Abstract
Members of the SMC (structural maintenance of chromosomes) protein family play a central role in higher-order chromosome dynamics from bacteria to humans. So far, studies of bacterial SMC proteins have focused only on unicellular rod-shaped organisms that divide by binary fission. The conversion of multigenomic aerial hyphae of the mycelial organism Streptomyces coelicolor into chains of unigenomic spores requires the synchronous segregation of multiple chromosomes. Here we focus on the contribution of SMC proteins to sporulation-associated chromosome segregation in S. coelicolor. Deletion of the smc gene causes aberrant DNA condensation and missegregation of chromosomes (7.5% anucleate spores). In vegetative mycelium, immunostained SMC proteins were observed sporadically, while in aerial hyphae about to undergo sporulation they appeared as irregularly spaced foci which accompanied but did not colocalize with ParB complexes. Our data demonstrate that efficient chromosome segregation requires the joint action of SMC and ParB proteins. SMC proteins, similarly to ParAB and FtsZ, presumably belong to a larger group of proteins whose expression is highly induced in response to the requirement of aerial hyphal maturation.
Higher-order chromosome structure is particularly critical in the segregation of replicated chromosomes during cell division in eubacteria, eukaryotes, and archaea. Recent studies have shown that members of the SMC (structural maintenance of chromosomes) protein family play a central role in higher-order chromosome dynamics, acting either as condensins or cohesins (15-17, 30, 32, 36, 40). SMC are large proteins (110 to 170 kDa) that form dimers. Their N- and C-terminal parts form a single head domain (see references 13, 15-17, and 38 for reviews). SMC proteins exhibit ATPase activity that is required for their dynamic interaction with DNA (17). The results of recent studies demonstrated that bacterial SMC protein functions in complexes with other proteins, i.e., ScpA and ScpB (Scp's are segregation and condensation proteins) (6, 29, 37).
In contrast to eukaryotes, in eubacteria the cell-cycle events, including replication, condensation, and segregation, take place simultaneously (16, 26). Thus, during the bacterial cell cycle, chromosomes must undergo dynamic changes. So far, studies regarding chromosome dynamics have focused on Bacillus subtilis, Caulobacter crescentus, and Escherichia coli. In these organisms, SMC proteins (or MukB, a functional homologue in E. coli) appear to play similar roles; they are involved in chromosome organization and segregation. The genes encoding these proteins are not essential, although their deletion results in temperature-sensitive growth (2, 9, 22, 31, 32, 38). In B. subtilis, smc deletion mutants exhibit complex phenotypes under permissive conditions, including poorly compacted nucleoids and a relatively large amount of anucleate cells (up to 15% of the cells were anucleate) (10). Interestingly, the inactivation of smc enhances the chromosome segregation defect of spo0J- or soj-null mutants (25). The proteins encoded by these genes, Soj (ParA, an ATPase) and Spo0J (ParB, a DNA-binding protein), form a nucleoprotein complex at the oriC region that ensures its proper segregation. Many attempts have been made to localize SMC proteins in bacterial cells. Despite some differences in SMC protein localization, comprehensive microscopic observations in B. subtilis revealed that SMC proteins form foci or a punctate pattern associated with the nucleoid (42). SMC protein colocalizes with its interacting partners ScpA and ScpB, and the specific localization of SMC protein depends on both Scp proteins, showing that all three components of the SMC complex are required for proper localization (41).
Here we extend the studies of SMC proteins, which have previously focused only on unicellular rod-shaped bacteria that divide by binary fission, to the more-complex bacterium Streptomyces coelicolor. Streptomycetes are gram-positive multicellular sporulating bacteria that grow by tip extension and hyphal branching to form a dense mycelial network of vegetative hyphae in which septation occurs at some distance from the tips (7). Compartments of the vegetative hyphae contain several copies of the large (8 to 9 Mbp), linear uncondensed chromosome (4). Older Streptomyces colonies differentiate to form aerial hyphae, which subsequently develop into chains of spores (3, 4). The rapid growth of the aerial hyphae is accompanied by intensive replication of the chromosomes, such that 50 or more uncondensed nonsegregated chromosomes may be present in one long tip compartment (33). After an aerial hypha has stopped growing, a large number of FtsZ rings form a regular ladder in the long tip compartment, giving rise to sporulation septa that delimit the prespore compartments (8). At this stage, dozens of chromosomes are condensed and uniformly aligned along the hyphal tip, ensuring that each prespore compartment receives a single copy (21).
Very little is known about the orchestration of major cell cycle processes, particularly the synchronization of condensation and the segregation of several dozen chromosomes during the formation of unigenomic spores. Here we focus on the contribution of SMC proteins to sporulation-associated chromosome segregation in S. coelicolor A3(2), the model organism for the Streptomyces species.
MATERIALS AND METHODS
DNA manipulation and bacterial strain growth conditions.
DNA manipulations were carried out according to standard protocols (34). Enzymes were supplied by Fermentas, New England BioLabs, or Roche; fluorescent reagents were from Invitrogen (Molecular Probes); and oligonucleotides were from The Institute of Biochemistry and Biophysics (Poland). The S. coelicolor and E. coli strains are listed in Table 1. Culture conditions, media, antibiotic concentrations, and transformation and conjugation methods followed the general procedures for E. coli (34) and Streptomyces (23). S. coelicolor was cultivated in Trypticase soy broth-yeast extract-malt extract (1:1) complex liquid medium or on mannitol soy flour agar plates unless otherwise stated.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory stock |
| BW25113/pIJ790 | K-12 derivative; ΔaraBAD ΔrhaBAD λ− Red(gam bet exo) cat araC rep101(Ts) | 12 |
| ET12567/pUZ8002 | dam-13::Tn9 dcm cat tet hsd zjj-201::Tn10 tra neo RP4 | 32a |
| BL21 | B F−dcm ompT hsdS(rB− mB−) gal | Stratagene |
| S. coelicolor M145 | ||
| SCP1− SCP2− | 1 | |
| ΔparB (J3305) | D. Jakimowicz, unpublished data | |
| ΔparA (J3306) | 21 | |
| parB-egfp (J3310) | 19 | |
| dnaN-egfp (J3337) | 33 | |
| attBfC31::KF41(ftsZ-egfp) (K202) | 8 | |
| smc-egfp acc(3)IV (RMD2) | 5a | |
| pIJ8641 | 39 | |
| Δsmc::apra | This study | |
| ΔscpAB | This study | |
| ΔparB Δsmc::apra | This study | |
| ΔparA Δsmc::apra | This study | |
| parB-egfp Δsmc::apra | This study |
Construction of strains carrying a deletion of smc or scpAB.
