Abstract
The cellular localization of a chimera formed by fusing a monomeric red fluorescent protein to the C terminus of the Klebsiella oxytoca type II secretion system outer membrane secretin PulD (PulD-mCherry) in Escherichia coli was determined in vivo by fluorescence microscopy. Like PulD, PulD-mCherry formed sodium dodecyl sulfate- and heat-resistant multimers and was functional in pullulanase secretion. Chromosome-encoded PulD-mCherry formed fluorescent foci on the periphery of the cell in the presence of high (plasmid-encoded) levels of its cognate chaperone, the pilotin PulS. Subcellular fractionation demonstrated that the chimera was located exclusively in the outer membrane under these circumstances. A similar localization pattern was observed by fluorescence microscopy of fixed cells treated with green fluorescent protein-tagged affitin, which binds with high affinity to an epitope in the N-terminal region of PulD. At lower levels of (chromosome-encoded) PulS, PulD-mCherry was less stable, was located mainly in the inner membrane, from which it could not be solubilized with urea, and did not induce the phage shock response, unlike PulD in the absence of PulS. The fluorescence pattern of PulD-mCherry under these conditions was similar to that observed when PulS levels were high. The complete absence of PulS caused the appearance of bright and almost exclusively polar fluorescent foci.
Over the past 10 years, fluorescence microscopy has been extensively used to study the cellular localization of large multiprotein bacterial membrane complexes in relation to their biological functions. For example, the Tat (28) protein export system is evenly distributed in the cell envelope. Helical patterns were reported for the Sec protein export system (33), the outer membrane porin LamB (11), and other outer membrane proteins, as well as newly synthesized lipopolysaccharide (10). Proteins that constitute the cell division machinery are located at midcell, the site of constriction (12). Polar localization has been demonstrated for flagellar (23), type IV pilus (4), type III (17) and type IV (18) secretion machineries, chemotaxis receptors (33), and some components of the type II secreton (8, 29, 30) and its site of action (30, 34). It seems probable that assembly, disassembly, and targeting of protein machineries are temporally and spatially coordinated processes.
We have addressed the correlation between localization and complex assembly of a prototypical type II secretion system (T2SS) machinery. Secretion via this pathway is a two-step process. Exoproteins, such as hydrolytic enzymes and toxins, are first translocated to the periplasm via the Sec (26) or Tat (36) protein export system. The second step is secretion to the external milieu by the T2SS machinery, the secreton, which is composed of at least 12 proteins that likely span the cell envelope as a large complex. An inner membrane platform composed of the GspE, GspF, GspL, and GspM proteins (27) is connected to the outer membrane exit channel, formed by the secretin GspD (15), by GspC (24). The pseudopilins might assemble into a periplasmic type IV pilus-like structure (the pseudopilus) that is directly involved in the secretion of substrates (19). Green fluorescent protein (GFP) chimeras of GspM (PulM) and GspL (PulL) of the Klebsiella oxytoca Pul secreton localized circumferentially in the Escherichia coli cell envelope, with occasional brighter foci when other secreton factors were present (3). The major pilin of this secreton, PulG, localized evenly throughout the inner membrane independently of other secreton components when fused to fluorescent reporter proteins (9). We proposed that the clustered membrane fluorescence patterns of GFP-PulL and GFP-PulM might represent nucleation sites for the assembly of part of the secreton, with protein secretion occurring upon complete assembly with motor proteins, the outer membrane secretin, and the pseudopilus (3). GspD and GspC determine secreton specificity (1, 32). One would expect their correct localization to be essential for assembly and function of the secreton. Localization of PulD (GspD) to the outer membrane requires a dedicated lipoprotein chaperone, the pilotin PulS (13, 15, 16), but the topological organization of PulD in the outer membrane has not been studied. In this work, we examined the cellular localization of the secretin PulD as a chimera with a stable variant of monomeric red fluorescent protein (RFP), mCherry (31), and studied the effect of its pilotin, PulS, and other secreton components on its membrane localization and organization. To validate the use of PulD-mCherry as a model for localizing PulD in the envelope, we also introduced the use of Sac7d-derived binding proteins (22), hereafter called affitins. GFP-tagged affitins can be used as alternatives for fluorescein-labeled antibodies in immunofluorescence-like tests.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains used in this study are listed in Table 1. Luria broth and agar were prepared as described previously (21). When appropriate, media were supplemented with ampicillin (100 μg/ml for plasmids and 25 μg/ml for chromosomal insertions) and kanamycin (30 μg/ml). Generalized transduction using P1 phage was performed as described previously (21). Stable introduction of gene fusions into at the λ attachment of the E. coli chromosome site was done using the λInCh integration vector (2). Strain PAP105 was used for cloning purposes.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| Strains | ||
| PAP105 | Δ(lac-pro) pAPIP501 [F′(lacIq ΔlacZM15 pro+ Tn10)] (Tcr) | Laboratory collection |
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb-5301 deoC ptsF25 rbsR | Laboratory collection |
| NS30 | MC4100 degP | Laboratory collection |
| PAP7232 | MC4100 malP::(pulS pulAB pulC to -O) F′ (lacIqpro+ Tn10) (Tcr) | 25 |
| PAP7447 | MC4100 malP::(pulS pulAB pulC to -O ΔpulD) F′ (lacIqpro+ Tn10) (Tcr) | 15 |
| PAP9123 | PAP7447Δ(λattL-lom)::bla lacIqp204-mCherry | This study |
| PAP9124 | PAP7447Δ(λattL-lom)::bla lacIqp204-pulD | This study |
| PAP9138 | PAP7447Δ(λattL-lom)::bla lacIqp204-pulD-mCherry | This study |
| PAP9210 | PAP105Δ(λattL-lom)::bla lacIqp204-pulD-mCherry | This study |
| PAP9312 | NS30Δ(λattL-lom)::bla lacIqp204-pulD | This study |
| PAP9313 | NS30Δ(λattL-lom)::bla lacIqp204-pulD-mCherry | This study |
| Plasmids | ||
| pCHAP3671 | pUC18-pulD | 14 |
| pDSW204 | Modified ptrc promoter in pTrc99A | 37 |
| pCHAP580 | pSU19-pulS | 14 |
| pCHAP7539 | pCHAP3671 AvrII | This study |
| pCHAP7542 | pCHAP3671-pulD-mCherry | This study |
| pCHAP7545 | pDSW204-mCherry | This study |
Plasmid construction.
