Abstract
Human herpesvirus 8 (HHV-8) is the etiological agent of Kaposi's sarcoma (KS). HHV-8 encodes an antiapoptotic viral Fas-associated death domain-like interleukin-1β-converting enzyme-inhibitory protein (vFLIP/K13). The antiapoptotic activity of vFLIP/K13 has been attributed to an inhibition of caspase 8 activation and more recently to its capability to induce the expression of antiapoptotic proteins via activation of NF-κB. Our study provides the first proteome-wide analysis of the effect of vFLIP/K13 on cellular-protein expression. Using comparative proteome analysis, we identified manganese superoxide dismutase (MnSOD), a mitochondrial antioxidant and an important antiapoptotic enzyme, as the protein most strongly upregulated by vFLIP/K13 in endothelial cells. MnSOD expression was also upregulated in endothelial cells upon infection with HHV-8. Microarray analysis confirmed that MnSOD is also upregulated at the RNA level, though the differential expression at the RNA level was much lower (5.6-fold) than at the protein level (25.1-fold). The induction of MnSOD expression was dependent on vFLIP/K13-mediated activation of NF-κB, occurred in a cell-intrinsic manner, and was correlated with decreased intracellular superoxide accumulation and increased resistance of endothelial cells to superoxide-induced death. The upregulation of MnSOD expression by vFLIP/K13 may support the survival of HHV-8-infected cells in the inflammatory microenvironment in KS.
Kaposi's sarcoma (KS) is a multifocal tumor that appears in four epidemiologically distinct forms: classical, iatrogenic, AIDS-associated, and African endemic KS (63). KS lesions evolve over time from patch- or plaque-like lesions at early stages to nodular lesions characteristic of later stages (for a review, see references 21 and 63). Histologic hallmarks that characterize all stages of KS include a prominent microvasculature and a large inflammatory compartment of CD8+ T cells and monocytes (for a review, see references 21 and 63). Especially in later stages of development, bundles of spindle-shaped cells become the dominant cellular feature. These so-called KS spindle cells (KSC) express markers of lymph vessel endothelial cells and are regarded as the tumor cells of KS (17, 76). As yet, it is unclear whether KSC are derived from lymph or blood vessel endothelial cells, or from both (32, 60, 73).
Human herpesvirus 8 (HHV-8), also known as KS-associated herpesvirus, is regarded as the etiological agent of KS (8). In addition, HHV-8 is associated with primary effusion lymphoma and multicentric Castleman's disease (1). In KS lesions, only a few lytically HHV-8-infected cells are detected (6), whereas more than 70% of KSC are latently infected (58, 59). During latency, only a few of the more than 80 viral genes are expressed (22, 53). These genes are crucial to establish latent infection and to protect infected cells from apoptosis (62). The latter is specifically important considering the inflammatory conditions present in KS, which may induce apoptosis in infected cells by cell-mediated cytotoxicity and/or the formation of high concentrations of reactive oxygen species (ROS) (56, 77).
A growing body of evidence suggests that the inhibition of cell death rather than increased proliferation may be key for the growth of the lesions. For example, KSC exhibit longer doubling times than normal endothelial cells in vitro (14, 57), which is in agreement with the fact that generally low proliferation rates of KSC are observed in all stages of KS in vivo (14, 57). Most importantly, the numbers of apoptotic cells in KS decrease from early to late stages, and this is inversely related to increasing numbers of HHV-8-infected cells (62).
The K13 gene of HHV-8 is a latent gene that is expressed in almost all KSC in vivo (62). This gene encodes a viral Fas-associated death domain-like interleukin-1β (IL-1β)-converting enzyme-inhibitory protein (vFLIP/K13) with homology to cellular FLIP (cFLIP) molecules (cFLIP-long, -short, and -R) and with certain caspases (67, 71, 78). It is well established that vFLIP/K13 has antiapoptotic activity (20, 64) and plays an important role in the pathogenesis of HHV-8-associated tumors, including KS, primary effusion lymphoma, and multicentric Castleman's disease (15, 28, 62). In contrast, the mechanisms of vFLIP/K13 antiapoptotic activities are controversial and continue to be debated.
vFLIP/K13 harbors a death effector domain, suggesting that it may bind to caspase 8 and inhibit its activation, which may protect cells from death receptor-induced apoptosis (9, 67). This hypothesis was initially supported by a report showing that vFLIP/K13 and caspase 8 can bind to each other (5). However, these results were questioned in subsequent studies (10, 24, 44). Presently, evidence is accumulating that vFLIP/K13 can interact with and activate the regulatory components of the IκB kinase complex (3, 23, 39). It has been shown that this activates NF-κB signaling (2, 9, 27, 44) and induces the expression of cellular proteins, including antiapoptotic factors, such as cellular inhibitor of apoptosis (cIAP) and cFLIP (28, 64). As yet, the effect of vFLIP/K13 on cellular-gene expression has been investigated only for selected genes. It is not clear whether the respective genes may also encode the proteins most abundantly induced by vFLIP/K13 and whether additional factors may mediate the antiapoptotic activity of vFLIP/K13 in endothelial cells.
Using two-dimensional difference gel electrophoresis (2D-DIGE) analysis, we identified manganese superoxide dismutase (MnSOD), a mitochondrial antioxidant and an important antiapoptotic enzyme, as the protein most strongly upregulated by vFLIP/K13 in primary human endothelial cells. Evidence is provided that MnSOD may be an important mediator of the antiapoptotic activity of vFLIP/K13.
MATERIALS AND METHODS
Cell culture and HHV-8 infection.
Primary human umbilical vein endothelial cells (HUVEC) were purchased from Cambrex Bio Science (Verviers, Belgium) and PromoCell (Heidelberg, Germany) and cultivated in EGM-2-MV medium (Cambrex) or ECGM (PromoCell), respectively. The cells from Cambrex were cultivated on 1.5% bovine skin gelatin type B (Sigma-Aldrich, Munich, Germany)-coated flasks and split routinely at a ratio of 1:4 (54, 74).
HEK 293 cells were infected with a recombinant HHV-8 encoding green fluorescent protein (GFP) (70) to produce high-titer HHV-8 supernatants and were selected for 10 days with 1 μg/ml puromycin. Afterwards, lytic replication of HHV-8 was induced by transfection (the calcium phosphate method) with an expression plasmid encoding RTA (open reading frame 50). The cells were washed once with 1× phosphate-buffered saline (PBS) 7 h after transfection. Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS), 1 mM sodium butyrate, and 20 ng/ml phorbol-12-myristate-13-acetate was added. The medium was replaced after 20 h with DMEM containing 1 mM sodium butyrate only; 48 h later, the supernatants were collected and debris was removed by centrifugation (300 × g; 10 min; 4°C). The supernatants were first filtered (0.45 μm) and then centrifuged (48,400 × g; 2 h; 4°C) to concentrate the virus particles. Finally, the supernatants were removed and the viral pellets were dissolved in 500 μl EGM-2-MV or ECGM containing 8 μg/ml Polybrene. Virus-containing supernatants were added immediately to HUVEC (passage 4.5), which had been plated in 12-well plates at a density of 50,000 cells/well 24 h before. The medium was replaced with fresh EGM-2-MV or ECGM after 48 h. After an additional 24 h, the cells were selected with 0.5 μg/ml puromycin for 10 days.
Plasmids.
The full-length K13 sequence was amplified from genomic viral DNA of BCBL-1 cells by PCR and was either tagged with a 3′ Myc/His epitope or left untagged. The K13 constructs were cloned into the Moloney murine leukemia virus-derived retroviral vector pBabe-Puro or into the vector pMCV 1.4(−) (Mologen AG, Berlin, Germany). The mutation in vFLIP/K13-58 was generated with a QuikChange Site-Directed Mutagenesis kit (Stratagene, Amsterdam, The Netherlands) and appropriate primers. In the resulting vFLIP/K13-58 protein, the amino acid residues 58 to 60 (Glu-Cys-Leu) of wild-type vFLIP/K13 were replaced with alanine residues, which fully abrogated the capability of vFLIP/K13-58 to activate NF-κB (66). The sequence of MC159 from the molluscum contagiosum virus was amplified by PCR and cloned as a construct encoding N-terminally vesicular stomatitis virus (VSV)-tagged vFLIP/MC159 into the pMCV 1.4(−) vector. The sequences of the PCR-amplified products were confirmed by sequencing them.
Retrovirus production and infection and transfection of HUVEC.
For retroviral transduction, 2.5 × 106 human embryonic kidney (HEK 293T) cells were seeded in 10-cm dishes and cultivated in 9 ml DMEM supplemented with 10% FCS. On the next day, the cells were transfected (by the calcium phosphate method) with 1 μg pVSV-G vector, 5 μg pGag-Pol vector, and 5 μg either K13-pBabe or pBabe vector. The cells were incubated for 24 h at 37°C and 5% CO2. Afterwards, the cells were washed once with 1× PBS, and 10 ml of Ultraculture medium (Cambrex) supplemented with 1% glutamine was added to the cells. After incubation for 24 h at 37°C and 5% CO2, the supernatants were filtered (0.45 μm) and the viruses were concentrated by centrifugation (48,400 × g; 2 h; 4°C). For infection, the viral pellets were resuspended in 300 μl EGM-2-MV or ECGM medium containing 5% FCS and 8 μg/ml Polybrene and added to HUVEC (passage 3) that had been seeded the day before (29, 47). The medium was changed 24 h after infection, and 48 h after infection, the cells were selected for 10 days with EGM-2-MV medium containing 5% FCS and 0.3 μg/ml puromycin. Three independent pools of vFLIP/K13-transduced HUVEC were generated and analyzed. One pool was transduced with an untagged vFLIP/K13, and two other pools, obtained from two different companies (Cambrex and PromoCell), were transduced with a Myc-tagged version of the protein. With every pool, identical results were obtained.
Transient transfection of HUVEC was performed with SuperFect (Qiagen, Hilden, Germany) as described previously (47). For cotransfection with small interfering RNA (siRNA) molecules, MnSOD or control siRNA was added to the transfection mixture containing the expression plasmid and SuperFect in a final concentration of 33 nM. MnSOD siRNA (5′-GGAACAACAGGCCUUAUUC-3′; ID8959) was purchased from Ambion/Applied Biosystems (Darmstadt, Germany). Control siRNAs were purchased from Ambion (negative control no. 1 siRNA) or Invitrogen (Block-iT control siRNA); both were tested and shown not to target any known gene. For inhibition of the NF-κB signaling pathway, cells were treated 6 h prior to transfection with 2 μM Bay 11-7082 (Calbiochem, Darmstadt, Germany) (26). The medium was changed 2 h after transfection. The new medium again contained 2 μM Bay 11-7082. After 48 h, cells were harvested for analysis.
Isolation of RNA.
A vector encoding vFLIP/K13 fused to a Myc tag at its C terminus for immunochemical detection (K13-EC) and a retroviral control vector (Ctrl-EC) were harvested using trypsin/EDTA (0.05%/0.02%) in 1× PBS (PAA Laboratories, Pasching, Austria). Total RNA was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The RNA concentration was determined photometrically (GeneQuant; GE-Healthcare, Munich, Germany) at a λ of 260 nm, and RNA integrity was controlled by nondenaturing agarose gel electrophoresis (1%) (Peqlab, Erlangen, Germany) and ethidium bromide (Roth, Karlsruhe, Germany) staining.