A PCR-targeted gene replacement strategy (11, 12) was used for the construction of deletions of the smc and scpAB genes. Deletion of smc was created by replacing the smc gene within cosmid 7A1 with apra-oriT, an apramycin resistance (Aprar) cassette flanked by FLP recombinase recognition target sites. The cassette was amplified from pIJ773 (12) with the pair of primers pDelsmc_f and pDelsmc_r (Table 2). The resulting construct, 7A1Δsmc::apra, was used to transform ET12567/pUZ8002, from which it was mobilized into S. coelicolor M145 or other S. coelicolor derivatives (Table 1). The Aprar exconjugants were screened for the loss of Kanr (the kan gene is a part of cosmid 7A1Δsmc::apra), indicating that there was double-crossover allelic exchange of the smc locus.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequencea | Application |
|---|---|---|
| pScpA_fw | GGATCCGTGCGGCTCGCCAACTTCGAGG (BamHI) | scpA gene amplification |
| pScpA_rv | CTCGAGCGCCTTCCGCTCCTCCTTCGG (XhoI) | |
| pScpB_fw | GGATCCGTGAGTGAGCGGATCACGGAGG (BamHI) | scpB gene amplification |
| pScpB_rv | CTCGAGAAATTCCGTCTTGTCGTCTGCGTC (XhoI) | |
| pDelspn_f | GACGACGGTGTCTTCAAGGTGCGGCTCGCCAACTTCGAGATTTAAATATTCCGGGGATCCGTCGACC (SwaI) | scpA and scpB (SCO1769 and SCO1770) deletion in cosmid StI51 |
| pDelspn_r | TTCCGTCTTGTCGTCTGCGTCCCCGGCATCCGGGGCGTCATTTAAATTGTAGGCTGGAGCTGCTTC (SwaI) | |
| pSmc_5_fw | CATATGCACCTGAAGGCCCTGACCCTC (NdeI) | smc gene fragment amplification |
| pSmc_5_rv | CTCGAGGGCGGCGTTCCTCCATGGCC (XhoI, NcoI) | |
| pSmc_ap_fw | CGGTGTGTCGAAGGTCATCAGCCAGCGGTTGCGTCAGCCCCTCGAGTGTAGGCTGGAGCTGCTTC (XhoI) | XhoI sites before codon stop smc gene in cosmid 7AI |
| pSmc_ap_rv | AGTCAAGAGGGCGAGAGCTGTGACTTTCAGGGGAGCTGAAATTCCGGGGATCCGTCGACC | |
| pDelsmc_f | TCGTGGAGTGAGACAGACCCGAAAGGTAGAGTCCGACGGCATTCCGGGGATCCGTCGACC | smc (SCO5577) deletion in cosmid 7A1 |
| pDelsmc_r | GCTGAAGTGGTTATGTGTTCAAGTCTTGAAGAACGAGGGTTGTAGGCTGGAGCTGCTTC |
Boldface indicates restriction sites, and italics indicate apra cassette complementary fragment.
An in-frame scpAB deletion was created by using the appropriate pair of primers, pDelspn_f and pDelspn_r. Cosmid StI51ΔscpAB::apra was digested with SwaI and religated. The amp gene in the SuperCos part of cosmid StI51ΔscpAB was subsequently exchanged for an apra-oriT cassette, yielding StI51ΔscpAB amp::apra-oriT. This construct was used to transform ET12567/pUZ8002 and for conjugation into S. coelicolor M145. The Aprar exconjugants were screened for the loss of both Aprar and Kanr, indicating a double-crossover allelic exchange of the ΔscpAB locus of M145.
Purification of SMC, ScpA, and ScpB proteins.
The vectors pET-21a and pGEX-6P-2 were chosen to overexpress the recombinant SMC protein and the fusion proteins glutathione-S-transferase (GST)-ScpA and GST-ScpB. Cosmid 7A1, which contains the entire coding region of the S. coelicolor smc gene, was used for subcloning the smc gene into pET-21a. The 5′ end (1,135 bp) of the smc gene was PCR amplified using the appropriate pair of primers, pSmc_5_fw and pSmc_5_rv (Table 2). The resulting DNA fragment was digested with NdeI and XhoI and inserted into the NdeI- and XhoI-restricted pET-21a. Afterward, the rest of the smc gene was subcloned into the expression vector as follows. An XhoI restriction site was created directly before the stop codon of smc on cosmid 7A1 by using PCR targeting directly, and then a 2,456-bp fragment of smc was cut out and subcloned into NcoI- and XhoI-restricted pET-21a carrying smc (1,135 bp). The S. coelicolor His6-SMC protein was isolated as described previously (43).
The scpA and scpB genes were PCR amplified with the appropriate pairs of primers, pScpA_fw and pScpA_rv and pScpB_fw and pScpB_rv (Table 2). The PCR products were cloned into pGEM-T Easy (Promega) and later recloned as BamHI-XhoI fragments into pGEX-6P-2. All the PCR-derived clones were analyzed by DNA sequencing to check their fidelity. The fusion proteins GST-ScpA and GST-ScpB were purified on glutathione-Sepharose 4B as described earlier (27). For removal of the GST part, the bound fusion proteins were treated with PreScission protease (Amersham-Pharmacia-Biotech) and ScpA or ScpB protein was released from the beads. The purified SMC, ScpA, and ScpB proteins were more than 95% pure, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (see Fig. S1A in the supplemental material).
