presents the structures and lengths of the constructed B/NS1_N derivatives that were fused to the coding sequences for β-Gal and GFP. MDCK cells were transfected with the indicated expression plasmids; 24 h posttransfection, the cells were fixed, permeabilized, and stained with SC35-specific antibody and secondary α-mouse IgG-Alexa 594 conjugate. The GFP and Alexa 594 signals were analyzed by CLSM, and fields with merged signals are shown. The white squares in the large micrographs indicate areas that are expanded on the right, with separate recordings of both channels shown. Scale bars = 10 μm.
Since the identified B/NS1 NLS at positions 46 to 57 alone was insufficient to mediate speckle association (Fig. 6C), we concluded that there were additional elements involved. Therefore, we analyzed N- or C-terminal truncation variants of the NS1 1-to-104 fragment that had all retained the NLS in the context of the β-Gal-GFP fusion protein (Fig. 8B). The smallest B/NS1 fragment that still interacted with nuclear speckles contained amino acids 1 to 90 (Fig. 8B). The other five fusion constructs, carrying NS1 amino acids 12 to 90, 12 to 104, 1 to 83, 1 to 73, and 1 to 65, respectively, entered the nucleus but failed to change the morphology of SC35-positive speckles (Fig. 8B). Thus, we conclude that the N-terminal 90 amino acids comprise a minimal sequence that is necessary for speckle association of the B/NS1 protein.
The B/NS1 protein is exported from the nucleus by a CRM1-independent pathway.
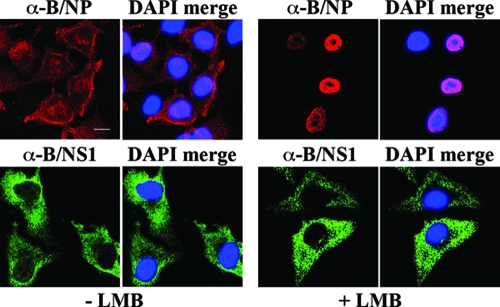
Having identified the elements that are important for nuclear import and speckle association of the B/NS1 protein, we addressed the question of how the protein is exported from the nucleus at late times of infection. CRM1 is the major export protein that recognizes leucine-rich nuclear export signals, binds to the cargo proteins, and mediates their export to the cytoplasm. The drug LMB specifically inhibits CRM1-dependent export (23, 57) and was previously shown to block the nucleocytoplasmic transport of the influenza A virus NP late in infection (21, 71, 72). MDCK cells were infected with influenza B virus in the absence or presence of LMB, which was added 3.5 h p.i., and stained for localization of the viral B/NP and B/NS1 proteins at 20 h p.i. A large fraction of the B/NP localized in the cytoplasm of control-treated cells (Fig. 9). In contrast, the addition of LMB led to pronounced nuclear B/NP staining, indicating that export of this protein also depends on the CRM1 pathway. Interestingly, LMB did not inhibit the translocation of the B/NS1 protein to the cytoplasm, demonstrating that its export is not regulated by the CRM1 pathway (Fig. 9).
FIG. 9.
Cytoplasmic relocalization of the B/NS1 protein late in infection is not inhibited by LMB. MDCK cells were infected with influenza B WT virus. Starting at 3.5 h p.i., cells were left untreated or treated with LMB. The cells were fixed at 20 h p.i. and stained as indicated for the viral nucleoprotein (B/NP; top) or the B/NS1 protein (B/NS1; bottom) with primary monoclonal α-B/NP antibody or α-B/NS1 rabbit antiserum, together with secondary α-rabbit IgG-Alexa 488 or α-mouse IgG-Alexa 594 conjugate, respectively. DAPI was used to stain the nuclei. The cells were analyzed by CLSM, and the DAPI signals were analyzed by fluorescence microscopy. For each field, separate micrographs with the signal of the viral proteins alone (α-B/NP and α-B/NS1) and with merged DAPI signals (DAPI merge) are shown. Scale bar = 10 μm.
