Abstract
The proteasome has an essential function in the intracellular degradation of protein in eukaryotic cells. We found that the dimeric quinone reductase Lot6 uses the flavin mononucleotide (FMN)-binding site to bind to the 20S proteasome with a 1:2 stoichiometry—that is, one 20S proteasome molecule can associate with two quinone reductases. Furthermore, reduction of the FMN cofactor by either NADH or light irradiation results in the binding of the b-Zip transcription factor Yap4 to the Lot6–proteasome complex, indicating that recruitment of the transcription factor depends on the redox state of the quinone reductase. Here, we show that binding of Yap4 to the complex not only protects it from ubiquitin-independent proteasomal degradation, but also regulates its cellular localization. In non-stressed wild-type cells, we did not detect any Yap4 in the nucleus, whereas Yap4 was present in the nuclei from quinone-stressed yeast cultures. Thus, the Lot6–proteasome complex can be regarded as a redox switch in which the quinone reductase acts as a sensor for oxidative stress.
Keywords: Lot6, redox state, 20S proteasome, Yap4, transcription factor
Introduction
Organisms have evolved cellular responses to cope with stress caused by reactive oxygen species (Halliwell & Gutteridge, 1984). As these harmful molecules are constantly being generated, all aerobically growing cells maintain an assemblage of antioxidants and enzymes to maintain a state of redox balance (Toone et al, 2001). In the yeast Saccharomyces cerevisiae, the b-Zip transcription factor Yap1 is controlled primarily by redox-sensitive nuclear export, regulating its nuclear accumulation on activation by H2O2 (Delaunay et al, 2000). However, Yap1 is not directly oxidized by H2O2; instead, a thiol peroxidase Gpx3 has been identified to promote oxidation of Yap1 (Delaunay et al, 2002). Other molecular mechanisms to sense oxidative stress involve iron–sulphur centres, haem or flavin (Bauer et al, 1999). In Klebsiella, Azotobacter and Enterobacter, the activity of NifA, a transcriptional activator of nitrogen fixation genes, is inhibited by the redox-sensitive protein NifL in response to elevated levels of oxygen and fixed nitrogen in vivo (Dixon, 1998). NifL is a flavoprotein that contains FAD as a redox-responsive cofactor, which inhibits NifA activity in vitro in the oxidized but not in the reduced form. Although details of the interaction of NifL with NifA are still unclear, this system so far represents the only example of flavin-dependent transcriptional regulation.
Several species have been shown to have flavin-dependent enzymes that allow the two-electron reduction of quinones to the hydroquinone form to avoid the generation of one-electron reduced semiquinone that is known to cause oxidative stress (Deller et al, 2008). One of the human NADPH:quinone reductases (NQOs) was recently shown to interact with the 20S proteasome. In the presence of NADH, the NQO1–proteasome complex recruits transcription factors p53 and p73 (Asher et al, 2001), presumably to protect them from proteasomal degradation (Asher & Shaul, 2005). Remarkably, degradation of p53 by the 20S proteasome is inhibited in the presence of excess NQO1 and NADH, suggesting that NQO1 directly regulates the proteasomal degradation of this protein. In vitro binding assays in the presence of the NQO1 cofactors NAD+, FAD or NADH showed that the binding of NQO1 to p53 increased in the presence of NADH (Asher et al, 2005). The involvement of NQO1 in the accumulation of p53 suggests that redox reactions controlled by oxidoreductases such as NQO1 might therefore be important in determining the intracellular level of p53. However, the molecular mechanism and the exact role of NADH in this process are so far unclear.
Here, we show that Lot6, the yeast orthologue of human NQO1, is physically associated with the 20S proteasome. Each proteasome barrel structure can bind to two Lot6 dimers, which might be associated with the α-ring system. Furthermore, recruitment of the yeast transcription factor Yap4, a member of the yeast activator protein (Yap) family, to the quinone reductase–20S proteasome complex is regulated by the redox state of the flavin cofactor. Association of the transcription factor with the pre-formed complex is shown to affect both proteasomal degradation and localization. Hence, we conclude that Lot6 is a redox-regulated switch that directly affects the physiological function of the proteasome, as well as the stability and localization of yeast transcription factor Yap4.
