Abstract
Recent high profile clinical trials demonstrate that microarray-based gene expression profiling has the potential to become an important tool for predicting prognosis in breast cancer. Earlier work in our laboratory using mouse models and human breast cancer populations has enabled us to demonstrate that metastasis susceptibility is an inherited trait. This same combined approach facilitated the identification of a number of candidate genes that when dysregulated, have the potential to induce prognostic gene expression profiles in human datasets. To investigate if these gene expression signatures were of somatic or germline origin, and to assess the contribution of different cell types to the induction of these signatures, we have performed a series of expression profiling experiments in a mouse model of metastatic breast cancer. These results demonstrate that both the tumor epithelium and invading stromal tissues contribute to the development of prognostic gene signatures. Furthermore, analysis of normal tissues and tumor transplants suggests that prognostic signatures result from both somatic and inherited components, with the inherited components being more consistently predictive.
Introduction
Microarray technology has become an important tool to define the mechanisms driving the most lethal forms of cancer: those that disseminate beyond the primary site and form distant malignancies. In the case of most solid tumors, these metastatic lesions are difficult to manage with currently available therapies (1-8), and a clearer understanding of metastatic progression is therefore necessary in order to develop more effective therapeutic strategies (9). The development of microarray-based systems for classifying individuals at higher or lower risk of developing metastatic disease is gaining more prominence in terms of breast cancer therapy (10, 11). One of the primary aims of utilizing this type of global expression based profiling as a prognostic tool in breast cancer is to identify those women who are more likely to develop secondary disease, which in turn would facilitate swift and aggressive initiation of adjuvant anti-metastatic therapy. Additionally, microarray-based prognostic assessment could spare women with gene expression profiles indicating a lower risk of metastatic disease from needless therapy.
Most of these investigations have been based on the assumption that the metastasis-predictive gene expression signatures are the result of early somatic mutation (12, 13). However, studies from our laboratory have demonstrated that inherited polymorphism also play a role in metastatic progression (14-18), and that this germline variation drives the establishment of gene expression signatures that distinguish tumors with varying propensities to metastasize (19). More recently we have identified a number of genes with differential functionality, presumably as a consequence of germline polymorphism, in recombinant inbred mice derived from founder strains with inherently different metastatic capacities (14, 20, 21). We subsequently demonstrated that ectopic expression of these genes could induce gene signatures in mouse tumor epithelium that predict outcome in human breast cancer clinical samples. These studies have provided some preliminary evidence to suggest that metastasis-predictive gene signatures may be induced by germline polymorphism of metastasis susceptibility genes. However, these initial studies do not enable dissection of the contribution of different cell types in the bulk tumor or the relative contribution of somatic mutation versus germline variation in the establishment of these expression patterns.
The aim of the current study is to gain a better understanding of the origins of the metastasis predictive gene expression profiles. To achieve this aim, we have utilized a mouse model system to define the factors driving the induction of metastasis-predictive gene expression signatures. Our studies suggest that the signatures are likely due to a combination of pre-existing signatures established by inherited factors present in all tissues as well as somatic mutations within the tumor epithelium.
Materials and Methods
Primary tissue extraction and processing for Affymetrix GeneChip analysis
F1 hybrids of differing metastatic propensities were generated by crossing the polyoma middle T (PyMT) mouse model of mammary tumorigenesis (FVB/NTgN(MMTV-PyVT)634Mul) to either the high metastatic potential AKR/J strain or the low metastatic potential DBA/2J strain (17). PyMT male animals were bred to female DBA/2J or AKR/J females to produce transgene-positive F1 hybrid female progeny. These virgin transgene-positive F1 hybrid females were euthanized at 100 days of age for tissue harvesting. Transgene-negative females were used for harvesting of normal tissues. RNA extraction and Affymetrix GeneChip analysis was performed as previously described (19).
Generation of mouse tissue gene signatures
Analysis of mouse tissue microarray data were performed using BRB-ArrayTools Version: 3.5.0 - Patch_1. Signatures distinguishing the tissues from the high- or low-metastatic genotypes were developed using the Class Comparison tool. The data were pre-filtered to include only probe sets whose log-ratio variation were p < 0.01, and included in the signature only if univariate analysis for differential expression between the genotypes was p< 0.001. For the spleen and thymus samples the univariate p-value thresholds were p < 0.0001 or p< 0.00001, respectively to truncate the number of probe sets included in the signature. Gene expression data from these studies can be accessed at the NCBI GEO database (22) under the accession GSE13231.
Tumor transplant assays
Two days before injection, highly metastatic Mvt-1 mouse mammary tumor cells (23) were passaged and permitted to grow to 80−90% confluence. The cells were then washed with PBS and trypsinized, collected, washed twice with cold PBS, counted in hemocytometer and resuspended at a concentration of 106 cells/ml. One hundred thousand cells (100 μl) were injected into the fourth mammary gland of 6 week old virgin FVB/NJ female mice. The mice were then aged for 28 days and euthanized by anesthetic overdose. The 28 day time point was selected based on previously observed tumor growth and metastatic capacities (18, 24). Tumors were dissected and weighed. Lungs were isolated and surface metastases enumerated using a dissecting microscope. These experiments were performed in compliance with the National Cancer Institute's Animal Care and Use Committee guidelines.
Generation of human gene signatures
Human gene signatures were generated using Affymetrix Netaffx tools (http://www.affymetrix.com/analysis/index.affx). Mouse tissue signature probe sets generated by the Class Comparison analysis of BRB Array Tools were used to query the database using the Batch Query tool of the Exon/Gene Array Expression toolset. Human probe sets corresponding to the individual mouse tissue signatures were identified using the Show Orthologs tool and the Human Genome U133 Plus 2.0 Array probe sets downloaded for further analysis. Generation of the Rosetta Hu25K signatures was performed by matching the mouse gene symbols to the human gene symbols in the Hu25K annotation data.
Analysis of human gene expression datasets
Analysis of human gene expression datasets was performed as previously described (14, 20, 25). Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam (http://linus.nci.nih.gov/BRB-ArrayTools.html). The GSE1456 (26), GSE2034 (1), GSE3494 (27) and GSE4922 (28) datasets were downloaded from the NCBI Gene Expression Omnibus website (www.ncbi.nlm.nih.gov/projects/geo/). Where samples were present in more than GEO submission (e.g. GSE1456 and GSE4922) duplicate samples were excluded from one or more of the datasets to ensure independence among the datasets. The Rosetta dataset (10) was downloaded from the Rosetta Inpharmatics website (http://www.rii.com/publications/2002/vantveer.html). Expression data were loaded into BRB ArrayTools using the Affymetrix GeneChip Probe Level Data option or the Data Import Wizard. The equivalent human tissue gene signatures used to filter the expression data using the Select Gene Subset tool to exclude any probe set that was not a component of the relevant tissue gene expression signature, and to eliminate any probe set whose expression variation across the dataset was p ≥ 0.01.