DISCUSSION
The present study reveals the dynamic intracellular trafficking of the influenza B virus NS1 protein during the course of infection, with unexpected early accumulation in nuclear speckle domains and a largely cytoplasmic localization at late time points of infection. Speckle localization did not require any other viral factor, demonstrating that this specific nuclear trafficking is an inherent property of the B/NS1 protein. Remarkably, we found that influenza B virus lacking the NS1 gene did not affect speckle morphology. Since this mutant virus is strongly attenuated even in hosts lacking an intact type I IFN system, this finding indicates that speckle association of the B/NS1 protein is involved in productive virus infection (15, 16). In fact, the recombinant viruses expressing full-length NS1 proteins with reduced or abrogated speckle localization were attenuated for replication, suggesting that this NS1 property contributes to influenza B virus replication.
The signals required for association of the B/NS1 protein with nuclear speckles were located within the N-terminal 90 amino acids. This domain is known to bind to single- and double-stranded RNAs with multiple basic amino acids at positions 47 and 50; 58, 60, and 64; and 77 and 78 being essential for double-stranded-RNA binding (18, 70). In addition, this B/NS1 domain can bind to the IFN-stimulated gene 15 product, which is part of its suppressive effects on innate immunity (41). Our study identified in this region a monopartite polybasic NLS at positions 46 to 56 (DRLHRLKRKLE). This signal sequence directed a fused β-Gal-GFP to the nucleus; it interacted with all members of the importin α receptor family, and mutations of the basic amino acids it contained eliminated nuclear accumulation. The apparent sequence similarity and the overlapping positions within the suggested spatial structures indicate that the B/NS1 signal corresponds to the N-terminal NLS1 of the influenza A virus NS1 protein (35-RLRRDQK-41) (29, 48, 76). Apparently, the B/NS1 protein lacks a second NLS found at the C termini of many A/NS1 proteins, which also functions as a nucleolar targeting signal (29, 48).
The NLSs of the influenza A and B virus NS1 proteins are located within their RNA-binding domains at the N-terminal 73 and 93 amino acids, respectively (48, 70). Structural analyses of these domains revealed a common unique six-helical chain fold that differs from cellular RNA-binding motifs (76). NLSs overlapping with an RNA-binding domain have been described for other viral regulatory proteins, including UL69 of human cytomegalovirus and human immunodeficiency virus type 1 Rev, that promote nuclear export of the respective viral mRNAs (35, 68). RNA binding and nuclear import are mutually exclusive in the Rev protein, thereby ensuring that exported viral mRNAs do not return immediately to the nucleus (35). Although it has not yet been determined whether the B/NS1 protein is involved in the transport of viral RNA, it is possible that this overlap indicates a similar regulatory mechanism (see below).
The NLS of the B/NS1 protein was necessary but not sufficient to mediate speckle association, which required the whole N-terminal domain, consisting of amino acids 1 to 90. Previously, several unrelated sequence elements as short as 37 amino acids were shown to target a heterologous protein to nuclear speckles. This included the R/S-rich domains of the SR splicing factors SC35, SRp20, and Tra (6, 32); two contiguous SR-rich regions in the splicing factor SRm160; the two RNA recognition motifs of the alternative splicing factor ASF/SF2 (6); a threonine-proline-rich domain in the splicing factor SF3b; and the RNA-binding “SELOR” module in the putative export protein MLN51 (6, 17, 19, 20, 69). The mechanistic basis for the specific subnuclear targeting activities of these domains is not well understood but may involve interactions with other speckle-resident factors or nuclear matrix proteins (6, 69). The B/NS1 protein does not contain an R/S repeat sequence, and there is no recognizable homology to a known cellular speckle protein, which could explain its specific trafficking (61). Interestingly, our study showed that mutation of the basic amino acids within the NLS at positions 46 to 56 eliminated not only nuclear entry, but also the specific subnuclear trafficking when the protein was imported into the nucleus via a heterologous NLS. One explanation for this phenotype is that speckle association requires RNA binding, since this activity is also affected by the NLS mutation (18). Alternatively, it is possible that the basic residues within the B/NS1 NLS mediate an interaction with a presently unknown speckle-resident factor. In both scenarios, the conservation of basic amino acids at positions 47, 50, 52, and 54 within the more than 179 NS1 sequences of natural influenza B viruses available in public databases indicates a functional importance of the B/NS1 karyophilic signal (1). Amino acids 1 to 45 and 57 to 90 flanking the NLS may be necessary to maintain a proper structure of the N-terminal domain.
