Abstract
The fungus, Candida albicans, and the bacterium, Pseudomonas aeruginosa, are opportunistic human pathogens that have been coisolated from diverse body sites. Pseudomonas aeruginosa suppresses C. albicans proliferation in vitro and potentially in vivo but it is the C. albicans hyphae that are killed while yeast cells are not. We show that hyphal killing involves both contact-mediated and soluble factors. Bacterial culture filtrates contained heat-labile soluble factors that killed C. albicans hyphae. In cocultures, localized points of hyphal lysis were observed, suggesting that adhesion and subsequent bacteria-mediated cell wall lysis is involved in the killing of C. albicans hyphae. The glycosylation status of the C. albicans cell wall affected the rate of contact-dependent killing because mutants with severely truncated O-linked, but not N-linked, glycans were hypersensitive to Pseudomonas-mediated killing. Deletion of HWP1, ALS3 or HYR1, which encode major hypha-associated cell wall proteins, had no effect on fungal susceptibility.
Keywords: Candida, cell wall, glycosylation, hypha, Pseudomonas aeruginosa
Introduction
Candida albicans is a fungus that is part of the normal human commensal microbial communities, but it is also an opportunistic pathogen that can cause life-threatening disseminated infections and superficial mucosal infections such as oral and vaginal thrush. The ability of the fungus to alter its morphology interchangeably between yeast, pseudohyphal and true hyphal growth forms is thought to contribute to pathogenicity (Liu, 2002; Saville et al., 2003; Zheng et al., 2004). Yeast, pseudohyphae and true hyphae have been observed together at infected sites and in biofilms formed by the fungus on the surface of invasive surgical devices (Baillie & Douglas, 1999; Gow, 2002; Netea et al., 2006).
At most body sites, C. albicans coexists alongside other microorganisms. The size of the fungal population can be manipulated by suppressing or supporting the growth of commensal enteric bacteria (Payne et al., 2003), suggesting that C. albicans proliferation in the gut is controlled by the bacterial microbial communities. Candida albicans and the opportunistic bacterial pathogen, Pseudomonas aeruginosa, have been coisolated from sputum from patients with cystic fibrosis (CF) (Bakare et al., 2003). Pseudomonas aeruginosa and C. albicans were also coisolated from patients undergoing surgery and growth of C. albicans was observed in sputum samples only after the bacterium had been eradicated (Kerr, 1994). In a study of C. albicans and P. aeruginosa coisolated from the lungs of CF patients, growth of the fungus was inhibited in vitro by the coisolated bacterial strain (Kerr, 1994), suggesting that the bacterium was able to directly suppress the growth of C. albicans.
Pseudomonas aeruginosa has an extensive repertoire of virulence determinants and is antagonistic to a wide range of microorganisms (Tan et al., 1999). In vitro studies of its antifungal activity showed that P. aeruginosa inhibited the growth of Aspergillus fumigatus and Candida spp. (Grillot et al., 1994; Kerr, 1994). The yeast form of C. albicans is however resistant to P. aeruginosa (Hogan & Kolter, 2002). In a study of direct interactions between a C. albicans monomorphic tup1 mutant that is constitutively hyphal, P. aeruginosa was found to form a biofilm on hyphae and to selectively kill them (Hogan & Kolter, 2002). Suppression of fungal growth has been correlated with production of the bacterial phenazine derivatives, pyocynanin and 1-hydroxyphenazine, in culture filtrates (Kerr et al., 1999). Conversely, pyocyanin and the quorum-sensing molecule, 3-oxo-C12 homoserine lactone, can promote growth of C. albicans yeast forms in vitro (Kerr et al., 1999; Hogan et al., 2004) leading to their escape from P. aeruginosa killing. To date it is unclear what mediates colonization and killing of C. albicans hyphae by P. aeruginosa. In this study, the role of cell-surface glycosyl groups and the presence of associated hypha-specific proteins were evaluated as potential facilitators of contact-mediated killing of C. albicans hyphae by P. aeruginosa.