Microarray analysis.
Preparation of cDNA probes (from 5 to 10 μg total RNA), fragmentation, hybridization of HG-U133 plus 2.0 microarray GeneChips (Affymetrix, Santa Clara, CA), washing, staining, and scanning were performed following the manufacturer's protocols (Affymetrix) with a commercial partner (L. Klein-Hitpass, Institute for Cell Biology, University of Essen, Essen, Germany) (46). Raw data derived from GeneChips was normalized by “global scaling” using the Affymetrix Microarray Suite Data Mining Tool. Comparison files were further filtered to detect differentially upregulated genes. The filter criteria included a change of “I” (increased), a change of >4-fold, a K13-EC-versus-Ctrl-EC P value change of <0.001, a K13-EC detection P value of <0.001, and an overall K13-EC signal intensity of >300.
Identification of statistically overrepresented gene ontology terms was done with the GOstat analysis tool (4) with the following settings: GO-gene-association database, goa_human; minimal length of considered GO paths, 3; maximal P value in GO output list, 0.3; cluster GOs, −1; and correct for multiple testing, false discovery rate (Benjamini).
2D-DIGE.
Cells were harvested with trypsin/EDTA and washed once with EGM-2-MV medium and twice with 0.5× PBS. The cells were lysed in labeling buffer (8 M urea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 30 mM Tris-HCl, pH 8.5). Proteins (50 μg in 20 μl lysate) were labeled with 400 pmol CyDyes DIGE Cy3 or Cy5 (GE-Healthcare) for 30 min on ice. The N-hydroxysuccinimidyl ester reactive group of the DIGE fluors covalently attaches to the ɛ-amino group of lysine residues in proteins by an amide linkage formation. Since the amount of dye added is limiting, this method is referred to as minimal labeling. An excess of lysine was added (0.5 μM final concentration) to stop the labeling reaction. The lysates were combined, and 160 μl of rehydration buffer (8 M urea, 4% CHAPS, 25 μl Pharmalyte, pH 3 to 10, 5 mg dithiothreitol) was added for isoelectric focusing. Using DryStrip gels (GE-Healthcare; pH 3 to 11 nonlinear; 11 cm) and the IPGphor3 focusing unit (GE-Healthcare), the proteins were separated according to their isoelectric points by applying a total of 12 kVh. The DryStrips were incubated in sodium dodecyl sulfate (SDS) equilibration buffer 1 (6 M urea, 30% glycerol, 2% SDS, 100 mM Tris-HCl, pH 8.0, 0.5% dithiothreitol) and equilibration buffer 2 (6 M urea, 30% glycerol, 2% SDS, 100 mM Tris-HCl, pH 8.0, 4.5% iodoacetamide) for 10 min each before polyacrylamide gel electrophoresis. The separation of the proteins was performed in a 13- by 18-cm 10% polyacrylamide gel (29:1; Bio-Rad, Munich, Germany). The gel was scanned with a FLA-5000 laser scanner (Fuji, Düsseldorf, Germany) after the run. Image analyses and quantification were done with Decodon 2D image software (Decodon, Greifswald, Germany). Before fusion of the gel images with the union-type program, amplitude rescaling and background removal were done. As spot detection criteria, the local background was set to a value of 39, the average spot size to a value of 13, and the sensitivity to a value of 15. Spot selection criteria were a relative volume of >0.003% on the fused gel image and >0.002% on the individual Cy3 and Cy5 gel images.
Nuclear/cytosol fractionation.
Fractionation was done with the Nuclear/Cytosol-Fractionation kit from BioVision (Wiesbaden, Germany) (52). The protein concentration was determined with a microplate reader (Bio-Rad) at 750 nm using the DC protein assay (Bio-Rad). All extracts were stored at −80°C until analysis was performed.
Western blotting.
Western blotting was performed using polyvinylidene difluoride membranes (Roth) and 5% nonfat milk as a blocking reagent, as described previously (29, 40). The following antibodies were used: anti-Myc (9B11, monoclonal antibody [MAb], mouse, 1:3,000; Cell Signaling, Frankfurt, Germany), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (MAb, mouse, 1:60,000; Chemicon, Hampshire, United Kingdom), anti-MnSOD (polyclonal antibody [PAb], rabbit, 1:1,000; Upstate/Millipore, Schwalbach, Germany), anti-VSV (MAb, mouse, 1:2,000; Sigma-Aldrich, Munich, Germany), anti-NF-κB (p65) (PAb, rabbit, 1:200; Zymed/Invitrogen, Karlsruhe, Germany), anti-lamin A/C (PAb, goat, 1:1,000; Santa Cruz, Heidelberg, Germany), and anti-vFLIP/K13 (clone 4C1, MAB, rat, 1:10) (52). Horseradish peroxidase-conjugated secondary antibodies were purchased from GE-Healthcare and were all diluted 1:5,000.
Flow cytometry analysis.
HUVEC (2 × 105) were treated with the specified concentrations of the superoxide inducer Ly83583 (Sigma-Aldrich) for 24 h. An Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences, Heidelberg, Germany) was used for the determination of apoptotic HUVEC. The cells were detached with trypsin/EDTA, washed in 1× PBS, and resuspended in binding buffer according to the manufacturer's protocol. Cells (1 × 105) in 100 μl buffer were stained with 5 μl annexin V-fluorescein isothiocyanate and 5 μl propidium iodide (PI) for 15 min at 4°C. Afterwards, the cells were analyzed using flow cytometry (FACSCalibur and CellQuest Pro Software; BD Biosciences). For determination of intracellular superoxide levels, 2 × 105 HUVEC were incubated for 24 h with 4 μM Ly83583 or left untreated. Then, dihydroethidium (DHE) was added to the medium to a final concentration of 10 μM. The cells were incubated for an additional 30 min at 37°C, washed twice with 1× PBS, detached with trypsin/EDTA, and washed again with 1× PBS. The cells were centrifuged and resuspended in 500 μl 1× PBS and analyzed by flow cytometry.
Immunofluorescence.
Cells were grown to confluence in chamber slides, fixed with 10% neutral buffered formaldehyde (Sigma-Aldrich) for 10 min, and permeabilized with 0.1% saponin for 30 min. MnSOD antibody (Upstate/Millipore) was diluted 1:100 and incubated for 1 h. Alexa 546-coupled secondary antibody (Molecular Probes/Invitrogen, Karlsruhe, Germany) was diluted 1:500 and incubated for 45 min (52, 69). The slides were mounted with fluorescent mounting medium (Dako, Hamburg, Germany). Staining was visualized using a Zeiss Axiovert 100 M (Zeiss, Oberkochen, Germany) confocal laser scanning microscope.
Immunocytochemistry.
Cells were grown to confluence in chamber slides and fixed with ice-cold ethanol for 20 min (54). MnSOD antibody (Upstate/Millipore) was diluted 1:100 and incubated for 1 h. Stains were visualized using the R.T.U. Vectastain Elite Kit Universal (Vector Laboratories). The stains were developed with NovaRed substrate (Vector) and counterstained with Haematoxylin Gill III (Merck, Darmstadt, Germany). The stained cells were photographed using an Aristoplan microscope (Leica, Wetzlar, Germany) equipped with a 40× objective and a 3CCD Exwave HAD color camera (Sony, Berlin, Germany).
Nucleotide sequence accession numbers.
The accession number for the vFLIP/K13 protein is AAB62643, and that for the vFLIP/MC159 protein is AAC55287.
Microarray data accession number.
The microarray experiment design, setup, and results are available through Array Express (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MEXP-1469.
RESULTS
vFLIP/K13 induces spindle cell morphology in HUVEC.
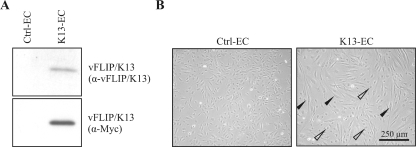
To assess potential changes in cellular-gene expression induced by vFLIP/K13, HUVEC were stably transduced with a retroviral control vector (Ctrl-EC) or a vector encoding for vFLIP/K13 fused to a Myc tag at its C terminus (K13-EC). Western blot analysis of the transduced cells with a specific monoclonal rat anti-vFLIP/K13 antibody (52) (Fig. 1A, top) or with an antibody directed against the Myc tag (Fig. 1A, bottom) demonstrated that the tagged vFLIP/K13 protein was expressed in HUVEC. vFLIP/K13 expression was also detectable in HUVEC stably expressing untagged vFLIP/K13 with the monoclonal vFLIP/K13 antibody in Western blot analysis (data not shown). Since the signal intensity with an anti-vFLIP/K13 antibody was significantly lower than with the anti-Myc antibody (Fig. 1A), the Myc-tagged protein was used in combination with the anti-Myc antibody for all subsequent experiments, except where otherwise indicated. Ctrl-EC exhibited the characteristic endothelial homogeneous cobblestone morphology (Fig. 1B, left), while in K13-EC cultures, numerous spindle-shaped cells appeared (Fig. 1B, right), which is consistent with previous reports (27, 44). In addition to spindle-shaped cells, cells with cobblestone morphology were also present in the K13-transduced cultures (Fig. 1B, right). This suggests that the spindle cell morphology was correlated with vFLIP/K13 expression levels in the individual cells. The same mixture of cell morphologies was also seen in cultures expressing untagged vFLIP/K13 (data not shown).
FIG. 1.
vFLIP/K13 expression induces spindle shape in HUVEC. (A) HUVEC were transduced with the control vector (Ctrl-EC) or a vector encoding vFLIP/K13 fused to a Myc tag at its C terminus for immunochemical detection (K13-EC). vFLIP/K13 expression could be detected using both an anti-vFLIP/K13-specific rat MAb (top) and an anti-Myc antibody (bottom). (B) Control vector-transduced cells showed the cobblestone morphology typical of endothelial cells (left), whereas in K13-transduced cultures, cells with an elongated spindle shape (right, filled arrowheads) and cobblestone morphology (open arrowheads) were detectable.
The vFLIP/K13-modulated transcriptome is characterized by increased expression of immune, defense, and inflammation genes.
In order to detect cellular genes induced by vFLIP/K13 on the transcript level, a cDNA microarray comparison between K13-EC and Ctrl-EC was performed (Table 1). Analysis of the cDNA microarray data with the GOstat (4) tool showed that the statistically most overrepresented gene ontology terms belonged to the groups immune system process (P value, 5.88 × 10−36), defense response (P value, 6.13 × 10−22), inflammatory response (P value, 2.87 × 10−19), and cytokine activity (P value, 8.56 × 10−19). The genes assigned to these groups are indicated in Table 1. More than one-fourth (26.6%) of all vFLIP/K13-upregulated genes (173 genes) were associated with one of these groups. Among them, several cytokines and their receptors and chemokines with known functions in the regulation of inflammation and response against pathogens were found, e.g., IL-1α, IL-1β, IL-8, IL18R, CCL5, CCL20, and CXCL2. In contrast, only a few of the vFLIP/K13-upregulated genes (9.8%) belonged to the gene ontology group of apoptosis regulation (Table 1). About half of them are known to possess antiapoptotic functions (e.g., BCL2A1, BIRC3, TNFSF13b, and SOD2 [encoding MnSOD]) (30, 45, 55, 72), whereas others are assigned to proapoptotic activities (e.g., RIPK2, TNFRSF19, and RASSF5) (19, 33, 50).