Significantly, we not only observed colocalization of the viral B/NS1 protein with SC35 speckles, we also found that these domains had a rounded and coalesced appearance. Not all of the analyzed B/NS1 mutant proteins associated with speckles. However, the variants that did also caused a morphological change of SC35 domains, indicating that the involved sequence elements overlap or are identical. Rounding and enlargement of nuclear speckles have been described during heat shock and inhibition of RNA polymerase II-mediated transcription (39). Under these circumstances, RNA processing and transport events are downregulated, leading to a redistribution of the involved factors from nucleoplasmic transcription sites to the interchromatin granule clusters. We noticed a striking similarity of the coalesced shapes of SC35 speckles during influenza B virus infection with the appearance of those domains after infection with herpes simplex virus type 1, Kaposi's sarcoma-associated herpesvirus, herpesvirus saimiri, and Epstein-Barr virus (11, 14, 44, 58, 62). This is caused by a conserved group of viral posttranscriptional regulator proteins comprising the herpes simplex virus type 1 ICP27, the herpesvirus saimiri open reading frame 57, the Kaposi's sarcoma-associated herpesvirus open reading frame 57, and the Epstein-Barr virus EB2 proteins that promote nuclear export of intronless viral mRNA. These viral regulators bind to viral mRNA transcripts and interact directly or indirectly with the speckle-associated nuclear export factor Aly/REF and thereby access the TAP-dependent major mRNA export pathway (5, 10, 33, 37, 45). In addition, they associate with RNA-processing factors and regulate pre-mRNA splicing (5, 31, 34, 44, 64). Thus, the redistribution of speckle factors reflects apparent intrusions of viral proteins in cellular RNA biogenesis, thereby supporting the expression of viral transcripts at the expense of host cell mRNA.
How might these findings relate to the transient speckle accumulation observed during influenza B virus infection? The B/NS1 protein has been reported to differ from the NS1 protein of influenza A viruses in not being able to interfere with the splicing and export of cellular mRNAs (24, 27, 63, 70). We were also unable to detect an effect of the B/NS1 protein on the splicing of an expressed model pre-mRNA (J. Schneider and T. Wolff, unpublished observations). The regulation of mRNA processing by the A/NS1 protein is consistent with the subtle alterations of nuclear speckles observed in influenza A virus-infected cells (25, 58, 74). However, there was no colocalization of A/NS1 and nuclear speckles (this study and references 25 and 74). If the transient association of the B/NS1 protein with speckles does not reflect inhibition of RNA processing, it is tempting to speculate that it functions at another level of gene expression. Although there is no recognizable sequence homology, this activity could well correspond to the recruitment functions of the speckle-modifying herpesvirus regulator proteins that engage the cellular export pathway. Possibly, the interaction with nuclear speckles reflects a similar activity of the B/NS1 protein involving the coupling of viral transcripts with cellular export factors. This suggestion is supported by our finding that the B/NS1 protein interacted with the RNA helicase UAP56, a protein involved in RNA export (28). The difference in A/NS1 and B/NS1 protein localizations may indicate that the two influenza virus types have evolved slightly different mechanisms to integrate viral transcripts into cellular pathways. Clearly, more experimental work is required to test these hypotheses.