pCHAP7539 containing pulD carrying a silent mutation in its 3′ end, resulting in an AvrII recognition site, was constructed by site-directed mutagenesis. The AvrII site was generated by PCR using the primers EndDAvr_for (5′-GCTATCGACGCGTTCAACCTAGGAGGCAATCTATG-3′) and EndDAvr_rev (5′-CATAGATTGCCTCCTAGGTTGAACGCGTCGATAGC-3′) with pCHAP3671 (14) as template DNA. PCR amplifications were performed for 18 cycles (30 s at 94°C, 1 min at 55°C, and 6 min at 72°C) with Pfu Turbo polymerase (Invitrogen) based on the Stratagene Quick Change protocol. Amplified DNA was digested with DpnI and transformed into competent PAP7447 cells. The gene encoding mCherry was amplified by PCR using the primers mCherry_Avr_for (5′-CGCCCTAGGAGTGAGCAAGGGCGAGGAG-3′) and mCherry_Avr_rev (5′-GCGCCTAGGTTCTTGTACAGCTCGTCCATGC-3′) with pRSETb-mCherry (31) as template DNA and inserted into pCHAP7539 as an AvrII (sequence underlined in primers) fragment, resulting in pCHAP7542. The constructs were verified by DNA sequencing (GATC). A plasmid with mCherry was constructed as a control for protein localization and degradation of fluorescent fusion proteins. The gene encoding mCherry was amplified by PCR using the primers 5′-NcoI_SC/YFP (5′-GCGCCATGGTGAGCAAGGGCGAG-3′) and 3′-EcoRI_SC/YFP (5′-CATGAATTCCTTGTACAGCTCGTCCATG-3′) with pRSETb-mCherry as template DNA using Taq DNA polymerase (Invitrogen). The PCR product was cloned in TOPO pCR2.1 (Invitrogen) and recloned as a NcoI-EcoRI fragment in pDSW204, resulting in pCHAP7545.
Cell fractionation and separation of inner and outer membranes by sucrose flotation gradient.
One hundred milliliters of cultures at an optical density at 600 nm of 0.8 to 1.0 were collected by centrifugation, and the pellet was resuspended in 10 ml of 25 mM HEPES (pH 7.4). Cells were disrupted by two passages through a French press (1,200 × 105 Pa), and the lysate was mixed with 10 μg/ml (each) of DNase I, pancreatic RNase A, and Pefabloc protease inhibitor and then centrifuged for 10 min at 5,000 × g to eliminate unbroken cells. Membranes were then collected by ultracentrifugation at 160,000 × g for 1 h at 4°C, resuspended, saturated with 60% sucrose in 200 μl of 25 mM HEPES (pH 7.4), and then placed at the bottom of the centrifuge tube. Steps (600 μl) were created using 56.2, 53.2, 50.2, 47.1, 44.2, 41.2, 38.1, and 35.9% sucrose solutions, and tubes were centrifuged in a swing-out rotor for 36 h at 230,000 × g at 10°C. Fractions (250 μl) were collected from the top of the tubes and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with appropriate antibodies. The concentration of the sucrose in each fraction was determined from the refraction index.
SDS-PAGE and immunoblotting.
Cells were treated with phenol to dissociate PulD multimers, and proteins were precipitated with acetone as described previously (15). Proteins were solubilized in loading buffer with 4 mM dithiothreitol, heated at 100°C for 5 min, and separated by SDS-PAGE in gels containing 10% or 12% acrylamide. After transfer of the gel including the stacking gel onto nitrocellulose membranes, proteins were detected by incubating the membranes with primary polyclonal rabbit antibodies against PulD or RFP (Chemicon International) and then with horseradish peroxidase-conjugated secondary antibodies to rabbit immunoglobulin G (GE Healthcare). Bound secondary antibodies were detected by enhanced chemiluminescence (Pierce).
Fluorescence microscopy.