Materials and methods
Organisms and media
The C. albicans strains used in this study are listed in Table 1. Candida albicans was maintained on solid YPD [1% (w/v) yeast extract (Oxoid, Unipath, Basingstoke, UK), 2% (w/v) mycological peptone (Oxoid), 2% (w/v) glucose, 2% (w/v) Agar Technical No. 3 (Oxoid)]. Candida albicans was grown overnight in SD minimal medium [yeast nitrogen base without amino acids, with ammonium sulfate (Bio101, Carlsbad, California), 2% (w/v) glucose] at 30 °C. Hyphal cells were induced by growth in RPMI-1640. A wild-type P. aeruginosa strain, ATCC27853, was used in all experiments and was grown in Luria–Bertani (LB) medium [1% (w/v) tryptone, 1% (w/v) NaCl, 0.5% (w/v) yeast extract] at 37 °C.
Table 1.
Survival of Candida albicans cell surface mutants cultured with Pseudomonas aeruginosa in hyphal growth conditions
| Strain | Genotype | Description | Mean survival time (days) | References |
|---|---|---|---|---|
| NGY152 | CAI4/CIp10-URA3 | Control strain | 3.7 ± 0.8 | Brand et al. (2004) |
| Ca44 | hyr1Δ | Mutant lacking a hypha-specific cell wall protein | 3.7 ± 0.6 | Bailey et al. (1996) |
| Ca86 | als3Δ | Mutant lacking a hypha-specific cell wall protein | 3.0 ± 0.0 | Hoyer et al. (1998) |
| CAH7-1A1E2 | hwp1Δ | Mutant lacking a hypha-specific cell wall protein | 4.0 ± 0.8 | Staab et al. (1999) |
| BCa2-10 | tup1Δ | Mutant of transcriptional repressor of hypha-specific genes | 1.8 ± 0.0* | Braun & Johnson (1997) |
| NGY337 | mnt1Δ/mnt2Δ | O-glycosylation double mutant | 2.3 ± 0.5* | Munro et al. (2005) |
| NGY158 | mnt1Δ | O-glycosylation mutant | 2.7 ± 0.6* | Munro et al. (2005) |
| NGY145 | mnt2Δ | O-glycosylation mutant | 2.7 ± 0.6* | Munro et al. (2005) |
| NGY204 | och1Δ | N-glycosylation mutant | 3.7 ± 0.6 | Bates et al. (2006) |
| DH15 | mnn4Δ | Glycosylation mutant lacking phosphomannan in N-linked glycan | 3.3 ± 0.6 | Hobson et al. (2004) |
| NGY355 | pmr1Δ | Golgi ATPase mutant, partially deficient in O- and N-glycosylation | 3.5 ± 0.5 | Bates et al. (2005) |
| NGY146 | mnt3Δ | N-linked glycosyl-transferase mutant | 4.0 ± 0.0 | Unpublished |
| NGY313 | mnt4Δ | N-linked glycosyl-transferase mutant | 3.0 ± 0.0 | Unpublished |
| NGY147 | mnt5Δ | N-linked glycosyl-transferase mutant | 3.7 ± 0.6 | Unpublished |
Candida albicans hyphae were mixed with P. aeruginosa ATCC27853 and the viable cell population determined by daily plating. All fungal strains were Ura+ (Bain et al., 2001). The mean survival time ± SD (n=3) in cocultures was defined as the time point at which the viable concentration of C. albicans was reduced to <0.1% of the original population.
Significant difference from the control (tup1ΔP=<0.001, mnt1ΔP=0.027, mnt2ΔP=0.027, mnt1Δ/mnt2ΔP=<0.001).