At about 8 h p.i., the B/NS1 protein started to progressively localize to the cytoplasm until only a minor fraction was detectable in the nuclear compartment at 16 h p.i. At this time point, the SC35 antigen was not stained in rounded or speckled structures but was more evenly distributed in the nucleoplasm, illustrating a loss of colocalization with B/NS1. The cytoplasmic localization of the B/NS1 protein at the late time point is in line with its previously described suppressive effects on the antiviral protein PKR and the signaling pathway leading to type I IFN induction (16). The cytoplasmic localization late in infection was unaffected by LMB, indicating that the protein enters the cytoplasm in a CRM1-independent manner, which is apparently accompanied by overruling of the B/NS1 karyophilic signal. Interestingly, the late relocalization was observed only during virus infection and did not occur in B/NS1-transfected cells, suggesting that additional virus-encoded proteins or RNAs trigger this process. The cytoplasmic accumulation of the B/NS1 protein may depend on the interaction with another transported protein and/or RNA or, alternatively, could be regulated by an exposed export signal. It will be interesting to distinguish between these possibilities.
In conclusion, our analysis of the influenza B virus NS1 protein revealed its highly dynamic intracellular localization during infection. This includes early accumulation in nuclear speckle domains and an intriguing coalescence of their appearance, indicating a role in viral- or cellular-RNA biogenesis that is important for efficient virus replication. Further analysis of this novel virus-host interaction not only may reveal how influenza viruses facilitate efficient gene expression, but also may lead to novel insights into the pathways of cellular-RNA maturation.
Acknowledgments
We are grateful to Elmar Wahle (Halle, Germany) for anti-PABP2 antiserum and to Thomas Stamminger (Erlangen, Germany), David Goodrich (Buffalo, NY), and Stuart Wilson (Sheffield, United Kingdom) for expression plasmids. We thank Anne Sadewasser, Gudrun Heins, and Andrea Zoehner for excellent technical support.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (Wo 554-3/2), the Virgil network of excellence funded within FP6 of the European Commission, and the influenza research program “FSI” of the German federal government.
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Bao, Y., P. Bolotov, D. Dernovoy, B. Kiryutin, L. Zaslavsky, T. Tatusova, J. Ostell, and D. Lipman. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17251-256. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 746203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernardi, R., and P. P. Pandolfi. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 81006-1016. [DOI] [PubMed] [Google Scholar]
- 5.Boyne, J. R., K. J. Colgan, and A. Whitehouse. 2008. Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front. Biosci. 132928-2938. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., T. Misteli, G. R. Screaton, D. L. Spector, and A. R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calado, A., and M. Carmo-Fonseca. 2000. Localization of poly(A)-binding protein 2 (PABP2) in nuclear speckles is independent of import into the nucleus and requires binding to poly(A) RNA. J. Cell Sci. 1132309-2318. [DOI] [PubMed] [Google Scholar]
- 8.Carmo-Fonseca, M., R. Pepperkok, M. T. Carvalho, and A. I. Lamond. 1992. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 1171-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca, M., R. Pepperkok, B. S. Sproat, W. Ansorge, M. S. Swanson, and A. I. Lamond. 1991. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 101863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 7612877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., G. Liao, M. Fujimuro, O. J. Semmes, and S. D. Hayward. 2001. Properties of two EBV Mta nuclear export signal sequences. Virology 288119-128. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 182273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colwill, K., T. Pawson, B. Andrews, J. Prasad, J. L. Manley, J. C. Bell, and P. I. Duncan. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15265-275. [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper, M., D. J. Goodwin, K. T. Hall, A. J. Stevenson, D. M. Meredith, A. F. Markham, and A. Whitehouse. 1999. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J. Gen. Virol. 801311-1316. [DOI] [PubMed] [Google Scholar]