Strains were grown overnight at 30°C, diluted 1:100 in fresh medium, and grown to an optical density at 600 nm of 0.2. Isopropyl-β-d-thiogalactopyranoside (IPTG) was omitted from the cultures, and basal levels of expression from the ptrc promoter at the λatt site integrated genes resulted in production of fluorescent fusion proteins. Maltose (0.4%) was added for strains carrying pul genes under control of the maltose-inducible promoter. The cells were incubated for 1 h at 30°C under continuous shaking. Live cells were immobilized on wet agarose-coated glass slides and analyzed using a Zeiss Axioplan2 fluorescence microscope mounted with a Hamamatsu charge-coupled device camera. Images were collected using the software program OpenLab and processed using the program Photoshop.
For localization of PulD using the purified Sac7*40-GFP fusion protein (22), cells were resuspended in phosphate-buffered saline (PBS) and immobilized on polylysine-coated glass coverslips at room temperature for 30 min. After the coverslips were washed two times with PBS, the cells were fixed in 2% formaldehyde in PBS for 30 min. Excess formaldehyde was removed by washing followed by quenching with 1 M Tris-HCl (pH 8) for 30 min. The cells were then partially permeabilized by incubation in 100 mM Tris-HCl (pH 8), 10 mM EDTA, and 100 μg/ml lysozyme at room temperature for 45 min to allow access to the periplasm. After washing and incubation with 2% bovine serum albumin in PBS for 30 min, the coverslips were incubated for 1 h in 10 μg/ml purified Sac7*40-GFP in PBS containing 2% bovine serum albumin. The coverslips were washed three times in PBS, incubated for another 30 min in PBS, washed again three times, and finally mounted in glycerol for examination by fluorescence microscopy.
RESULTS AND DISCUSSION
PulD-mCherry forms multimers and is functional.
The outer membrane insertion of the multimeric secretin PulD depends on its pilotin protein, PulS (13, 15, 16). In the presence of PulS and the other Pul factors, PulD forms an active secreton that promotes pullulanase (PulA) secretion. In the absence of PulS, PulD multimers insert into the inner membrane and induce the phage shock response (13, 15, 16). To determine whether PulD is localized at specific sites in the cell envelope, it was fused to a bright, stable variant of monomeric RFP, mCherry (31). mCherry could be readily fused to the carboxy terminus of PulD, leaving the PulS binding domain (5) intact. Attempts to fuse mCherry to the amino terminus of PulD were unsuccessful. The pulD-mCherry gene fusion was inserted into the E. coli chromosome at the λ attachment site (λatt) (2) in strain PAP7447, which carries all pul genes (the maltose-inducible pulC to -O operon with a pulD deletion, the maltose-inducible pulAB operon, which includes the pulA gene, encoding the surface-bound secreted protein pullulanase [PulA], and the constitutively expressed pulS gene), as described previously for gfp-pulL and gfp-pulM (3), to give the strain PAP9138. An untagged pulD gene was inserted at the same site in the control strain, PAP9124.
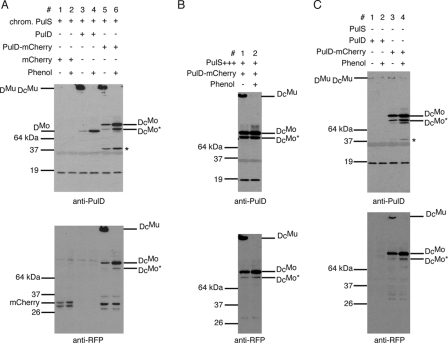
PulD-mCherry was analyzed for its ability to multimerize and to promote PulA secretion in PAP9138 (24). Like PulD, PulD-mCherry formed SDS-resistant multimers that were dissociated into monomers (95 kDa) by phenol (15) (Fig. 1A, lanes 5 and 6). Full-length PulD-mCherry multimers (DcMu) and monomers (DcMo) were recognized by antibodies against both PulD and RFP (Fig. 1A, lanes 5 and 6). Small amounts of mCherry, a 45-kDa PulD degradation product (15), and a truncated form of PulD-mCherry monomer (DcMo*), presumably resulting from degradation of the chimera, were observed under conditions of low-level PulS production (PAP9138, without maltose induction, in which chromosomal pulS is constitutively expressed from its own promoter (6, 7). Upon phenol treatment, an increase in PulD-mCherry monomers (DcMo) was observed under noninducing conditions (Fig. 1A, lane 6). These monomers reacted with antibodies against PulD and RFP, suggesting that PulD-mCherry multimers are mainly composed of full-length PulD-mCherry subunits. High levels of PulS protected PulD-mCherry from degradation, so that mCherry and the 45-kDa degradation product were no longer detected (Fig. 1B). The amount of truncated PulD-mCherry monomer (DcMo*) was unaffected, however. PulD-mCherry monomers and some multimers were detected in the absence of the entire secreton (in strain PAP9313 lacking DegP) (Fig. 1C, lanes 3 and 4), whereas PulD encoded by wild-type pulD integrated at the same chromosomal location as pulD-mCherry (PAP9312) was rapidly degraded, even in the absence of the periplasmic protease DegP, which degrades unassembled PulD (13) (Fig. 1C, lanes 1 and 2). This result suggests that the C-terminal mCherry extension protects PulD from degradation (see below).
FIG. 1.
