Abstract
Cytoplasmic RNA receptors are important in the detection of and response to viral infections. We analyzed ligand recognition by the retinoic acid-inducible protein I (RIG-I) protein in biochemical assays and in transiently transfected cells and characterized the requirements for both single- and double-stranded RNA agonists for RIG-I activation of signaling. RIG-I mutants such as K270A and T409A/S411A that were defective in signaling with triphosphorylated single-stranded RNAs were perfectly capable of signaling with dsRNAs. Furthermore, phosphorothioated oligodeoxynucleotides were found to antagonize RIG-I signaling. Both agonists and antagonist bind purified RIG-I protein and a truncated RIG-I protein that lacked the signaling domain. The agonists were necessary to activate RIG-I ATPase activity in vitro, whereas antagonist inhibited ATPase activity. Differential scanning fluorometry showed that RIG-I bound to agonists, and antagonists have different denaturation properties, suggesting a difference in protein conformations. Last, single particle reconstruction was used to generate three-dimensional models of the RIG-I dimers in complex with an agonist and an antagonist. The two complexes exhibited dramatically different structures.
Viral RNAs can bind to a subset of several innate immunity receptors and induce the signal transduction cascade to result in the production of cytokines and chemokines that can regulate the responses of the infected cells and communicate the infection to uninfected cells (1-5). Understanding how the receptors recognize ligands that can agonize and/or antagonize signaling is, thus, important in being able to intervene in viral infections.
Single- and double-stranded RNAs (ssRNA and dsRNA, respectively)2 RNAs are recognized by the membrane-associated Toll-like receptors 3, 7, and 8 as well as by cytoplasmic receptors that contain the DEXD/H box helicase motifs such as retinoic acid-inducible protein I (RIG-I), MDA5, and LGP2 (5-8).
The RIG-I-mediated antiviral response can target RNA viruses from the Flaviviridae, Paramyxoviridae, Orthomyxoviridae, Rhabdoviridae (5, 9, 10). A mutation (T55I) in the RIG-I gene resulted in a cell line, Huh7.5, that increased HCV replication (9). Furthermore, the HCV-encoded protease NS3-4A can cleave IPS-1 (variously called IPS-1, MAVS, VISA, and Cardif), an adaptor that mediates the communication between RIG-I and the activation of transcription factors IRF3 and NF-κB (11-16). IPS-1 cleavage can lead to its release from the mitochondrial membrane and disrupts the RIG-I pathway by preventing downstream activation of IRF3 and NF-κB (17). These studies unambiguously demonstrate the importance of the RIG-I pathway in viral infection and pathogenesis.
RIG-I encodes two N-terminal caspase recruitment domains (CARD) that function in signaling, a middle portion containing DEXD/H RNA helicase motifs that can bind dsRNA, and a C-terminal regulatory domain that interacts with the helicase domain and inhibits signaling by the CARD domain (18). Intramolecular and intermolecular interactions are critical to regulate the conformational changes upon ligand binding that are a part of the activation of signaling. In the presence of an agonist, RIG-I is thought to undergo a conformational change to expose the CARD domain that can trigger multimerization and cell signaling (18). The structure for the regulatory domain (amino acids 800-925) of RIG-I was elucidated recently (19, 20). The regulatory domain (RD) is sufficient to bind RNA with a 5′-triphosphate with nanomolar affinity (19, 20). RNA binding was also shown to induce dimerization of the recombinant RIG-I protein (19).
The goal of this study is to better define ligand recognition by the RIG-I protein using cell-based and biochemical assays. A number of ligands were found to serve as agonists and also antagonists for RIG-I.
EXPERIMENTAL PROCEDURES
Expression Constructs—For nomenclature, the full-length RIG-I protein is simply named RIG-I. The deletion of the CARD domain (deleting amino acids 2-228) is named by the prefix “R” to designate RIG-I followed by single letters for the helicase and regulatory domains; that is, R-HR. Where a GST tag is present, it is denoted with a lowercase “g,” as in R-gHR.
The cDNA of RIG-I cloned in pUNO (pRIGI) was purchased from InvivoGen. Mutations within RIG-I were generated by site-directed mutagenesis using the QuikChange kit and the protocol recommended by the manufacturer (Stratagene). The presence of the designed mutations and the lack of spurious mutations were checked by DNA sequencing using the Big-Dye® Terminator Version 3.1 Cycle Sequencing kits (Applied Biosystems).
Full-length RIG-I, R-HR, and their GST-tagged versions were cloned into the baculovirus transfer vector pBAC1 (Novagen Inc.). The expression constructs were cotransfected with linearized Baculovirus DNA (BD Biosciences) into Sf9 cells to produce recombinant virus. The virus was amplified in Sf9 cells and used for protein expression.
RIG-I Expression and Purification—Sf9 (Spodoptera frugiperda 9) cells were cultured in HyQ® SFX-Insect™ media and grown at 27 °C in flasks.
