Abstract
We present a Monte Carlo sidechain sampling procedure and apply it to assessing the flexibility of protein binding pockets. We implemented a multiple “time step” Monte Carlo algorithm to optimize sidechain sampling with a surface generalized Born implicit solvent model. In this approach, certain forces (those due to long-range electrostatics and the implicit solvent model) are updated infrequently, in “outer steps”, while short-range forces (covalent, local nonbonded interactions) are updated at every “inner step”. Two multistep protocols were studied. The first protocol rigorously obeys detailed balance, and the second protocol introduces an approximation to the solvation term that increases the acceptance ratio. The first protocol gives a 10-fold improvement over a protocol that does not use multiple time steps, while the second protocol generates comparable ensembles and gives a 15-fold improvement. A range of 50–200 inner steps per outer step was found to give optimal performance for both protocols. The resultant method is a practical means to assess sidechain flexibility in ligand binding pockets, as we illustrate with proof-of-principle calculations on six proteins: DB3 antibody, thermolysin, estrogen receptor, PPAR-γ, PI3 kinase, and CDK2. The resulting sidechain ensembles of the apo binding sites correlate well with known induced fit conformational changes and provide insights into binding pocket flexibility.
Introduction
Sidechain sampling and optimization algorithms, mostly based on a rotamer approximation,1–5 have been used extensively in modeling proteins, including homology modeling,6,7 and predicting conformational changes due to ligand binding.8–10 We have been interested in developing sampling methods for protein sidechains (and, in other work, loops) that generate thermodynamic ensembles of conformations, in contrast to locating the global energy minimum.11,12 Minimization methods implicitly neglect the effect of entropy on sidechain conformations, and generally cannot distinguish whether sidechains will adopt a single well-defined conformation, or a distribution of conformations. For the many sidechains that are tightly packed in the core of a protein, minimization is an effective approach. For less tightly packed sidechains that display some degree of flexibility, a thermodynamic ensemble becomes a more appropriate description.
Sidechain conformational heterogeneity is important to protein–ligand binding. The ability to accurately predict the flexibility/rigidity of binding site residues would be useful in structure-based drug design.10,13 For example, a recent paper by Sherman et al.8 describes a computational method to predict “induced fit” effects upon ligand binding which relies on some advanced knowledge of which sidechains may adopt different conformations upon ligand binding, e.g., from multiple cocrystal structures. We demonstrate here that thermodynamic ensembles of sidechain conformations in apo proteins correlate well with known induced fit conformational changes in various well studied drug targets.
In principle, molecular dynamics sampling methods10,14 can be used to obtain thermodynamic ensembles for protein binding sites. The main disadvantage is that the timescales required to observe large changes in sidechain conformations can be long relative to the ∼1 fs timesteps employed in atomically detailed molecular dynamics simulations; transitions between sidechain rotamers can take up to μs, which is a known difficulty in binding affinity calculations.14–16 Monte Carlo sampling17 can lead to more efficient generation of the complete thermodynamic ensemble, if the trial moves are constructed carefully.
For macromolecules, which contain complex, heterogeneous, and densely packed atomic configurations, construction of efficient trial moves can be a substantial challenge. A variety of both rigorous and nearly rigorous methods have been used12,18–23 to address this challenge. One common idea among these involves decomposing the degrees of freedom into subspaces that are more manageable, both computationally and conceptually. The most natural decomposition for proteins is between backbone and sidechain degrees of freedom. Future work will incorporate backbone motions, but the current emphasis is on the sidechain sampling.
Another common decomposition is between solvent (water) and solute (protein) degrees of freedom. Here we use an implicit solvent model, which makes it possible to efficiently sample large sidechain conformational changes. By contrast, in explicit solvent, large changes (e.g., across rotamers) are difficult to sample with good acceptance rates because of steric clashes between waters and the sidechain, and the need for the solvent to relax around any new trial conformation. The same steric issues have motivated the use of implicit solvent in molecular dynamics studies as well.24–26 For this work, the electrostatic solvation term is evaluated with the SGB model27,28 and the nonpolar solvation energy with the nonpolar (NP) model.29 The solvation model here was developed for use with the all atom OPLS-AA 2001 forcefield30 and is implemented in the Protein Local Optimization Program.31,32 While this model is chosen as a compromise between efficiency and accuracy, it remains the most computationally expensive portion of the energy evaluations. The current effort is to develop a general sampling scheme which allows optimal use of an implicit solvation model in the context of a Monte Carlo scheme. The present application is to sidechain sampling, but can be extended to backbone sampling strategies in a straightforward manner.
The major innovation here in terms of computational methods is the implementation of a multiscale strategy, analogous to methods such as RESPA,33,34 used in molecular dynamics, to accelerate convergence toward the thermodynamic ensemble. In this approach, certain forces (primarily those due to long-range electrostatics and the implicit solvent model) are updated infrequently, in “outer steps”, while short-range forces (covalent, local nonbonded interactions) are updated at every “inner step”. The theory underlying this approach has been presented previously,35 and is only briefly reviewed here. The application of a multiscale Monte Carlo approach to sampling proteins in implicit solvent has been presented by Michel et al.,36 with different implementation details and approximations introduced. Other algorithmic details crucial for speed, including the rapid elimination of conformations with steric clashes, are also described. The resultant method is a practical means to assess sidechain flexibility in ligand binding pockets, as we illustrate with proof-of-principle calculations on six proteins.
Theory and Methods
Configuration Integral
The implicitly solvated37 macromolecular ensembles of interest can be represented by the following configuration integral:
| (1) |
where R is the set of all Cartesian coordinates of the macromolecule of interest, and
| (2) |
where A(R) is the sum of the forcefield energy, U(R), and the implicit solvation energy, G(R). The solvation energy is dependent on the Born radii, which are a function of the coordinate state of the macromolecule. In the SGB implementation we use, the Born radii α(R) are computed using surface integrals, and thus are dependent on the global coordinate state R of the protein. This calculation can take much longer (roughly 100 times longer in cases studied) than the pairwise energy terms. Some improvements have been gained by updating only local regions of the surface area as needed, and efforts are ongoing in this area to improve the efficiency and accuracy of this model.38,39
In general, however, any attempt to optimize sampling would benefit most from evaluating the solvation energy less frequently. While this approach is motivated by computational efficiency, a physical argument can also be made. The Born radii generally vary slowly for relatively small, local conformational changes. The sampling strategies presented are intended to make the best use of these ideas while still generating meaningful ensembles.