PulD-mCherry forms multimers and is more stable than PulD in the absence of PulS. (A) Multimerization of PulD and PulD-mCherry in the presence of low levels of PulS. Strains used were PAP9123 (lanes 1 and 2), PAP9124 (lanes 3 and 4), and PAP9138 (lanes 5 and 6). Two protein bands representing mCherry were detected on the anti-RFP immunoblot (lanes 1 and 2) due to the presence of two translational start codons in pCHAP7545. (B) Protection of PulD-mCherry by plasmid-encoded PulS in strain PAP9210. (C) Stability of PulD and PulD-mCherry in the absence of PulS. The strains used were PAP9312 (lanes 1 and 2) and PAP9313 (lanes 3 and 4). All fractions loaded were derived from the same volume of bacterial suspension. Stacking gels were transferred onto nitrocellulose for detection of PulD multimers, and samples were treated with phenol to resolve multimers into monomers. Immunoblots were developed with antibodies against PulD and against RFP. DMu and DMo, PulD multimers and monomers, respectively; DcMu and DcMo, PulD-mCherry multimers and monomers, respectively. The PulD degradation product of 45 kDa is indicated by an asterisk.
PulA (pullulanase) secretion to the cell surface was quantified by comparing its enzyme activity in whole cells to that in lysed cells (surface-exposed protein and the sum of surface-exposed and nonsecreted protein, respectively) (20). PulD-mCherry in maltose-induced PAP9138 promoted pullulanase secretion to the same extent as PulD in PAP9124 (70 to 90% of the active enzyme was accessible to the substrate in intact cells; data not shown), suggesting that the C-terminal extension on PulD does not interfere with its function. Similar levels of pullulanase secretion were observed in the presence of high levels of PulS. A slight increase in DcMo was observed upon phenol treatment, while DcMo* was present in similar amounts, suggesting that multimers are composed of full-length PulD-mCherry (Fig. 2). These results also indicate that a few PulD-mCherry multimers in the outer membrane (see below) fully support pullulanase secretion.
FIG. 2.
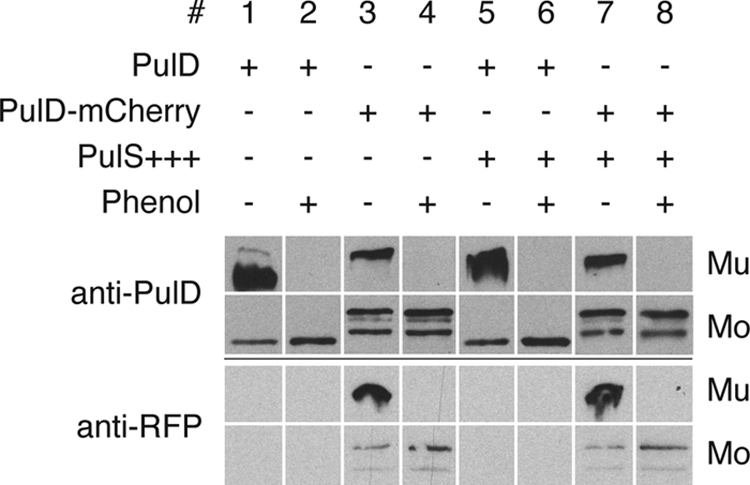
PulD-mCherry multimers composed of full-length PulD-mCherry subunits support pullulanase secretion. Strains PAP9124 without or with pCHAP580 (PulS+++) (lanes 1 and 2 and lanes 5 and 6, respectively) and PAP9138 without or with pCHAP580 (lanes 3 and 4 and lanes 7 and 8, respectively) were grown in minimal medium in the presence of 0.4% maltose to induce all Pul factors in order to monitor secretion of pullulanase. For other details, see the legend to Fig. 1.
PulS targets PulD-mCherry to the outer membrane.
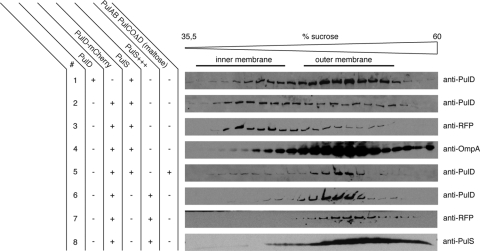
The observations that PulD-mCherry is able to form multimers and that it apparently functions in PulA secretion suggest that it is located in the outer membrane. PulD-mCherry in cells with PulS was exclusively located in the particulate (membrane) fraction of cells disrupted in a French press. These membranes were fractionated by sucrose flotation gradients. In the presence of low (chromosome-encoded) levels of PulS, PulD-mCherry was mainly located in the inner membrane fractions near the top of the gradient, whereas in control experiments, wild-type PulD was found almost exclusively in the outer membrane under these conditions (Fig. 3), as expected (13). PulS itself was found in the outer membrane fractions, irrespective of the presence of PulD or PulD-mCherry (not shown). Increasing the amount of PulS (in cells carrying pCHAP580) resulted in outer membrane localization of both PulD-mCherry and PulS (Fig. 3, lanes 6 to 8). At least 50 times more PulS was produced under these conditions (Fig. 4B, lanes 7 and 8). These results suggest that the S domain of PulD is partially masked by the mCherry protein or that its affinity for PulS is reduced, so that more PulS is required for efficient binding and targeting to the outer membrane.