Unsupervised clustering of each dataset was performed using the Samples Only clustering option of BRB ArrayTools. Clustering was performed using average linkage, the centered correlation metric and center the genes analytical option. Samples were assigned into two groups based on the first bifurcation of the cluster dendogram, and Kaplan-Meier analysis performed using the Survival module of the software package Statistica version 7.1 (StatSoft, Inc). Significance of outcome analyses was performed using the Cox F-test. Hazard ratios for the genes in the tissue gene expression signatures that correlated with outcome in the human datasets were identified using the Find Genes Correlated with Survival tool in the Survival Analysis toolset of BRB ArrayTools.
Survival analysis was performed using the publicly available outcome data. Where available, distant metastasis free survival was used (GSE2034 and Rosetta datasets). Since death by breast cancer is associated primarily with metastatic disease rather than primary tumors or local regional relapse, overall survival or death due to breast cancer was used for GSE1456 and GSE3494 datasets, respectively as a surrogate for metastatic disease. For GSE4922, only the relapse data, including both local and regional was available.
Pathway and functional category analysis
Pathway and biological functional category analysis was performed using the Ingenuity Pathways Analysis program (Ingenuity IPA 6.3−1402).
Results
Differences in tumor gene expression from high- versus low-metastatic genotype mouse tumors predicts outcome of human breast cancer
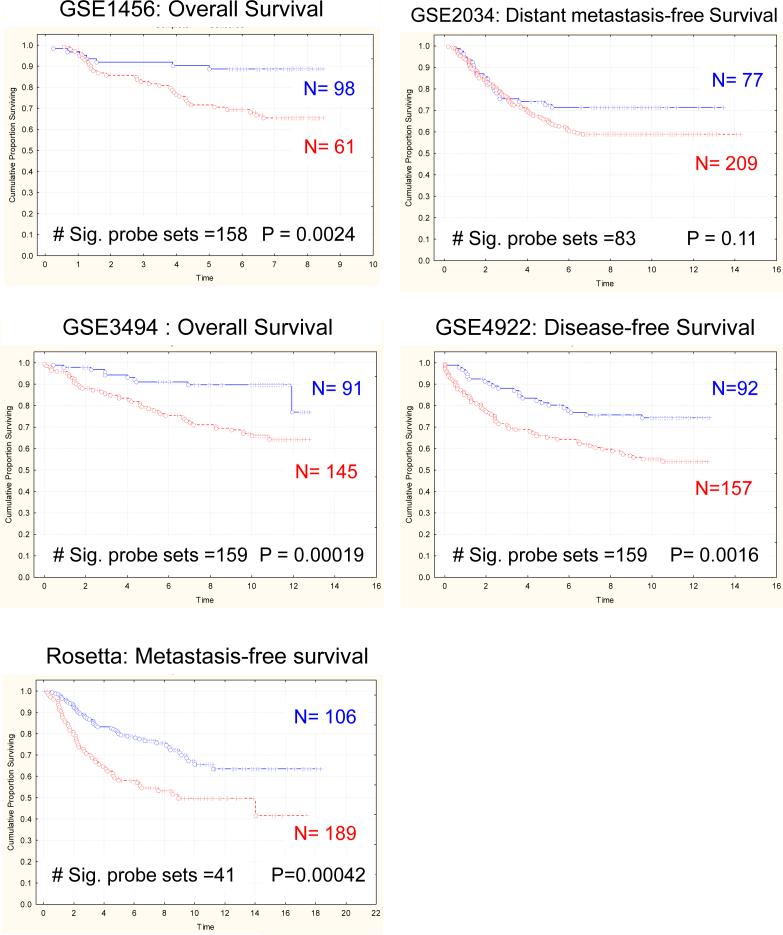
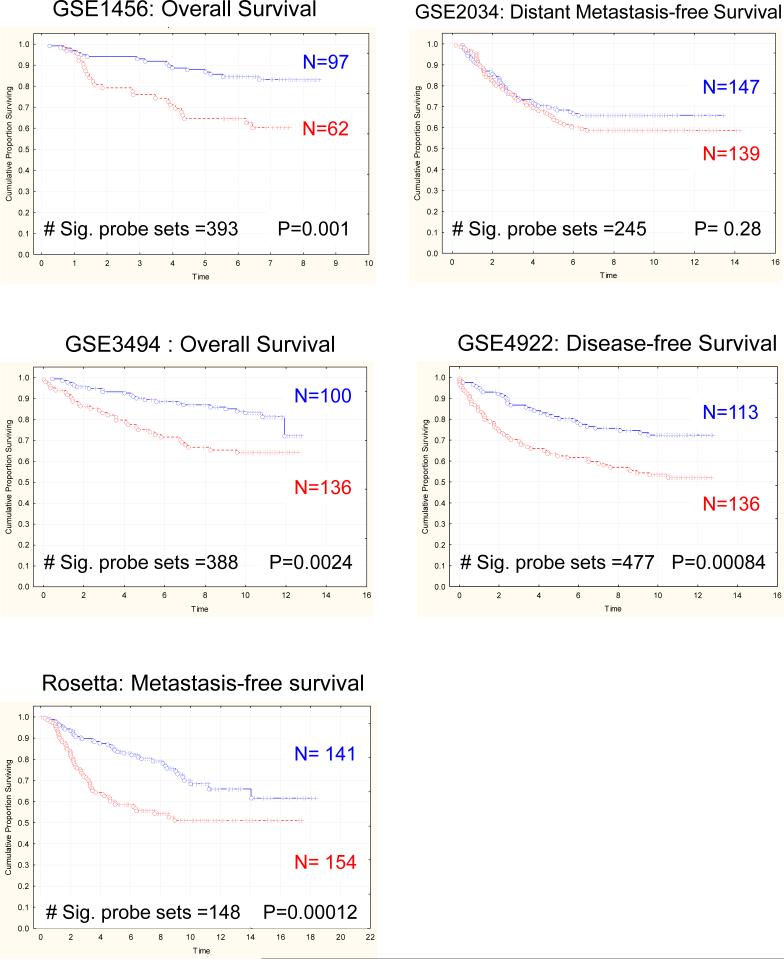
To determine whether mouse mammary tumor gene expression patterns would predict outcome in human breast cancer, gene expression profiles from high- versus low-metastatic genotype PyMT-induced tumors (17) were generated. PyMT-male animals were bred to either AKR/J (higly metastatic) or DBA/2J (poorly metastatic) females to generate tumors. Tumors from 3 independent animals of each genotype were arrayed and compared to generate a gene signature that distinguished the tumors (supplementary table S1). Probe sets in each mouse signature were converted to the orthologous human probe sets as described above (Supplemental table S2). Subsequently, signature gene expression was analyzed in publicly available gene expression datasets by performing unsupervised clustering of patient samples into two groups based on the first bifurcation of the resulting dendogram (14, 20). Kaplan-Meier analysis was then performed to determine whether steady state gene expression resulting from the genetic backgrounds upon which the primary tumor arose was sufficient to predict relapse or disease-free survival in five independent human datasets. The signature derived from differences in gene expression in tumors derived from high and low metastatic phenotype mice accurately predicted outcome in four of the five datasets (Rosetta, GSE1456, GSE3494 and GSE4922; Figure 1 & Table 1). These results indicate that mouse gene expression signatures derived from strains of different metastatic propensities were sufficient to distinguish human breast cancer patients of different outcomes.