Preparation of antisera.
Antisera were obtained by immunization of rabbits with the purified SMC and ScpA proteins mixed with the adjuvant monophosphoryl lipid A (a derivative of lipid A from Hafnia alvei PCM 1200) (5). Serum samples were taken 10 days after the second booster injection. Cellular particles were removed by centrifugation, and the serum was stored at −20°C. Anti-SMC and anti-ScpA antibodies were purified by solid-phase chromatography (14).
SDS-PAGE and Western blotting.
Proteins were separated by SDS-PAGE (10% or 12% acrylamide) as described by Laemmli (24) and transferred to a nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with TBST (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.05% Tween 20) containing 3% bovine serum albumin (BSA) and subsequently incubated with polyclonal anti-SMC antibody. Afterwards the membrane was incubated with a goat anti-rabbit secondary antibody conjugated with alkaline phosphatase. The membrane was stained with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium.
Dot blot overlay assay.
SMC or ScpB protein was spotted onto the nitrocellulose membrane strips. After drying, the strips were placed in a blocking solution (0.5% BSA, 0.25% gelatin, 0.5% Nonident P-40, 100 mM NaCl, 50 mM Tris-HCl, pH 7.5) and subsequently incubated with ScpA protein. The strips were exhaustively washed with TBST buffer containing 3% BSA. Afterwards, ScpA protein was detected by a polyclonal anti-ScpA antibody as described above.
Microscopy.
Strains used for fluorescence microscopic observations were inoculated into the acute-angle junction of coverslips inserted at an angle of 45° into minimal medium containing 1% mannitol (23) and cultured for 24 to 72 h. The staining procedures were described previously (21, 35). Briefly, mycelium was fixed for 10 min with a paraformaldehyde-glutaraldehyde mixture, digested for 2 min with 2 mg ml−1 lysozyme, washed with phosphate-buffered saline (PBS), and blocked with 2% BSA. For immunostaining, samples were incubated with purified antibody against SMC (1:2,500 dilution) overnight, washed six times with PBS, and then incubated for 1 h with secondary antibody (antirabbit) conjugated with Alexa Fluor 546. For DNA staining, samples were incubated with 0.1 to 1 μg ml−1 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes), and for cell wall visualization with 10 μg ml−1 wheat germ agglutinin (WGA)-Texas Red or Alexa Fluor 350 conjugate (Molecular Probes). After five washes with PBS, the coverslips were mounted in Vectashield (Vector Laboratories) antifade reagent. Fluorescence microscopy was carried out using a Zeiss Axio Imager Z1 microscope equipped with a 100× objective. The relative intensities of the ParB foci (maximal pixel value with the background signal subtracted) in images of aerial hyphae of the M145 Δsmc parB-egfp and M145 parB-egfp (expressing enhanced green fluorescent protein) strains taken with the same camera settings were measured.
RESULTS
Identification of the Streptomyces coelicolor smc, scpA, and scpB homologues.
Sequence analysis identified smc, scpA, and scpB genes in all the Streptomyces chromosomes sequenced so far (http://www.genomesonline.org/). The smc, scpA, and scpB genes encode putative proteins with calculated molecular masses of 128, 30, and 24 kDa, respectively. Sequence analysis of the S. coelicolor chromosome indicates that the smc gene is presumably independently transcribed, while scpA and scpB may be cotranscribed as part of a larger cluster of genes (scpA being the first gene of the operon, with scpB and four other genes, encoding putative pseudouridine synthase, putative DNA hydrolase, putative glycohydrolase, and a hypothetical protein, downstream of this gene) (1). The SMC, ScpA, and ScpB proteins from S. coelicolor share extensive homology with chromosomally encoded orthologues from other bacteria (sequence similarities to those of B. subtilis are 47%, 47%, and 44%, respectively). All the SMC signature motifs (Walker A and B, ABC signature) and domain organization (coiled-coil domains, hinge) are conserved in the predicted Streptomyces SMC protein.
In order to test whether ScpA interacts with SMC/ScpB, as the corresponding proteins do in B. subtilis, a dot blot overlay experiment was performed (see Fig. S1B in the supplemental material). Purified SMC and ScpB proteins (see Materials and Methods and see Fig. S1A in the supplemental material) were spotted onto a nitrocellulose membrane, followed by incubation with ScpA. The membrane was then washed extensively and probed with anti-ScpA antibodies. Immunoreactive spots were observed for the two proteins but not for the negative controls BSA or binding buffer alone, indicating that ScpA interacts with SMC and ScpB. These interactions presumably exist in vivo as well.
Deletion of smc or scpAB genes causes aberrant condensation and missegregation of chromosomes.
In B. subtilis and C. crescentus, deletion of the smc gene is not lethal, but it does cause defects in DNA compaction and chromosome segregation. In order to evaluate the importance of the smc and scpAB genes for viability, growth, and chromosome segregation in Streptomyces, deletion mutants were constructed.
Since the smc gene is supposed to be independently expressed, the insertion-deletion mutant was constructed by replacing the entire gene with the apramycin resistance cassette (see Materials and Methods). The deletion was confirmed by Southern hybridization (data not shown) and Western blotting (Fig. 1B). The resulting Δsmc mutant strain, S. coelicolor, showed no obvious defect in growth or morphology when cultured in liquid or on solid medium. However, visualization of the DNA (DAPI stained) and cell walls (stained with WGA conjugated with Texas red) of sporulating hyphae revealed missegregation of chromosomes; approximately 8% of prespore compartments were anucleate. Since both the smc and partitioning gene (parA and parB) deletion mutants have defects in chromosome segregation, we tested the phenotypes of double mutants Δsmc ΔparA and Δsmc ΔparB. The double mutants showed no obvious defects in morphology. The number of anucleate prespores resulting from Δsmc ΔparB mutations was approximately equal to the sum of the anucleate prespores observed with the two single mutations (Table 3). Interestingly, the total amount of anucleate prespores resulting from the double Δsmc ΔparA mutation was approximately the same as in the ΔparA mutant strain (Table 3).