TABLE 1.
Cellular genes upregulated in K13-EC compared to Ctrl-EC
| Gene symbola | UniGene ID | Induction | Gene title | Reference |
|---|---|---|---|---|
| CCL5 | Hs.514821 | 929.3 | Chemokine (C-C motif) ligand 5 (RANTES) | 27 |
| C15orf48 | Hs.112242 | 424.6 | Chromosome 15 open reading frame 48 | |
| TRPA1 | Hs.137674 | 418.8 | Transient receptor potential cation channel, subfamily A, member 1 | |
| TNC | Hs.143250 | 319.6 | Tenascin C (hexabrachion) | |
| CSF3 | Hs.2233 | 252.5 | Colony-stimulating factor 3 (granulocyte) | |
| TNFAIP6 | Hs.437322 | 212.3 | Tumor necrosis factor, alpha-induced protein 6 | |
| DNER | Hs.234074 | 186.1 | Delta-notch-like EGF repeat-containing transmembrane | |
| CCL3 | Hs.514107 | 162.0 | Chemokine (C-C motif) ligand 3 | |
| WT1 | Hs.591980 | 115.4 | Wilms' tumor 1 | |
| SEMA3C | Hs.269109 | 107.6 | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted | |
| WNT5A | Hs.561260 | 102.5 | Wingless-type MMTV integration site family, member 5A | |
| HSD11B1 | Hs.195040 | 96.3 | Hydroxysteroid (11-beta) dehydrogenase 1 | |
| PRRX1 | Hs.632475 | 92.4 | Paired related homeobox 1 | |
| VNN1 | Hs.12114 | 85.6 | Vanin 1 | |
| SERPINB2 | Hs.514913 | 81.6 | Serpin peptidase inhibitor, clade B (ovalbumin), member 2 | |
| PLA1A | Hs.437451 | 78.8 | Phospholipase A1 member A | |
| IL1B | Hs.126256 | 75.6 | IL-1β | |
| LUM | Hs.406475 | 69.1 | Lumican | |
| PAPPA | Hs.494928 | 66.3 | Pregnancy-associated plasma protein A, pappalysin 1 | |
| RGS7 | Hs.130171 | 60.1 | Regulator of G-protein signaling 7 | |
| CXCL5 | Hs.89714 | 58.9 | Chemokine (C-X-C motif) ligand 5 | |
| CCL20 | Hs.75498 | 58.1 | Chemokine (C-C motif) ligand 20 (MIP-3α) | 27 |
| TNFRSF11B | Hs.81791 | 56.9 | TNF receptor superfamily, member 11b (osteoprotegerin) | |
| KYNU | Hs.470126 | 51.6 | Kynureninase (l-kynurenine hydrolase) | |
| IL1A | Hs.1722 | 48.8 | IL-1α | |
| C12orf39 | Hs.130692 | 48.8 | Chromosome 12 open reading frame 39 | |
| STC1 | Hs.25590 | 47.2 | Stanniocalcin 1 | |
| EBI3 | Hs.501452 | 45.3 | Epstein-Barr virus-induced gene 3 | |
| IFI30 | Hs.14623 | 44.3 | Gamma interferon-inducible protein 30 | |
| CXCL10 | Hs.632586 | 44.0 | Chemokine (C-X-C motif) ligand 10 | |
| CSF2 | Hs.1349 | 39.7 | Colony-stimulating factor 2 (granulocyte-macrophage) | |
| TNFSF13B | Hs.525157 | 39.1 | TNF (ligand) superfamily, member 13b | |
| UBD | Hs.44532 | 38.3 | Ubiquitin D | |
| INDO | Hs.840 | 35.3 | Indoleamine-pyrrole 2,3 dioxygenase | |
| LOC129607 | Hs.7155 | 32.0 | Hypothetical protein LOC129607 | |
| IL-6 | Hs.512234 | 30.9 | IL-6 (interferon, beta 2) | 27, 44 |
| C1S | Hs.458355 | 30.3 | Complement component 1, s subcomponent | |
| MX1 | Hs.517307 | 26.7 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 | |
| MGAT4C | Hs.126195 | 25.8 | mannosyl (α-1,3-)-glycoprotein β-1,4-N-acetylglucosaminyltransferase, isozyme C | |
| C3 | Hs.529053 | 25.3 | Complement component 3 | |
| BCL2A1 | Hs.227817 | 24.9 | BCL2-related protein A1 | |
| CX3CL1 | Hs.531668 | 24.1 | Chemokine (C-X3-C motif) ligand 1 | |
| MRGPRX3 | Hs.380177 | 23.6 | MAS-related GPR, member X3 | |
| TNIP3 | Hs.208206 | 22.9 | TNFAIP3-interacting protein 3 | |
| TNFRSF9 | Hs.193418 | 22.2 | TNF receptor superfamily, member 9 | |
| TLR2 | Hs.519033 | 21.4 | Toll-like receptor 2 | |
| CCL8 | Hs.271387 | 19.0 | Chemokine (C-C motif) ligand 8 | |
| NKX3-1 | Hs.55999 | 18.4 | NK3 transcription factor related, locus 1 (Drosophila) | |
| CXCL11 | Hs.632592 | 18.3 | Chemokine (C-X-C motif) ligand 11 | |
| CXCL3 | Hs.89690 | 17.9 | Chemokine (C-X-C motif) ligand 3 | 44 |
| C6orf58 | Hs.226268 | 17.9 | Chromosome 6 open reading frame 58 | |
| PELO | Hs.593754 | 16.9 | Pelota homolog (Drosophila) | |
| IL7R | Hs.591742 | 16.7 | IL-7 receptor | |
| BIRC3 | Hs.127799 | 15.8 | Baculoviral IAP repeat-containing 3 | |
| MMP1 | Hs.83169 | 14.9 | Matrix metallopeptidase 1 (interstitial collagenase) | |
| CTSS | Hs.181301 | 13.5 | Cathepsin S | |
| GBP4 | Hs.409925 | 13.5 | Guanylate binding protein 4 | |
| AMPD3 | Hs.501890 | 13.3 | AMP deaminase (isoform E) | |
| SERPINB4 | Hs.123035 | 13.3 | Serpin peptidase inhibitor, clade B (ovalbumin), member 4 | |
| GFPT2 | Hs.30332 | 12.8 | Glutamine-fructose-6-phosphate transaminase 2 | |
| HEY1 | Hs.234434 | 12.6 | Hairy/enhancer-of-split related with YRPW motif 1 | |
| OAS2 | Hs.414332 | 12.1 | 2′-5′-Oligoadenylate synthetase 2, 69/71 kDa | |
| SLCO4A1 | Hs.235782 | 12.1 | Solute carrier organic anion transporter family, member 4A1 | |
| GREM1 | Hs.40098 | 12.0 | Gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) | |
| C1orf24 | Hs.518662 | 11.7 | Chromosome 1 open reading frame 24 | |
| IFI44L | Hs.389724 | 11.3 | Interferon-induced protein 44-like | |
| CA8 | Hs.491813 | 11.3 | Carbonic anhydrase VIII | |
| RSAD2 | Hs.17518 | 11.2 | Radical S-adenosyl methionine domain containing 2 | |
| SELE | Hs.89546 | 10.6 | Selectin E (endothelial adhesion molecule 1) | |
| RELN | Hs.558371 | 10.4 | Reelin | |
| LOC387763 | Hs.530443 | 10.1 | Hypothetical LOC387763 | |
| IL-8 | Hs.624 | 9.8 | IL-8 | 27, 44 |
| MSC | Hs.442619 | 9.8 | Musculin (activated B-cell factor-1) | |
| TRAF1 | Hs.531251 | 9.6 | TNF receptor-associated factor 1 | |
| FLJ21986 | Hs.189652 | 9.6 | Hypothetical protein FLJ21986 | |
| BIC | Hs.388313 | 9.1 | BIC transcript | |
| CD74 | Hs.591258 | 9.0 | CD74 molecule, major histocompatibility complex, class II invariant chain | |
| TNFRSF19 | Hs.149168 | 8.9 | TNF receptor superfamily, member 19 | |
| CPM | Hs.484551 | 8.8 | Carboxypeptidase M | |
| LAMC2 | Hs.591484 | 8.6 | Laminin, gamma 2 | |
| KIAA1217 | Hs.445885 | 8.6 | KIAA1217 | |
| ISG20 | Hs.459265 | 8.2 | Interferon-stimulated exonuclease gene, 20 kDa | |
| G0S2 | Hs.432132 | 8.2 | G0/G1switch 2 | |
| EPSTI1 | Hs.546467 | 8.0 | Epithelial stromal interaction 1 (breast) | |
| TFPI2 | Hs.438231 | 7.9 | Tissue factor pathway inhibitor 2 | |
| PSTPIP2 | Hs.567384 | 7.8 | Proline-serine-threonine phosphatase-interacting protein 2 | |
| TNFAIP3 | Hs.591338 | 7.7 | TNF-α-induced protein 3 | |
| PTGES | Hs.146688 | 7.7 | Prostaglandin E synthase | |
| MMP10 | Hs.2258 | 7.6 | Matrix metallopeptidase 10 (stromelysin 2) | |
| PTGS2 | Hs.196384 | 7.6 | Prostaglandin-endoperoxide synthase 2 (Cox-2) | 44 |
| C1QTNF1 | Hs.201398 | 7.4 | C1q and TNF-related protein 1 | |
| TNFAIP2 | Hs.525607 | 7.1 | TNF-α-induced protein 2 | |
| OAS1 | Hs.524760 | 7.1 | 2′,5′-Oligoadenylate synthetase 1, 40/46 kDa | |
| FLJ20701 | Hs.409352 | 6.9 | Hypothetical protein FLJ20701 | |
| NLF1 | Hs.202656 | 6.8 | Nuclear-localized factor 1 | |
| CLEC4E | Hs.236516 | 6.8 | C-type lectin domain family 4, member E | |
| RGS7 | Hs.130171 | 6.7 | Regulator of G-protein signaling 7 | |
| GCH1 | Hs.86724 | 6.5 | GTP cyclohydrolase 1 (dopa-responsive dystonia) | |
| NFKBIZ | Hs.319171 | 6.5 | Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta | |
| ZC3H12A | Hs.471918 | 6.3 | Zinc finger CCCH-type containing 12A | |
| PRR16 | Hs.157461 | 6.2 | Proline rich 16 | |
| AFF3 | Hs.444414 | 6.2 | AF4/FMR2 family, member 3 | |
| SERPINE2 | Hs.38449 | 6.2 | Serpin peptidase inhibitor, clade E, member 2 | |
| PPM1H | Hs.435479 | 6.2 | Protein phosphatase 1H (PP2C domain containing) | |
| EDNRB | Hs.82002 | 6.1 | Endothelin receptor type B | |
| CD69 | Hs.208854 | 5.9 | CD69 molecule | |
| PRRG4 | Hs.471695 | 5.9 | Proline-rich Gla (G-carboxyglutamic acid) 4 (transmembrane) | |
| ITGB8 | Hs.592171 | 5.9 | Integrin, beta 8 | |
| VCAM1 | Hs.109225 | 5.9 | Vascular cell adhesion molecule 1 | 27 |
| RIPK2 | Hs.103755 | 5.8 | Receptor-interacting serine-threonine kinase 2 | |
| ANK2 | Hs.620557 | 5.8 | Ankyrin 2, neuronal | |
| PAPLN | Hs.509909 | 5.7 | Papilin, proteoglycan-like sulfated glycoprotein | |
| PTHLH | Hs.591159 | 5.7 | Parathyroid hormone-like hormone | |
| SOD2 | Hs.487046 | 5.6 | Superoxide dismutase 2, mitochondrial (MnSOD) | |
| ICAM1 | Hs.515126 | 5.5 | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 35 |
| CXCL2 | Hs.590921 | 5.5 | Chemokine (C-X-C motif) ligand 2 (GRO) | 27 |
| MYO1B | Hs.439620 | 5.5 | Myosin IB | |
| RASSF5 | Hs.497579 | 5.5 | Ras association (RalGDS/AF-6) domain family 5 | |
| ATP13A3 | Hs.529609 | 5.4 | ATPase type 13A3 | |
| PSMB9 | Hs.132682 | 5.4 | Proteasome subunit, beta type, 9 (large multifunctional peptidase 2) | |