Figure 1.
Kaplan-Meier analysis reveals that the gene signature distinguishing high- and low-metastatic spontaneous PyMT-induced mammary tumors predicts outcome in five different human breast cancer datasets. P-values were determined by log-rank analysis.
Table 1.
Univariate Analysis of Mouse Signatures on Human Breast Cancer Gene Expression Datasets
| Tissue | GSE1456 | GSE2034* | GSE3494 | GSE4922 | Rosetta | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Risk Ratio | 95% Conf. Int | Risk Ratio | 95% Conf. Int | Risk Ratio | 95% Conf. Int | Risk Ratio | 95% Conf. Int | Risk Ratio | 95% Conf. Int | |
| Spontaneous Tumor | 3.2 | 1.4 − 7.2 | - | - | 2.5 | 1.4 − 4.4 | 2.0 | 1.3 − 3.3 | 2.0 | 1.3 − 2.9 |
| Mvt-1 transplant | 4.8 | 1.9 − 12.4 | - | - | 1.7 | 1.1 − 2.9 | 1.8 | 1.2 − 2.9 | 2.2 | 1.4 − 3.5 |
| Lung | 3.1 | 1.5 − 6.3 | 1.45 | 1.0−2.2 | 1.8 | 1.1 − 2.9 | 1.7 | 1.1 − 2.6 | 1.5 | 1.0 − 2.3 |
| Spleen | 3.9 | 1.6 − 9.2 | - | - | 1.8 | 1.1 − 2.9 | 1.7 | 1.1 − 2.6 | 1.7 | 1.1 − 2.5 |
| Thymus | 1.9 | 2.4 − 19.2 | - | - | 1.9 | 1.1 − 3.1 | 2.0 | 1.3 − 3.2 | 2.0 | 1.3 − 3.0 |
| Met-1 vs DB-7 | 2.7 | 1.4 − 5.0 | - | - | 1.8 | 1.1 − 2.9 | 2.0 | 1.3 − 3.1 | 2.1 | 1.4 − 3.1 |
| Lung w/o proliferation genes | 2.8 | 1.3−6.0 | - | - | 1.9 | 1.2−3.0 | 2.1 | 1.3−3.3 | 1.6 | 1.1−2.3 |
Non-significant results for GSE2034 were not analyzed by Cox Regression
Stromal tissues contribute significantly to the induction of prognostic gene expression signatures
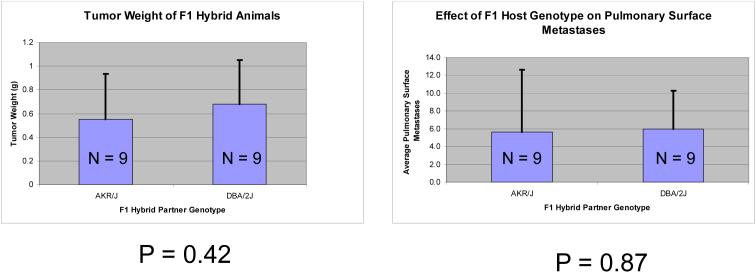
Transplant experiments were performed using the highly metastatic Mvt-1 mouse mammary tumor cell line to investigate whether the differential metastatic susceptibilities observed between AKR/J and DBA/2J mice were due to differences in the tumor epithelium, the normal stromal components, or a combination of the two.. Mvt-1 cells, which are an FVB/NJ derived epithelial line, were implanted into the mammary fat pad of the F1 progeny of FVB/NJ males bred to either the high metastatic AKR/J or the low metastatic DBA/2J females. Animals were euthanized following a 28 days incubation period, surface pulmonary metastases enumerated and primary tumors harvested for gene expression analysis. No significant differences in primary tumor weight or pulmonary metastasis were observed (Figure 2) suggesting the genetic polymorphism in the tumor epithelium, rather than the invading stroma is primarily responsible for differences in metastatic susceptibility, at least in this model system.
Figure 2.
Comparison of tumor weights and surface pulmonary metastasis counts 28 days after implantation of the highly metastatic Mvt-1 cell line into either the high metastatic susceptibility AKR/J strain or the low metastatic susceptibility DBA/2J strain.