Results And Discussion
The yeast gene lot6 (YLR011w) encodes a 21-kDa protein with a non-covalently bound flavin mononucleotide (FMN) cofactor. The protein adopts a flavodoxin-like fold and forms a dimer in solution (Liger et al, 2004). We have recently shown that the protein has quinone reductase activity that is similar to its mammalian orthologues (Sollner et al, 2007). Although a role of the enzyme in detoxification of quinones has been shown, the exact cellular function of the enzyme during oxidative stress is still unclear. In an approach to explain its cellular role, we performed pull-down assays for the isolation of potential protein interaction partners of Lot6. Initially, Lot6 with a carboxy-terminal hexahistidine tag was incubated with crude extracts of wild-type yeast cells and subsequently applied to a nickel-chelating sepharose column. After extensive washing, the tagged protein was eluted and protein fractions were analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE). As shown in Fig 1A, additional bands (with similar mobilities) were observed in the Lot6 fraction, which were identified by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry and amino-terminal sequencing as several subunits of the 20S proteasome. To confirm this finding, we isolated the proteasome from yeast, repeated the pull-down assay with the purified proteasome (Groll & Huber, 2005), and subjected the pull-down fractions to Western blotting using both Lot6 and 20S proteasome antibodies. Association of Lot6 with the yeast 20S proteasome was clearly detected (Fig 1B). Interestingly, Western blot analysis using Lot6 antibody showed the presence of Lot6 traces even in the affinity-purified 20S proteasome fraction (Fig 1B, lane 1). Furthermore, subjecting a yeast crude extract to size-exclusion chromatography (Fig 1C) and subsequent Western blotting showed that Lot6 coelutes with the 20S proteasome, indicating that a stable Lot6–20S proteasome protein complex forms in vivo.
Figure 1.
Lot6 is found associated with the yeast 20S proteasome. (A) Analysis of interaction partners of Lot6 (oxidized) by pull-down assays using Lot6–His6 as bait. (B) Purified yeast 20S proteasome (1), purified Lot6–His6 (2), and the first elution fraction of a pull-down assay using Lot6 and purified yeast 20S proteasome (3) were analysed by immunoblotting using both 20S antibody (upper panel) and Lot6 antibody (lower panel). (C) Gel filtration of crude yeast extract. Upper panel, original trace from experiment. Fractions were precipitated and analysed using Lot6 antibody (upper panel) and 20S antibody (lower panel). Std, standard.
To assess the importance of the FMN cofactor for the interaction with the proteasome, freshly prepared apo-Lot6 was used for the pull-down assay. The results revealed that apo-Lot6 is unable to form a complex with the 20S proteasome, suggesting that the flavin cofactor is required for the formation of the complex (Fig 2, lane 5). This prompted us to take a closer look at the potential role of the cofactor. As shown previously, Lot6 catalyses the reduction of a series of quinones at the expense of both NADH and NADPH (Sollner et al, 2007). Interestingly, reoxidation of reduced Lot6 by molecular oxygen is several orders of magnitudes slower compared with quinones (estimated kox=1.1 M−1 s−1). Similar rates of reaction were determined for the human quinone reductase NQO1 (Tedeschi et al, 1995). Thus, it can be concluded that oxygen is not a relevant oxidizing agent for reduced Lot6 in vivo.
Figure 2.
Protein–protein interactions depend on the presence and redox state of the flavin cofactor. (1) Lot6 control as used for all pull-down experiments. (2) Oxidized (ox) Lot6 interacts with 20S proteasome from crude yeast extract. (3, 4) Reduced (red) Lot6 binds to not only the 20S proteasome but also Yap4. (5) Lot6 lacking the cofactor (apo-Lot6) is unable to bind to both proteasome and Yap4. (6) When a yap4 deletion strain is used for the interaction studies, no other interaction partner is found. All lanes are showing 12.5% SDS–PAGE gels of pull-down experiments. SDS–PAGE, SDS–polyacrylamide gel electrophoresis; wt, wild type.