- 15.Dauber, B., G. Heins, and T. Wolff. 2004. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J. Virol. 781865-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dauber, B., J. Schneider, and T. Wolff. 2006. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J. Virol. 8011667-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degot, S., H. Le Hir, F. Alpy, V. Kedinger, I. Stoll, C. Wendling, B. Seraphin, M. C. Rio, and C. Tomasetto. 2004. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J. Biol. Chem. 27933702-33715. [DOI] [PubMed] [Google Scholar]
- 18.Donelan, N. R., B. Dauber, X. Wang, C. F. Basler, T. Wolff, and A. Garcia-Sastre. 2004. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 7811574-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dye, B. T., and J. G. Patton. 2001. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp. Cell Res. 263131-144. [DOI] [PubMed] [Google Scholar]
- 20.Eilbracht, J., and M. S. Schmidt-Zachmann. 2001. Identification of a sequence element directing a protein to nuclear speckles. Proc. Natl. Acad. Sci. USA 983849-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elton, D., M. Simpson-Holley, K. Archer, L. Medcalf, R. Hallam, J. McCauley, and P. Digard. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75408-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagerlund, R., L. Kinnunen, M. Kohler, I. Julkunen, and K. Melen. 2005. NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 28015942-15951. [DOI] [PubMed] [Google Scholar]
- 23.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 24.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortes, P., A. I. Lamond, and J. Ortin. 1995. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 761001-1007. [DOI] [PubMed] [Google Scholar]
- 26.Fu, X. D., and T. Maniatis. 1990. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343437-441. [DOI] [PubMed] [Google Scholar]
- 27.Garaigorta, U., and J. Ortin. 2007. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 354573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 111716-1721. [DOI] [PubMed] [Google Scholar]
- 29.Greenspan, D., P. Palese, and M. Krystal. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 623020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall, L. L., K. P. Smith, M. Byron, and J. B. Lawrence. 2006. Molecular anatomy of a speckle. Anat. Rec. A 288664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 687790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedley, M. L., H. Amrein, and T. Maniatis. 1995. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. USA 9211524-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278335-342. [DOI] [PubMed] [Google Scholar]
- 34.Hiriart, E., H. Gruffat, M. Buisson, I. Mikaelian, S. Keppler, P. Meresse, T. Mercher, O. A. Bernard, A. Sergeant, and E. Manet. 2005. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 28036935-36945. [DOI] [PubMed] [Google Scholar]
- 35.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365186-191. [DOI] [PubMed] [Google Scholar]
- 36.Huang, S., T. J. Deerinck, M. H. Ellisman, and D. L. Spector. 1994. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 126877-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause, S., S. Fakan, K. Weis, and E. Wahle. 1994. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 21475-82. [DOI] [PubMed] [Google Scholar]
- 39.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4605-612. [DOI] [PubMed] [Google Scholar]
- 40.Lange, A., R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine, and A. H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2825101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. T. Virgin. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 1041371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, Y., X. Wang, X. Zhang, and D. W. Goodrich. 2005. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol. Cell. Biol. 254023-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 81817-1828. [DOI] [PubMed] [Google Scholar]
- 44.Majerciak, V., K. Yamanegi, E. Allemand, M. Kruhlak, A. R. Krainer, and Z. M. Zheng. 2008. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 822792-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 27933001-33011. [DOI] [PubMed] [Google Scholar]
- 46.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416499-506. [DOI] [PubMed] [Google Scholar]
- 47.Matera, A. G., and K. B. Shpargel. 2006. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 18317-324. [DOI] [PubMed] [Google Scholar]
- 48.Melen, K., L. Kinnunen, R. Fagerlund, N. Ikonen, K. Y. Twu, R. M. Krug, and I. Julkunen. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 815995-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Min, J. Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363236-243. [DOI] [PubMed] [Google Scholar]