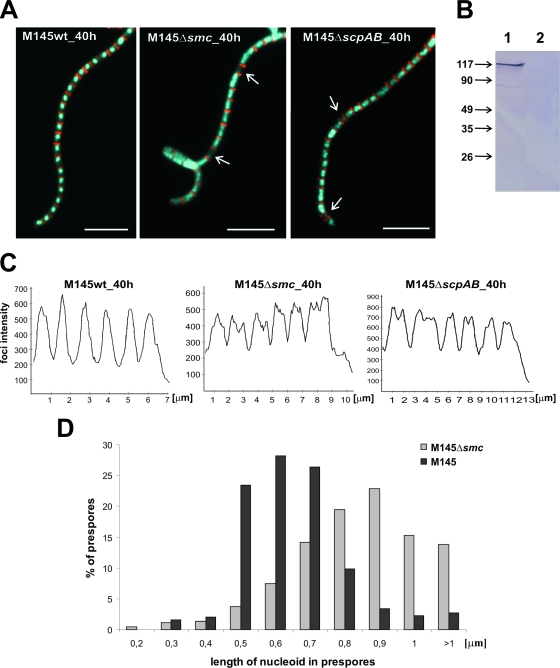
FIG. 1.
Influence of smc or scpAB deletion on chromosome condensation and segregation. (A) Examples of images showing hyphae with cell walls stained with WGA (red) and DNA stained with DAPI (blue). Anucleate compartments are indicated by white arrows. Scale bar, 5 μm. (B) Western blotting. Total proteins (20 μg) were separated by SDS-PAGE, and then SMC protein was identified by using antibodies raised against the recombinant SMC protein. Lanes: 1, wild type; 2, Δsmc mutant. Molecular sizes in kDa are shown on the left. (C) DNA signal intensity in spore compartments: fluorescent intensity of DNA stained with DAPI, presented in arbitrary units, was measured along a line drawn through the middle region of the spore compartments. (D) Percentage of prespores exhibiting different degrees of DNA compaction (length of nucleoid in prespores). wt, wild type; h, hour.
TABLE 3.
Segregation defects in Δsmc and ΔscpAB strains
| Strain or genotype | Anucleate prespores (%)a |
|---|---|
| Wild-type M145 | 1.4 |
| Δsmc | 7.5 |
| ΔscpAB | 6.3 |
| ΔparA | 24.1 |
| ΔparB | 19.5 |
| Δsmc ΔparB | 24.5 |
| Δsmc ΔparA | 24.7 |
Calculated for 400 to 700 prespores of each mutant and the wild type.
Deletion of smc also perturbed chromosome condensation in comparison with the chromosome condensation of the wild type; the Δsmc mutant's nucleoids filled almost all of a prespore's volume and did not stain very brightly with DAPI (Fig. 1A, C, and D), whereas the nucleoids of the wild type became smaller and stained very brightly during the formation of prespores (Fig. 1A, C, and D). This suggests that the nucleoids of the Δsmc mutant are less condensed during the early stages of sporulation (the smaller volume of mature spores made it difficult to assess chromosome condensation at later stages).
The scpA and scpB genes are supposed to be part of a larger operon. Indeed, disruption of the scpAB genes with the insertion of the Apra cassette caused markedly delayed growth (data not shown), suggesting perturbation in the transcription of genes that are located downstream of scpAB (including sco1768, encoding a putative pseudouridine synthase). To avoid polar effects of scpAB deletion on these genes, a strain carrying a nonpolar, unmarked, in-frame deletion of scpAB was constructed (see Materials and Methods). The scpAB unmarked deletion was verified by Southern hybridization. The S. coelicolor ΔscpAB mutant showed a phenotype similar to that of the S. coelicolor Δsmc mutant, including aberrations in chromosome condensation and segregation (Fig. 1A and C), although slightly fewer anucleate prespores were observed (6.3%).
In conclusion, these observations show that the SMC, ScpA, and ScpB proteins are involved in DNA condensation and influence chromosome segregation during sporulation.
In aerial hyphae, SMC forms irregularly spaced foci before septation.
In Streptomyces, dozens of linear chromosomes have to condense and segregate accurately and synchronously during sporulation to ensure that each spore receives a single copy of the chromosome.
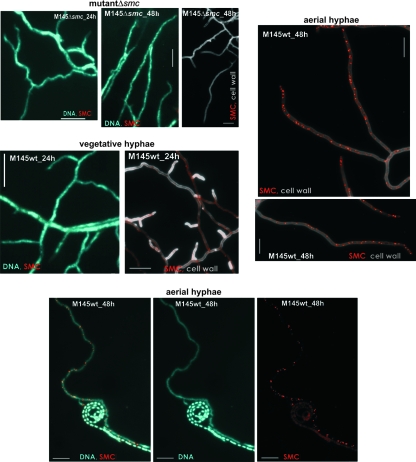
To localize SMC protein in the hyphae of a sporulating colony, rabbit polyclonal antibodies raised against the purified SMC protein (see Materials and Methods) were used in immunofluorescence microscopy. The affinity-purified anti-SMC antibodies reacted specifically with a protein of approximately 130 kDa that was present in a whole-cell extract of wild-type S. coelicolor, but they did not detect any protein when the S. coelicolor Δsmc mutant strain was tested (Fig. 1B). In agreement with the Western blot analysis, immunofluorescent signals (foci) were observed in the wild-type strain but not in the smc deletion mutant (Fig. 2). In the vegetative mycelium, sporadic immunostaining of SMC was observed (Fig. 2), while in the aerial hyphae, the number of foci increased as hyphae became close to full-length, reaching a maximum level before septation. At this stage, SMC protein was visualized as clusters of irregularly spaced foci. The abundance of foci was approximately seven foci/10 μm (Fig. 2). They were observed over uncondensed DNA; however, when the DNA was condensed in prespores, SMC foci were no longer detectable (Fig. 2). The foci disappeared after septum formation (see Fig. 4 and 6).