Constraints on various degrees of freedom can be introduced to generate a configuration integral q0 over a smaller subspace by identifying fixed (F) and sampled (S) degrees of freedom, such that dR = dR<F> dR<S>, and imposing a rigid constraint on the fixed degrees of freedom, yielding
| (3) |
Following the formulation of Deem,20 the transformation from Cartesian to torsional coordinates can be made with a Jacobian of unity, if bond lengths and angles are preserved. For the current work, the backbone torsions will be constrained to an initial value of φ0, and the fixed sidechains, to an initial value of χ0<F>. The resulting integral can be recast as
| (4) |
where χ<S> is the set of sidechain torsional coordinates that are sampled. The integral of interest over the subspace can be recast by letting dr = dR<S> and A(r) = A(χ<F>) = A(R<S>|R0<F>), yielding the more compact expression:
| (5) |
Generation of Trial Configurations
To generate a reversible trial move, a single sidechain i is chosen at random from the list of sampled sidechains and the updated set of torsions is assigned according to
| (6) |
where χi′ and χi are the trial and previous set of dihedral coordinates, respectively, for sidechain i, and ξ is a vector of uniform random variates of the same dimension, for which each value is drawn from the domain [−d/2, d/2]. To account for local fluctuations as well as larger fluctuations, the domain size d is assigned a value of either 360° or 18° with equal probability. The idea behind the heterogeneous move set is to alternate between large dihedral trial moves that cross local χ wells, and small trial moves, which sample the local χ basin. For the present work, selections from a rotamer library are not incorporated as a trial move, as slight nonuniformities in the distribution of the χ angles of the rotamer library have a quantitative effect on the distributions. As a practical matter, however, a mixture of rotamer and random moves could conceivably be implemented if quantitative energy distributions are not required.
For residues with rotatable polar hydrogen groups (Cys, Ser, Thr, Tyr), the torsional angle that places the hydrogen is also selected randomly when the rotamer state is assigned. Also, the torsions of the amine hydrogens of lysines are sampled. Torsions for methyl hydrogens are not currently sampled.
A hard sphere approximation is invoked, which vastly improves sampling efficiency, while preserving much of the essential physics of the system. This has been shown in liquid systems40,41 as well as proteins. For the current work, pairs of atoms that are closer than 0.7 times the sum of the Lennard-Jones radii are considered to be sterically disallowed. That is, no energy is computed for sterically disallowed states, because the steric clash will result in high energies and small acceptance probabilities. Cell lists (linked lists) further accelerate the identification of steric clashes, by only checking for clashes between atoms known to be proximal. A series of dihedral perturbations is generated as described until a configuration that is sterically allowed is generated. The resulting configuration is treated as a trial move. For the systems studied, the average number of sterically disallowed moves ranges from 0.5 to 0.75 (see Table 2), which is roughly a 2–4-fold improvement in sampling efficiency, because the CPU time per steric clash evaluation is negligible relative to the energy evaluation.
Table 2.
Binding Pockets Studieda
| label | protein | RB | RA | LA | no. residues | <NC> |
|---|---|---|---|---|---|---|
| A | db3 Antibody | 1dba | 1dbb | progesterone | 30 | 0.54 |
| B | thermolysin | 1kr6 | 1kjo | Z-D glutamic acid | 41 | 0.74 |
| C | estrogen receptor | 1err | 3ert | raloxifene | 73 | 0.65 |
| D | PPAR-γ | 1fm9 | 2prg | GI262570 | 65 | 0.75 |
| E | PI3 kinase | 2chx | 2chw | PIK-039 | 45 | 0.65 |
| F | CDK2 | 1buh | 1dm2 | hymenialdisine | 46 | 0.73 |
RB is the receptor used in the simulation (without ligand), and RA is a reference receptor with LA bound to it. <NC> is defined as the total number of steric clashes divided by the number of sterically allowed steps.
Multiple Time Step Monte Carlo (MTS-MC)
A sampling procedure known as multiple time step Monte Carlo,35 which was originally developed for Ewald sum calculations,42 can be used to optimally sample against a potential that can be decomposed into additive components. These components are typically, but not necessarily, short- and long-range contributions to the energy. The algorithm relies on the assumption that the short-range term varies rapidly with respect to the move set, while the long-range term varies more slowly. A related formalism is presented using approximate potentials.43 Many algorithms use similar ideas, including both molecular dynamics integrators33,34 and minimization algorithms.44 Some applications using algorithms that are similar in spirit involve evaluating Ewald sums less frequently in fluid simulations with periodic boundary conditions, sampling of polar fluids,45 and polarizable water sampling.46
While the formalisms in these approaches vary, they can all be thought of as relying on some decomposition of the overall potential to be sampled. The natural choice of decomposition, in general, is into short- and long-range terms, which we denote by subscripts S and L, respectively
| (7) |
The details of the nature of the decomposition of interactions into long and short-range can vary from system to system. A more detailed description of the decomposition for the present case, with proof of detailed balance, is given in the Appendix.
Using the above decomposition, detailed balance can be maintained using the following sampling protocol:
-
Starting with the configuration ri, generate a number NI of inner loop steps, where each step consists of a trial configuration rk′ that is generated reversibly (such as the trial configurations described by eq 6) and accepted according to the following short-range acceptance criterion:
(8) -
Take the final configuration from the inner loop to be the trial configuration rj for the outer loop and apply the long-range acceptance criterion:
(9)
It is important to note that any statistical quantities of interest can only be computed using the outer loop configurations. In all cases where the ratio of acceptance probabilities are given, the Metropolis acceptance criterion is used in practice.
Recasting MTS-MC to Account for Infrequent Born Radii Updates
For the present case, the most costly term to evaluate in the energy is the solvation term, which is due largely to the time intensive step of computing the Born radii, α(R), and we develop a strategy such that the Born radii are not updated in the inner steps. To motivate this method, it is helpful to express the potential in the following form:
| (10) |
where rn is nth configuration of the subset of sampled coordinates, α(Rm) is the set of Born radii which are evaluated based on the coordinates of the mth coordinate state Rm of the entire protein, U(rn), and G(α(Rm), rn) is the solvation energy evaluated at the given states. We can further express the energy deviation from the “true” potential, where the Born radii are synchronous with the current coordinate state, in terms of an error potential ε(α(Rm), rn):
| (11) |
Thus, the inner loop configurations are evaluated according to an approximate short-range potential AS(α(Rm), rn), where the Born radii are held at a previous or “latent” state. The relation to the true short-range potential can similarly be written in terms of a short-range error potential εS(α(Rm),rn):
| (12) |
where the coordinate state is rn, and the latent Born radii, α(Rm) are calculated from a previous step. Likewise, the true long-range potential can be described in terms of long-range error potential:
| (13) |
For simplicity, these energies can be expressed in terms of the state indices only:
| (14) |
Where, n is the index of the current coordinate state and m is the index of the Born radii held at a previous state. We can simply recast the decomposition as
| (15) |
where the index of the coordinate state is first argument in each of the functions, and the index of the Born radii state is the second argument. While the error potential described in eq 14 contains both long and short-range terms, the idea of the sampling protocols is to treat the all of error potential terms as long-range terms. Using this new decomposition, we can define two different sampling protocols:
-
In both protocols, start with the configuration Ri, generate a number NI of inner loop steps, where each trial configuration rk is generated using eq 6. The Born radii are held at a latent state i, such that the short-range acceptance criterion is the following:
(16) -
Take the final configuration from the inner loop to be the trial configuration rj for the outer loop and apply either of two acceptance criteria:
-
With error correction
(17) -
Without error correction
(18)
-
Protocol A rigorously obeys detailed balance, while protocol B is an approximation introduced to improve computational efficiency. It should be noted that the Born radii are completely updated in every outer loop calculation, regardless of protocol. The ideal error potential term would be narrowly distributed about a mean of zero, so that the distribution generated by neglecting the term would be nearly equivalent to the true distribution. The effect of the modification will be discussed in detail in the results section.