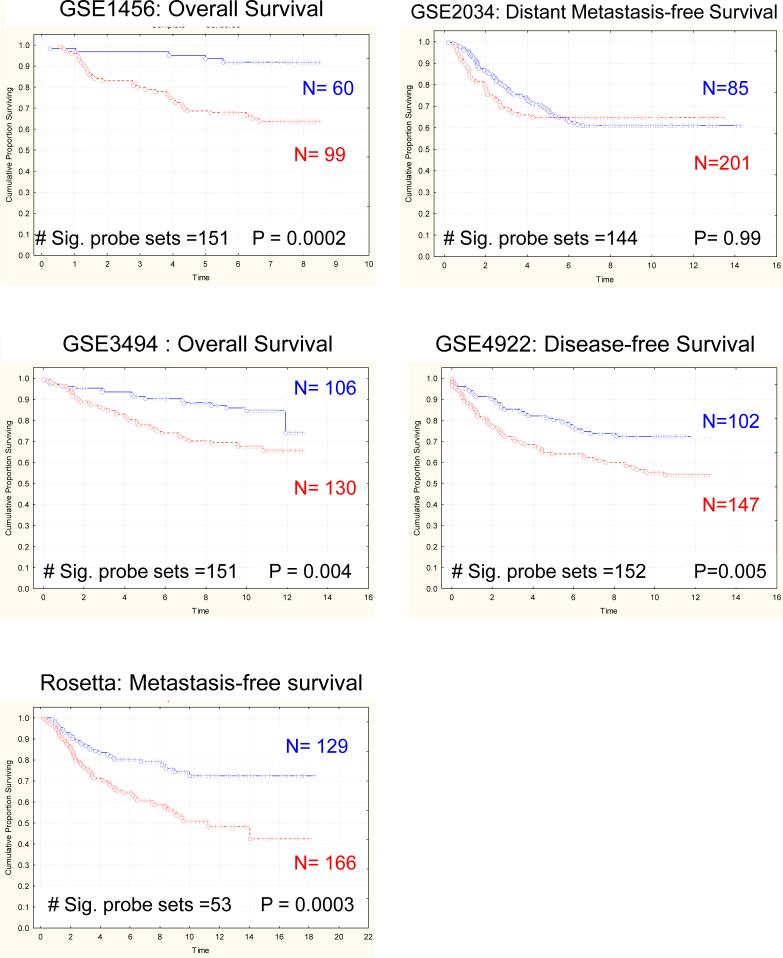
Primary tumors derived from implantation of this highly metastatic cell line into high and low metastatic genotype mice were then used to derive a gene expression signature indicative of the differences in tumor gene expression between strains (N=3 for each genotype; supplemental table S3). Since the epithelium of tumors from both strains originated from the Mvt-1 cell line, any differences in gene expression would most likely be due to either inherent differences in gene expression within host tissue components of the primary tumor, a differential response of the Mvt-1 cells to the different host genetic backgrounds, or a combination of the two. Kaplan-Meier analysis of the five human breast cancer datasets revealed that the resulting gene expression signature was capable of accurately predicting outcome in four of the human breast cancer datasets (GSE1456, GSE3494, GSE4922, Rosetta; Figure 3; supplemental table S4). These data suggest that a substantial fraction of the prognostic gene expression profiles derived tumors from the high and low metastatic potential may have their origins in the normal tissue surrounding the tumor epithelium, rather than just the invading tumor epithelium.
Figure 3.
Kaplan-Meier analysis of the gene signature derived from tumors induced by implantation of the Mvt-1 cell line into either high- or low- metastatic susceptibility mice.
Non-neoplastic tissue gene expression profiles from inbred mice strains with differing metastatic propensities can predict breast cancer outcome
There are a number of explanations as to how stromal components of the tumor contribute to prognostic gene expression signatures. One possibility is that it is solely the result of differential stromal reaction to tumor tissue (29-31). An alternative hypothesis is that some fraction of the predictive gene expression signatures might pre-exist in normal tissues prior to the onset of oncogenesis. This pre-existing difference in gene expression presumably results from the presence of constitutional polymorphisms that establish both the expression patterns and physiologic metastatic propensity.
To test these possibilities, gene expression analysis was performed using normal, non-neoplastic tissues isolated from transgene-negative highly metastatic AKR/J × FVB/NJ or low-metastatic DBA/2J × FVB/NJ F1 animals (supplemental tables S5-S9). Tissues were selected based on their presence in the primary tumor (whole blood, bone marrow), metastatic target organ and representative non-proliferative epithelial tissue (lung) and source of invading immunological cells (spleen and thymus). Due to the high adipose content of mouse mammary which is not represented in most human tumor samples used for gene expression, this tissue was excluded from the analysis. Signatures derived from expression differences from spleen and thymus of mice of differing metastatic capacities accurately predicted outcome in four of the five human breast cancer datasets analyzed in this study (Figure S1 & S2, respectively; supplemental tables 10-14). Furthermore, the gene expression signature derived from normal lung accurately predicted outcome in all five breast cancer datasets (Figure 4) consistent with the hypothesis that human breast cancer predictive signature profiles are driven, at least partially, by inherited, rather than acquired factors. However, no consistent outcome effects were observed for the gene expression signatures of whole blood or bone marrow (Supplemental Figures 3,4) suggesting that these tissues do not significantly contribute to the prognostic gene signatures derived from human tumor samples.
Figure 4.
Kaplan-Meier analysis demonstrates that the gene expression signature derived from comparison of normal lung tissue from high- and low-metastatic mouse strains accurately predicts outcome in all five breast cancer datasets.