FIG. 2.
Immunolocalization of SMC protein in the vegetative and aerial mycelium of the wild-type strain and in the Δsmc mutant. Immunolabeled SMC (red), cell wall stained with WGA (gray), and DNA stained with DAPI (blue) in the mycelium of strain grown for 24 to 48 h on solid minimal medium. Scale bar, 5 μm. wt, wild type; h, hour.
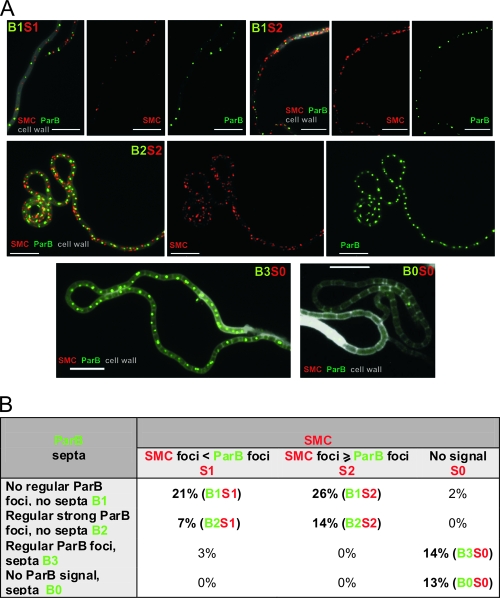
FIG. 4.
Localization of SMC and ParB proteins in the aerial hyphae. (A) Immunolocalization of SMC protein in a strain expressing ParB-EGFP fusion protein. The strain was grown for 44 h on solid minimal medium. SMC immunofluorescence is visible as red, ParB-EGFP as green, and cell walls in grey scale. The alphanumerical abbreviations indicate developmental stages as shown in panel B. Scale bar, 5 μm. (B) Developmental stages. Table shows percentages of aerial hyphae falling into specific developmental subclasses (100% = 102 apical compartments). Boldface indicates the most frequently observed stages.
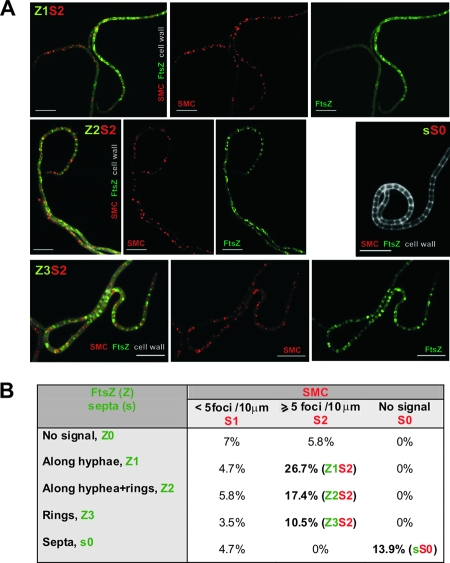
FIG. 6.
Localization of SMC and FtsZ in the aerial hyphae. (A) Immunolocalization of SMC protein in a strain expressing FtsZ-EGFP fusion protein. The strain was grown for 44 h on solid minimal medium. SMC immunofluorescence is visible as red, FtsZ-EGFP as green, and cell walls in grey scale. The alphanumerical abbreviations indicate developmental stages as shown in panel B of the figure. Scale bar, 5 μm. (B) Developmental stages. Table showing percentages of aerial hyphae falling into specific developmental subclasses. (100% = 105 apical compartments). Boldface indicates the most frequently observed stages.
Correlation of SMC localization with replication and segregation.
In rod-shaped bacteria dividing by binary fission, chromosome replication, condensation, and segregation take place simultaneously; the nascent origin regions are condensed and segregated toward the cell poles soon after replication starts (26, 36, 40). In Streptomyces, several dozen chromosomes need to be condensed and uniformly segregated along the hyphal tip, ensuring that each prespore compartment receives a single copy (21). In order to correlate SMC localization with particular cell-cycle events (replication and segregation) and stages of aerial hyphal development, SMC protein was immunolocalized in strains which allow the monitoring of replication (DnaN-EGFP) (33) and formation of partitioning complexes (ParB-EGFP) (19).
SMC and DnaN-EGFP foci formations are presumably spatially restricted.
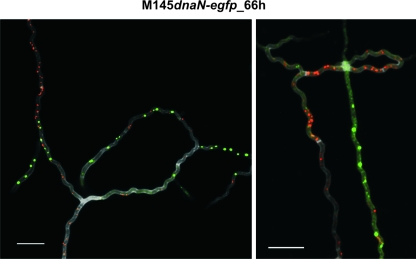
Rapid growth of aerial hyphae is accompanied by intensive replication of the chromosomes, such that 50 or more nonsegregated chromosomes may be present in one long apical compartment (33). To correlate the appearance of SMC foci with ongoing replication and aerial hyphal maturation, the SMC protein was immunostained. The examination of more than 100 apical compartments (n = 112) of the strain expressing DnaN-EGFP revealed that SMC foci did not generally colocalize with replisomes (Fig. 3). Instead, replisomes and/or SMC protein were frequently detected as separated arrays of foci representing, respectively, zones of intensive replication and/or condensation (Fig. 3). Thus, intensive replication and the formation of SMC foci appear to be mutually exclusive, presumably due to temporal separation of both processes, with the former preceding the latter.
FIG. 3.
Immunolocalization of SMC protein in the aerial hyphae of a strain expressing DnaN-EGFP fusion protein. The strain was grown for 66 h on solid minimal medium. SMC immunofluorescence is visible as red, DnaN-EGFP as green, and cell wall in grey scale. Scale bar, 5 μm. h, hour.