| BAMBI | Hs.533336 | 5.4 | BMP and activin membrane-bound inhibitor homolog (X. laevis) | |
| PDE5A | Hs.370661 | 5.3 | Phosphodiesterase 5A, cGMP-specific | |
| IL18R1 | Hs.469521 | 5.2 | IL-18 receptor 1 | |
| LPXN | Hs.125474 | 5.2 | Leupaxin | |
| IRF1 | Hs.436061 | 5.2 | Interferon regulatory factor 1 | |
| FST | Hs.9914 | 5.1 | Follistatin | |
| CXCL6 | Hs.164021 | 5.1 | Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 27 |
| CD44 | Hs.502328 | 5.1 | CD44 molecule (Indian blood group) | |
| LYPD6 | Hs.21929 | 5.1 | LY6/PLAUR domain containing 6 | |
| IFI6 | Hs.523847 | 5.1 | Interferon, alpha-inducible protein 6 | |
| IFIT3 | Hs.47338 | 5.0 | Interferon-induced protein with tetratricopeptide repeats 3 | |
| SLC25A37 | Hs.122514 | 5.0 | Solute carrier family 25, member 37 | |
| IFI35 | Hs.632258 | 5.0 | Interferon-induced protein 35 | |
| TLR1 | Hs.575090 | 4.9 | Toll-like receptor 1 | |
| FXYD6 | Hs.504031 | 4.9 | FXYD domain containing ion transport regulator 6 | |
| SLC7A2 | Hs.448520 | 4.9 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | |
| KIAA0146 | Hs.381058 | 4.9 | KIAA0146 | |
| PPAP2B | Hs.405156 | 4.9 | Phosphatidic acid phosphatase type 2B | |
| OAS3 | Hs.528634 | 4.8 | 2′-5′-Oligoadenylate synthetase 3, 100 kDa | |
| TNFAIP8 | Hs.271955 | 4.7 | TNF-α-induced protein 8 | |
| CYB5R2 | Hs.414362 | 4.7 | Cytochrome b5 reductase 2 | |
| TAP1 | Hs.352018 | 4.7 | Transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) | |
| C1R | Hs.567497 | 4.7 | Complement component 1, r subcomponent | |
| FSD1L | Hs.136901 | 4.7 | Fibronectin type III and SPRY domain containing 1-like | |
| C6orf155 | Hs.368337 | 4.6 | Chromosome 6 open reading frame 155 | |
| IRAK2 | Hs.449207 | 4.6 | IL-1 receptor-associated kinase 2 | |
| C9orf150 | Hs.445356 | 4.6 | Chromosome 9 open reading frame 150 | |
| LOC54103 | Hs.186649 | 4.6 | Hypothetical protein LOC54103 | |
| LOH11CR2A | Hs.152944 | 4.5 | Loss of heterozygosity, 11, chromosomal region 2, gene A | |
| MYLK | Hs.556600 | 4.4 | Myosin, light polypeptide kinase | |
| C8orf1 | Hs.436445 | 4.4 | Chromosome 8 open reading frame 1 | |
| WTAP | Hs.446091 | 4.3 | Wilms' tumor 1-associated protein | |
| CTSK | Hs.632466 | 4.3 | Cathepsin K (pycnodysostosis) | |
| BIRC4BP | Hs.441975 | 4.3 | XIAP-associated factor-1 | |
| HIST2H2AA3 | Hs.530461 | 4.3 | Histone 2, H2aa3 | |
| SLC22A4 | Hs.310591 | 4.3 | Solute carrier family 22 (organic cation transporter), member 4 | |
| DUSP16 | Hs.536535 | 4.3 | Dual-specificity phosphatase 16 | |
| PDLIM4 | Hs.424312 | 4.3 | PDZ and LIM domain 4 | |
| RP1-93H18.5 | Hs.381220 | 4.3 | Hypothetical protein LOC441168 | |
| CXCL1 | Hs.789 | 4.3 | Chemokine (C-X-C motif) ligand 1 | |
| ACSL1 | Hs.406678 | 4.3 | Acyl-coenzyme A synthetase long-chain family member 1 | |
| TNIP1 | Hs.543850 | 4.2 | TNFAIP3-interacting protein 1 | |
| GK | Hs.1466 | 4.2 | Glycerol kinase | |
| SOCS1 | Hs.50640 | 4.2 | Suppressor of cytokine signaling 1 | |
| CDKN2B | Hs.72901 | 4.2 | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | |
| ATF3 | Hs.460 | 4.2 | Activating transcription factor 3 | |
| SQRDL | Hs.511251 | 4.1 | Sulfide quinone reductase-like (yeast) | |
| BMP2 | Hs.73853 | 4.1 | Bone morphogenetic protein 2 | |
| NRG1 | Hs.453951 | 4.1 | Neuregulin 1 | |
| CSF1 | Hs.591402 | 4.1 | Colony-stimulating factor 1 (macrophage) | |
| CLIC2 | Hs.632837 | 4.1 | Chloride intracellular channel 2 | |
| SART2 | Hs.486292 | 4.0 | Squamous cell carcinoma antigen recognized by T cells 2 | |
| C13orf31 | Hs.210586 | 4.0 | Chromosome 13 open reading frame 31 |
Genes with fourfold and higher upregulation are included. Genes indicated in boldface have also been shown by others (27, 35, 44) to be upregulated by vFLIP/K13. Genes in italics are assigned to the statistically most overrepresented gene ontology terms: immune system process (P value, 5.88 × 10−36), defense response (P value, 6.13 × 10−22), inflammatory response (P value, 2.87 × 10−19), and cytokine activity (P value, 8.56 × 10−19). Underlined genes are assigned to the gene ontology term apoptosis (P value, 1.67 × 10−06). Analysis was done with GOstat (4). MnSOD (SOD2) is set off by spaces.
MnSOD is the major cellular protein upregulated by vFLIP/K13 in HUVEC.
In order to investigate the vFLIP/K13-induced changes at the protein level, K13-EC and Ctrl-EC were subjected to 2D-DIGE analysis. Comparison of protein signals identified a protein that was specifically and most intensively upregulated (25.1-fold) in K13-EC compared to Ctrl-EC (Fig. 2A). In addition to this spot, 13 additional protein candidates were detected as upregulated within a range of 4.0- to 9.4-fold in K13-EC versus Ctrl-EC. Isolation of the strongest spot from the gel and subsequent matrix-assisted laser desorption ionization mass spectrometry sequence analysis identified the protein as MnSOD (accession number AAP34407; gene name, SOD2). Western blot analysis with an MnSOD-specific antibody confirmed strong expression of MnSOD in K13-EC, whereas MnSOD was hardly detectable in Ctrl-EC (Fig. 2B). A comparable upregulation of MnSOD to the same extent was seen when HUVEC stably expressing untagged vFLIP/K13 were compared to Ctrl-EC (data not shown), suggesting that the attachment of the C-terminal Myc tag did not alter this activity of vFLIP/K13. MnSOD was selectively detected by immunocytochemical analysis in K13-EC but not in Ctrl-EC (Fig. 2C). MnSOD-positive cells showed an elongated spindle shape and a distinct MnSOD staining pattern that was similar to the staining pattern previously described for mitochondrion-associated proteins in HUVEC (11). This is in agreement with the mitochondrial expression of MnSOD (75).
FIG. 2.
MnSOD is induced in vFLIP/K13-expressing HUVEC and in HHV-8-infected HUVEC. (A) Protein lysates from Ctrl-EC or K13-EC were labeled with either Cy5 (Ctrl-EC) or Cy3 (K13-EC). The lysates were subjected to 2D-DIGE. Signals were detected by laser scanning and quantified with analysis software. The spot with the highest surplus in K13-EC-derived extracts (arrow) was isolated after Coomassie staining and subjected to mass spectroscopic analysis (Toplab GmbH, Marinsried, Germany). This identified MnSOD. (B) Western blot analysis confirmed increased MnSOD expression in K13-EC. vFLIP/K13 expression was detected with an anti-Myc antibody; MnSOD and GAPDH were detected with specific antibodies directed against the proteins. (C) Immunocytochemical staining of MnSOD in Ctrl-EC and K13-EC showed that MnSOD was selectively detectable in K13-transduced cells (red; arrowheads). The cells were counterstained by hematoxylin (light blue). (D) HUVEC were infected with a recombinant HHV-8 encoding GFP (HHV-8-EC). Phase-contrast (PhaCo) analysis of cell density and fluorescence microscopy (FM) for detection of HHV-8-infected cells confirmed efficient infection of HUVEC 72 h postinfection. After puromycin selection, the infected cells (HHV-8-EC) were subjected to Western blot analysis. Comparison of uninfected cells (HUVEC), Ctrl-EC, and K13-EC revealed upregulation of MnSOD in HHV-8-infected cells. GAPDH detection was used as a loading control.
As the next step, HUVEC were infected with recombinant HHV-8, which encodes GFP (70), to determine whether MnSOD protein expression was also increased by HHV-8 infection. The high efficiency of HUVEC infection was shown by GFP fluorescence (Fig. 2D, left, compare phase-contrast with GFP fluorescence). Western blot analysis showed that MnSOD protein is clearly induced in HHV-8-infected cells (Fig. 2D, right, HHV-8-EC). HUVEC transfected with a control plasmid and subjected to the same selection procedure as HHV-8-infected cells (at least 10 days of puromycin) (Fig. 2D, Ctrl-EC) and untransfected HUVEC (Fig. 2D, HUVEC) were used as negative controls. HUVEC transfected with the vFLIP/K13 expression plasmid and also subjected to puromycin selection for at least 10 days (Fig. 2D, K13-EC) served as a positive control. GAPDH was used as a loading control (Fig. 2D). These results identified MnSOD as the cellular protein most strongly induced by vFLIP/K13 in endothelial cells and confirmed that MnSOD is also induced in endothelial cells after infection with HHV-8.
vFLIP/K13 induces MnSOD expression via NF-κB activation in HUVEC.