Predictive gene signatures are likely due to a combination of inherited and somatic factors
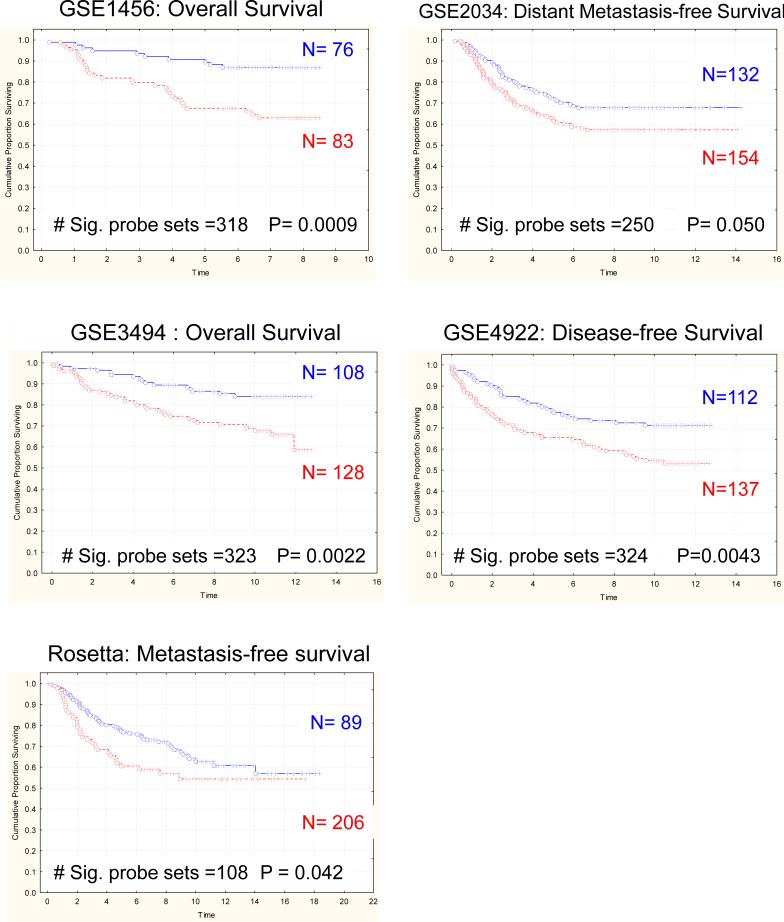
The previous results, while consistent with the hypothesis that germline variation induces an inherent susceptibility to metastasis, do not lessen the potential importance of somatic mutation in tumor progression. To attempt to evaluate the relative role of inherited versus somatic events in this model we investigated the Met-1 and DB-7 cell lines, which are derived from PyMT-driven mammary tumors from mice of an FVB/NJ genetic background (24). Met-1 is a highly metastatic tumor cell line derived from the original PyMT transgenic animal. DB-7, however, is a low metastatic potential cell line derived from a mutant PyMT construct that eliminates the activation of the Akt pathway. To investigate whether this type of somatic variation in identical genetic backgrounds can induce gene expression signatures with similar prognostic ability as those described above, these cell lines were implanted into the mammary fat pad of FVB/NJ virgin females and microarray analysis performed on the resulting tumors (supplemental table S15). As can be observed in Figure 5, the signature derived from the Met-1/DB-7 comparison was predictive in four of the five datasets (supplemental table S16), consistent with the presence of a significant somatically acquired component of the metastasis predictive gene expression profiles.
Figure 5.
Kaplan-Meier analysis demonstrates that the gene expression signature derived from comparison of tumors derived from the highly metastatic Met-1 cell line and the low metastatic DB-7 cell line accurately predicts outcome in the GSE1456, 3494, 4922 and Rosetta breast cancer datasets.
Signature probe set overlap and common network analysis of the mouse gene signatures
Since the polymorphisms modulating the gene expression patterns are present in all of the mouse tissues the possibility exists that the gene signatures derived from the various mouse tissues might be identifying different subsets of the same molecular network (32). To assess this possibility, the overlaps between the different signatures were assessed. As can be observed in supplemental tables S17-S18, the number of shared probes between signatures derived between different tissues varied significantly. To further investigate the potential overlap of these signatures, a combined analysis of the six signatures derived from polymorphic normal tissues was performed using the Ingenuity Pathway Analysis suite. Tumor samples (AKR vs DBA tumors, Mvt-1 transplant tumors) were excluded from this analysis to avoid any potential confounds due to somatic mutations within the tumor epithelium. A significant fraction of the genes in the individual tissue signatures could be assembled into a large network, consistent with the hypothesis that the overall basal transcription of the tissues from the two mouse inbred strains was likely altered due to constitutional polymorphisms. However, the inability of all tissues to predict outcome in the human datasets despite the interconnectivity of the network and substantial overlap of probe sets (supplemental tables S17-S18) suggests that only specific subsets and biological functions were relevant for prognosis. These subsets may be differentially expressed in different tissues. Thus, although there are interconnections in the global network diagrams, which represent averages across all tissue types, only specific tissues may harbor the appropriate transcriptional program relevant to disease outcome.
Network and biological function analysis of human datasets
To gain a better understanding of the genes and networks associated with prognosis in the human datasets the Ingenuity Pathway Analysis was performed on the orthologous human probe sets. For this analysis, only those probe sets that significantly varied (p<0.001; supplemental tables S10-14) in one or more of the datasets was included in the analysis. Similar to the mouse data, genes from each of the tissue signatures could be assembled into a large network consistent with the possibility of a common underlying mechanism.
Individual gene signatures were then analyzed for the biological functions significantly over-represented. Consistent with analysis of human gene signatures (33, 34) genes associated with cell growth and proliferation were among the most significant (figure S5A). In contrast, in the Mvt-1 transplant tumors, genes associated with cell growth or cell cycle were not the most significant biological functions (figure S5B), although they were present within the signature. Analysis of the normal tissue gene signatures also revealed the universal presence of growth associated genes in all of the profiles (figure S6).
The presence of proliferation-associated genes in all of the signature profiles, including those that did not consistently predict outcome, suggests that either specific subsets of proliferation-associated genes are important in predicting outcome or other biological networks present in some of the tissue profile but not others are also associated with outcome. To test the later possibility, probe sets associated with the biological functions of cell cycle, cell growth and proliferation, and cellular assembly and organization were removed from the non-proliferative adult lung gene signature and the human datasets re-analyzed using the truncated profile. As can be observed in figure S7 and table 1, the proliferation-truncated gene signature was still capable of discriminating outcome in four out of the five human datasets. This result is consistent with the possibility that other pathways in addition to cellular proliferation are capable of contributing to prognostic gene expression profiles. However, at this time we cannot rule out the possibility that proliferation-associated genes remain in the lung signature but were not identified due to incomplete annotation or because the genes in the lung signature have proliferation-associated functions that have not yet been identified.
Discussion
The discovery that gene expression profiles could predict breast cancer outcome has initiated widespread use of the technology for the development of expression profiles to improve individualized medicine for patients. It also reignited a debate in the literature as to the molecular origins of metastatic capacity (12, 35, 36). The prevailing theory of metastasis, the somatic evolution theory, predicted that only a small subset of tumor cells within the bulk tumor mass would acquire all of the capabilities required to successfully colonize a distant site. The ability of bulk tumor tissue to predict outcome however, suggested that on average the majority of primary tumor cells had to express the molecular signature of metastasis, which appeared potentially incompatible with the somatic evolution hypothesis. As a result some investigators offered a new hypothesis suggesting that metastatic potential might be encoded early within the tumor, potentially by the original transforming mutations themselves (12, 13). Simultaneously, work in our laboratory demonstrated that the propensity to metastasize was at least in part due to inherited susceptibility (16, 17). This led to an additional hypothesis that enabled the reconciliation of the data supporting both somatic evolution and early oncogenesis models. If a significant fraction of the prognostic gene signatures were encoded by inherited germline polymorphism, rather than somatic mutation, then the predictive gene signatures would be present throughout the tumor and metastasis-inducing somatic evolution could subsequently occur in susceptible individuals resulting in disseminating disease(37).