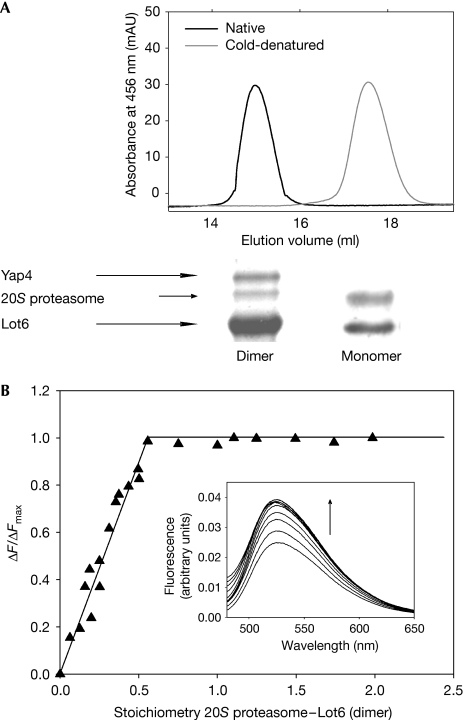
To probe the binding of reduced Lot6 to the proteasome, the enzyme was reduced in two different ways: by the addition of excess NADH or by light irradiation. By using one of these two procedures, reduced Lot6 was again incubated with yeast crude extract, and potential binding partners were analysed as described above. It was found that the interaction of Lot6 with the proteasome is independent of the redox state of the flavin (Fig 2, lanes 2–4). However, when Lot6 is present in its reduced state another band with a slightly lower mobility was detected (Fig 2, lanes 3 and 4). Subjecting this band to in-gel tryptic digestion and analysis by MALDI led to the identification of the b-Zip transcription factor Yap4, a member of the Yap family (Nevitt et al, 2004a, 2004b). When the pull-down experiments were repeated with protein extract from a yap4 knockout yeast strain, the Yap4 band was absent confirming our assignment (Fig 2, lane 6). The occurrence of this additional binding partner is independent of the flavin reduction method and hence does not rely on the presence of a reduced pyrimidine nucleotide but on the redox state of the cofactor. Conversely, Lot6 in its native dimeric state is essential for the binding of Yap4 to the complex. Cold denaturation of Lot6 yields a stable monomer, which in its reduced form still binds to the 20S proteasome but is no longer able to recruit Yap4 to the complex (Fig 3A). Dissociation of Lot6 dimers into monomers does not affect the catalytic properties of the enzyme with regard to quinone reduction (kcat=21.1±0.9 and 20.8±0.5 s−1, KM=2.7±0.4 and 2.4±0.3 μM for native and monomeric Lot6, respectively). Therefore, dimerization of Lot6 is not a prerequisite for enzymatic activity, but is essential for formation of the protein complex.
Figure 3.
Stoichiometry of the 20S proteasome–Lot6–Yap4 complex. (A) Upper panel, overlay of two gel filtrations of native dimeric (left) and monomeric (right) Lot6. Lower panel, pull-down assays under reduced conditions with both dimeric and monomeric Lot6. Reduced dimeric Lot6 is able to interact with both the 20S proteasome and Yap4, whereas reduced monomeric Lot6 can only bind to proteasome. (B) Fluorescence determination of the stoichiometry of binding of the 20S proteasome to Lot6. Lot6 (1 μM) was titrated with 20S proteasome at 25°C. The results of the titration are plotted as a fraction of maximum change (ΔF/ΔFmax at 530 nm) compared with the molecular stoichiometry of 20S proteasome–Lot6 (dimer). The insert shows titration spectra with 20S proteasome concentrations ranging from 0 to 2.5 μM.
To determine the stoichiometry of the Lot6–20S proteasome complex, we titrated dimeric Lot6 with 20S proteasome while monitoring the fluorescence emission of the FMN cofactor. As shown in Fig 3B, the titration yielded a 2:1 (Lot6:20S) stoichiometry, suggesting that two quinone reductases are bound per 20S proteasome. Furthermore, the titration shows that binding of Lot6 to the proteasome is tight, as indicated by the sharp titration end point.
We also investigated the effect of complex formation on enzymatic activity of Lot6 and the 20S proteasome. Quinone reductase activity was five times lower in the presence of a 10-fold excess of 20S proteasome (data not shown). This is consistent with the assumption that the 20S proteasome binds to or near the FMN site of Lot6. Similarly, chymotrypsin-like activity of the proteasome was 10 times lower in the presence of Lot6 (Fig 4A). As substrate access is considered to be the rate-limiting step, it is possible that Lot6 binds to the α-ring of the proteasome, thereby hampering access to the catalytic chamber and decreasing proteasomal activity (Groll et al, 1997).
Figure 4.