It has been reported that vFLIP/K13 activates NF-κB (9). Cell fractionation of K13-EC and Ctrl-EC confirmed NF-κB activation in K13-EC as demonstrated by nuclear translocation of the NF-κB p65 protein (Fig. 3A). Only minimal amounts of nuclear p65 were detectable in Ctrl-EC, which might have resulted from slight contamination of the nuclear fraction with remaining cytosolic protein (note the faint GAPDH signal in the nuclear fraction of Ctrl-EC). GAPDH served as a marker protein for the cytosolic fraction and lamin A/C for the nuclear fraction. In agreement with previous findings by us and others, vFLIP/K13 localized in both the cytoplasm and nucleus (43, 52), whereas MnSOD was strictly cytoplasmic (Fig. 3A). In order to determine whether vFLIP-induced NF-κB activation was necessary for the increased expression of MnSOD, endothelial cells were transiently transfected with either a control vector, a vector encoding vFLIP/K13, or a vector encoding vFLIP/MC159. MC159 is the vFLIP molecule of molluscum contagiosum virus and is a structural homologue of vFLIP/K13 that does not activate NF-κB (25, 68). vFLIP/K13, but not vFLIP/MC159, induced MnSOD expression in HUVEC, as detected by Western blotting (Fig. 3B, top), suggesting that the capability of vFLIP/K13 to activate NF-κB may be responsible for the induction of MnSOD expression. The lack of vFLIP/K13-induced MnSOD expression in the presence of the NF-κB inhibitor Bay 11-7082 confirmed this hypothesis (Fig. 3B, bottom). The inhibitor completely blocked MnSOD expression in the vFLIP/K13-expressing cells, while the expression of the two vFLIP molecules was not affected (Fig. 3B, bottom). In order to demonstrate that MnSOD induction depends on vFLIP/K13-mediated NF-κB activation, the mutant vFLIP/K13-58 (K13-58) was used. In K13-58, 3 amino acids (Glu58, Cys59, and Leu60) are replaced by alanines, which abrogates its effect on NF-κB (66). This mutant did not induce MnSOD expression in HUVEC (Fig. 3C). In conclusion, these experiments demonstrated that vFLIP/K13 induced MnSOD expression via activation of NF-κB.
FIG. 3.
vFLIP/K13 induces MnSOD via NF-κB activation. (A) Nuclear (Nuc) and cytoplasmic (Cyto) fractions of Ctrl-EC and K13-EC were analyzed by Western blotting using antibodies against p65, MnSOD, Myc, GAPDH, or lamin A/C. Nuclear localization of p65 was detectable in K13-EC. In agreement with previous findings (43, 52), vFLIP/K13 was also partially located in the nucleus, while MnSOD was strictly cytoplasmic. GAPDH served as a marker protein for the cytosolic fraction and lamin A/C for the nuclear fraction. (B) HUVEC were transfected with control vector (Ctrl) or with a vector encoding either VSV-tagged vFLIP/MC159 (MC159) or Myc-tagged vFLIP/K13 (K13). The cells were left untreated (top) or were treated 6 h before transfection with 2 μM of the NF-κB inhibitor Bay 11-7082 (Bay) (bottom). After 48 h, Western blot analysis was performed with antibody against MnSOD, vFLIP/K13, VSV, or GAPDH. MC159 and GAPDH were detected on the same blot by adding anti-VSV and anti-GAPDH antibodies simultaneously. Specifically, vFLIP/K13 induced MnSOD expression (top), while vFLIP/MC159 (no NF-κB activating function) did not. Addition of the NF-κB inhibitor blocked vFLIP/K13-induced MnSOD expression (bottom). (C) HUVEC were transfected with vectors encoding either VSV-tagged vFLIP/MC159 (MC159), Myc-tagged vFLIP/K13 (K13), Myc-tagged vFLIP/K13-58 (K13-58), or control vector (Ctrl). Western blot analysis was performed 48 h after transfection with antibody against MnSOD, vFLIP/K13, VSV, or GAPDH. Specifically, vFLIP/K13 induced MnSOD expression (top), while vFLIP/MC159 and vFLIP/K13-58 (no NF-κB-activating functions) did not.
vFLIP/K13 induces MnSOD by cell-intrinsic mechanisms in HUVEC.
It has been shown previously (27, 44) and confirmed in the microarray study discussed above that vFLIP/K13 can induce the expression of a variety of cytokines and chemokines in endothelial cells. These may also induce MnSOD expression by paracrine mechanisms in cells that do not express vFLIP/K13 themselves. In order to test this hypothesis, endothelial cells constitutively expressing GFP (GFP-EC) were cocultivated for 72 h with Ctrl-EC or K13-EC (Fig. 4A). Immunocytochemical staining showed that MnSOD was not expressed in GFP-EC/Ctrl-EC cocultures (Fig. 4A, left). Instead, MnSOD was selectively expressed in GFP-EC/K13-EC cocultures (Fig. 4A, right). In these cultures, MnSOD was exclusively expressed in the GFP-negative cells (Fig. 4A, right). To ensure that GFP-EC were able to produce MnSOD, the cells were treated with tumor necrosis factor alpha (TNF-α) (1,000 U/ml; 24 h). This resulted in strong upregulation of MnSOD (data not shown), as described previously (31). Finally, HUVEC were cultivated in conditioned medium obtained from K13-EC (K13-EC-CM) or Ctrl-EC (Ctrl-EC-CM). MnSOD was not induced in HUVEC under these conditions (Fig. 4B, first and second lanes from left). Again, TNF-α was used as a control and induced MnSOD expression in HUVEC (Fig. 4B, fourth lane from left). In addition, the K13-EC cultures that were used to obtain K13-EC-CM expressed MnSOD strongly (Fig. 4B, right lane). Taken together, these results showed that MnSOD was predominantly induced by cell-intrinsic mechanisms in K13-EC and not secondarily by paracrine mechanisms through cytokines released from the cells.
FIG. 4.
MnSOD is induced via cell-intrinsic effects by vFLIP/K13. (A) HUVEC constitutively expressing GFP (GFP-EC) were mixed and cocultivated with either Ctrl-EC or K13-EC (ratio, 2:1) on chamber slides. The cells were stained 72 h after being seeded with an anti-MnSOD antibody and an Alexa-546-coupled anti-rabbit antibody. The cells were analyzed using laser scanning microscopy. MnSOD was detected only in K13-EC/GFP-EC cultures (right), but not in Ctrl-EC/GFP-EC cultures (left). In K13-EC/GFP-EC cultures, MnSOD was specifically observed in the GFP-negative cells (arrow). (B) K13-EC and Ctrl-EC were cultivated for 48 h with EGM-2-MV medium without puromycin. Conditioned media (K13-EC-CM and Ctrl-EC-CM) were filtered (0.22 μm) and mixed with fresh EGM-2-MV (1:2). The respective mixture of conditioned and fresh media was directly applied to HUVEC, which had been seeded 12 h before at a density of 1.0 × 104/cm2. The HUVEC were lysed after 48 h and analyzed together with the previously collected lysates of K13-EC and Ctrl-EC. As a positive control, HUVEC were incubated with 1,000 U/ml TNF-α for 48 h. As a negative control, HUVEC were incubated in pure EGM-2-MV for 48 h. Induction of MnSOD was detectable in K13-EC lysates and the positive control only.
vFLIP/K13 inhibits superoxide-induced cell death in endothelial cells.
MnSOD converts superoxide into oxygen and hydrogen peroxide and thereby protects cells against the damage caused by superoxide (12). To investigate whether vFLIP/K13-induced MnSOD expression protects against superoxide-induced cell death, K13-EC and Ctrl-EC were treated with increasing concentrations (1 to 8 μM) of the superoxide inducer Ly83583. Severe cytotoxic effects, such as cell deformation and detachment, were visible in Ctrl-EC after 48 h (Fig. 5A, upper right; 4 μM Ly83583), while K13-EC were only slightly affected at the same concentration of Ly83583 (Fig. 5A, lower right; 4 μM Ly83583). Interestingly, while in untreated cultures of K13-EC a mixture of spindle-shaped cells and cells with cobblestone morphology was detectable (Fig. 5A, lower left), treatment of K13-EC with Ly83583 seemed to select those cells with a spindle shape (Fig. 5A, lower right). As we described above, these spindle-shaped cells are selectively induced in vFLIP/K13-transduced cultures and express MnSOD at very high levels (Fig. 2C, right).
FIG. 5.
vFLIP/K13 inhibits superoxide-induced cell death in HUVEC. (A) Phase-contrast images of Ctrl-EC and K13-EC treated for 48 h with the superoxide inducer Ly83583 (4 μM) or left untreated. Severe cell damage was detectable in Ly83583-treated Ctrl-EC (upper right). Ly83583-treated K13-EC showed only moderate effects (lower right). (B) Ctrl-EC and K13-EC were incubated with DHE for 30 min at 37°C after the cells were treated with Ly83583 (4 μM) for 24 h or left untreated. The cells were analyzed by fluorescence microscopy (excitation, 546 nm; emission, 590 nm). A significant amount of intracellular superoxide was detected by red staining of the nuclei in Ly83583-treated Ctrl-EC (top). DHE fluorescence was quantified using flow cytometry; 95% of untreated Ctrl-EC (R1) (upper graph, filled) were DHE negative, while 94.2% of Ly83583-treated Ctrl-EC were DHE positive (R2) (upper graph, open). Treated K13-EC remained DHE negative (91.0% in R3) (lower graph, open), comparable to untreated K13-EC (92.1% in R3) (lower graph, filled). (C) Ctrl-EC and K13-EC were treated with the indicated concentrations of Ly83583. The cells were harvested and stained with annexin V and PI after the addition of Ly83583 for 24 h. Afterwards, the cells were analyzed using flow cytometry. Annexin V- and PI-negative cells were regarded as viable cells. The numbers of treated cell were calculated as relative percentages compared to untreated control cells (no Ly83583 treatment). The data are given as mean plus standard deviation; n = 4. A comparison between two groups (K13-EC versus Ctrl-EC treated with the same concentration of Ly83583) was done using a paired two-tailed Student's t test with the SPSS 15.0 software package. ***, P = 0.001; **, P = 0.010; *, P = 0.034.
Superoxide formation in Ly83583-treated cells was detected by fluorescence microscopy using DHE staining. In the presence of superoxide, this compound is converted into a red fluorescent dye (2-hydroxyethidium) that intercalates into the DNA. In Ly83583-treated Ctrl-EC, almost all nuclei were stained, indicating the presence of large amounts of superoxide in the cells (Fig. 5B, top), while untreated Ctrl-EC showed low background (Fig. 5B, upper left). No nuclear accumulation of the dye above background was detectable in K13-EC even after treatment with Ly83583 (Fig. 5B, bottom). For quantification, stained cells were detected by flow cytometry analysis (Fig. 5B, right). Untreated control cells were DHE negative (95.0% in R1) (Fig. 5B, top graph), and 94.2% became DHE positive upon Ly83583 treatment (R2) (Fig. 5B, top graph). In contrast, K13-EC stayed DHE negative after Ly83583 treatment (91.0% of treated and 92.1% of untreated K13-EC in R3) (Fig. 5B, bottom graph). Together, these findings demonstrated that vFLIP/K13 potently prevented the accumulation of superoxide in the cells, most likely via the upregulation of MnSOD expression.