- 51.Molenaar, C., A. Abdulle, A. Gena, H. J. Tanke, and R. W. Dirks. 2004. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J. Cell Biol. 165191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1991-1000. [DOI] [PubMed] [Google Scholar]
- 53.Norton, G. P., T. Tanaka, K. Tobita, S. Nakada, D. A. Buonagurio, D. Greenspan, M. Krystal, and P. Palese. 1987. Infectious influenza A and B virus variants with long carboxyl terminal deletions in the NS1 polypeptides. Virology 156204-213. [DOI] [PubMed] [Google Scholar]
- 54.O'Keefe, R. T., A. Mayeda, C. L. Sadowski, A. R. Krainer, and D. L. Spector. 1994. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Opitz, B., A. Rejaibi, B. Dauber, J. Eckhard, M. Vinzing, B. Schmeck, S. Hippenstiel, N. Suttorp, and T. Wolff. 2007. IFNβ induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 9930-938. [DOI] [PubMed] [Google Scholar]
- 56.Palese, P., and M. Shaw. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647-1690. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 57.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6187-198. [DOI] [PubMed] [Google Scholar]
- 58.Phelan, A., M. Carmo-Fonseca, J. McLaughlan, A. I. Lamond, and J. B. Clements. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 909056-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Politz, J. C., R. A. Tuft, K. V. Prasanth, N. Baudendistel, K. E. Fogarty, L. M. Lifshitz, J. Langowski, D. L. Spector, and T. Pederson. 2006. Rapid, diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Mol. Biol. Cell 171239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sacco-Bubulya, P., and D. L. Spector. 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saitoh, N., C. S. Spahr, S. D. Patterson, P. Bubulya, A. F. Neuwald, and D. L. Spector. 2004. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 153876-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 696063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzweig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 1041853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith, R. W., P. Malik, and J. B. Clements. 2005. The herpes simplex virus ICP27 protein: a multifunctional post-transcriptional regulator of gene expression. Biochem. Soc. Trans. 33499-501. [DOI] [PubMed] [Google Scholar]
- 65.Sorg, G., and T. Stamminger. 1999. Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to beta-galactosidase. BioTechniques 26858-862. [DOI] [PubMed] [Google Scholar]
- 66.Spector, D. L., X. D. Fu, and T. Maniatis. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 103467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Summers, M. D., and G. E. Smith. 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. Tex. Agric. Exp. Stn. Bull. 15551-57. [Google Scholar]
- 68.Toth, Z., P. Lischka, and T. Stamminger. 2006. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 341237-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner, S., S. Chiosea, and J. A. Nickerson. 2003. The spatial targeting and nuclear matrix binding domains of SRm160. Proc. Natl. Acad. Sci. USA 1003269-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, W., and R. M. Krug. 1996. The RNA-binding and effector domains of the viral NS1 protein are conserved to different extents among influenza A and B viruses. Virology 22341-50. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe, K., N. Takizawa, M. Katoh, K. Hoshida, N. Kobayashi, and K. Nagata. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 7731-42. [DOI] [PubMed] [Google Scholar]
- 72.Whittaker, G., M. Bui, and A. Helenius. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J. Virol. 702743-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams, B. J., J. R. Boyne, D. J. Goodwin, L. Roaden, G. M. Hautbergue, S. A. Wilson, and A. Whitehouse. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolff, T., R. E. O'Neill, and P. Palese. 1998. NS1-binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 727170-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeakley, J. M., H. Tronchere, J. Olesen, J. A. Dyck, H. Y. Wang, and X. D. Fu. 1999. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 145447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin, C., J. A. Khan, G. V. Swapna, A. Ertekin, R. M. Krug, L. Tong, and G. T. Montelione. 2007. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J. Biol. Chem. 28220584-20592. [DOI] [PubMed] [Google Scholar]
- 77.Zeng, C., E. Kim, S. L. Warren, and S. M. Berget. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 161401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407401-405. [DOI] [PubMed] [Google Scholar]