The physiological role of Yap4. (A) Chymotrypsin-like (CL) activity was measured in the presence of both oxidized (ox) and reduced (red) Lot6 and compared with 20S proteasome only. (B) In vitro study showing that Yap4 is protected from ubiquitin-independent proteasomal degradation through association with reduced Lot6. Yap4 was incubated with yeast 20S proteasome, Lot6 (third and fourth panels from top) and NADH (second and fourth panels from top). Samples were taken after 0, 30, 60 and 90 min, and immunoblotted using Yap4 and Lot6 antibodies (fifth panel from top). (C) Immunoblot of homogenate (hom), cytosol (cyt) and nucleus (nuc) preparations from non-stressed (1) and quinone-stressed (2) yeast cultures. On treatment with naphthoquinone, Yap4 relocalizes from the cytosol to the nucleus. As a control, the distribution of Sir2 (3), a nuclear histone deacetylase, is shown.
Previous results using the human system have suggested that association of a transcription factor with the quinone reductase–proteasome complex prevents ubiquitin-independent proteasomal degradation (Asher et al, 2001, 2005; Asher & Shaul, 2005; Garate et al, 2008). For the yeast system, we also found that Yap4 is protected from proteasomal degradation when bound to the Lot6–20S proteasome complex (Fig 4B). However, the fact that the localization of Yap1 is regulated by a redox-sensitive mechanism prompted us to investigate whether the association of Yap4 with protein interaction partners might also have implication for its localization. Thus, we prepared homogenate, cytosol and nuclei from stressed and non-stressed cultures. Immunoblotting revealed the presence of Yap4 in the nucleus after induction of stress, whereas no Yap4 was detected in the nuclei from a non-stressed culture (Fig 4C). Hence, we propose that stress leads not only to the dissociation of Yap4 from the Lot6–proteasome complex, but subsequently Yap4 relocalizes to the nucleus.
Sequestration of a transcription factor by a quinone reductase–proteasome complex discovered here in a unicellular eukaryote is similar to findings in mammalian cells (Asher et al, 2001). In humans, NQO1 interacts with the 20S proteasome and is proposed to recruit the transcription factor p53 in an NADH-dependent manner (Asher et al, 2001, 2005). Similarly, the transcription factor Yap4 reported here is recruited to the (reduced) quinone reductase–20S proteasome complex of yeast. However, it shows only low sequence similarity to human p53 (sequence identity of 22.5%) and hence it is unclear whether Yap4 is a functional precursor of p53.
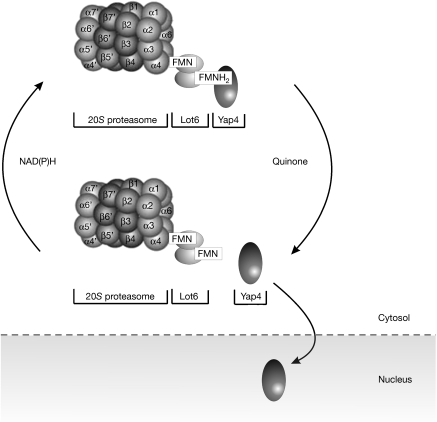
Our data indicate that the redox state of the flavin cofactor controls the recruitment of Yap4 to the 20S proteasome in yeast irrespective of the mode of reduction. Therefore, we propose that the generation of the reduced state of the flavin cofactor results in the recruitment of Yap4 to the Lot6–proteasome complex. This would suggest that the stability and localization of the transcription factor Yap4 is under direct redox control of the quinone reductase–proteasome system. Under reducing conditions, Yap4 is bound to the Lot6–proteasome complex and thus is retained in the cytosol (Fig 5). In the presence of quinones, Yap4 is released from the Lot6–proteasome complex and rapidly relocalizes to the nucleus to avoid ubiquitin-independent proteasomal degradation (Asher et al, 2006). A similar redox controlled mechanism might regulate the stabilization and localization of p53 and related transcription factors in mammalian cells.
Figure 5.
Proposed model for redox-dependent binding and release of Yap4 from the Lot6–20S proteasome complex. Under conditions in which the Lot6 flavin is reduced and does not get reoxidized by quinone species, Yap4 is bound to the Lot6–proteasome complex and thus is retained in the cytosol. In the presence of quinones, Yap4 is released from the complex and relocalizes to the nucleus to induce the transcription of target genes.
Although it seems obvious that NADPH acts as a physiological reducing agent for the quinone reductase, the molecular nature of the reoxidizing agent remains elusive. The biochemical data indicate that molecular oxygen is an unlikely candidate and suggests that this role is performed by cellular quinones. In this context, it is important to note that quinone reductases have been found in a wide range of eukaryotic organisms from yeast to mammals (Deller et al, 2008), and it is therefore possible that these enzymes have evolved as redox sensors of the proteasome, recruiting and stabilizing transcription factors that are crucial for the response to oxidative stress.