Next, we investigated whether the vFLIP/K13-mediated inhibition of superoxide accumulation protects against programmed cell death. To this end, K13-EC and Ctrl-EC were treated with Ly83583 for 24 h and subsequently stained with annexin V and PI. Compared to Ctrl-EC, K13-EC showed a statistically significantly increased percentage of viable cells (annexin V and PI negative) (Fig. 5C). The most significant differences in Ly83583 resistance between K13-EC and Ctrl-EC were seen at 2 μM (P = 0.001) and 4 μM (P = 0.010) Ly83583 concentrations. These results showed that superoxide-induced apoptosis in HUVEC can be prevented by vFLIP/K13.
Depletion of MnSOD expression renders vFLIP/K13-expressing HUVEC sensitive to superoxide-induced cell death.
Finally, we analyzed the biological role of vFLIP/K13-mediated MnSOD upregulation in ROS accumulation and inhibition of cell death with an siRNA approach. MnSOD siRNA efficiently inhibited MnSOD expression but did not affect vFLIP/K13 expression in HUVEC transiently transfected with a vFLIP/K13 expression plasmid (Fig. 6A). Treatment with Ly83583 (4 μM; 24 h) resulted in a clearly greater superoxide accumulation in MnSOD siRNA-treated cells than in control siRNA-treated cells (Fig. 6B). Quantitative analysis by fluorescence-activated cell sorting showed that 70% of MnSOD siRNA-treated cells (Fig. 6C, R1), but only 44% of control siRNA-treated cells (Fig. 6C, R1) accumulated elevated concentrations of superoxide. Cell death was clearly increased in HUVEC, in which MnSOD expression was inhibited by siRNA after prolonged incubation (72 h) with 4 μM Ly83583 (Fig. 6D).
FIG. 6.
Knockdown of MnSOD sensitizes vFLIP/K13-expressing cells to superoxide-induced cell death. (A) HUVEC were transiently cotransfected with a K13-encoding plasmid and MnSOD siRNA or with a K13-encoding plasmid and control (Ctrl) siRNA. After 48 h, Western blot analysis was performed with antibodies against MnSOD, Myc, or GAPDH. Cotransfection of K13 with MnSOD-specific siRNA led to significant reduction of the MnSOD protein level (top, left lane). Cells cotransfected with K13 and control siRNA still showed strong induction of MnSOD (top, middle lane) compared to untreated HUVEC (top, right lane). (B) HUVEC transiently cotransfected with a K13-encoding plasmid and MnSOD siRNA or with a K13-encoding plasmid and control siRNA, as described for panel A, were treated 24 h after transfection with 4 μM Ly83583. After an additional 24 h, the cells were stained with 10 μM DHE at 37°C for 30 min. The cells were analyzed by fluorescence microscopy (excitation, 546 nm; emission, 590 nm). A larger amount of intracellular superoxide was detected in Ly83583-treated cells transfected with K13 plus MnSOD siRNA (right). (C) DHE fluorescence was quantified for the cells described in panel B using flow cytometry; 70% of the cells cotransfected with K13 plasmid and MnSOD siRNA became DHE positive (R1) (open graph), while only 44% of K13- and control siRNA-transfected cells became DHE positive (R1) (filled graph) upon Ly83583 treatment. (D) Phase-contrast images of HUVEC transiently cotransfected with a K13-encoding plasmid and MnSOD siRNA or with a K13-encoding plasmid and control siRNA and cultured for 72 h after Ly83583 treatment (4 μM). Cotransfection of MnSOD siRNA resulted in more cell damage (right).
DISCUSSION
We showed that vFLIP/K13 inhibits superoxide-induced apoptosis in endothelial cells via upregulation of MnSOD expression. The induction of MnSOD required NF-κB activation, which is a known consequence of vFLIP/K13 action.
It is well established that vFLIP/K13 has antiapoptotic activity, but there is an ongoing controversy as to whether the protein is antiapoptotic by inhibition of caspase 8 activation (5), by activation of NF-κB (20, 64) and subsequent induction of antiapoptotic proteins (28, 64), or both. A cDNA microarray analysis showed that vFLIP/K13 activates the expression of many NF-κB-induced cytokines, such as IL-6, IL-8, and RANTES (CCL5), in endothelial cells (Table 1). This was in agreement with previous findings (27, 44) and confirmed the activity of the vFLIP/K13 used in our study. In addition, this cDNA microarray analysis showed that vFLIP/K13-induced genes are involved in inflammation and defense responses, suggesting that vFLIP/K13 may actively contribute to the Th-1-like immune reaction that has commonly been observed in KS (21, 63). However, this analysis did not provide new hints about how vFLIP/K13 regulates apoptosis. Only a few genes with a known function in apoptosis regulation were found to be subject to vFLIP/K13 transcription regulation, and both anti- and proapoptotic genes were among them. Therefore, it would be difficult to predict if induction or inhibition of apoptosis would be the net outcome of vFLIP/K13 expression just by considering the data from the cDNA array.
As a novel approach to identify factors that may mediate the antiapoptotic activity of vFLIP/K13, we used 2D-DIGE analysis in order to compare the proteomes of K13-EC and Ctrl-EC. Although limited in resolution, this technique has the advantage that strongly expressed proteins that are differentially regulated can be detected. Using this technique, we identified MnSOD as the most strongly upregulated protein in K13-EC. MnSOD was also found to be upregulated in the cDNA microarray (SOD2), though the differential regulation at the RNA level (5.6-fold) was much weaker than at the protein level (25.1-fold). This indicated the advantage of the proteome analysis in this setting.
Due to the low level of expression of vFLIP/K13, we were not able to demonstrate coexpression of MnSOD and vFLIP/K13 in the same cells with immunocytochemical techniques. However, we provided several lines of evidence that MnSOD is induced in endothelial cells by vFLIP/K13 via cell-intrinsic mechanisms. This was surprising, since it has been shown that vFLIP/K13 induces the expression of several cytokines and chemokines, such as IL-1β and IL-6, at the transcription level in endothelial cells (Table 1) (27, 65). IL-1β and IL-6 have been shown to induce MnSOD expression in endothelial cells and hepatocytes, respectively (16, 51). However, cell culture supernatants obtained from K13-EC did not induce MnSOD expression in HUVEC that did not express vFLIP/K13 (Fig. 4B). IL-6 was detected in high concentrations, up to 340 pg/ml, after 24 h in the cell culture supernatants of K13-transfected HUVEC by enzyme-linked immunosorbent assay, whereas IL-1β was not detected (data not shown). This may be due to the fact that IL-1β secretion requires the activation of the inflammasome (34, 42), which may not be activated by K13 expression in endothelial cells. The fact that IL-6 did not increase MnSOD expression may be explained by a recent report showing that HUVEC do not express the IL-6 receptor (36), which is in agreement with our own findings that the IL-6 receptor was not detected in KSC in vivo (61). Based on these negative results, we decided that the most relevant test of whether an MnSOD-activating activity is released from K13-EC may be the cocultivation experiment shown in Fig. 4A. With this approach, paracrine and juxtacrine MnSOD-inducing activity can be detected. Also, in this case, MnSOD was selectively expressed in K13-EC and not in vFLIP/K13-negative cells. This demonstrated that vFLIP/K13 induces MnSOD exclusively in a cell-intrinsic manner.
In agreement with previous findings (27, 44), we demonstrated that vFLIP/K13 expression activates NF-κB constitutively in HUVEC (Fig. 3). This activation of NF-κB was necessary for the induction of MnSOD. Treatment of K13-expressing endothelial cells with an NF-κB inhibitor blocked the induction of MnSOD. Furthermore, the expression of a vFLIP/K13 mutant (vFLIP/K13-58) and the vFLIP/K13-homologous vFLIP/MC159, both unable to activate NF-κB (9, 25, 66, 68), did not induce MnSOD expression. The induction of MnSOD seems to be specific for vFLIP/K13, since vFLIP/K13 is the only viral FLIP homologue that activates NF-κB (9). The caspase 8-inhibitory function of vFLIP/K13 was likely not involved in the induction of MnSOD expression. First, caspase 8 was not activated in HUVEC under our experimental conditions (data not shown), and second, cotransfection of vFLIP/K13 with vFLIP/MC159, which can inhibit caspase 8 activation but does not activate NF-κB (68), did not affect MnSOD induction by vFLIP/K13 (data not shown).
MnSOD is an important antioxidant molecule localized in the mitochondria (75). It eliminates toxic mitochondrial superoxide by converting it into hydrogen peroxide and oxygen (12). MnSOD levels in endothelial progenitor cells are correlated with increased resistance against superoxide-induced cell damage compared to mature endothelial cells (7, 13, 31). We showed that vFLIP/K13-induced upregulation of MnSOD in HUVEC similarly prevented the accumulation of superoxide and protected HUVEC against cell damage. Downregulation of MnSOD sensitized K13-expressing cells to superoxide-induced cell death, showing that MnSOD is the mediator of K13-induced protection against superoxide-induced death.
It is well established that inflammation as characteristically observed in KS (21, 63) establishes high concentrations of ROS (56). In addition, activation of both of the major apoptotic signaling pathways (the intrinsic stress-induced pathway [37, 38, 48] and the extrinsic receptor-mediated pathway [18, 49]) leads to increased intracellular concentrations of ROS (Fig. 7). This indicated that the protection against ROS may provide an important survival advantage for HHV-8-infected KSC in the lesions. In fact, we found that infection of endothelial cells with HHV-8 induces expression of MnSOD. Additionally, increased expression of MnSOD was previously reported in AIDS KS lesions (41). These results support the relevance of our findings for the situation in KS.
FIG. 7.
Model of vFLIP/K13-induced prevention of cell death. Extrinsic (e.g., FasL and TNF-α) and intrinsic (stress-induced) death signals lead to the accumulation of superoxide (O2·−) in the mitochondria. The latent protein vFLIP/K13 possesses two major antiapoptotic functions. (I) It can inhibit caspase 8 activation by direct interaction with the molecule. (II) It activates NF-κB, which results in upregulation of several antiapoptotic molecules, including cFLIP and cIAP, with MnSOD being the most strongly induced protein. Upregulation of the antioxidant MnSOD prevents accumulation of O2·− in the mitochondria and subsequent damage to the organelle (left). Additionally, vFLIP/K13 can inhibit the activation of caspase 8 and of downstream effector caspases (via direct interaction and induction of antiapoptotic proteins, like cFLIP and cIAPs) (right). Thus, vFLIP/K13 can inhibit both extrinsic and intrinsic death signaling pathways in this way, providing comprehensive protection for the cell.