Survival assays
Candida albicans cells from overnight cultures were diluted to 1 × 106 cells mL−1 in 10 mL RPMI-1640 with 25 mM HEPES and NaHCO3 (Sigma-Aldrich, Dorset, UK), supplemented with 0.3 mg mL−1l-glutamine, and grown at 37 °C for 3 h to induce hyphal growth. Pseudomonas aeruginosa cells from an overnight culture were inoculated into 50 mL 1 : 3 LB : RPMI-1640 (described above) and grown to an OD650 nm of 1.25 at 37 °C. A 25 mL sample was passed through a 0.2 μm PES filter unit (Nalge Nunc International, New York) to generate sterile conditioned medium. Samples consisting of 2.5 mL aliquots of P. aeruginosa culture or sterile conditioned medium were added to hyphal cells of C. albicans in shake flasks or sterile tubes. The concentration of viable C. albicans cells was determined as mean CFUs by plating triplicate 5 μL samples on YPD plates containing tetracycline (60 μg mL−1; Progen Industries Ltd, Queensland, Australia), gentamicin (30 μg mL−1; Duchefa, Haarlem, the Netherlands) and chloramphenicol (30 μg mL−1; Duchefa) to suppress the growth of P. aeruginosa. As CFUs decreased, 50 μL aliquots were spread on plates. The limit of detection was c. 20 CFU mL−1. Plates were incubated for 24–48 h at 30 °C. Results were expressed as the mean time point at which the viable cell concentration decreased to <0.1% of the initial population. Three independent samples were analyzed per strain in each experiment and experiments were carried out on three independent occasions. To compare the survival of cells grown as yeasts, C. albicans from an overnight culture was inoculated into YPD, instead of RPMI-1640, and grown at 30 °C for 3 h before addition of P. aeruginosa. The anti-Candida activity of medium supernatants was determined using 4-day cocultures. Media were filtered with a 0.2-μm PES filter unit, divided into aliquots and C. albicans hyphae were inoculated, with or without P. aeruginosa, into filtered or autoclaved–filtered media. For controls, C. albicans hyphae were inoculated into filtered media from 4-day cultures of the fungus alone. The Dunnett t-test in the spss software package (SPSS, Woking, UK) was used for statistical analyses.
Microscopy
The wells of a sterile Lab-Tek four-well glass slides (Scientific Laboratory Supplies, Nottingham, UK) were coated with fetal bovine serum (Biosera) at 37 °C for 24 h and washed with 0.9 mL phosphate-buffered saline (PBS) (Invitrogen, Paisley, UK). Candida albicans yeast cells (1 × 105 cells mL−1per well) were incubated at 37 °C for 3 h to induce hypha formation and adhesion. Wells were washed with PBS and 0.9 mL of an exponential-phase P. aeruginosa culture was added. Slides were incubated at 37 °C with shaking for up to 3 h. For fluorescence microscopy, 1 μL 10 mM FUN1 LIVE/DEAD stain (Molecular Probes, Leiden, the Netherlands) was added and incubation was continued in the dark for 30 min. Wells were washed with PBS and the well-housing and gasket were removed. Images were captured using an Axioplan 2 microscope (Carl Zeiss Ltd, UK) with a Hamamatsu CCD camera and analyzed with openlab 3.0.9 (Improvision Ltd, Coventry, UK). For light microscopy, images were captured using an Olympus BX50 microscope fitted with an Olympus DP11 camera. For scanning electron microscopy, hyphae and bacteria were cocultured for 48 h. Cultures were filtered through 25-mm polycarbonate filters with 12 μm pores (Costar, High Wycombe, UK) and the cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4. Cells were postfixed with 1% osmium tetroxide and dehydrated in 70%, 90%, 95% and 100% ethanol and critical point dried in CO2. After sputter-coating with gold, cells were viewed in a JEOL35CF Scanning electron microscope at a voltage of 10 kV.
Pyocyanin assay
The presence of pyocyanin was measured spectrophotometrically by the method of Essar et al. (1990). Briefly, 1 mL of a 6-day culture was extracted in triplicate in 2 mL chloroform and then 1 mL 0.2 M HCl. The OD520 nm was determined and concentration calculated using the pyocyanin molar extinction coefficient. Results were expressed as micrograms of pyocyanin per milliliter of culture (MacDonald, 1967).