FIG. 3.
Flotation sucrose gradient analysis of multimers of PulD and PulD-mCherry in the presence of chromosomal (PulS) or plasmid levels of PulS (PulS+++). pulD and pulD-mCherry were expressed from the chromosome at the λatt site without IPTG induction in PAP9124 (lane 1) and PAP9138 (lanes 2 to 4), respectively. Induction of the pul operon was obtained with 0.4% maltose in PAP9138 (lane 5). PulD-mCherry was analyzed in the presence of elevated PulS levels in PAP9138 (containing pCHAP580; lanes 6 to 8). Proteins in samples collected from the gradients were dissolved in SDS and examined by SDS-PAGE and immunoblotting with anti-PulD, anti-RFP, anti-OmpA, and anti-PulS antibodies.
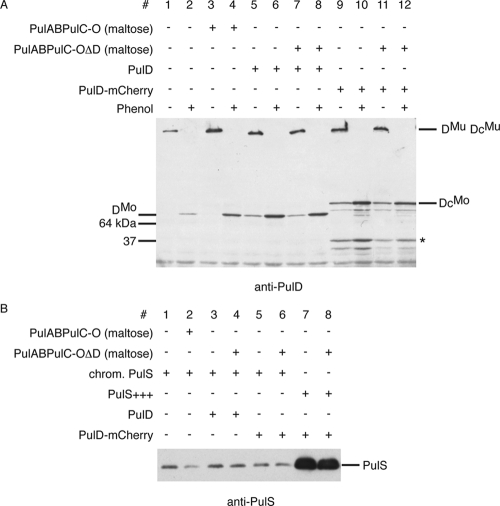
FIG. 4.
Levels of the PulD protein encoded by the maltose-inducible pul operon are comparable to PulD and PulD-mCherry levels obtained by expression of integrated genes at the λatt site and are independent of other Pul factors. (A) Comparison of protein levels of PulD (PAP9124, lanes 5 to 8) and PulD-mCherry (PAP9138, lanes 9 to 12) as multimers (DMu and DcMu, respectively) and as monomers (DMo and DcMo, respectively) (after treatment with phenol) compared to PulD expressed from the pul operon (PAP7232, lanes 1 to 4) with noninduced or induced (maltose) expression of pul genes. The 45-kDa PulD degradation product is indicated by an asterisk. (B) Comparison of chromosomal and plasmid-encoded levels of PulS. The amount of PulS encoded by IPTG-induced pCHAP580 (lanes 7 and 8) is at least 50 times higher than PulS from the constitutively expressed pulS gene in PAP7232 (lanes 1 and 2). Chromosomal levels of PulS are similar in PAP7232 (lanes 1 and 2), PAP9124 (lanes 3 and 4), and PAP9138 (lanes 5 and 6) and are independent of the presence of other Pul factors (maltose).
In the presence of the entire Pul secreton (in strain PAP9138 grown in the presence of maltose), PulD-mCherry multimers were detected in the outer membrane fractions (Fig. 3, lane 5). The amount of chromosome-encoded PulS was the same irrespective of maltose induction of the pulC to -O operon (7) (Fig. 4B, lanes 5 and 6). The levels of PulD-mCherry and PulD multimers derived from expression of the encoding genes from the λatt sites were similar to PulD levels from the pulC to -O operon upon maltose induction (Fig. 4A). These results suggest that other secreton factors besides PulS play a role in the outer membrane localization of PulD-mCherry.
PulD-mCherry forms foci in both the inner and outer membrane.
Unless the other Pul factors are induced, PulD-mCherry inserts into the inner membrane when PulS levels are low. Unlike PulD, however, PulD-mCherry does not induce the phage shock response in the absence of PulS (Fig. 5, lane 5). PspA levels were similar when pulD was expressed from the λatt site (Fig. 5, lane 3), when PulD multimers are located mainly in the outer membrane fractions (Fig. 3). A small decrease in PspA levels was observed when other secreton components were present (Fig. 5, lane 6), which is in agreement with its outer membrane localization under these conditions. As seen previously with PulD, PulD-mCherry could not be dissociated from inner membrane vesicles by 5 M urea (data not shown). Thus, the amount of PulD-mCherry, which is comparable to untagged PulD in the inner membrane (Fig. 3, lanes 1 and 2), might be too low to cause a phage shock response. Alternatively, PulD-mCherry might insert into the inner membrane in a conformation in which the C-terminal mCherry extension interferes with the ability of PulD to form the (hypothetical) channel that induces the phage shock response.
FIG. 5.
PulD-mCherry does not induce the phage shock response when inserted in the inner membrane in the presence of low levels of PulS. Production of PspA was determined by immunoblotting with antibodies against PspA in strain PAP9138 without and with induction of the pulC to -O operon by 0.4% maltose (lanes 5 and 6, respectively). PAP7232 without (lane 1) or with maltose (lane 2) and PAP9124 grown without or with maltose (lanes 3 and 4, respectively) were used as controls. Multimers of PulD and PulD-mCherry were detected by antibodies against PulD and RFP.