ParB and SMC cooperate at the stage of chromosome segregation.
In young aerial hyphae, ParB-EGFP is visible as irregularly spaced and rather weak foci. However, during subsequent steps of aerial hyphal development, ParB forms bright and regularly distributed foci (19). They appear before DNA condensation and disassemble after septum formation. To define the timing of SMC foci formation with respect to ParB complexes, a substantial number of presporulation aerial hyphae were examined (n = 102). Apical compartments containing ParB-EGFP foci were classified as described earlier (Fig. 4B) (21). It was found that SMC foci appeared before the assembly of regular ParB complexes (Fig. 4; B1S1, 21%). During later developmental stages, the number of SMC foci increased (B1S2, 26%), reaching a “maximum” level when the regular ParB complexes were formed (B2S2, 14%). Interestingly, SMC foci disappeared after septum formation, while ParB foci were still visible (Fig. 4; B3S0 14%). Taken together, SMC is observed as irregularly spaced foci which accompany but do not colocalize with ParB complexes in aerial hyphae about to undergo sporulation. SMC foci disappeared earlier than ParB complexes.
Deletion of smc influences the formation of ParB segregation complexes.
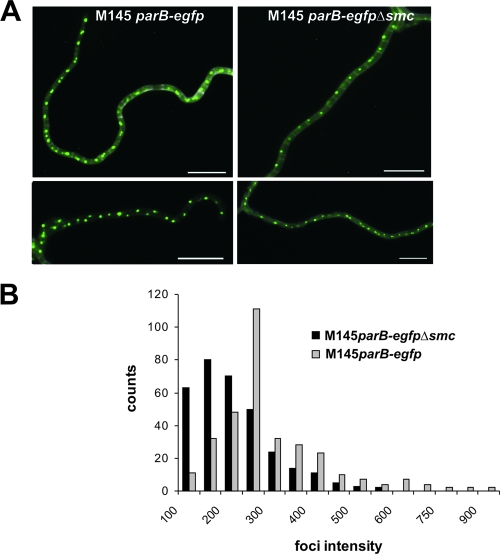
To determine whether SMC protein affects the positioning and/or formation of segregation complexes, we analyzed ParB localization in an smc deletion strain. The smc deletion was introduced into a strain which expressed ParB-EGFP protein instead of the wild-type ParB. The resulting strain, S. coelicolor parB-egfp Δsmc, was used to monitor microscopically the formation of ParB-EGFP complexes. As in the wild type, we observed arrays of regularly spaced ParB complexes, but they appeared smaller and less intensely fluorescent; the average signal intensity of the ParB foci in the Δsmc mutant was about 70% of that in the wild type (see Materials and Methods and Fig. 5). The results indicate that SMC protein may facilitate the formation of large segregation complexes.
FIG. 5.
Influence of smc deletion on ParB complex formation. (A) Examples of merged images showing ParB-EGFP (green) and cell walls stained with WGA (gray). Scale bar, 5 μm. (B) Frequency of occurrence of ParB foci with indicated signal intensity (in arbitrary units) in aerial hyphae of strain M145 parB-egfp and parB-egfp Δsmc mutants.
SMC foci appear shortly before FtsZ spiral formation.
During sporulation, the key cell division protein FtsZ forms spiral-shaped filaments along hyphae that are gradually replaced by regularly spaced rings (8). In order to correlate SMC protein localization more precisely in relation to particular stages of cytokinesis, we immunostained SMC protein in strain K202, in which, in addition to FtsZ encoded by the normal chromosomal copy, FtsZ-EGFP fusion protein is expressed from an integrated plasmid (8). Several compartments had SMC signals but no FtsZ, implying that SMC aggregation marginally preceded FtsZ aggregation (Fig. 6B; Z0S1 7.0%, and Z0S2, 5.8%). Compartments exhibiting FtsZ signals were grouped into three subclasses as described previously (21), and the abundance of SMC foci in each subclass was estimated (Fig. 6B). A substantial number of compartments contained both SMC foci and FtsZ spirals (Z1S2, 26.7%). SMC protein was also observed in compartments with some spirals and some irregular Z-rings (Z2S2, 17.4%) and in compartments where regular Z-rings were already present (Z3S2, 10.5%). The foci and the Z-rings disappeared rapidly after septum formation (sS0, 13.9%).
DISCUSSION
In eukaryotes, major events of the chromosome cycle, i.e., replication, cohesion, condensation, and segregation, are temporally separated and occur at discrete stages of the cell cycle. In contrast, replication, condensation, and chromosome segregation take place simultaneously in rod-shaped bacteria. Not much is known about the coupling of replication, condensation, and segregation in multinucleoid compartments of filamentous multicellular bacteria, such as Streptomyces. Here we studied the influence of smc, scpA, and scpB deletions on the condensation of Streptomyces chromosomes and attempted to relate this process to their replication and segregation during the differentiation of sporulating aerial hyphae.
Genes encoding the SMC, ScpA, and ScpB proteins are present in the sequenced Streptomyces species S. coelicolor, S. avermitilis, and S. scabies. As for other bacteria, the deletion of smc revealed that the product of this gene is not essential to S. coelicolor. However, smc deletion affects DNA condensation (Fig. 1) and correct chromosome segregation during sporulation (Table 3). The scpAB deletion mutant displays a phenotype similar to that of Δsmc (Table 3). In addition, we demonstrated in vitro that the ScpA protein interacts with the ScpB and SMC proteins. Thus, the ScpA, ScpB, and SMC proteins presumably act in concert in vivo. Probably, SMC/ScpA/ScpB protein complexes interact randomly with DNA, with subsequent interactions between complexes bringing about condensation.
Developmental-stage-specific assembly of irregularly spaced SMC complexes in aerial mycelium.