Results
Pseudomonas aeruginosa ATCC27853 kills wild-type C. albicans hyphae, but not yeast
It was reported previously that P. aeruginosa killed a constitutively hyphal C. albicans mutant (tup1Δ) within 72 h in M63 medium at 30 °C (Hogan & Kolter, 2002). We found that CAI4/CIp10, a prototrophic control strain in the genetic background used to create most C. albicans mutants (Fonzi & Irwin, 1993; Brand et al., 2004), grew as yeasts in these conditions and were not killed by P. aeruginosa (data not shown). In order to test wild-type hyphae and a range of C. albicans mutants (i.e. non-tup1Δ), we grew CAI4/CIp10 in RPMI-1640 at 37 °C. This medium induces the formation of true, nonconstricted hyphae. No reversion to yeast growth was observed over the duration of the experiments, even after 4 days' coculture in the presence of high bacterial densities. Pseudomonas aeruginosa grew slowly in RPMI-1640 in the absence of C. albicans (data not shown). We compared the viability of the C. albicans control strain under hypha-inducing conditions during culture with and without P. aeruginosa. Without P. aeruginosa, C. albicans cultures reached high cell densities and tended to aggregate; hence, the viable cell concentration was likely to be underestimated. On mixing with P. aeruginosa, hyphal aggregation was abolished, thereby facilitating CFU counts by plating. However, hyphae consist of multiple cellular compartments; hence, hyphal CFUs are an underestimate of the total number of viable cells present.
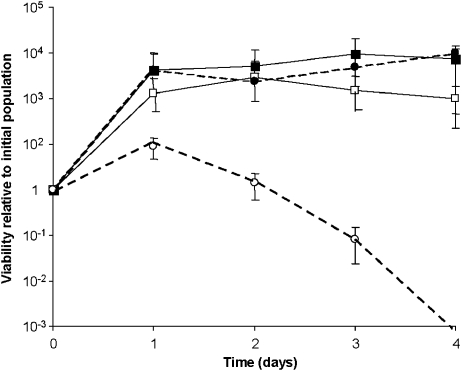
In RPMI-1640, CFUs of the control strain rose and remained in the region of 108 cells mL−1 by day 4. When incubated with P. aeruginosa ATCC27853, the mean survival time of hyphae was 3.7 days (Fig. 1). The growth rate of P. aeruginosa in YPD at 30 °C was twice that in RPMI at 37 °C (data not shown); yet the viable population of C. albicans yeast cells, grown with or without the bacterium, increased 60–100-fold within 48 h. By day 4, viable yeast cell concentrations in cocultures declined by a third (Fig. 1). This trend was observed for all C. albicans strains growing in the yeast form. The decline of yeast cell viability in YPD cocultures coincided with the appearance of pyocyanin pigment, which reached a mean concentration of 2.2±0.36 μg mL−1 (data not shown) by day 4. In contrast, the mean pyocyanin concentration in RPMI-1640 by day 4 was 0.05±0.01 μg mL−1. When grown in yeast-inducing conditions, the tup1Δ mutant, which is constitutively filamentous, followed the same growth profile as the control strain. Under hypha-inducing conditions, tup1Δ was hypersensitive to P. aeruginosa (Table 1), consistent with previously reported findings using M63 medium (Hogan & Kolter, 2002).
Fig. 1.
Pseudomonas aeruginosa kills Candida albicans hyphae, but not yeast. Candida albicans yeast cells (1 × 106 cells mL−1) were inoculated into 10 mL RPMI-1640 (circles) or YPD (squares) and incubated for 3 h at 37 or 30°C, to produce hyphae or yeast cells, respectively. Candida albicans was then cultured for 4 days at 37 or 30°C with (open symbols) or without (closed symbols) the addition of P. aeruginosa cells. The fungal viable cell population was determined daily by plating on YPD solid medium containing antibacterial agents. Error bars are SD (n=3).