Methods
Plasmids, strains and growth conditions. The S. cerevisiae yeast strains Y00000 (BY4741 Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and Y01804 (BY4741 Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yap4Δ∷KanMX4) were obtained from the Euroscarf strain collection. Strains were grown under aerobic conditions at 30°C on YPD medium.
Preparation of cell extracts. Yeast cells were collected at late logarithmic phase and protein extracts were prepared by using glass beads. Protein was quantified using a method described previously (Lowry et al, 1951).
Yap4 cloning and expression. Cloning was performed using standard techniques. The gene was integrated into pET21a (Novagen, San Diego, CA, USA) using NdeI and XhoI restriction sites for expression of the protein with an N-terminal hexahistidine tag. Yap4 was coexpressed with the vector pTf16 containing the tig chaperone (Takara Bio, Otsu, Shiga, Japan) in Escherichia coli BL21(DE3) and purified as described for Lot6.
Pull-down assays and photoreduction. Native Lot6–His6 was expressed and purified as described previously (Sollner et al, 2007). For interaction studies, 1.5 ml of cell extract (2 mg/ml protein) was incubated with 800 μg of recombinant Lot6–His6 for 5 min. The protein solution was subjected to Ni-NTA resin (1 ml HiTrap Chelating HP columns; GE Healthcare, Uppsala, Sweden). Fractions were precipitated using 15% trichloroacetic acid and applied to SDS–PAGE. Interaction partners of reduced Lot6 were investigated either by reduction with an excess of NADH or by photoreduction as outlined previously (Sollner et al, 2007). Subsequently, reduced Lot6 was transferred to a nitrogen-filled glove bag (AtmosBag; Sigma-Aldrich, St Louis, MO, USA) and pull-down assays were performed as described above. Fractions were analysed by Western blotting using a primary rabbit antibody against Lot6 (Haid & Suissa, 1983). When pull-down assays with Lot6 and purified 20S proteasome were conducted, yeast 20S proteasome was expressed and purified as described previously (Groll & Huber, 2005).
Gel filtration. For the detection of the Lot6–20S proteasome complex, 3 ml of crude yeast cell extract (2 mg/ml protein) was loaded onto a HiLoad™ 16/60 Superdex™ 200 prep grade (Amersham Biosciences, Uppsala, Sweden) column and eluted at a flow rate of 1 ml/min. Fractions were precipitated with 15% (w/v) ice-cold trichloroacetic acid and subjected to Western blotting using both a primary antibody against Lot6 and a 20S proteasome rabbit antibody (Biomol International, Exeter, UK).
Rapid reaction studies. Stopped-flow experiments were performed at 4°C using a Hi-Tech Scientific KinetAsyst SF-61 DX2 stopped-flow spectrometer. Enzyme and substrate solutions were made anaerobic in glass tonometers by repeated cycles of evacuation and flushing with purified argon. Oxidative half-reactions were carried out using 20 μM Lot6 (reduced with dithionite) in 50 mM N-[tris(hydroxymethyl)methyl]-3-aminopropane sulfonic acid, pH 7.4. The apparent rate constant for oxygen was determined by mixing reduced enzyme solution with air-saturated buffer (21% oxygen) at 25°C.
Enzyme activity assays. Lot6 quinone reductase assays were conducted as described previously (Sollner et al, 2007). For the generation of monomeric Lot6, the buffer of recombinant Lot6 was changed to 50 mM phosphate, 10 mM β-mercaptoethanol and 150 mM NaCl, pH 7.0. The protein solution was then flash-frozen with liquid nitrogen, slowly thawed and subjected to a HiLoad 16/60 Superdex prep grade (Amersham Biosciences) column. Fractions corresponding to monomeric Lot6 were concentrated by ultracentrifugation (CentriPrep; Amicon, Millipore, Billerica, MA, USA; 10 kDa molecular weight cut off). When Lot6 quinone reductase assays were performed in the presence of 20S proteasome, a 10-fold excess (50 nM) of native yeast 20S proteasome was added.
20S proteasome activity assays were performed using native 20S proteasome (Groll & Huber, 2005). For the determination of proteasomal activity in the absence or presence of Lot6, the chymotrypsin-like activity against chromogenic substrate was tested as described previously (Groll & Huber, 2005). When reduced conditions were measured, an excess of NADH (500 μM) was added.
Fluorescence titration. Fluorescence measurements were performed with a Shimadzu RF-5301PC spectrofluorophotometer at 25°C. Emission was recorded from 480 to 650 nm at an excitation wavelength of 370 nm. The band width for excitation was 5 nm and for emission it was 10 nm.