Our results provide further evidence that vFLIP/K13 is an important antiapoptotic molecule encoded by HHV-8 (Fig. 7). According to the literature, it possesses two major antiapoptotic functions. First, it can inhibit caspase 8 activation by direct interaction with the molecule (Fig. 7I) (5). Second, it activates NF-κB, which results in upregulation of several antiapoptotic molecules, including cFLIP and cIAP (28), with MnSOD being the most strongly induced protein (Fig. 7II). Both functions may be required to protect cells from apoptosis induced by death receptor activation, which can cause both increased intracellular ROS concentrations and activation of caspases. Altogether, vFLIP/K13 possesses manifold antiapoptotic functions that may protect HHV-8-infected cells against a broad spectrum of different death stimuli, such as Fas and TNF-α receptor signaling (5, 9, 15), growth factor withdrawal (64), and anoikis (20). The induction of MnSOD may establish an important protection shield for HHV-8-infected KSC specifically in the inflammatory ROS-generating microenvironment of KS.
Acknowledgments
We thank Mahimaidos Manoharan and Melanie Nurtsch (both Division of Molecular and Experimental Surgery, University of Erlangen-Nuremberg.) for excellent technical assistance, Werner Hohenberger (Department of Surgery, University of Erlangen-Nuremberg) for his continuous support, Matthew Miller for critically reading the manuscript, and Christine Hohenadl (Austrianova Biotechnology GmbH and Research Institute of Virology and Biomedicine, University of Veterinary Medicine, Vienna, Austria) for providing the pVSV-G and pGag-Pol vectors. We are grateful to Jeffrey Vieria (Department of Laboratory Medicine, University of Washington) for providing the rKSHV.219-producing cell line.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG-SPP 1130, DFG-GK 1071, and DFG 317/2-1), the German Cancer Aid (Deutsche Krebshilfe, Apoptose-Schwerpunktprogramm), and the Interdisciplinary Centre for Clinical Research (IZKF, D8/D9) of the University of Erlangen-Nuremberg to Michael Stürzl and a tandem project grant of the IZKF (project B11) to Michael Stürzl and Frank Neipel. Mathias Thurau was supported by a grant from the ELAN program (AZ 05.11.13.1) of the University of Erlangen-Nuremberg. Margot Thome is supported by grants from the Swiss National Science Foundation and the Swiss Cancer League (Oncosuisse). Thomas F. Schulz is also supported by the EU Integrated Project INCA (LSHC-CT-2005-018704).
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Ablashi, D. V., L. G. Chatlynne, J. E. Whitman, Jr., and E. Cesarman. 2002. Spectrum of Kaposi's sarcoma-associated herpesvirus, or human herpesvirus 8, diseases. Clin. Microbiol. Rev. 15439-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, J., Y. Sun, R. Sun, and M. B. Rettig. 2003. Kaposi's sarcoma-associated herpesvirus encoded vFLIP induces cellular IL-6 expression: the role of the NF-κB and JNK/AP1 pathways. Oncogene 223371-3385. [DOI] [PubMed] [Google Scholar]
- 3.Bagneris, C., A. V. Ageichik, N. Cronin, B. Wallace, M. Collins, C. Boshoff, G. Waksman, and T. Barrett. 2008. Crystal structure of a vFlip-IKKγ complex: insights into viral activation of the IKK signalosome. Mol. Cell 30620-631. [DOI] [PubMed] [Google Scholar]
- 4.Beissbarth, T., and T. P. Speed. 2004. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics 201464-1465. [DOI] [PubMed] [Google Scholar]
- 5.Belanger, C., A. Gravel, A. Tomoiu, M. E. Janelle, J. Gosselin, M. J. Tremblay, and L. Flamand. 2001. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J. Hum. Virol. 462-73. [PubMed] [Google Scholar]
- 6.Blasig, C., C. Zietz, B. Haar, F. Neipel, S. Esser, N. H. Brockmeyer, E. Tschachler, S. Colombini, B. Ensoli, and M. Stürzl. 1997. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J. Virol. 717963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, H., P. Gehrig, T. M. Scott, R. Zimmermann, R. Schlapbach, and A. H. Zisch. 2006. MnSOD marks cord blood late outgrowth endothelial cells and accompanies robust resistance to oxidative stress. Biochem. Biophys. Res. Commun. 350364-369. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhary, P. M., A. Jasmin, M. T. Eby, and L. Hood. 1999. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 185738-5746. [DOI] [PubMed] [Google Scholar]
- 10.Chugh, P., H. Matta, S. Schamus, S. Zachariah, A. Kumar, J. A. Richardson, A. L. Smith, and P. M. Chaudhary. 2005. Constitutive NF-κB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc. Natl. Acad. Sci. USA 10212885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, T. J., M. J. Berridge, P. Lipp, and M. D. Bootman. 2002. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 211616-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culotta, V. C., M. Yang, and T. V. O'Halloran. 2006. Activation of superoxide dismutases: putting the metal to the pedal. Biochim. Biophys. Acta 1763747-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dernbach, E., C. Urbich, R. P. Brandes, W. K. Hofmann, A. M. Zeiher, and S. Dimmeler. 2004. Antioxidative stress-associated genes in circulating progenitor cells: evidence for enhanced resistance against oxidative stress. Blood 1043591-3597. [DOI] [PubMed] [Google Scholar]
- 14.De Thier, F., T. Simonart, P. Hermans, J. Andre, J. P. Van Vooren, and J. C. Noel. 1999. Early- and late-stage Kaposi's sarcoma lesions exhibit similar proliferation fraction. Am. J. Dermatopathol. 2125-27. [DOI] [PubMed] [Google Scholar]
- 15.Djerbi, M., V. Screpanti, A. I. Catrina, B. Bogen, P. Biberfeld, and A. Grandien. 1999. The inhibitor of death receptor signaling, FLICE-inhibitory protein defines a new class of tumor progression factors. J. Exp. Med. 1901025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dougall, W. C., and H. S. Nick. 1991. Manganese superoxide dismutase: a hepatic acute phase protein regulated by interleukin-6 and glucocorticoids. Endocrinology 1292376-2384. [DOI] [PubMed] [Google Scholar]
- 17.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 964546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dussmann, H., D. Kogel, M. Rehm, and J. H. Prehn. 2003. Mitochondrial membrane permeabilization and superoxide production during apoptosis. A single-cell analysis. J. Biol. Chem. 27812645-12649. [DOI] [PubMed] [Google Scholar]
- 19.Eby, M. T., A. Jasmin, A. Kumar, K. Sharma, and P. M. Chaudhary. 2000. TAJ, a novel member of the tumor necrosis factor receptor family, activates the c-Jun N-terminal kinase pathway and mediates caspase-independent cell death. J. Biol. Chem. 27515336-15342. [DOI] [PubMed] [Google Scholar]
- 20.Efklidou, S., R. Bailey, N. Field, M. Noursadeghi, and M. K. Collins. 2008. vFLIP from KSHV inhibits anoikis of primary endothelial cells. J. Cell Sci. 121450-457. [DOI] [PubMed] [Google Scholar]
- 21.Ensoli, B., M. Stürzl, and P. Monini. 2001. Reactivation and role of HHV-8 in Kaposi's sarcoma initiation. Adv. Cancer Res. 81161-200. [DOI] [PubMed] [Google Scholar]
- 22.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 766213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Field, N., W. Low, M. Daniels, S. Howell, L. Daviet, C. Boshoff, and M. Collins. 2003. KSHV vFLIP binds to IKK-gamma to activate IKK. J. Cell Sci. 1163721-3728. [DOI] [PubMed] [Google Scholar]
- 24.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1273-296. [DOI] [PubMed] [Google Scholar]
- 25.Gil, J., J. Rullas, J. Alcami, and M. Esteban. 2001. MC159L protein from the poxvirus molluscum contagiosum virus inhibits NF-κB activation and apoptosis induced by PKR. J. Gen. Virol. 823027-3034. [DOI] [PubMed] [Google Scholar]
- 26.Grimm, T., S. Schneider, E. Naschberger, J. Huber, E. Guenzi, A. Kieser, P. Reitmeir, T. F. Schulz, C. A. Morris, and M. Stürzl. 2005. EBV latent membrane protein-1 protects B cells from apoptosis by inhibition of BAX. Blood 1053263-3269. [DOI] [PubMed] [Google Scholar]
- 27.Grossmann, C., S. Podgrabinska, M. Skobe, and D. Ganem. 2006. Activation of NF-κB by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J. Virol. 807179-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guasparri, I., S. A. Keller, and E. Cesarman. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J. Exp. Med. 199993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Guenzi, E., K. Töpolt, C. Lubeseder-Martellato, A. Jörg, E. Naschberger, R. Benelli, A. Albini, and M. Stürzl. 2003. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 223772-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, B., A. Chadburn, E. Jou, E. J. Schattner, D. M. Knowles, and A. Cerutti. 2004. Lymphoma B cells evade apoptosis through the TNF family members BAFF/BLyS and APRIL. J. Immunol. 1723268-3279. [DOI] [PubMed] [Google Scholar]
- 31.He, T., T. E. Peterson, E. L. Holmuhamedov, A. Terzic, N. M. Caplice, L. W. Oberley, and Z. S. Katusic. 2004. Human endothelial progenitor cells tolerate oxidative stress due to intrinsically high expression of manganese superoxide dismutase. Arterioscler. Thromb. Vasc. Biol. 242021-2027. [DOI] [PubMed] [Google Scholar]
- 32.Hong, Y. K., K. Foreman, J. W. Shin, S. Hirakawa, C. L. Curry, D. R. Sage, T. Libermann, B. J. Dezube, J. D. Fingeroth, and M. Detmar. 2004. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat. Genet. 36683-685. [DOI] [PubMed] [Google Scholar]
- 33.Inohara, N., L. del Peso, T. Koseki, S. Chen, and G. Nunez. 1998. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem. 27312296-12300. [DOI] [PubMed] [Google Scholar]
- 34.Keller, M., A. Ruegg, S. Werner, and H. D. Beer. 2008. Active caspase-1 is a regulator of unconventional protein secretion. Cell 132818-831. [DOI] [PubMed] [Google Scholar]
- 35.Lagos, D., M. W. Trotter, R. J. Vart, H. W. Wang, N. C. Matthews, A. Hansen, O. Flore, F. Gotch, and C. Boshoff. 2007. Kaposi sarcoma herpesvirus-encoded vFLIP and vIRF1 regulate antigen presentation in lymphatic endothelial cells. Blood 1091550-1558. [DOI] [PubMed] [Google Scholar]