Defective C. albicans O-glycosylation increases susceptibility to P. aeruginosa
Because yeast cells were resistant to P. aeruginosa, we investigated whether components of the hypha cell wall influenced susceptibility to the bacterium. Hypha-specific cell wall proteins of C. albicans are involved in adhesion and aggregation (Bailey et al., 1996; Hoyer et al., 1998; Staab et al., 1999; Phan et al., 2007) and mannan components of the cell surface have been implicated in adhesion of C. albicans to several cell types (Timpel et al., 2000; Dalle et al., 2003; Munro et al., 2005; Bates et al., 2006). Therefore, we tested a range of mutants that lacked hypha-specific cell wall mannoproteins and others that lacked specific glycosyl epitopes. Mutant C. albicans strains that lacked the hypha-specific proteins Hyr1p, Hwp1p and Als3p or enzymes involved in N-glycosylation (Och1p, Mnn4p, Mnt3p, Mnt4p, and Mnt5p) of surface glycoproteins, were assayed for altered rates of killing by P. aeruginosa but none showed increased resistance or sensitivity to the bacterium (Table 1). However, the survival of mnt1Δ, mnt2Δ and mnt1Δ/mnt2Δ mutants with truncated O-linked mannan was significantly reduced in the presence of P. aeruginosa as compared with the control strain, suggesting that O-mannan is protective against the P. aeruginosa killing activity. Survival of the yeast form of the O-glycosylation mutants was the same as the control strain (not shown).
Medium filtrates have anti-C. albicans activity
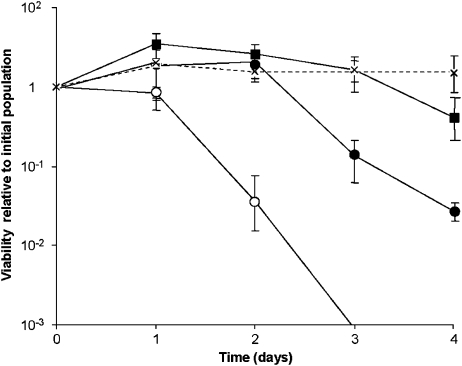
We assessed whether the presence of P. aeruginosa in the culture medium was required for the killing of C. albicans and whether secreted antifungal factors contributed to fungal cell death. Candida albicans was inoculated with or without P. aeruginosa into filtered conditioned medium, obtained by coculturing the two organisms in RPMI-1640 for 4 days (Pa–Ca CM). Candida albicans was also inoculated into the same filtrate that had been heat treated by autoclaving (Pa–Ca HCM) or into a medium in which the fungus alone had been cultured for 4 days (Ca-only CM), as a control. Viable C. albicans hyphae were not detectable by day 3 in Pa–Ca CM containing P. aeruginosa (Fig. 2) and in the same medium, but without P. aeruginosa, C. albicans viability declined by 97% by day 4. When inoculated into the same medium that had been heat treated (Pa–Ca HCM), viability fell by 59% by day 4. In contrast, the CFUs for C. albicans inoculated into Ca-only conditioned medium increased by 45% over 4 days. The decline in viability in coculture filtrate was therefore likely to be linked to P. aeruginosa-specific factors and not to nutrient deprivation. The rate of C. albicans cell death was therefore highest in the presence of the bacterium but cell-free filtrates of Pa–Ca CM and Pa–Ca HCM also significantly affected viability (P=0.01 and 0.033, respectively).
Fig. 2.
The susceptibility of Candida albicans to the presence of Pseudomonas aeruginosa, its heat-labile and heat-stable secreted factors. Candida albicans hyphae were inoculated into 10 mL cell-free conditioned medium from a 4-day C. albicans–P. aeruginosa coculture (Pa–Ca CM). Hyphae were incubated either with (open circles) or without (closed circles) the addition of fresh P. aeruginosa. Hyphae were also inoculated into the same filtrate that had been heat treated to denature proteins (Pa–Ca HCM – closed squares) or into filtrate generated from a 4-day C. albicans-only culture as a control (Ca-only CM – broken line). Error bars are SD (n=3).