As a control, a “standard” Monte Carlo trajectory, or protocol S, was also studied. For the standard Monte Carlo protocol, the same trial move set was used, including steric screening, but with the Born radii updated at every step, with no decomposition of potentials. For every step, the acceptance criterion is simply:
| (19) |
Estimation of the Time to Convergence and Improvement Ration
To estimate the optimal number of inner steps, we express the total processor time T to compute a trajectory as
| (20) |
where <dt/dNO> is the expectation value of the time required to generate an outer step. This is not a fixed value, since the innermost sampling loop samples an arbitrary number of configurations until a sterically allowed configuration is obtained. NO,T is the total number of outer steps, which includes the both the nonequilibrated steps, nO, and equilibrated steps, NO. This can also be expressed as
| (21) |
where ts is the average time required to generate a single (sterically allowed) trial coordinate and evaluate the short-range potential. The rate tL is the time required to evaluate the long-range potential, which includes the long-range energies and the time required to update the Born radii. This quantity does not need to be averaged, since there is no dependence on the number of steric clashes. NI is the number of inner steps that are set for the simulation. Since statistics can only be gathered on the equilibrated outer steps, we can express NO in terms of the standard error:
| (22) |
where σ is the variance of the energy over the entire equilibrated portion of the trajectory, ε is the desired error in the estimate of the energy, and g(NI) is the correlation interval, or distance between uncorrelated snapshots. This quantity is measured from the simulation, and will vary with the number of inner steps for a given system with all other conditions held constant. It is closely related to other measures of quality of Monte Carlo trajectories, such as acceptance ratio, and a low correlation interval often corresponds to a high acceptance ratio.
Since the number of steps required to equilibrate depends strongly on the initial condition, we shall overestimate this quantity by assuming that nO = NO. This varies in practice from a few correlation intervals to less than half of the number of outer steps. As long as the equilibration time is proportional to the number of equilibrated steps, it will cancel out in the improvement ratio calculation. Using this assumption, the estimated CPU time required for a converged trajectory is
| (23) |
where the number of inner steps can be adjusted to locate the optimal computing time. As a measure of sampling efficiency, the following quantity can be expressed:
| (24) |
where I is the improvement, and Ts is the time required for a converged trajectory in a standard Monte Carlo protocol.
Convergence Determination and Error Estimation
Determination of the number of steps required for equilibration and the correlation interval was performed iteratively. Initially, the number of steps required for equilibration was estimated very approximately as 3000 for the standard trajectory, 1000 for NI = 1, 50, 100, 200, 300, and 400 for the remaining inner step settings. To estimate the correlation time, an autocorrelation function of the energy was computed, and the correlation interval g was identified as the first place that the autocorrelation function crosses zero. This initial estimate is expected to overestimate the true correlation time since the trajectory may include nonequilibrated regions, which contain slow fluctuations toward the equilibrium state that would not be present in the stationary distribution. Using this initial estimate, a blocksize was assigned to have a value of g. A block standard deviation σB is computed at each point (using the points preceding the point of interest), and the trajectory was deemed to be converged if the block standard deviation was less than a nominal value σB = 15kBT.
With this new estimate of the equilibrated region of the trajectory, another estimate of the correlation time was applied. To improve the estimate, the autocorrelation function was fit to a simple exponential exp(−τ/τD) where τD is the decay constant, or correlation time. For this procedure, a least-squares fit was performed where the sum of the squares of the errors between the function and the data points are weighted according to the inverse of error at that point. The error in the autocorrelation function is given by45
| (25) |
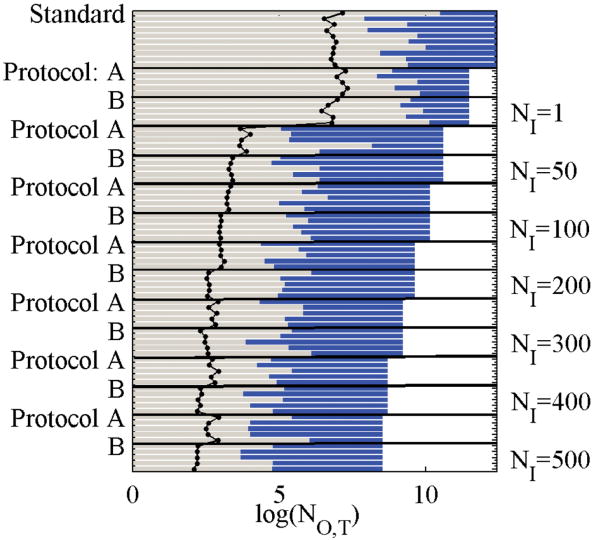
where g = 1 + 2τD is the correlation interval, or the number of steps between uncorrelated snapshots. Once a correlation time is obtained, the reverse cumulative averaging (RCA) method was used to obtain a better estimate of the location of the equilibrated region,47 with the blocksize set to g. A confidence level of 85% was used to reject the hypothesis that the block averaged samples came from a normal distribution, according to the Shapiro–Wilk Test.48,49 The location of the equilibrated portion of the trajectory depends heavily on the value of the blocksize, and vice versa, so 30 iterations of the blocksize and RCA convergence calculation were run. See Figure 1 for the convergence times, correlation intervals, and total simulation lengths for each simulation.
Figure 1.
Summary statistics for validation data set. Bars represent the log of simulation lengths, and black dots connected with lines represent the correlation interval for that simulation. All simulations are run at 600 K. The blue portion of each bar is the unequilibrated portion, and the green portion is equilibrated. Different values are given for different runs, which are trajectories using the same settings, including initial condition, but assigned different random seeds. The natural log of the number of total steps, NO,T, appears on the x-axis.
Preparation of Unbound receptors
The proteins studied are listed in Table 2. A few of the proteins had missing sidechains or loops, outside of the binding sites (> 15Å) being studied. These were reconstructed in arbitrary configurations free of steric clashes using standard routines in the protein local optimization program. The sidechains to be sampled in the Monte Carlo were defined as those within 8 Å of any atom of the ligand in the holo structure. All calculations were performed in the absence of the ligand.
Composite Energy Histograms
In order to represent multiple simulations of the same sampling protocol as a single histogram, a superposition of individual energy histograms was computed. This is done to obtain better statistics so that detailed balance may be demonstrated for protocol A.
For each trajectory histogram, an error εB = √(gnB) was assigned at each bin point, where nB is the number of entries in each bin. To generate the composite histograms for protocols A and B, each of the trajectory histograms for each protocol were superimposed with a weight proportional to the number of uncorrelated entries in each bin of each trajectory. The errors are computed a superposition of square of the errors of each trajectory, with the same weights used to compute the composite histograms. It should be noted that the sampling protocols produce the same distribution of energies, independent of number of inner steps chosen. The data from all ranges of inner steps can therefore be combined to form a single histogram. Since the error is computed using the autocorrelation times, the fact that the distributions fall within error suggest also that the correlation times are correctly estimated.