- 36.Larsson, P., E. Ulfhammer, L. Karlsson, M. Bokarewa, K. Wahlander, and S. Jern. 2008. Effects of IL-1β and IL-6 on tissue-type plasminogen activator expression in vascular endothelial cells. Thromb. Res. doi: 10.1016/j.thromres.2008.03.013. [DOI] [PubMed]
- 37.Li, A. E., H. Ito, I. I. Rovira, K. S. Kim, K. Takeda, Z. Y. Yu, V. J. Ferrans, and T. Finkel. 1999. A role for reactive oxygen species in endothelial cell anoikis. Circ. Res. 85304-310. [DOI] [PubMed] [Google Scholar]
- 38.Lieberthal, W., V. Triaca, J. S. Koh, P. J. Pagano, and J. S. Levine. 1998. Role of superoxide in apoptosis induced by growth factor withdrawal. Am. J. Physiol. 275F691-F702. [DOI] [PubMed] [Google Scholar]
- 39.Liu, L., M. T. Eby, N. Rathore, S. K. Sinha, A. Kumar, and P. M. Chaudhary. 2002. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the IκB kinase complex. J. Biol. Chem. 27713745-13751. [DOI] [PubMed] [Google Scholar]
- 40.Lubeseder-Martellato, C., E. Guenzi, A. Jörg, K. Töpolt, E. Naschberger, E. Kremmer, C. Zietz, E. Tschachler, P. Hutzler, M. Schwemmle, K. Matzen, T. Grimm, B. Ensoli, and M. Stürzl. 2002. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am. J. Pathol. 1611749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallery, S. R., P. Pei, D. J. Landwehr, C. M. Clark, J. E. Bradburn, G. M. Ness, and F. M. Robertson. 2004. Implications for oxidative and nitrative stress in the pathogenesis of AIDS-related Kaposi's sarcoma. Carcinogenesis 25597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinon, F., K. Burns, and J. Tschopp. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10417-426. [DOI] [PubMed] [Google Scholar]
- 43.Matta, H., V. Punj, S. Schamus, L. Mazzacurati, A. M. Chen, R. Song, T. Yang, and P. M. Chaudhary. 2008. A nuclear role for Kaposi's sarcoma-associated herpesvirus-encoded K13 protein in gene regulation. Oncogene 275243-5253. [DOI] [PubMed] [Google Scholar]
- 44.Matta, H., R. M. Surabhi, J. Zhao, V. Punj, Q. Sun, S. Schamus, L. Mazzacurati, and P. M. Chaudhary. 2007. Induction of spindle cell morphology in human vascular endothelial cells by human herpesvirus 8-encoded viral FLICE inhibitory protein K13. Oncogene 261656-1660. [DOI] [PubMed] [Google Scholar]
- 45.Mohr, A., C. Buneker, R. P. Gough, and R. M. Zwacka. 2008. MnSOD protects colorectal cancer cells from TRAIL-induced apoptosis by inhibition of Smac/DIABLO release. Oncogene 27763-774. [DOI] [PubMed] [Google Scholar]
- 46.Naschberger, E., R. S. Croner, S. Merkel, A. Dimmler, P. Tripal, K. U. Amann, E. Kremmer, W. M. Brueckl, T. Papadopoulos, C. Hohenadl, W. Hohenberger, and M. Stürzl. 2008. Angiostatic immune reaction in colorectal carcinoma: impact on survival and perspectives for antiangiogenic therapy. Int. J. Cancer. 1232120-2129. [DOI] [PubMed] [Google Scholar]
- 47.Naschberger, E., C. Lubeseder-Martellato, N. Meyer, R. Gessner, E. Kremmer, A. Gessner, and M. Stürzl. 2006. Human guanylate binding protein-1 is a secreted GTPase present in increased concentrations in the cerebrospinal fluid of patients with bacterial meningitis. Am. J. Pathol. 1691088-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orrenius, S., V. Gogvadze, and B. Zhivotovsky. 2007. Mitochondrial oxidative stress: implications for cell death. Annu. Rev. Pharmacol. Toxicol. 47143-183. [DOI] [PubMed] [Google Scholar]
- 49.Pardo, M., J. A. Melendez, and O. Tirosh. 2006. Manganese superoxide dismutase inactivation during Fas (CD95)-mediated apoptosis in Jurkat T cells. Free Radic. Biol. Med. 411795-1806. [DOI] [PubMed] [Google Scholar]
- 50.Park, S. J., D. Lee, C. Y. Choi, and S. Y. Ryu. 2008. Induction of apoptosis by NORE1A in a manner dependent on its nuclear export. Biochem. Biophys. Res. Commun. 36856-61. [DOI] [PubMed] [Google Scholar]
- 51.Rogers, R. J., J. M. Monnier, and H. S. Nick. 2001. Tumor necrosis factor-alpha selectively induces MnSOD expression via mitochondria-to-nucleus signaling, whereas interleukin-1β utilizes an alternative pathway. J. Biol. Chem. 27620419-20427. [DOI] [PubMed] [Google Scholar]
- 52.Sander, G., A. Konrad, M. Thurau, E. Wies, R. Leubert, E. Kremmer, H. Dinkel, T. F. Schulz, F. Neipel, and M. Stürzl. 2008. Intracellular localization map of HHV-8 proteins. J. Virol. 821908-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 721005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schellerer, V. S., R. S. Croner, K. Weinländer, W. Hohenberger, M. Stürzl, and E. Naschberger. 2007. Endothelial cells of human colorectal cancer and healthy colon reveal phenotypic differences in culture. Lab. Investig. 871159-1170. [DOI] [PubMed] [Google Scholar]
- 55.Simmons, M. J., G. Fan, W. X. Zong, K. Degenhardt, E. White, and C. Gelinas. 2008. Bfl-1/A1 functions, similar to Mcl-1, as a selective tBid and Bak antagonist. Oncogene 271421-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon, H. U., A. Haj-Yehia, and F. Levi-Schaffer. 2000. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 5415-418. [DOI] [PubMed] [Google Scholar]
- 57.Simonart, T., C. Degraef, R. Mosselmans, P. Hermans, Y. Lunardi-Iskandar, J. C. Noel, J. P. Van Vooren, D. Parent, M. Heenen, and P. Galand. 2001. Early- and late-stage Kaposi's sarcoma-derived cells but not activated endothelial cells can invade de-epidermized dermis. J. Investig. Dermatol. 116679-685. [DOI] [PubMed] [Google Scholar]
- 58.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stürzl, M., C. Blasig, A. Schreier, F. Neipel, C. Hohenadl, E. Cornali, G. Ascherl, S. Esser, N. H. Brockmeyer, M. Ekman, E. E. Kaaya, E. Tschachler, and P. Biberfeld. 1997. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int. J. Cancer 7268-71. [DOI] [PubMed] [Google Scholar]
- 60.Stürzl, M., H. Brandstetter, and W. K. Roth. 1992. Kaposi's sarcoma: a review of gene expression and ultrastructure of KS spindle cells in vivo. AIDS Res. Hum Retrovir. 81753-1763. [DOI] [PubMed] [Google Scholar]
- 61.Stürzl, M., H. Brandstetter, C. Zietz, B. Eisenburg, G. Raivich, D. P. Gearing, N. H. Brockmeyer, and P. H. Hofschneider. 1995. Identification of interleukin-1 and platelet-derived growth factor-B as major mitogens for the spindle cells of Kaposi's sarcoma: a combined in vitro and in vivo analysis. Oncogene 102007-2016. [PubMed] [Google Scholar]
- 62.Stürzl, M., C. Hohenadl, C. Zietz, E. Castanos-Velez, A. Wunderlich, G. Ascherl, P. Biberfeld, P. Monini, P. J. Browning, and B. Ensoli. 1999. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 911725-1733. [DOI] [PubMed] [Google Scholar]
- 63.Stürzl, M., C. Zietz, P. Monini, and B. Ensoli. 2001. Human herpesvirus-8 and Kaposi's sarcoma: relationship with the multistep concept of tumorigenesis. Adv. Cancer Res. 81125-159. [DOI] [PubMed] [Google Scholar]
- 64.Sun, Q., H. Matta, and P. M. Chaudhary. 2003. The human herpes virus 8-encoded viral FLICE inhibitory protein protects against growth factor withdrawal-induced apoptosis via NF-kappa B activation. Blood 1011956-1961. [DOI] [PubMed] [Google Scholar]
- 65.Sun, Q., H. Matta, G. Lu, and P. M. Chaudhary. 2006. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-κB activation. Oncogene 252717-2726. [DOI] [PubMed] [Google Scholar]
- 66.Sun, Q., S. Zachariah, and P. M. Chaudhary. 2003. The human herpes virus 8-encoded viral FLICE-inhibitory protein induces cellular transformation via NF-κB activation. J. Biol. Chem. 27852437-52445. [DOI] [PubMed] [Google Scholar]
- 67.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386517-521. [DOI] [PubMed] [Google Scholar]
- 68.Thurau, M., H. Everett, M. Tapernoux, J. Tschopp, and M. Thome. 2006. The TRAF3-binding site of human molluscipox virus FLIP molecule MC159 is critical for its capacity to inhibit Fas-induced apoptosis. Cell Death Differ. 131577-1585. [DOI] [PubMed] [Google Scholar]
- 69.Tripal, P., M. Bauer, E. Naschberger, T. Mörtinger, C. Hohenadl, E. Cornali, M. Thurau, and M. Stürzl. 2007. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 2744-52. [DOI] [PubMed] [Google Scholar]
- 70.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325225-240. [DOI] [PubMed] [Google Scholar]
- 71.Walczak, H., and P. H. Krammer. 2000. The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp. Cell Res. 25658-66. [DOI] [PubMed] [Google Scholar]
- 72.Wang, C. Y., M. W. Mayo, R. G. Korneluk, D. V. Goeddel, and A. S. Baldwin, Jr. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 2811680-1683. [DOI] [PubMed] [Google Scholar]
- 73.Wang, H. W., M. W. Trotter, D. Lagos, D. Bourboulia, S. Henderson, T. Makinen, S. Elliman, A. M. Flanagan, K. Alitalo, and C. Boshoff. 2004. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat. Genet. 36687-693. [DOI] [PubMed] [Google Scholar]
- 74.Weinländer, K., E. Naschberger, M. H. Lehmann, P. Tripal, W. Paster, H. Stockinger, C. Hohenadl, and M. Stürzl. 2008. Guanylate binding protein-1 inhibits spreading and migration of endothelial cells through induction of integrin α4 expression. FASEB J. doi: 10.1096/fj.08-107524. [DOI] [PubMed]
- 75.Weisiger, R. A., and I. Fridovich. 1973. Mitochondrial superoxide simutase. Site of synthesis and intramitochondrial localization. J. Biol. Chem. 2484793-4796. [PubMed] [Google Scholar]
- 76.Weninger, W., T. A. Partanen, S. Breiteneder-Geleff, C. Mayer, H. Kowalski, M. Mildner, J. Pammer, M. Stürzl, D. Kerjaschki, K. Alitalo, and E. Tschachler. 1999. Expression of vascular endothelial growth factor receptor-3 and podoplanin suggests a lymphatic endothelial cell origin of Kaposi's sarcoma tumor cells. Lab. Investig. 79243-251. [PubMed] [Google Scholar]
- 77.Wesche-Soldato, D. E., C. S. Chung, S. H. Gregory, T. P. Salazar-Mather, C. A. Ayala, and A. Ayala. 2007. CD8+ T cells promote inflammation and apoptosis in the liver after sepsis: role of Fas-FasL. Am. J. Pathol. 17187-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, J. K. 2008. FLIP as an anti-cancer therapeutic target. Yonsei Med. J. 4919-27. [DOI] [PMC free article] [PubMed] [Google Scholar]