Pseudomonas aeruginosa adhere to live C. albicans hyphae and cause localized cell lysis
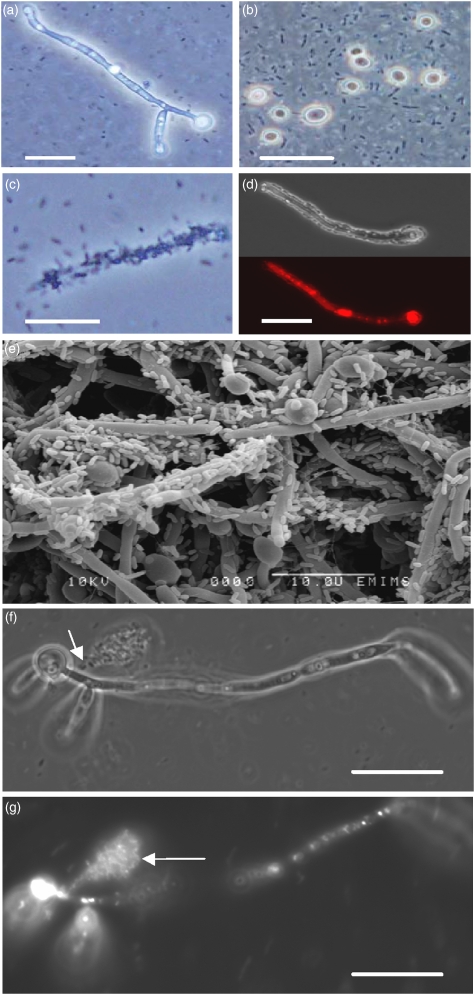
The interactions between P. aeruginosa and C. albicans wild-type yeast and hyphae were studied by microscopy. Motile bacteria did not immediately associate with hyphae upon inoculation of P. aeruginosa into a hyphal culture of C. albicans in RPMI-1640. After 1, 2, 3 and 48 h coincubation in this medium, hyphae were observed with and without attached bacteria (Fig. 3a, c and e). Pseudomonas aeruginosa adhered preferentially to specific hyphae at all time points. There appeared to be no preferred attachment site relative to the length of the hypha, the presence or absence of a branch or any other morphological parameter. The vital fluorescent stain FUN1 LIVE/DEAD, applied after colonization, showed that hyphal cell death did not occur before the development of a substantial biofilm on the surface of hyphae (Fig. 3d). Release of intracellular material was observed from sites of bacterial foci on the cell wall (Fig. 3f), suggesting that hyphal lysis occurred as a result of exposure to P. aeruginosa. Fluorescent staining of the fungal cytosol confirmed that exudate was released from hyphae into the medium (Fig. 3g). Consistent with previous studies (Hogan & Kolter, 2002), no bacteria were seen adhering to C. albicans yeast cells, even after 7 days' coculture (Fig. 3b).
Fig. 3.
Candida albicans hyphae – adhesion, colonization and lysis by Pseudomonas aeruginosa. Adhered hyphae were viewed by light microscopy after 3 h coincubation with P. aeruginosa in RPMI-1640. Most hyphae and all yeast cells (b) had no observable adherent bacteria (a, b); yet some hyphae were completely colonized by P. aeruginosa (c, d). After 48 h, yeast cells and some hyphae were not colonized by P. aeruginosa (e). Treatment after colonization with FUN1 LIVE/DEAD red vacuolar stain revealed that hyphae were alive before adhesion by bacteria (d). Cytosol appeared to be released from points on the hyphal surface (arrow, f). The exudate released into the medium (arrow, g) from the point of lysis was confirmed as cytosolic material by FUN1 staining. Scale bar=10 μm.