Timings
Since simulations were run on a variety of machines, smaller trajectories were collected to estimate the average time per outer step (see Table 1). Timings of the simulations were measured on a Linux machine, using a single CPU from a dual AMD Opteron CPU running at 2.2 GHz.
Table 1.
Simulation Data for Model Systema
| NI | NO,T | <dt/dNO,T> (s) | <E> – <E>STD (RT) | σ | ε | NO (all) |
|---|---|---|---|---|---|---|
| S | 250000 | 6.02 | 0.00 | 5.65 | 0.37 | 2366928 |
| A-1 | 95000 | 6.29 | −0.19 | 5.88 | 0.76 | 421025 |
| A-50 | 40000 | 12.02 | −0.01 | 5.83 | 0.21 | 195235 |
| A-100 | 25000 | 16.37 | 0.13 | 5.83 | 0.19 | 122849 |
| A-200 | 15000 | 26.08 | 0.17 | 5.86 | 0.22 | 74035 |
| A-300 | 10000 | 34.37 | 0.25 | 5.82 | 0.24 | 48860 |
| A-400 | 6000 | 43.44 | 0.12 | 5.83 | 0.31 | 29354 |
| A-500 | 5000 | 51.48 | 0.13 | 5.91 | 0.37 | 24195 |
| B-1 | 95000 | 6.20 | 1.77 | 5.96 | 0.28 | 393587 |
| B-50 | 40000 | 11.11 | 1.58 | 6.07 | 0.07 | 198311 |
| B-100 | 25000 | 16.01 | 1.79 | 6.12 | 0.08 | 123416 |
| B-200 | 15000 | 24.72 | 1.89 | 6.12 | 0.08 | 73904 |
| B-300 | 10000 | 32.86 | 1.71 | 6.19 | 0.10 | 48913 |
| B-400 | 6000 | 41.40 | 1.69 | 6.09 | 0.11 | 29440 |
| B-500 | 5000 | 50.47 | 1.83 | 6.18 | 0.12 | 24568 |
Data shown summarizes the results for 10 simulations of each protocol and inner step setting. For the leftmost column, NI is the number of inner steps. S indicates a standard protocol (no inner steps). For the remaining columns, protocol and number of inner steps are given. (A-50 represents protocol A using 50 inner steps). NO,T is the total number of steps simulated, including nonequilibrated portions of the trajectory. dt/dNO,T is the average time to generate an outer step, as described in the text. <E> – <E>STD (RT) is the average equilibrium energy minus the standard measurement, σ and ε are the standard deviation and standard error of the equilibrated energies. The rightmost column is the total number of equilibrated steps (across all simulations at the designated setting) used for the calculation.
Results and Discussion
Comparison of Protocols Using Antibody DB3
To optimize the number of inner steps and other parameters of the algorithm, the binding pocket of apo antibody DB3 (1dba)50,51 was selected as a model system. A total of three sampling protocols were explored, as defined in Theory and Methods. To compare the effect of neglecting the short-range error in the Born updates, identical simulations were run using protocols A (rigorous) and B (approximate). A single set of 10 trajectories using protocol S was also generated. The number of inner steps (NI) was set to 1, 50, 100, 200, 300, 400, and 500. For each inner step setting, five trajectories were collected, starting from the same (nonequilibrium) initial condition with different random seeds. Since the backbone is held fixed, room temperature simulations tend to exhibit frustrated dynamics. To obtain better statistics, especially for protocol S, all simulations were run at 600 K. The goals of these simulations are twofold: (1) to generate sufficient statistics to demonstrate detailed balance and (2) to study the effect of adjusting the number of inner steps and protocol. A total of 80 separate trajectories were collected for the analysis. Figure 1 summarizes the pertinent information on these trajectories.
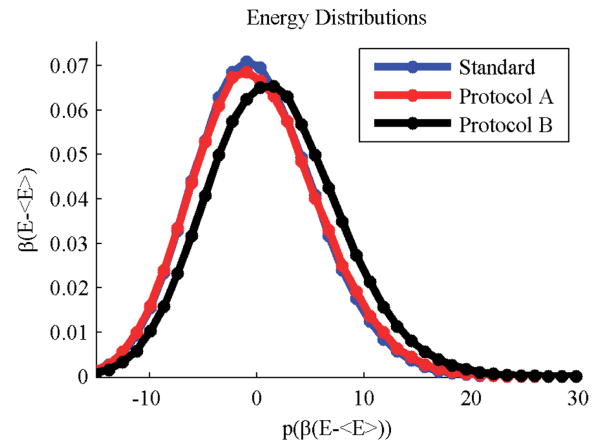
The average energies and standard errors of each simulation are in Table 2, and Figure 2 shows histograms of equilibrated energies for each sampling protocol. The energy distributions of protocols A and S (standard) appear to be equivalent. While error bars are not shown for clarity, the histograms superimpose to well within the estimated error. The energy distribution of protocol B is offset by roughly 1.75 RT, and is clearly from a different distribution than protocol A. The standard deviation of protocol B is larger by roughly 0.3 RT. The broader distribution and higher mean value is due to the more permissive approximation, which increases the number of states that are accepted.
Figure 2.
Protocol A distributions superimpose with standard energy histograms, and protocol B generates a similar approximate distribution. All simulations were run at 600 K, under the conditions summarized in Figure 1. Dimensionless energy is plotted on the x-axis, with the mean of the energies of the standard simulation <E> subtracted from the energy (see Table 1). On the y-axis is the probability of observing that energy.
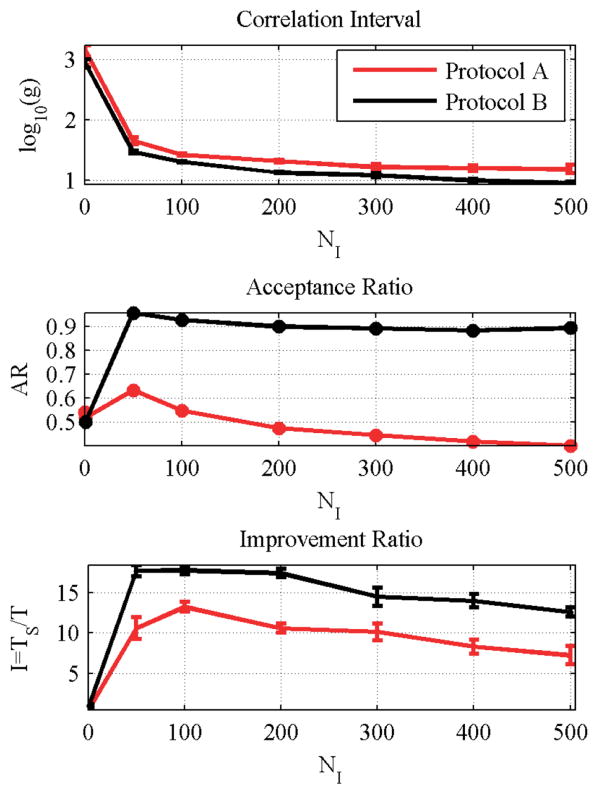
The correlation interval is shown in Figure 3. A sharp decrease is observed from NI = 50–200, which steadily decreases over the remaining inner step settings. The acceptance ratio shows an initially sharp increase, since a smaller number of inner steps helps to generate better trial moves for the outer loop. As the number of inner steps increase however, the inner loop becomes less efficient at generating trial configurations. This effect is more prominent in protocol A, which is the rigorous approach. Figure 3c shows the relative improvement over protocol S (no inner steps). Optimal values are in the range NI = 50–200. For both protocols A and B, a broad optimal range is observed, which suggests that this optimal range should hold for a wide variety of proteins.