This hypothesis makes several predictions. The most important is that if the predictive gene signatures are due in part to inherited polymorphism, it would suggest that the signatures should be detectable in normal, preneoplastic tissue in susceptible individuals. The aim of this study was therefore to test this hypothesis and to evaluate the ability to translate the results of our mouse genetic model system of breast cancer progression to human clinical samples. To do so we performed a series of gene expression array analyses to ask the following questions: 1) do gene expression profiles from mouse models of inherited metastasis susceptibility predict outcome in human breast cancer; 2) what are the cellular origin(s) of prognostic gene expression signatures; 3) does germline variation contribute to the induction of prognostic expression patterns in human breast cancer; and 4) if there is indeed an inherited component to such signatures, what are the relative contributions of somatic and inherited factors in the establishment of the predictive expression profiles?
The strategy we employed was to examine spontaneous tumors, transplant tumors and normal tissues in mouse strains with different genetic susceptibility to metastatic progression for the presence of gene signatures that were able to discriminate outcomes in human breast cancer datasets. Our previous studies suggested that like mice, humans also exhibit an inherited genetic susceptibility to metastasis (14, 15, 20). This in turn implied that the prognostic gene expression profiles observed in human breast cancer datasets might be at least partially the result of inherited factors (14, 20, 21). In the current study, we provide further support for the hypothesis that metastasis susceptibility is a complex heritable trait. More significantly, we provide evidence supporting our hypothesis that metastasis-predictive microarray gene expression signatures, which are currently being evaluated as potential prognostic tools in the clinical setting, may be partially driven by host germline polymorphism.
To investigate this, we performed microarray analysis to derive a gene expression signature indicative of the differences in gene expression between primary spontaneous mammary tumors from mice with a 20-fold difference in metastatic propensity (17). The resulting gene expression signature accurately predicted outcome in four of the five human breast cancer datasets examined. Additionally, non-neoplastic tissues from five other organs involved in the process of tumorigenesis were analyzed to investigate the relative cellular contributions to signatures derived from complex, bulk human tumors. Whole blood, spleen and thymus were chosen to investigate the contribution of hematologically-derived cells present within the primary tumor mass. Additionally, we characterized gene expression patterns in bone marrow since these cells have recently been demonstrated to promote metastasis in both the primary tumor (38, 39) and secondary site (40). Finally, lungs were selected for gene expression analysis since the majority of metastatic lesions in this model system form at this site.
Several important conclusions can be drawn from these experiments. First, as predicted by the genetic predisposition hypothesis, metastasis-predictive gene expression signatures could be derived from a variety of normal, non-neoplastic tissues. Specifically, normal lung, spleen and thymus derived from mice of differing metastatic propensities exhibited gene expression signatures that could predict outcome in breast cancer. No consistent predictive signal was observed for the circulating whole blood or bone marrow, supporting the conclusion that the contribution of these tissues to metastatic phenotype, while potentially critical to the clinical phenotype, may not contribute a large fraction of the expression patterns of most bulk primary tumors. The ability of the lung, spleen and thymus to distinguish patient outcomes suggests that both basal epithelial and lymphocyte signals may comprise the majority of the signal observed in bulk tumor tissue.
The cellular origins of the inherited components of the predictive gene signatures were further investigated using a transplant strategy. Previously published analyses and earlier work in our laboratory demonstrated that genes associated with stromal tissues and the immune compartments are frequently dysregulated in tumors more prone to metastasizing (10, 13, 41, 42). We therefore sought to investigate the relative contribution of these tissues to signatures by removing a major source of genetic heterogeneity: the tumor epithelium. This was achieved by implanting a malignant highly metastatic mouse mammary tumor cell line into the mammary fat pad of mice with differing metastasis susceptibilities. The resulting primary tumors were therefore composed of identical tumor epithelium, but contained different infiltrating host components from the two mouse genotypes. Thus, any gene expression differences between tumors from different hosts would result directly from host tissue germline polymorphism and/or the reaction of tumor cells to the differing microenvironments.
Based on the presence of numerous host-derived, non-epithelial transcripts in the prognostic signatures, we anticipated that both the spontaneous and transplant tumors would be able to discriminate patient outcome. Indeed, we did observe that this was the case. However, no difference was observed in the metastatic capacity of this tumor cell line in spite of the previously observed twenty-fold difference in metastatic susceptibility of the host genotypes (17). The one possible explanation for this lies in the highly malignant properties of the Mvt-1 cell line. It may be that the influence that host germline polymorphism exerts upon the tumor epithelium is too subtle to be detected by in vivo orthotopic transplantation assays using a cell line selected for high malignant potential (23). Microarray analysis is, however, a very sensitive means of detecting changes in gene expression. Therefore, the observed prognostic gene expression signature in the Mvt-1 implant tumors likely reflects the subtle changes in gene expression resulting from interaction with the different hosts. Alternatively, it is possible that the effect of inherited polymorphisms on metastatic capacity is a tumor autonomous effect and the prognostic gene expression profile from the transplant tumors is due entirely from the infiltrating host tissues. Thus, although the prognostic signature is apparent in the bulk tumor, the presence of the same highly malignant cell line in both hosts results in equivalent metastatic capacity. Additional work will be necessary to resolve these two scenarios.
Significant variation in the number of significant probe sets and the discriminatory ability of the tissue signatures was also observed across the human datasets. We believe that this reflects the underlying heterogeneity of the human populations represented in each dataset, which are comprised of mixtures of different molecular subtypes and stages. Previously bioinformatic investigation into gene expression signatures demonstrated that subsets of predictive genes would be identified based on the particular subset of patients analyzed (43, 44). As a result, the different sets of patients included within each dataset, as well as different experimental variation introduced during array analysis, would be expected to generate different significant subsets of each tissue signature. Despite these fluctuations, all of these large datasets in the analysis to increase the probability that any results that were observed was due to a general phenomenon, rather than a dataset specific effect, or due to false-positives from analyzing only one of a limited number of datasets.