Discussion
The fungus, C. albicans, and the bacterium, P. aeruginosa, are frequently coisolated from the sputum of CF patients (Kerr, 1994) and it is therefore possible that the two populations interact directly in the respiratory tract (Hogan & Kolter, 2002). Resistance of the yeast form and susceptibility of the hyphal form of C. albicans to killing by P. aeruginosa has been reported (Hogan & Kolter, 2002). Induced reversion to the yeast form by P. aeruginosa secretion of 3-oxo-C12-homoserine lactone in media that otherwise favored hyphal growth resulted in survival of C. albicans (Hogan et al., 2004). Initial studies of this interaction used the transcription factor tup1 mutant to generate constitutive pseudohyphal growth and thereby avoid the reversion to yeast. We induced hypha formation in a wild-type background in RPMI-1640 medium and were able to cultivate true hyphae that grew without reversion to yeast or pseudohyphal growth for a minimum of 5 days. While the tup1Δ mutant was hypersensitive to killing by P. aeruginosa under hypha-inducing conditions, it was almost as resistant to killing as the control strain under yeast-inducing conditions. This suggests that yeast-inducing conditions are protective for the constitutively filamentous tup1Δ mutant. Under these conditions, P. aeruginosa may not produce key antifungal factors, or alternatively, a hypha-specific property required for susceptibility is not induced in the tup1Δ mutant. In this case, the susceptibility of C. albicans to P. aeruginosa is due to a Tup1-independent factor.
Upon inoculation, bacteria did not immediately associate with hyphae and hyphae were not all evenly colonized with bacteria. Instead, bacteria formed foci on selected hyphae. This pattern of selective adhesion by bacterial cells is markedly different from the rapid adhesion of C. albicans to host buccal epithelial cells (Watts et al., 1998). Complete colonization of the hyphal surface was observed within 3 h of coincubation. Live/dead staining of hyphae showed that they were viable before colonization. Microscopy revealed points of localized lysis on hyphae where intracellular material escaped into the surrounding medium. Taken together, these observations suggest an active role for P. aeruginosa in affecting the integrity of the C. albicans cell wall. Cell wall lysis may therefore occur due to the localized accumulation of cytotoxic molecules or degradative enzymes within bacterial foci. The formation of foci of killing by P. aeruginosa has also been reported in investigations of adhesion to confluent mammalian cells, suggesting that bacterial aggregation and biofilm formation is important for efficient killing (Apodaca et al., 1995).
Adhesion of P. aeruginosa to C. albicans is likely to be mediated by the outer, glycoprotein-rich layer of the fungal cell wall. It has been reported that P. aeruginosa requires glycoproteins as adhesion sites on host kidney cells (Apodaca et al., 1995). Candida glycans are also known ligands for recognition by pattern recognition receptors of the immune system (Netea et al., 2006, 2008). Loss of the hypha-specific proteins Hyr1p, Hwp1p and Als3p or truncation of the wild-type N-linked glycan structure did not alter resistance or susceptibility to P. aeruginosa. In contrast, mutant hyphae that were deficient in O-glycosylation were killed significantly faster than the control strain and the severity of O-mannan truncation correlated with increased susceptibility, suggesting that there is a specific role for O-glycans in resistance to P. aeruginosa. The truncation of O-glycans may also result in the exposure of a high-affinity adhesion site for the bacterium or may cause the loss, mislocalization or misfolding of specific surface proteins that are required for fungal cell wall integrity during colonization of P. aeruginosa. Therefore, both soluble secreted and insoluble surface factors participate in the selective killing of C. albicans hyphae by P. aeruginosa.
Acknowledgments
This work was funded by the BBRSC (BB/E008372/1), Wellcome Trust (080088) and a University of Aberdeen-NCIMB studentship award to N.A.R.G. and F.C.O. to support J.D.B.
Statement
OnlineOpen articles are made available in accordance with the terms of the Creative Commons Deed, Attribution 2.5 (further details available from http://www.creativecommons.org), which allows Open Access dissemination of the article, but does not permit commercial exploitation or the creation of derivative works.