Figure 3.
Approximate protocol provides slightly better performance, and optimal performance of both protocols is in the range of NI = 50–200. (a) Logarithm of correlation interval. (b) Acceptance ratio. (c) Improvement ratio, as given by eq 24.
Binding Pocket Studies
As a first application, we investigate the flexibility of sidechains in protein binding pockets. As a test set, we consider several proteins from Sherman et al.,8 as well as PI3K.52 The assumption of this work is that sidechains that show more flexibility in our ensembles will be capable of undergoing rearrangements upon binding ligands. Table 2 lists the binding pockets studied. For all trajectory data which is displayed, individual sidechains conformations were filtered such that no two conformations are less than an rmsd of 0.05 Å from one another.
Protocols A and B were used to generate sidechain ensembles, at a variety of temperatures. Temperatures >300 K were explored for three primary reasons. First, our goal is to predict conformational changes that could occur upon binding a ligand. In the limit of pure “conformational selection”, the bound conformation of the protein would be populated significantly, or at least measurably, at ambient temperature. However, there can also be some additional conformational rearrangement of the 1protein to accommodate the ligand (“induced fit”), derived from the free energy of ligand binding. Here, we have essentially postulated that ligand binding can “induce” conformational changes that may not be observable with a room temperature thermal ensemble. It has been observed that sidechain rearrangements within binding pockets can be cost up to 4 kcal/mol of free energy.15,16
Another reason for considering higher temperature distributions of 600 K is related to limitations of the energy function. In particular, it has been widely reported that generalized Born solvent models can overstabilize hydrogen bonds and salt bridge interactions.39,53 This known limitation of the implicit solvent model will tend to result in reduced flexibility of charged residues at ambient temperatures.
Finally, the use of a rigid backbone will also reduce sidechain flexibility. The test cases were chosen in part because ligand binding does not induce large changes in backbone conformation; clearly, further algorithmic development, which will be reported in due course, is needed to deal with backbone fluctuations. When there is reason to believe that backbone changes are likely to be small, simply using a higher temperature may help to reduce artifacts due to the rigid backbone.
Ultimately, from the standpoint of identifying “flexible” sidechains in a binding site, we view the choice of temperature as a user-definable parameter; in practice, performing simulations with multiple values of the temperature may be advisable. Note that, since the backbone is held fixed, the protein will not denature during the simulation, which provides considerable freedom in the choice of temperature and simulation protocol.
Antibody DB3.50,51
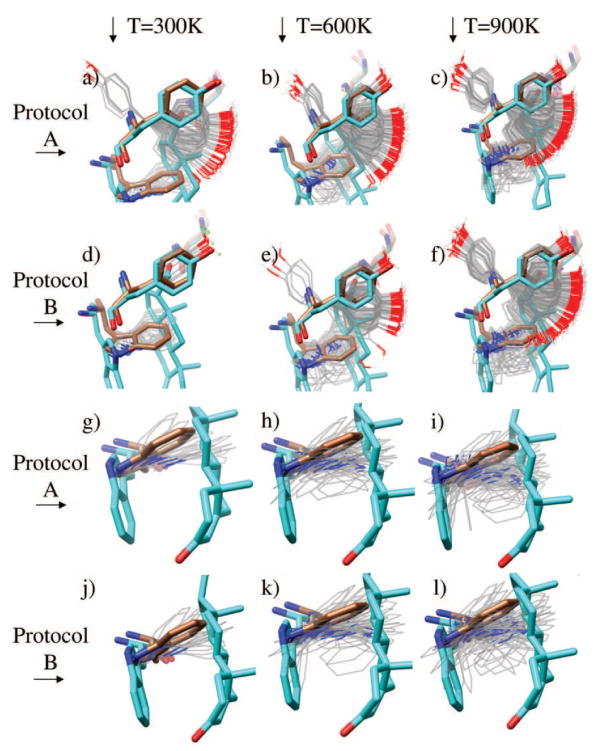
For the DB3 antibody (Figures 4 and 5a), the primary conformational change between the two structures is the large movement of the Trp100 sidechain to accommodate 4-hydroxytamoxifen. We studied this system with both protocols A and B at T = 300, 600, and 900 K, with NI = 200 (the upper end of the optimal range). It is encouraging to observe that the large conformational change in Trp100 is observed in the Monte Carlo simulations, performed without a ligand present, at 600 K using protocol B and at 900 K using protocol A. Two conformational states of Trp100 are observed: a low-population state where the sidechain is in a similar conformation as the holo structure and a high-population state where it is similar to the apo structure, although significant fluctuation is observed. Intermediate conformations are not observed suggesting a high energy barrier for the rotation.
Figure 4.
Distribution of sidechain configurations for Tyr97 and Trp100 of 1dba. Brown configurations are from the native structure, and cyan configurations are from the holo structure. Grey sidechains are distinct configurations from a sidechain trajectory at the given conditions. (a) Tyr97 at 300 K, protocol A. (b) Tyr97 at 600 K, protocol A. (c) Tyr97 at 900 K, protocol A. (d) Tyr97 at 300 K, protocol B. (e) Tyr97 at 600 K, protocol B. (f) Tyr97 at 900 K, protocol B. (g) Trp100 at 300 K, protocol A. (h) Trp100 at 600 K, protocol A. (i) Trp100 at 900 K, protocol A. (j) Trp100 at 300 K, protocol B. (k) Trp100 at 600 K, protocol B. (l) Trp100 at 900 K, protocol B.
Figure 5.