In Streptomyces hyphae, linear chromosomes are uncondensed during both vegetative and aerial growth and remain so until sporulation septation is completed. Using in situ immunodetection, we demonstrated that SMC foci occur sporadically in vegetative hyphae. In contrast, irregularly spaced foci are detectable over uncondensed DNA in aerial hyphae; their number increases during aerial hyphal development, reaching a maximum before septation. At this stage, the number of SMC foci was about equal to or slightly exceeded the number of ParB complexes (Fig. 4). Since each ParB complex is bound to a single chromosome (21), we can assume that statistically there is one SMC focus per nucleoid. In contrast to ParB complexes, the SMC foci are not regularly positioned. Like other SMC proteins, S. coelicolor SMC protein presumably does not exhibit any sequence specificity. Thus, we assume that SMC molecules bind randomly to a few distinct regions and, by bringing them into close proximity, form a condensation center, i.e., a condensome. The foci presumably reflect ongoing DNA condensation. Indeed, the lack of SMC protein causes inefficient condensation of chromosomes in prespores (Fig. 1).
The presence of SMC foci is presumably a direct consequence of increased SMC protein production (see Fig. S2 in the supplemental material) in aerial hyphae; smc transcription is probably developmentally controlled. This is consistent with the idea that there is a need for a greater amount of SMC protein at the time of sporulation; several dozen chromosomes have to be condensed simultaneously in order to fit into tiny prespore compartments. SMC complexes are transiently localized at the time of sporulation, when DNA has to be compacted and segregated. Thus, SMC, like FtsZ (8) and ParAB (19, 20), seems to belong to the larger group of proteins whose expression is highly induced in response to the requirement of aerial hyphal maturation. The SMC foci, like FtsZ and ParAB, disappear after septation. In B. subtilis, when the cells enter the stationary phase, SMC protein is subject to induced proteolysis (28). A future question will be to identify the agents of this presumed proteolysis during spore maturation, as well as their mechanisms of activation.
Efficient chromosome segregation requires the joint action of ParB and SMC.
In addition to the effect of smc and scpAB deletion on DNA condensation, our observations suggest a role for SMC/ScpA/ScpB complexes in accurate chromosome segregation: the elimination of any of these genes resulted in the relatively frequent formation of anucleate prespores (about 7.5%) compared with their rate of formation in the wild-type strain (1.4%). Moreover, in the Δsmc mutant, the ParB foci were smaller and less intensive, although the ParB-EGFP protein was produced at about wild-type levels (data not shown). In S. coelicolor, ParB binds to numerous parS sites scattered over a 400-kbp chromosomal segment containing oriC and assembles into a large nucleoprotein complex that is necessary for proper chromosome segregation (18, 19). Probably, condensation brought about by SMC/ScpA/ScpB protein complexes is needed for the effective formation of segregosomes, perhaps by bringing parS sequences closer together by condensing the DNA around the oriC region. The resulting nucleoprotein complexes may enhance ParB oligomerization. Condensomes and segregosomes do not colocalize, but they appear at nearly the same time, at the beginning of aerial hyphal maturation. In contrast to B. subtilis (31), deletion of the smc gene does not influence the positioning of the regularly spaced ParB complexes. Interestingly, in S. coelicolor the Z-rings and, consequently, septa are also not mispositioned in the Δsmc mutant—measurements did not reveal a statistically significant difference in spacing between Z-rings and septa compared with that in the wild type (data not shown). In S. coelicolor, ParA provides a scaffolding for the proper distribution of ParB complexes and controls the synchronized segregation of several dozen chromosomes, possibly mediating a segregation and septation checkpoint. On the basis of our current and previous results (21), we suggest that the formation of the ParA scaffolding precedes chromosome condensation and segregation events, and therefore, deletion of the smc gene in the ΔparA mutant does not significantly increase the number of anucleate prespores. In contrast to Streptomyces, the deletion of B. subtilis smc unmasked a role for Soj in chromosome segregation; a soj-null mutation dramatically enhanced the production of anucleate cells in an smc-null mutant (25). In S. coelicolor, ParA probably promotes ParB binding to parS sites, possibly by enhancing oligomerization (21), while SMC-dependent chromosome condensation may help in clustering ParB binding sites.
Sequential stages of aerial hyphal development with respect to the replication, condensation, and segregation of chromosomes.
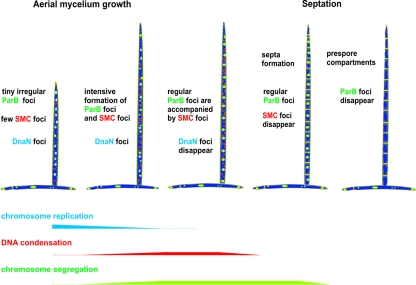
In rod-shaped bacteria, such as B. subtilis and E. coli, SMC protein (MukB in E. coli) is present within defined bipolar foci, one in each cell half, while the replication machinery is located mid-cell. Newly replicated DNA, which moves from the central part of the cell toward opposite poles, is probably condensed within SMC centers. In contrast, in vegetative as well as in young and short aerial hyphae of Streptomyces, chromosome replication is not accompanied by condensation: the chromosomes remain uncondensed until the aerial hyphae are converted into prespores. The rapid growth of the aerial hyphae requires intensive chromosome replication, and our results suggest that at this stage of growth there is no chromosome condensation machinery (we could observe separate zones of intensive replication and condensomes). A decrease in replication intensity and an increase in the formation of condensomes was accompanied by the formation of segregation complexes and could be observed before Z-ring and septum formation (Fig. 6). Thus, in Streptomyces, all three processes, chromosome replication, condensation, and segregation, only partially overlap during aerial hyphal maturation, and they all precede Z-ring formation (Fig. 7).
FIG. 7.
Sequential stages of aerial hypha development with respect to replication, condensation, and segregation of chromosomes.
Addendum.
In related work, Dedrick et al. (5a) have obtained similar results for the segregation phenotype of an smc mutant and SMC localization.