References
- Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D, Mostov KE, Wiener-Kronish J. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DA, Feldmann PJF, Bovey M, Gow NAR, Brown AJP. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999;48:671–679. doi: 10.1099/00222615-48-7-671. [DOI] [PubMed] [Google Scholar]
- Bain JM, Stubberfield C, Gow NAR. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol Lett. 2001;204:323–328. doi: 10.1111/j.1574-6968.2001.tb10905.x. [DOI] [PubMed] [Google Scholar]
- Bakare N, Rickerts V, Bargon J, Just-Nübling G. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses. 2003;46:19–23. doi: 10.1046/j.1439-0507.2003.00830.x. [DOI] [PubMed] [Google Scholar]
- Bates S, MacCallum D, Bertram G, Munro CA, Hughes HB, Buurman ET, Brown AJ, Odds FC, Gow NA. Candida albicans Pmr1p: a secretory pathway Ca2+/Mn2+ P-type ATPase required for glycosylation and virulence. J Biol Chem. 2005;280:23408–23415. doi: 10.1074/jbc.M502162200. [DOI] [PubMed] [Google Scholar]
- Bates S, Hughes HB, Munro CA, et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJP, Gow NAR, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Dalle F, Jouault T, Trinel PA, Esnault J, Mallet JM, d'Athis P, Poulain D, Bonnin A. beta-1,2- and alpha-1,2-linked oligomannosides mediate adherence of Candida albicans blastospores to human enterocytes in vitro. Infect Immun. 2003;71:7061–7068. doi: 10.1128/IAI.71.12.7061-7068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR. Cell biology and cell cycle of Candida. In: Calderone RA, editor. Candida and Candidiasis. Washington, DC: ASM Press; 2002. pp. 145–158. [Google Scholar]
- Grillot R, Portmann-Coffin V, Ambroise-Thomas P. Growth inhibition of pathogenic yeasts by Pseudomonas aeruginosa in vitro: clinical implications in blood cultures. Mycoses. 1994;37:343–347. doi: 10.1111/myc.1994.37.9-10.343. [DOI] [PubMed] [Google Scholar]
- Hobson RP, Munro CA, Bates S, MacCallum DM, Cutler JE, Heinsbroek SE, Brown GD, Odds FC, Gow NA. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J Biol Chem. 2004;279:39628–39635. doi: 10.1074/jbc.M405003200. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. Pseudomonas–Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Kerr JR. Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J Clin Microbiol. 1994;32:525–527. doi: 10.1128/jcm.32.2.525-527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JR, Taylor GW, Rutman A, Hoiby N, Cole PJ, Wilson R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol. 2002;292:299–311. doi: 10.1078/1438-4221-00215. [DOI] [PubMed] [Google Scholar]
- MacDonald JC. Pyocyanine. In: Gottlieb D, Shaw PD, editors. Antibiotics. New York: Springer-Verlag; 1967. pp. 52–65. [Google Scholar]
- Munro CA, Bates S, Buurman ET, et al. Mnt1p and Mnt2p of Candida albicans are functionally redundant α-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J Biol Chem. 2005;280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Gow NAR, Munro CA, et al. Immune sensing of Candida albicans: cooperative recognition of mannans and glucan by lectin and Toll-like receptors. J Clin Invest. 2006;6:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg B-J, Gow NAR. An integrated model of recognition of Candida albicans in innate immunity. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- Payne S, Gibson G, Wynne A, Hudspith B, Brostoff J, Tuohy K. In vitro studies on colonization resistance of the human gut microbiota to Candida albicans and the effects of tetracyclin and Lactobacillus plantarum LPK. Curr Issues Intest Microbiol. 2003;4:1–8. [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1537. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausebel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpel C, Zink S, Strahl-Bolsinger S, Schroppel K, Ernst JF. Morphogenesis, adhesive properties and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J Bacteriol. 2000;182:3063–3071. doi: 10.1128/jb.182.11.3063-3071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts HJ, Cheah FHS, Hube B, Sanglard D, Gow NAR. Altered adherence in strains of Candida albicans with null mutations in secreted aspartic proteinase genes. FEMS Microbiol Lett. 1998;159:129–135. doi: 10.1111/j.1574-6968.1998.tb12851.x. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang Y, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]