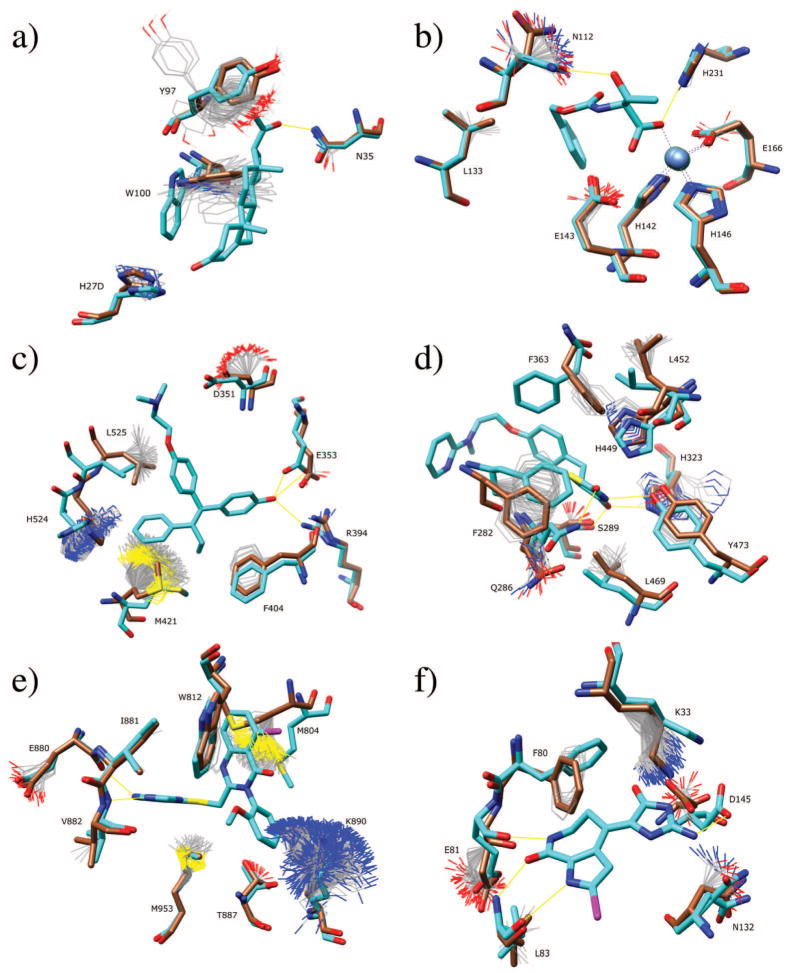
Binding pocket ensembles. Simulations are carried out in the absence of ligand at 600 K, with protocol B (no error correction). Ligand and bound (holo) structures are shown in cyan. Unbound native sidechains in starting configurations are shown in brown. The computed ensemble is shown as thin lines. The ligand from the holo structure is shown for reference. (a) DB3 antibody and progesterone. (b) Thermolysin and Z-D glutamic acid. (c) Estrogen receptor and raloxifene. (d) PPAR-γ and GI262570. (e) PI3 kinase and ligand PIK-039. (f) CDK2 and hymenialdisine.
The residues His27D and Asn35 show less flexibility in the simulations and also little conformational change between the apo and holo structures (Figure 5a). Tyr97, in contrast, appears to fluctuate in multiple basins. This is because it is mostly solvent exposed, and there is very little steric hindrance. The sidechain adopts similar conformations in the apo and holo structures. This does not necessarily imply a failure of the computational prediction, however. It is possible that this sidechain could adopt different conformations in complex with other ligands.
The magnitudes of fluctuations observed using protocols A and B for Trp100 and Tyr97 are similar (Figure 4). Since protocol B is slightly more efficient and appears to provide similar configurational diversity, it was used for the data presented for all the remaining binding pockets in Figure 5. In addition, we have chosen to use T = 600 K for the remainder of the test cases, because it provides a balance between sampling alternative conformations that may be important in ligand binding, but not so much diversity as to be uninformative. We reiterate that we view temperature as a user-adjustable parameter, and using multiple temperatures, as with this test case, may be advisable.
Thermolysin.54
The residues His142, His146, and Glu66, which coordinate the Zn ion are correctly predicted to be rigid (Figure 5b). For this simulation, the zinc ion was included. The hydrogen bonding network of His231 is correctly preserved. Asn112 is predicted to be very flexible, and in fact rotates significantly upon ligand binding.
Estrogen Receptor.55,56
Residues Leu525, Met421, and His524 all show significant flexibility in the simulations, and also undergo significant rearrangements upon binding 4-hydroxytamoxifin (Figure 5c). Glu353 and Arg394 display less flexibility due to the strong salt bridge. These show small conformational rearrangements upon binding the ligand due to formation of hydrogen bonds to it. Backbone rearrangements observed upon ligand binding, such as those seen in His524 and Leu525, are of course not captured by the sidechain MC simulations. As a rough guide, however, the ensemble correlates well with observed rearrangements.
PPAR-γ.57
The hydrophobic residues Phe282, Leu452, and Leu469 display flexibilities that correspond to structural rearrangements upon ligand binding (Figure 5d). Phe363 fails to sample the bound configuration, and is the first of only two false negative cases from the entire data set (see CDK2). It is likely that this is due to the fact that the rigid backbone occupies a region which occludes the possibility of sampling an alternative state. His449 displays a narrow range of flexibility which corresponds to the displacement in the target structure. Tyr473 samples alternative solvent exposed configurations, similar to Tyr97 in the DB3 antibody. Gln286 displays flexibility and appears to sample some conformations similar to the holo conformation, to the extent that the slightly different backbone configurations permit.
PI3 Kinase.52,58
All residues which do not undergo significant rearrangement upon ligand binding are predicted to be rigid in the simulations (Figure 5e). Glu880 and Lys890 display conformational diversity in the simulations which encompasses the observed apo and holo conformations. Met804 displays significant flexibility in the sidechain ensemble which encompasses the apo and holo conformations. The movement of this sidechain is critical for opening a hydrophobic pocket that is critical for ligand binding and specificity.
CDK2.59–61
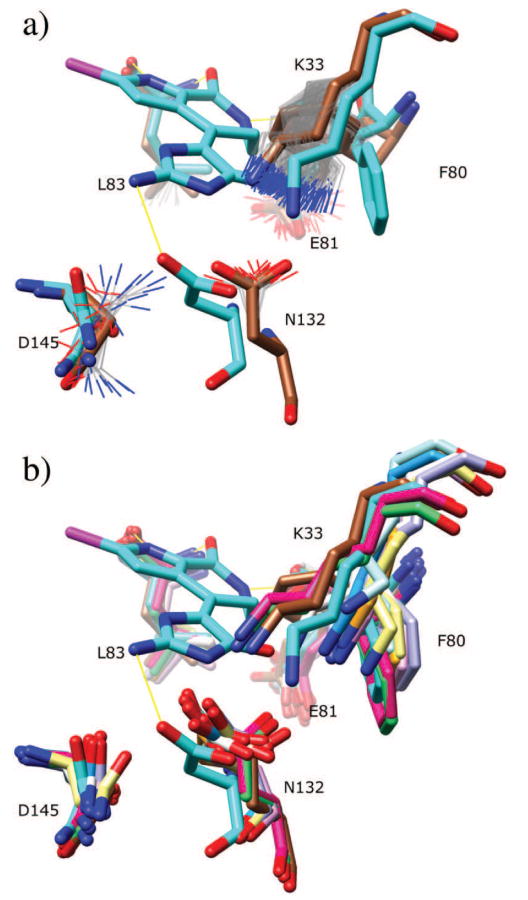
Residues Glu81, Leu83, and Asn132 each appear to display conformational diversity commensurate with the observed changes between the apo and holo structures (Figure 5f), while Phe80 is the second false negative of the data set. Lys33 displays flexibility, although it does not quite sample the bound configuration. Instead, in the absence of ligands, it forms a salt bridge with Asp145, which is disrupted by the hymenialdiside interaction in the bound form.