In addition, differences in the clinical characteristics of each patient set may also contribute significantly to the probe set selection and discriminatory ability of each dataset. The dataset from Wang et al. (GSE2034)(1), for example, consists of only untreated lymph node-negative patients, while the other datasets contain a mixture of node-positive, node-negative and adjuvant therapy treated patients. The GSE2034 dataset therefore represents the natural progression of node-negative breast cancer since there is no confound due to adjuvant therapy to account for. The Rosetta dataset, in contrast, was designed to develop a discriminatory assay for younger patients (10). The differences observed for the prognostic ability of our samples between the datasets may therefore be potentially explained by these confounding variables. Of note, however, is the fact that the lung expression profile had prognostic value in all of the datasets, regardless of these confounding clinical differences. Since GSE2034 represents the natural progression of node-negative patients this results supports our hypothesis that germline encoded transcriptional differences may in fact account for some measurable fraction of the prognostic gene signatures.
Finally, investigations over the past few years into the factors underlying the metastasis predictive expression profiles have suggested that all of the prognostic gene signatures may be sampling the same underlying network (32), most commonly thought to be cell cycle and proliferation (33, 34). The data presented here are consistent with these being important biological functions associated with progression. The signature profile derived from the spontaneous PyMT-induced tumors from (AKR × PyMT)F1 and (DBA × PyMT)F1 mice was capable of discriminating outcome in four of the five human datasets, and was trending toward significance in the GSE2034 dataset (figure 1). Removal of potential differences in proliferative capacity of the tumor epithelium resulting from constitutional polymorphism by implanting the same cell line into non-transgenic hosts eliminated any trend in GSE2034 (figure 3) and somewhat reduced the risk ratio in both GSE3494 and GSE4922 (table 1). Similar results were observed when proliferation associated genes were stripped out of the lung gene expression signature (figure S7 and table 1).
The ability of Mvt-1 transplant and truncated lung signatures to predict outcome in the datasets other than GSE2034, however, raises the possibility that other biological networks may also be predictive of breast cancer outcome. There are several possibilities that would need to be considered. First, these other pathways may not be causative factors predicting outcome. It is possible that the same polymorphic differences that are driving the predictive proliferation-associated gene sets may also be impacting the other networks as a bystander effect. Second, they may be causative factors, but have not been detected as a common mechanism in analysis of the human datasets because of the dominant effect of the cell proliferation pathway and/or effects only in subsets of the human population. Third, it is possible that genes remaining in the Mvt-1 and truncated lung profiles are in fact members of the proliferation network but have not been so annotated either because their functional significance in cell growth is as of yet unrealized, or that the current annotations are incomplete. While it is not possible to definitely distinguish between these possibilities at this time, we favor the first two possibilities. Previous studies have demonstrated that expression profiles are an independent predictive factor compared to standard clinical measures, including mitotic index. This suggests that the signatures either are a much more accurate measure of proliferation compared to standard immunohistochemistry, or that they are measuring factors in addition to cellular growth. However, additional studies will be necessary to investigate and definitely address these possibilities.
In summary, these results provide additional evidence for the role of inherited factors in human breast cancer progression. In addition, they suggest that the prognostic gene signatures currently in clinical trial likely result from a complex mixture of somatic and inherited factors present not only in the tumor epithelium, but also infiltrating non-neoplastic cells. Further investigations will hopefully improve our current understanding of the relationship between these various factors not only in the tumor epithelium itself, but also in the infiltrating non-neoplastic tissues, with a goal of improving not only the current prognostic tools but also developing more effective therapeutic strategies for therapeutic intervention.
Supplementary Material
Table S10: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA whole blood comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S2: Probe sets with significant variation in human datasets from mouse AKR vs DBA tumors. Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S3: Mouse expression signature from Mvt-1 transplant tumors in AKR versus DBA F1 hosts
Table S4: Probe sets with significant variation in the human datasets from the MVt-1 AKR vs DBA host comparison. Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S5: Mouse AKR vs DBA whole blood gene list
Table S6: Mouse AKR vs DBA bone marrow gene list
Table S7: Mouse AKR vs DBA lung gene list
Table S8: Mouse AKR vs DBA spleen gene list
Table S90: Mouse AKR vs DBA thymus gene list
Supplemental Tables
Table S1: Mouse AKR vs DBA tumor gene list
Supplemental Figure Legends
Figure S1: Kaplan-Meier analysis demonstrates that the gene expression signature derived from comparison of normal spleen tissue from high- and low-metastatic mouse strains accurately predicts outcome in the GSE1456, 3494, 4922 and Rosetta breast cancer datasets.
Figure S2: Kaplan-Meier analysis demonstrates that the gene expression signature derived from comparison of normal thymus tissue from high- and low-metastatic mouse strains accurately predicts outcome in the GSE1456, 3494, 4922 and Rosetta breast cancer datasets.
Figure S3: Kaplan-Meier analysis of the gene expression signature derived from comparison of normal whole blood from high- and low-metastatic mouse strains. P-values were determined by log-rank analysis.
Figure S4: Kaplan-Meier analysis of the gene expression signature derived from comparison of normal bone marrow from high- and low-metastatic mouse strains.
Figure S5: Ingenuity biological function analysis of the gene signatures derived from comparison of mouse tumors. For clarity only the top 12 categories are shown. A) Comparison of the signature derived from spontaneous tumors from (AKR × PyMT )F1 and (DBA × PyMT)F1 animals. B) Comparison of the signature derived from tumors derived from implantation of the Mvt-1 cell line into non-transgenic (AKR × FVB)F1 and (DBA × FVB)F1 animals.
Figure S6: Ingenuity biological function analysis of the comparison of normal tissues derived from non-transgenic (AKR × FVB)F1 and (DBA × FVB)F1 animals. For clarity only the top 12 categories are shown.
Figure S7: Kaplan-Meier of the gene expression signature derived from comparison of lung tissue signature after truncation of genes associated with proliferation.
Table S11: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA bone marrow comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S12: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA lung comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S13: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA spleen comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S14: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA thymus comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S15: Mouse Met1 versus DB7 tumor gene list
Table S16: Probe sets with significant variation in human datasets from the Met1 versus DB7 tumor comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S17: Number of overlapping probe sets between mouse gene signatures
Table S18: Probe sets shared betweeen mouse tissue samples Probe sets are listed in the rows, mouse tissue datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam.