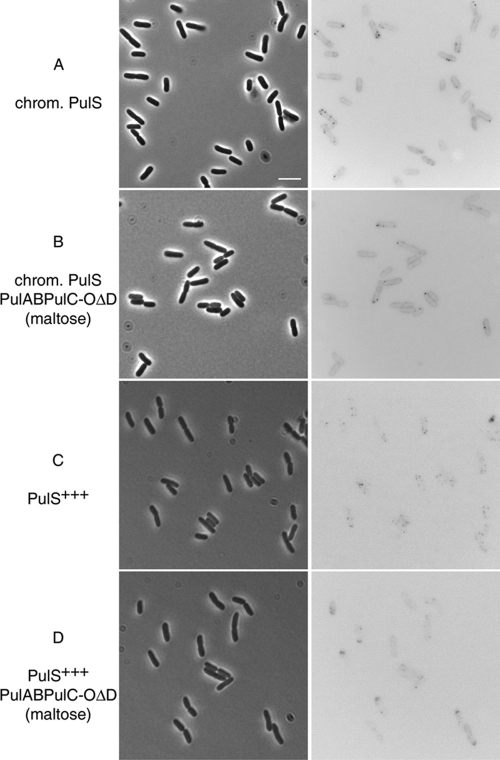
Under these conditions, one to four bright fluorescent foci were observed (Fig. 6A) in 44% of the cells, while the remaining 56% of cells did not have detectable foci (n = 239). In addition, the cell circumference could be discriminated in the fluorescence image, possibly due to the presence of unassembled PulD-mCherry monomers associated with the membrane or mCherry in the periplasm. A similar localization pattern was observed in the presence of the full complement of Pul components (Fig. 6B), indicating that PulD-mCherry multimers appear as foci irrespective of whether they are located in the inner or outer membrane.
FIG. 6.
PulD-mCherry localizes as bright fluorescent foci in the inner membrane (low levels of PulS) and in the outer membrane (high levels of PulS). The localization of fluorescent PulD-mCherry is shown on the right, and the corresponding phase-contrast images are shown on the left. (A) Microscopy of cells producing PulD-mCherry in the presence of chromosomal levels of PulS in strain PAP9138. (B) Localization of PulD-mCherry in the presence of chromosomal levels of PulS with pulC to -O operon expression (strain PAP9138 with maltose). (C) Localization of PulD-mCherry in the presence of high levels of PulS (PAP9138 with pCHAP580). (D) Localization of PulD-mCherry in the presence of high levels of PulS and other Pul factors (PAP9138 with pCHAP580 and maltose). Bar, 5 μm.
Fluorescent foci were numerous (up to six foci per cell) when PulS was sufficiently abundant to promote complete outer membrane association of PulD-mCherry (strain PAP9138 with pCHAP580) (Fig. 6C). The majority of the cells (70%) had detectable foci; the rest of the cells (30%) were not in the same focal plane but probably did have PulD-mCherry foci (n = 172). The number of PulD-mCherry foci increased with increasing levels of PulS, corresponding to more PulD-mCherry multimers located in the outer membrane. PulD-mCherry foci were located at random cellular positions, irrespective of the level of PulS, but occasionally polar foci were observed when PulS levels were high (Fig. 6C and D). The presence of other secreton factors did not affect the general distribution of PulD-mCherry, but some arc-shaped polar foci could be detected (strain PAP9138 with pCHAP580 and maltose) (Fig. 6D).
In strain PAP7232 (carrying the maltose-inducible pulC to -O operon, maltose-inducible pulAB, and the constitutively expressed pulS gene), the number of PulD molecules was estimated to be between 120 and 300 per cell, equivalent to 10 and 25 dodecameric multimers under maltose-induced conditions. The levels of PulD and PulD-mCherry attained by expressing the genes integrated at the λatt site were comparable to these levels (Fig. 4), suggesting that each focus represents at least one multimer of PulD. The existence of clusters of multimers would be in line with previous results showing PulD is present in clusters in a small proportion of vesicles when it is located in the inner membrane (in the absence of PulS) (13).
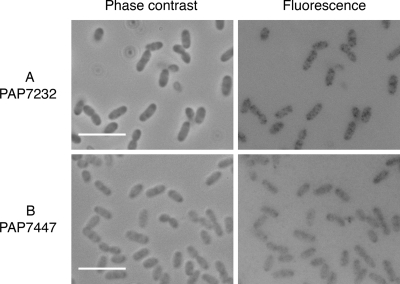
To confirm that PulD in strain PAP7232 is present at comparable levels and shows the same distribution pattern as PulD-mCherry, we examined the localization of wild-type PulD by indirect fluorescent detection using Sac7*40-GFP as a reporter. Sac7*40 is an artificial protein (affitin) that binds specifically and with high affinity to an unidentified epitope in the N-terminal half of PulD (22). Incubation of fixed and permeabilized PAP7232 cells with Sac7*40-GFP followed by fluorescence microscopy showed the presence of zero to four bright foci per cell and a weaker fluorescence of the whole cell body. Foci were not detected in a strain lacking the pulD gene (PAP7447), although some background fluorescence was present (Fig. 7). This background fluorescence was not PulD dependent and is therefore nonspecific. Furthermore, fluorescent foci were not detected when Sac7*40-GFP was used on nonpermeabilized cells (not shown), suggesting that the epitope recognized by this affitin in the N domain of PulD faces the periplasm. The very similar number and distribution on fluorescent foci observed in this experiment and in strain PAP9138 was taken as validation of PulD-mCherry as a model of wild-type PulD. These results extend the range of applications for affitins to include in situ localization of the proteins they recognize by fluorescence microscopy when they are GFP tagged. Other approaches, including localization in thin-sectioned cells using affitins bound to gold beads, would represent another potential application.