Figure 6a shows a closeup of the salt bridge which is transiently disrupted in the 600 K simulation. Figure 6b shows a superposition of multiple structures of CDK2 which display a similar structural diversity.
Figure 6.
CDK2 salt bridge interaction. (a) Binding pocket ensemble and representation which is identical to Figure 5f, but from a different perspective. (b) Sidechains from CDK2 structures 1h24, 1h25, 1h26, 1h27, 1h28, 1hc1, 1pw2, 1w98, and 2jgz.
Conclusions and Future Directions
A novel application of the MTS-MC algorithm has been applied to sampling sidechain degrees of freedom in implicit solvent. Relative to a “simple” Monte Carlo algorithm without the use of inner steps, the multiscale approach increases the convergence by a factor of 10–15. Rapid steric screening provides an additional factor of 2–4 speed up, and other algorithmic details (rapid updates of energies when only a portion of the protein is moving) also contribute to efficiency. Applications to small molecule ligand binding sites in proteins demonstrate that the method can be used to efficiently sample large changes in sidechain conformations and identifies sidechains that may undergo conformational changes upon ligand binding.
Additional degrees of freedom can be incorporated into this approach in a straightforward manner. For example, local changes in backbone conformation can be included using analytical loop closure62,63 methods with an appropriate Jacobian.64 Such a method, which is under development, could be an efficient means of sampling conformational changes such as those that have been observed in the kinase DFG motif, or in loop latching as in TIM barrels,65 in a way that obeys detailed balance and thus can capture entropy differences between states.
Acknowledgments
We thank Kevan Shokat (UCSF) for pointing out the PI3K system. All molecular figures were generated with UCSF Chimera.66 This work was supported by NSF grant MCB-0346399, NIH grant GM081710, and the Sandler Program in Basic Science (UCSF). M.J. is a consultant to Schrödinger, Inc. Special thanks to Katarzyna Bernacki, Dave Mobley, Arjun Narayanan, and Chris Mc-Clendon for helpful discussions.
Appendix: Proof of Detailed Balance with a Short Range Cutoff
A more detailed accounting of the short- and long-range decompositions is presented. These details are omitted from the body of the text for clarity.
The use of a short- and long-range cutoff is a common way of improving calculation efficiencies. The advantage gained is in the infrequent updating of the long-distance interactions. To explicitly track the updating of the short- and long-range cutoffs, eqs 14 and 15 can be re-expressed as follows:
| (26) |
Where S(l) is a “switching function” of the coordinate state l, which divides the space over which the potential A(m, n), as expressed in eq 15, is the potential at Born state m and coordinate state n. When the Born radii are evaluated based on the current coordinate state, the short- and long-range potentials can be expressed in terms of the current coordinate (and Born radii) state n, and latent cutoff state l:
| (27) |
Since S(l) is a function of the complete set of coordinates, a full update of the distances must be computed. The idea behind the use of the cutoff is to limit the number of times the full distance matrix is computed, as well as the full potential.
To this end, an efficient Monte Carlo protocol will update the switching function infrequently, while maintaining detailed balance or very nearly doing so. For the updating scheme that is used for the present work, detailed balance is rigorously maintained with regard to the short and long-range evaluations. The simplest form that the switching function can take is a simple distance cutoff, but more complicated forms, such as cell neighbor lists and other types of additive decompositions can be used. For this work, atoms are treated as short-range if any single atom within a sidechain is within a cutoff distance of another sidechain. Default settings that were developed for an optimal minimization strategy were used.44 The cutoffs vary according to type of interaction. Each sidechain is identified as either charged or nonpolar. All atoms in the given sidechain are labeled as such. For nonpolar atoms interacting with nonpolar atoms, the cutoff is 15 Å. For charged–nonpolar interactions, the cutoff is 20 Å, and for charged–charged interactions, the cutoff is 30 Å. The updating scheme used for the current work is to update the switching function at the beginning of the each outer iteration of the sampling loop.
While the proof of detailed balance for the switching function updating scheme is independent of the Born radii updating scheme, the full bookkeeping of all latent states is presented here for completeness. Re-expressing the short- and long-range potentials in eq 13 with the short-range state made explicit gives the following:
| (28) |
The resulting (unnormalized) probability distributions are
| (29) |
where q is given by eq 5. Expressing the probability of a single state in terms of the decomposed states gives the following:
| (30) |
Following the derivations presented in refs 35 and 43, the required detailed balance condition is
| (31) |
where T(j|i) is the probability of transitioning from coordinate state i to j. Expanding this expression gives:
| (32) |
where α(j|i) and accL(j|i) are the selection and acceptance probabilities of outer state j from state i. Following the MTS-MC derivation,35 the probability of selecting state j from state i is given by the following:
| (33) |
where the above transition probability is the product of the individual transition probabilities of the inner loop
| (34) |
In the short-range, or inner loop of sampling, neither the switching function nor the Born radii are updated, so that each step obeys the following detailed balance relation:
| (35) |
The transition between outer states j and i obeys the following detailed balance relation:
| (36) |
Combining eqs 32–35 and solving for the ratio of acceptance probabilities gives the following:
| (37) |
Protocols A and B follow the same updating scheme for the switching functions. The acceptance probability for protocol A is expressed in eq 17 as follows:
| (38) |
The ratio of eqs 37 and 38 is unity
| (39) |
and therefore, the sampling scheme described by eqs 37 and 38 rigorously obeys detailed balance. For all equations in the body of the text, the state of the switching function is not shown, but is updated according to the scheme described. It should be noted, however that the “standard” protocol is not updated according to this scheme, since there is no need to express the energies in terms of the latent states.