References
- 1.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy S, Tamayo P, Rifkin R, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A. 2001;98(26):15149–54. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 5.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102(10):3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 7.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Smid M, Wang Y, Klijn JG, et al. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24(15):2261–7. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 9.Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nat Clin Pract Oncol. 2008;5(4):206–19. doi: 10.1038/ncponc1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 11.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 12.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 13.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 14.Crawford NP, Walker RC, Lukes L, Officewala JS, Williams RW, Hunter KW. The Diasporin Pathway: a tumor progression-related transcriptional network that predicts breast cancer survival. Clin Exp Metastasis. 2008 doi: 10.1007/s10585-008-9146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford NP, Ziogas A, Peel DJ, Hess J, Anton-Culver H, Hunter KW. Germline polymorphisms in SIPA1 are associated with metastasis and other indicators of poor prognosis in breast cancer. Breast Cancer Res. 2006;8(2):R16. doi: 10.1186/bcr1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter KW, Broman KW, Voyer TL, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 2001;61(24):8866–72. [PubMed] [Google Scholar]
- 17.Lifsted T, Le Voyer T, Williams M, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77(4):640–4. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Park YG, Zhao X, Lesueur F, et al. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet. 2005;37(10):1055–62. doi: 10.1038/ng1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Crawford N, Lukes L, Finney R, Lancaster M, Hunter KW. Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis. 2005;22(7):593–603. doi: 10.1007/s10585-005-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford NP, Qian X, Ziogas A, et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3(11):e214. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford NPS, Alsarraj J, Lukes L, et al. Bromodomain 4 activation predicts breast cancer survival. PNAS. 2008;105(17):6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–5. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei XF, Noble MS, Davoli MA, et al. Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In Vitro Cell Dev Biol Anim. 2004;40(12):14–21. doi: 10.1290/1543-706X(2004)40<14:ECOPMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 24.Borowsky AD, Namba R, Young LJ, et al. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22(1):47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- 25.Crawford NP, Alsarraj J, Lukes L, et al. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105(17):6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 29.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 30.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiyama Y, Farrow B, Murillo C, et al. Analysis of differential gene expression patterns in colon cancer and cancer stroma using microdissected tissues. Gastroenterology. 2005;128(2):480–6. doi: 10.1053/j.gastro.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 33.Dai H, van't Veer L, Lamb J, et al. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005;65(10):4059–66. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- 34.Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genomics. 2008;1:11. doi: 10.1186/1755-8794-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernards R, Weinberg RA. Metastasis: objections to the same-gene model. Nature. 2002;419(6907):560. doi: 10.1038/419560a. [DOI] [PubMed] [Google Scholar]
- 36.Fidler IJ, Kripke ML. Genomic analysis of primary tumors does not address the prevalence of metastatic cells in the population. Nat Genet. 2003;34:23. doi: 10.1038/ng0503-23a. [DOI] [PubMed] [Google Scholar]
- 37.Hunter KW, Welch DR, Liu ET. Genetic background is an important determinant of metastatic potential. Nat Genet. 2003;34:23–4. doi: 10.1038/ng0503-23b. [DOI] [PubMed] [Google Scholar]
- 38.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 39.Kitamura T, Kometani K, Hashida H, et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat Genet. 2007;39(4):467–75. doi: 10.1038/ng1997. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan RN, Riba RD, Zacharoulis S, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438(7069):820–7. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergamaschi A, Tagliabue E, Sorlie T, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol. 2008;214(3):357–67. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, Rouse J, Lukes L, et al. Caffeine suppresses metastasis in a transgenic mouse model: a prototype molecule for prophylaxis of metastasis. Clin Exp Metastasis. 2005;21(8):719–35. doi: 10.1007/s10585-004-8251-4. [DOI] [PubMed] [Google Scholar]
- 43.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21(2):171–8. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 44.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103(15):5923–8. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S10: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA whole blood comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S2: Probe sets with significant variation in human datasets from mouse AKR vs DBA tumors. Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S3: Mouse expression signature from Mvt-1 transplant tumors in AKR versus DBA F1 hosts
Table S4: Probe sets with significant variation in the human datasets from the MVt-1 AKR vs DBA host comparison. Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S5: Mouse AKR vs DBA whole blood gene list
Table S6: Mouse AKR vs DBA bone marrow gene list
Table S7: Mouse AKR vs DBA lung gene list
Table S8: Mouse AKR vs DBA spleen gene list
Table S90: Mouse AKR vs DBA thymus gene list
Supplemental Tables
Table S1: Mouse AKR vs DBA tumor gene list
Supplemental Figure Legends
Figure S1: Kaplan-Meier analysis demonstrates that the gene expression signature derived from comparison of normal spleen tissue from high- and low-metastatic mouse strains accurately predicts outcome in the GSE1456, 3494, 4922 and Rosetta breast cancer datasets.
Figure S2: Kaplan-Meier analysis demonstrates that the gene expression signature derived from comparison of normal thymus tissue from high- and low-metastatic mouse strains accurately predicts outcome in the GSE1456, 3494, 4922 and Rosetta breast cancer datasets.
Figure S3: Kaplan-Meier analysis of the gene expression signature derived from comparison of normal whole blood from high- and low-metastatic mouse strains. P-values were determined by log-rank analysis.
Figure S4: Kaplan-Meier analysis of the gene expression signature derived from comparison of normal bone marrow from high- and low-metastatic mouse strains.
Figure S5: Ingenuity biological function analysis of the gene signatures derived from comparison of mouse tumors. For clarity only the top 12 categories are shown. A) Comparison of the signature derived from spontaneous tumors from (AKR × PyMT )F1 and (DBA × PyMT)F1 animals. B) Comparison of the signature derived from tumors derived from implantation of the Mvt-1 cell line into non-transgenic (AKR × FVB)F1 and (DBA × FVB)F1 animals.
Figure S6: Ingenuity biological function analysis of the comparison of normal tissues derived from non-transgenic (AKR × FVB)F1 and (DBA × FVB)F1 animals. For clarity only the top 12 categories are shown.
Figure S7: Kaplan-Meier of the gene expression signature derived from comparison of lung tissue signature after truncation of genes associated with proliferation.
Table S11: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA bone marrow comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S12: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA lung comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S13: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA spleen comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S14: Probe sets with significant variation in human datasets from the Mouse AKR vs DBA thymus comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S15: Mouse Met1 versus DB7 tumor gene list
Table S16: Probe sets with significant variation in human datasets from the Met1 versus DB7 tumor comparison Probe sets are listed in the rows, human breast cancer datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.
Table S17: Number of overlapping probe sets between mouse gene signatures
Table S18: Probe sets shared betweeen mouse tissue samples Probe sets are listed in the rows, mouse tissue datasets are in the columns. Filled in cells indicate the presence of the associated probe set in the dataset.