Supplementary Material
Acknowledgments
We thank Rebekah M. Dedrick and Joseph R. McCormick for communicating their results to us prior to publication and for S. coelicolor strain RMD2 expressing SMC-EGFP. We are grateful to Keith Chater for his critical reading of the manuscript.
This work was supported by the Ministry of Science and Higher Education (grant no. N301 2035 33). A.K. and J.C.-Z. gratefully acknowledge financial support received from the Foundation for Polish Science (Start and MISTRZ Programmes).
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 2.Britton, R. A., D. C. Lin, and A. D. Grossman. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 121254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47685-713. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4667-673. [DOI] [PubMed] [Google Scholar]
- 5.Chodaczek, G., M. Zimecki, J. Lukasiewicz, and C. Lugowski. 2006. A complex of lactoferrin with monophosphoryl lipid A is an efficient adjuvant of the humoral and cellular immune response in mice. Med. Microbiol. Immunol. 195207-216. [DOI] [PubMed] [Google Scholar]
- 5a.Dedrick, R. M., H. Wildschutte, and J. R. McCormick. 2009. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J. Bacteriol. 191320-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fennell-Fezzie, R., S. D. Gradia, D. Akey, and J. M. Berger. 2005. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. EMBO J. 241921-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flardh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6564-571. [DOI] [PubMed] [Google Scholar]
- 8.Grantcharova, N., U. Lustig, and K. Flardh. 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 1873227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graumann, P. L. 2001. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie 8353-59. [DOI] [PubMed] [Google Scholar]
- 10.Graumann, P. L. 2000. Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions. J. Bacteriol. 1826463-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gust, B., C. Bruton, T. Yuqing, D. Jakimowicz, G. Chandra, and K. F. Chater. 2004. λ red mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54107-128. [DOI] [PubMed] [Google Scholar]
- 12.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 1001541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9773-788. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press. New York, NY.
- 15.Hirano, M., and T. Hirano. 2006. Opening closed arms: long distance activation of SMC ATPase by hinge-DNA interaction. Mol. Cell 21175-186. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16399-414. [DOI] [PubMed] [Google Scholar]
- 17.Hirano, T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7311-322. [DOI] [PubMed] [Google Scholar]
- 18.Jakimowicz, D., K. F. Chater, and J. Zakrzewska-Czerwińska. 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 451365-1377. [DOI] [PubMed] [Google Scholar]
- 19.Jakimowicz, D., B. Gust, J. Zakrzewska-Czerwińska, and K. F. Chater. 2005. Developmental stage-specific assembly of the partitioning protein ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 1873572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakimowicz, D., S. Mouz, J. Zakrzewska-Czerwińska, and K. F. Chater. 2006. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 1881710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakimowicz, D., P. Żydek, A. Kois, J. Zakrzewska-Czerwińska, and K. F. Chater. 2007. Helical ParA scaffolding for ParB complexes in S. coelicolor aerial hyphae. Mol. Microbiol. 65625-641. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. USA 9610661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 24.Laemmli, U. K. 1970. Claevage of structural proteins during assembly of the head of the bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lee, P. S., and A. D. Grossman. 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60853-869. [DOI] [PubMed] [Google Scholar]
- 26.Leonard, T. A., J. Moller-Jensen, and J. Lowe. 2005. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majka, J., D. Jakimowicz, W. Messer, H. Schrempf, M. Lisowski, and J. Zakrzewska-Czerwińska. 1999. Interactions of the Streptomyces lividans initiator protein DnaA with its target. Eur. J. Biochem. 260325-335. [DOI] [PubMed] [Google Scholar]
- 28.Mascarenhas, J., A. V. Volkov, C. Rinn, J. Schiener, R. Guckenberger, and P. Graumann. 2005. Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biol. 296-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascarenhas, J., J. Soppa, A. V. Strunnikov, and P. L. Graumann. 2002. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 213108-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melby, T., E. Ciampaglio, G. Briscoe, and H. P. Erickson. 1998. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 1421595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriya, S., E. Tsujikawa, A. K. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29179-187. [DOI] [PubMed] [Google Scholar]
- 32.Niki, H., A. Jaffé, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruban-Ośmiałowska, B., D. Jakimowicz, A. Smulczyk-Krawczyszyn, K. F. Chater, and J. Zakrzewska-Czerwińska. 2006. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 1887311-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J. F., E. F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor LaboratoryPress., New York, NY.
- 35.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25847-858. [DOI] [PubMed] [Google Scholar]
- 36.Sherratt, D. J. 2003. Bacterial chromosome dynamics. Science 301780-785. [DOI] [PubMed] [Google Scholar]
- 37.Soppa, J., K. Kobayashi, M. F. Noirot-Gros, D. Oesterhelt, S. D. Ehrlich, E. Dervyn, N. Ogasawara, and S. Moriya. 2002. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 4559-71. [DOI] [PubMed] [Google Scholar]
- 38.Strunnikov, A. V. 2006. SMC complexes in bacterial chromosome condensation and segregation. Plasmid. 55135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun, J., G. H. Kelemen, J. M. Fernández-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology. 1452221-2227. [DOI] [PubMed] [Google Scholar]
- 40.Thanbichler, M., and L. Shapiro. 2006. Chromosome organization and segregation in bacteria. J. Struct. Biol. 156292-303. [DOI] [PubMed] [Google Scholar]
- 41.Volkov, A., J. Mascarenhas, C. Andrei-Selmer, H. D. Ulrich, and P. L. Graumann. 2003. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol. Cell. Biol. 235638-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, L. J. 2004. Structure and segregation of the bacterial nucleoid. Curr. Opin. Gen. Dev. 1426-132. [DOI] [PubMed] [Google Scholar]
- 43.Zawilak, A., A. Kois, G. Konopa, A. Smulczyk-Krawczyszyn, and J. Zakrzewska-Czerwinska. 2004. Mycobacterium tuberculosis DnaA initiator protein: purification and DNA binding requirements. Biochem. J. 382247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.