FIG. 7.
In situ detection of PulD by PulD-specific affitin Sac70*40-GFP. After fixation and permeabilization, PulD was detected by fluorescence microscopy using the PulD-binding artificial protein Sac7*40-GFP as a fluorescent probe. The fluorescence image is shown in the right panels; the corresponding phase-contrast image is shown on the left. (A) Microscopy of cells expressing pulAB, pulC to -O, and pulS at chromosomal levels (PAP7232 with maltose). (B) Microscopy of an isogenic strain lacking pulD (PAP7447 with maltose). Bar, 5 μm.
PulD-mCherry forms bright polar foci in the complete absence of PulS.
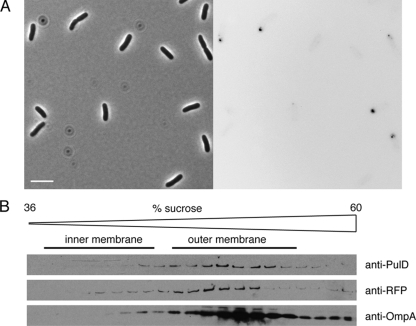
PulS both protects PulD from degradation and targets it to the outer membrane. In the complete absence of PulS, PulD is degraded by periplasmic and membrane proteases, of which the most active is DegP (13). Unlike the case with PulD, monomers and some multimers of PulD-mCherry made without PulS could be detected in whole-cell lysates of a strain lacking DegP (Fig. 1C, lanes 3 and 4), suggesting that fusing mCherry to the C terminus increases PulD stability. Under these conditions, PulD-mCherry formed bright, round, polar foci of various sizes (Fig. 8A). This localization pattern was clearly different from that observed in cells in which PulD-mCherry was targeted to the outer membrane when PulS levels were high and when other Pul factors were present, when numerous foci and some polar arc-shaped patches were observed (Fig. 6). PulD-mCherry multimers from cells lacking PulS fractionated with the outer membrane in flotation sucrose gradients (Fig. 8B), but this material is likely to be small, membrane-associated aggregates. Indeed, this localization pattern resembles the polar foci observed when GFP-PulL or GFP-PulM was produced from high-copy-number plasmids (3).
FIG. 8.
PulD-mCherry forms small membrane-associated aggregates in the complete absence of PulS. (A) Microscopy of cells expressing pulD-mCherry in the absence of PulS (PAP9313). Note the bright foci at the cell poles in the fluorescence panel (right). Bar, 5 μm. (B) Flotation sucrose gradient analysis of PulD-mCherry multimers from cells of the same culture. Proteins were analyzed by immunoblotting using antibodies against PulD, RFP, and OmpA. For other details, see the legend to Fig. 3.
Concluding remarks.
PulD-mCherry displays many characteristics of the type II secretin PulD: it forms heat-resistant multimers, it can function as a secretin to secrete pullulanase, and its outer membrane location is dependent on the pilotin PulS. Thus, it can be used as a reporter for PulD localization. The bright fluorescent foci that it forms when PulS levels are high presumably represent multimers of secretin in the outer membrane, whereas those formed at low PulS levels presumably represent nonfunctional PulD-mCherry multimers in the inner membrane. In wild-type cells carrying the complete secreton, these sites presumably represent positions in the cell envelope where PulA is translocated and where artificially elongated PulG pseudopili (resulting from increased PulG production) are anchored in the cell envelope (35).
We reported previously that GFP-PulL and GFP-PulM distribute over the entire inner membrane, with occasional brighter foci. The formation of GFP-PulL foci was dependent on other components of the secreton and not only on PulM (3). GFP-PulG and PulG-RFP were localized evenly throughout the inner membrane without foci (9). These data clearly show that components of the K. oxytoca T2SS are not localized at the cell poles in E. coli. However, we note that while GFP-PulM and GFP-PulL appeared to be mostly uniformly distributed throughout the inner membrane, occasional brighter foci were also observed. One intriguing possibility is that these sites are the same as those at which PulD dodecamers are located and that they represent the location of fully assembled secreton complexes that span the inner membrane, the periplasm and peptidoglycan, and the outer membrane. These conjectures underscore the need to correlate localization with function when determining the location of envelope-associated nanomachines, such as the secreton.
Acknowledgments
We thank Ingrid Guilvout for her constant interest in this work and other members of the Molecular Genetics Unit of the Institut Pasteur for their support. We thank Ghislaine Behar for technical support. Roger Tsien kindly provided the plasmid pRSETb-mCherry.
The work was supported in part by a grant from the French ANR (ANR-05-0307-01).