The acceptance probabilities for protocol B, as given in 18, are as follows:
| (40) |
The ratio of the true acceptance probabilities is equivalent to the acceptance probabilities given in eq 37, and the ratio is given simply as follows:
| (41) |
References
- 1.Dunbrack RL, Jr, Karplus M. Nat Struct Biol. 1994;1:334. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 2.Dunbrack RL, Jr, Cohen FE. Protein Sci. 1997;6:1661. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Z, Honig B. J Mol Biol. 2001;311:421. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- 4.Kuhlman B, Baker D. Proc Natl Acad Sci USA. 2000;97:10383. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, Kuhlman B, Kortemme T, Baker D. Proteins. 2005;58:893. doi: 10.1002/prot.20347. [DOI] [PubMed] [Google Scholar]
- 6.Fiser A, Do RK, Sali A. Protein Sci. 2000;9:1753. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiser A, Sali A. Methods Enzymol. 2003;374:461. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 8.Sherman W, Day TJ, Jacobson M, Friesner RA, Farid R. J Med Chem. 2006;49:534. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 9.Meiller J, Baker D. Proteins. 2006;65:538. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari AM, Wei B, Constantino L, Shoichet BK. J Med Chem. 2004;47:5076. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voigt CA, Gordon DB, Mayo SL. J Mol Biol. 2000;299:789. doi: 10.1006/jmbi.2000.3758. [DOI] [PubMed] [Google Scholar]
- 12.Jain T, Cerutti DS, McCammon JA. Protein Sci. 2006;15:2029. doi: 10.1110/ps.062165906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. J Mol Biol. 1982;161:269. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 14.Schlick T. Molecular Modeling and Simulation. Springer-Verlag; New York: 2002. [Google Scholar]
- 15.Mobley D. J Chem Theory Comput. 2007;3:1231. doi: 10.1021/ct700032n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobley D, Graves A, Chodera JD, McReynolds A, Shoichet BK, Dill KA. J Mol Biol. 2007;371:1118. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller A, Teller E. J Chem Phys. 1953;21:1087. [Google Scholar]
- 18.Rosenbluth MN, Rosenbluth AW. J Chem Phys. 1955;23:356. [Google Scholar]
- 19.Deem MW. J Chem Phys. 1999;111:6625. [Google Scholar]
- 20.Deem MW. Mol Phys. 1999;97:559. [Google Scholar]
- 21.Dinner AR. J Comput Chem. 2000;21:1132. [Google Scholar]
- 22.Ulmschneider JP, Jorgensen WL. J Am Chem Soc. 2004;126:1849. doi: 10.1021/ja0378862. [DOI] [PubMed] [Google Scholar]
- 23.Li ZQ, Scheraga HA. Proc Natl Acad Sci USA. 1987;84:6611. doi: 10.1073/pnas.84.19.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorin EJ, Engelhardt MA, Herschlag D, Pande VS. J Mol Biol. 2002;317:493. doi: 10.1006/jmbi.2002.5447. [DOI] [PubMed] [Google Scholar]
- 25.Pande VS, Baker I, Chapman J, Elmer SP, Khaliq S, Larson SM, Rhee YM, Shirts MR, Snow CD, Sorin EJ, Zagrovic B. Biopolymers. 2003;68:91. doi: 10.1002/bip.10219. [DOI] [PubMed] [Google Scholar]
- 26.Felts AK, Gallicchio E, Wallqvist A, Levy RM. Proteins. 2002;48:404. doi: 10.1002/prot.10171. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh A, Rapp CS, Friesner RA. J Phys Chem B. 1998;102:10983. [Google Scholar]
- 28.Still W Clark, T A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127. [Google Scholar]
- 29.Gallicchio E, Zhang LY, Levy RM. J Comput Chem. 2002;23:517. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- 30.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474. [Google Scholar]
- 31.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. Proteins. 2004;55:351. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson MP, Friesner RA, Xiang Z, Honig B. J Mol Biol. 2002;320:597. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 33.Tuckerman M, Berne BJ. J Chem Phys. 1990;94:1465. [Google Scholar]
- 34.Tuckerman M, Berne BJ, GJ M. J Chem Phys. 1992;97:1990. [Google Scholar]
- 35.Hetenyi B, Bernacki K, Berne BJ. J Chem Phys. 2002;117:8203. doi: 10.1063/1.1755195. [DOI] [PubMed] [Google Scholar]
- 36.Michel J, Taylor R, Essex J. J Chem Theory Comput. 2006;2:732. doi: 10.1021/ct600069r. [DOI] [PubMed] [Google Scholar]
- 37.Roux B. Implicit Solvent Models. Marcel Dekker; New York: 2001. [Google Scholar]
- 38.Yu Z, Jacobson MP, Friesner RA. J Comput Chem. 2005;27:72. doi: 10.1002/jcc.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobson M. J Phys Chem B. 2004;108:6643. [Google Scholar]
- 40.Verlet L. Phys Rev. 1968;165:201. [Google Scholar]
- 41.Weeks JD, Chandler D, Andersen HC. J Chem Phys. 1971;55:5422. [Google Scholar]
- 42.Bernacki K, Hetenyi B, Berne BJ. J Chem Phys. 2004;121:44. doi: 10.1063/1.1755195. [DOI] [PubMed] [Google Scholar]
- 43.Gelb LD. J Chem Phys. 2003;118:7747. [Google Scholar]
- 44.Zhu K, Friesner RA, Jacobson MP. J Comput Chem. 2006 doi: 10.1002/jcc.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frenkel D, S B. Understanding Molecular Simulation: From Algorithms to Applications. Academic Press; Boston: 2002. [Google Scholar]
- 46.Chen B, Siepmann JI. Theor Chem Acc. 1999;103:87. [Google Scholar]
- 47.Yang Wei, Karplus Martin, R BP. J Chem Phys. 2004;120 doi: 10.1063/1.1738106. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro MBW, Chen HJ. 1968;63:1343. [Google Scholar]
- 49.Shapiro S, Wilk MB. Biometrika. 1965;52:591. [Google Scholar]
- 50.Arevalo JH, Hassig CA, Stura EA, Sims MJ, Taussig MJ, Wilson IA. J Mol Biol. 1994;241:663. doi: 10.1006/jmbi.1994.1543. [DOI] [PubMed] [Google Scholar]
- 51.Arevalo JH, Stura EA, Taussig MJ, Wilson IA. J Mol Biol. 1993;231:103. doi: 10.1006/jmbi.1993.1260. [DOI] [PubMed] [Google Scholar]
- 52.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. Cell. 2006;125:733. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roe DR, Okur A, Wickstrom L, Hornak V, Simmerling C. J Phys Chem B. 2007;111:1846. doi: 10.1021/jp066831u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senda M, Senda T, Ogi T, Kidokoro S, Stihle R, Boroni E, Hennig M. Acta Crystallogr. 2002;58:C278. [Google Scholar]
- 55.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. Cell. 1998;95:927. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 56.Pike AC, Brzozowski AM, Walton J, Hubbard RE, Bonn T, Gustafsson JA, Carlquist M. Biochem Soc Trans. 2000;28:396. [PubMed] [Google Scholar]
- 57.Gampe RT, Jr, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE. Mol Cell. 2000;5:545. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 58.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Nature. 1998;395:137. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 59.Meijer L, Thunnissen AM, White AW, Garnier M, Nikolic M, Tsai LH, Walter J, Cleverley KE, Salinas PC, Wu YZ, Biernat J, Mandelkow EM, Kim SH, Pettit GR. Chem Biol. 2000;7:51. doi: 10.1016/s1074-5521(00)00063-6. [DOI] [PubMed] [Google Scholar]
- 60.Bourne Y, Watson MH, Hickey MJ, Holmes W, Rocque W, Reed SI, Tainer JA. Cell. 1996;84:863. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 61.Groban ES, Narayanan A, Jacobson MP. PLoS Comput Biol. 2006;2:e32. doi: 10.1371/journal.pcbi.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coutsias EA, Seok CL, Jacobson MP, Dill KA. J Comput Chem. 2004;25:510. doi: 10.1002/jcc.10416. [DOI] [PubMed] [Google Scholar]
- 63.Go N, Scheraga HA. Macromolecules. 1969;3:178. [Google Scholar]
- 64.Dodd LR, Boone TD, Theodorou DN. Mol Phys. 1993;78:961. [Google Scholar]
- 65.Wong S, Jacobson MP. Proteins. 2008;71:153. doi: 10.1002/prot.21666. [DOI] [PubMed] [Google Scholar]
- 66.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]