Preparation of yeast organelles. Nuclei were prepared from yeast wild-type BY4741 cells with or without 6 h of treatment with naphthoquinone (10 μM) in the stationary phase. Cytosol was prepared as described previously (Zinser & Daum, 1995). Nuclei were enriched by sucrose density gradient centrifugation (Hurt et al, 1988; Aris & Blobel, 1991) and immunoblotted using Yap4 antibody. Sir2 was detected using a rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
MALDI–time of flight MS. We analysed MALDI–time of flight (MALDI–TOF) with a Micro MX (Waters, Milford, MA, USA) TOF instrument using standard procedure as in gel digestion (Shevchenko et al, 1996).
Acknowledgments
We thank the Austrian Science Foundation (FWF) for the support through the Doktoratskolleg Molecular Enzymology (W901-B05) to P.M. and the US Public Health Service for support of B.A.P. through National Institutes of Health (NIH) grant R01GM61807.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aris JP, Blobel G (1991) Isolation of yeast nuclei. Methods Enzymol 194: 735–749 [DOI] [PubMed] [Google Scholar]
- Asher G, Shaul Y (2005) p53 proteasomal degradation: poly-ubiquitination is not the whole story. Cell Cycle 4: 1015–1018 [DOI] [PubMed] [Google Scholar]
- Asher G, Lotem J, Cohen B, Sachs L, Shaul Y (2001) Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci USA 98: 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Tsvetkov P, Kahana C, Shaul Y (2005) A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev 19: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Reuven N, Shaul Y (2006) 20S proteasomes and protein degradation ‘by default'. Bioessays 28: 844–849 [DOI] [PubMed] [Google Scholar]
- Bauer CE, Elsen S, Bird TH (1999) Mechanisms for redox control of gene expression. Annu Rev Microbiol 53: 495–523 [DOI] [PubMed] [Google Scholar]
- Delaunay A, Isnard AD, Toledano MB (2000) H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19: 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB (2002) A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481 [DOI] [PubMed] [Google Scholar]
- Deller S, Macheroux P, Sollner S (2008) Flavin-dependent quinone reductases. Cell Mol Life Sci 65: 141–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R (1998) The oxygen-responsive NIFL–NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in γ-proteobacteria. Arch Microbiol 169: 371–380 [DOI] [PubMed] [Google Scholar]
- Garate M, Wong RP, Campos EI, Wang Y, Li G (2008) NAD(P)H quinone oxidoreductase 1 inhibits the proteasomal degradation of the tumour suppressor p33(ING1b). EMBO Rep 9: 576–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Huber R (2005) Purification, crystallization, and X-ray analysis of the yeast 20S proteasome. Methods Enzymol 398: 329–336 [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Haid A, Suissa M (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Methods Enzymol 96: 192–205 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM (1984) Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J 219: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC, McDowall A, Schimmang T (1988) Nucleolar and nuclear envelope proteins of the yeast Saccharomyces cerevisiae. Eur J Cell Biol 46: 554–563 [PubMed] [Google Scholar]
- Liger D, Graille M, Zhou CZ, Leulliot N, Quevillon-Cheruel S, Blondeau K, Janin J, van Tilbeurgh H (2004) Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J Biol Chem 279: 34890–34897 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Nevitt T, Pereira J, Azevedo D, Guerreiro P, Rodrigues-Pousada C (2004a) Expression of YAP4 in Saccharomyces cerevisiae under osmotic stress. Biochem J 379: 367–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt T, Pereira J, Rodrigues-Pousada C (2004b) YAP4 gene expression is induced in response to several forms of stress in Saccharomyces cerevisiae. Yeast 21: 1365–1374 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M (1996) Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA 93: 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner S, Nebauer R, Ehammer H, Prem A, Deller S, Palfey BA, Daum G, Macheroux P (2007) Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification. FEBS J 274: 1328–1339 [DOI] [PubMed] [Google Scholar]
- Tedeschi G, Chen S, Massey V (1995) DT-diaphorase. Redox potential, steady-state, and rapid reaction studies. J Biol Chem 270: 1198–1204 [DOI] [PubMed] [Google Scholar]
- Toone WM, Morgan BA, Jones N (2001) Redox control of AP-1-like factors in yeast and beyond. Oncogene 20: 2336–2346 [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11: 493–536 [DOI] [PubMed] [Google Scholar]