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J. Mol. Biol. 27205-219. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buddelmeijer, N., O. Francetic, and A. P. Pugsley. 2006. Green fluorescent chimeras indicate nonpolar localization of pullulanase secreton components PulL and PulM. J. Bacteriol. 1882928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang, P., M. Habash, and L. L. Burrows. 2005. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daefler, S., I. Guilvout, K. R. Hardie, A. P. Pugsley, and M. Russel. 1997. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol. Microbiol. 24465-475. [DOI] [PubMed] [Google Scholar]
- 6.d'Enfert, C., and A. P. Pugsley. 1989. Klebsiella pneumoniae pulS gene encodes an outer membrane lipoprotein required for pullulanase secretion. J. Bacteriol. 1713673-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.d'Enfert, C., A. Ryter, and A. P. Pugsley. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 63531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorsey, F. C., J. F. Fischer, and J. M. Fleckenstein. 2006. Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell Microbiol. 81516-1527. [DOI] [PubMed] [Google Scholar]
- 9.Francetic, O., N. Buddelmeijer, S. Lewenza, C. A. Kumamoto, and A. P. Pugsley. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 1891783-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, A. S., and K. D. Young. 2005. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 1871913-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs, K. A., D. D. Isaac, J. Xu, R. W. Hendrix, T. J. Silhavy, and J. A. Theriot. 2004. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 531771-1783. [DOI] [PubMed] [Google Scholar]
- 12.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15R514-R526. [DOI] [PubMed] [Google Scholar]
- 13.Guilvout, I., M. Chami, A. Engel, A. P. Pugsley, and N. Bayan. 2006. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 255241-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilvout, I., K. R. Hardie, N. Sauvonnet, and A. P. Pugsley. 1999. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol. 1817212-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15978-988. [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie, K. R., A. Seydel, I. Guilvout, and A. P. Pugsley. 1996. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol. 22967-976. [DOI] [PubMed] [Google Scholar]
- 17.Jaumouille, V., O. Francetic, P. J. Sansonetti, and G. Tran Van Nhieu. 2008. Cytoplasmic targeting of IpaC to the bacterial pole directs polar type III secretion in Shigella. EMBO J. 27447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Judd, P. K., R. B. Kumar, and A. Das. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55115-124. [DOI] [PubMed] [Google Scholar]
- 19.Köhler, R., K. Schäfer, S. Müller, G. Vignon, K. Driedrichs, A. Philippsen, P. Ringler, A. P. Pugsley, A. Engel, and W. Welte. 2004. Structure and assembly of pseudopilin PulG. Mol. Microbiol. 54647-664. [DOI] [PubMed] [Google Scholar]
- 20.Michaelis, S., C. Chapon, C. d'Enfert, A. P. Pugsley, and M. Schwartz. 1985. Characterization and expression of the structural gene for pullulanase, a maltose-inducible secreted protein of Klebsiella pneumoniae. J. Bacteriol. 164633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Mouratou, B., F. Schaeffer, I. Guilvout, D. Tello-Manigne, A. P. Pugsley, P. M. Alzari, and F. Pecorari. 2007. Remodeling a DNA-binding protein as a specific in vivo inhibitor of bacterial secretin PulD. Proc. Natl. Acad. Sci. USA 10417983-17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obuchowski, P. L., and C. Jacobs-Wagner. 2008. PflI, a protein involved in flagellar positioning in Caulobacter crescentus. J. Bacteriol. 1901718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Possot, O., G. Vignon, N. Bomchil, F. Ebel, and A. P. Pugsley. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 1822142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugsley, A. P. 1993. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol. Microbiol. 9295-308. [DOI] [PubMed] [Google Scholar]
- 26.Pugsley, A. P., M. G. Kornacker, and I. Poquet. 1991. The general secretion pathway is directly required for extracellular pullulanase secretion in Escherichia coli. Mol. Microbiol. 5343-352. [DOI] [PubMed] [Google Scholar]
- 27.Py, B., L. Loiseau, and F. Barras. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2244-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray, N., A. Nenninger, C. W. Mullineaux, and C. Robinson. 2005. Location and mobility of twin-arginine translocase subunits in the Escherichia coli plasma membrane. J. Biol. Chem. 28017961-17968. [DOI] [PubMed] [Google Scholar]
- 29.Scott, M., Z. Dossani, and M. Sandkvist. 2001. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc. Natl. Acad. Sci. USA 9413978-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senf, F., J. Tommassen, and M. Koster. 2008. Polar secretion of proteins via the Xcp type II secretion system in Pseudomonas aeruginosa. Microbiology 1543025-3032. [DOI] [PubMed] [Google Scholar]
- 31.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 32.Shevchik, V. E., J. Robert-Badouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO. J. 163007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiomi, D., M. Yoshimoto, M. Homma, and I. Kawagishi. 2006. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 60894-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tinker, J. K., J. L. Erbe, and R. K. Holmes. 2005. Characterization of fluorescent chimeras of cholera toxin and Escherichia coli heat-labile enterotoxins produced by use of the twin arginine translocation system. Infect. Immun. 733627-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vignon, G., R. Köhler, E. Larquet, S. Giroux, M.-C. Prévost, P. Roux, and A. P. Pugsley. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization and surface presentation of peptides. J. Bacteriol. 1853416-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L.-F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO. J. 206735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]