At 48 and 72 h after infection with the recombinant viruses, the cells were pelleted by centrifugation for 5 min at 680 × g. The cell pellets were resuspended in ice-cold lysis buffer (200 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm PMSF phenylmethylsulfonyl fluoride, and 1% Nonidet P-40), and the lysate was clarified with a 15-min centrifugation at 30,600 × g. The supernatant was added to nickel-nitrilotriacetic acid beads equilibrated in the lysis buffer. The proteins bound to the beads were washed 4 times with lysis buffer amended with 10 mm imidazole followed by repeated washes by centrifugation at 1 min and 3400 × g. Finally, the desired proteins were eluted with the lysis buffer amended with 250 mm imidazole. The protein was then concentrated and loaded onto a Superdex200 10/300 GL column and eluted with TS buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl). The peak of protein was dialyzed with storage buffer (50 mm Tris, pH 7.5, 300 mm NaCl, 40% glycerol, and 2 mm β-mercaptoethanol) and stored in aliquots. gRIG-I and R-gRH were purified by glutathione resin before elution with buffer E (50 mm Tris, pH 7.5, 150 mm NaCl, 5% glycerol, and 2 mm β-mercaptoethanol). The peak of protein was dialyzed with the storage buffer and stored in aliquots at -70 °C. Removal of the GST tag was performed by incubating the protein for 30 min with thrombin. The cleaved reaction was then purified again using the GST resin.
Size-exclusion Chromatography—Wild type or mutant protein samples (∼100 μg) were filtered through a Superdex 200 column using a GE Healthcare fast protein liquid chromatography system calibrated with the molecular mass standards blue dextran (2000 kDa), thyroglobulin (660 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (140 kDa), BSA (67 kDa), and chymotrypsinogen (25 kDa) according to the protocol described in Guarino et al. (21).
Cell-based Assay for RIG-I Activity—Actively growing HEK293T cells was plated in CoStar White 96-well plates at 4.4 × 104/ml for transfection. Cells were between 60 to 80% confluent and were transfected with a mixture of Lipofectamine 2000 reagent (Invitrogen) and plasmids pNiFty Luc (15 ng, InvivoGen), phRL-TK (5 ng, Promega Corp., Madison, WI), and pRIG1 (0.5 ng) that, respectively, code for the firefly luciferase reporter, the Renilla luciferase transfection control, and plasmids coding for either wild-type or mutant RIG-I. The cells were incubated for 24 h to allow expression from the plasmids. Ligands were then transfected into the cells at 0.5 μm unless otherwise specified. After another incubation for 12 h, the cells were assayed using the Dual Glo Luciferase Assay System reagents (Promega), quantifying luminescence with the FLUOstar OPTIMA Plate Reader (BMG Labtech, Inc).
ATPase Assay—ATPase assay was performed in a buffer containing 20 mm Tris, pH 7.5, 5 mm MgCl2, 8 mm dithiothreitol, 30 mm NaCl, and 4% glycerol. The reaction contained ligand RNA (1 μm) or pIC (500 ng) and 1 mm cold ATP along with trace amounts of [γ-32P]ATP. Specified proteins were added, and samples were incubated at 37 °C for 30 min. Reaction products were then separated on thin-layer chromatography using cellulose polyethyleneimine sheets. Relative ATPase activity was calculated as the amount of 32P released relative to a reaction where RIG-I was not exposed to a ligand.
Reversible Protein-RNA Cross-linking and Peptide Mapping—N-hydroxy succinimide-Sepharose (20 μl) was washed with 0.1 m sodium tetraborate, pH 8.5, and mixed with 20 μl of 50 μm oligodeoxynucleotide 2006 (ODN2006) containing an amine group attached to the 3′ end. The mixture was incubated overnight followed by washes with buffer A (20 mm Hepes, 30 mm NaCl, 5 mm MgCl2, and 2 mm dithiothreitol) and resuspended in the same buffer. RIG-I (5 μg) was added to it and incubated at room temperature for 15 min followed by the addition of formaldehyde to a final concentration of 0.1% in a 50-μl reaction. The cross-linking reaction was terminated by the addition of glycine at a 0.2 m final concentration. After 5 min, sequencing grade trypsin (Trypsin Gold; Promega) was added at a protease: substrate ratio 1:50 (w/w) in 100 mm NH4HCO3, pH 7.8, at 37 °C and allowed to digest overnight. The released the ODN2006-peptide conjugates were purified and subjected to incubation at 70 °C for 1 h to reverse the cross-links. The samples were centrifuged at 3000 × g for 5 min, and the supernatants containing the peptides were desalted using a Ziptip (Millipore, Bedford, MA). The bound peptides were eluted in 2.5 μl of 70% acetonitrile and 0.1% trifluoroacetic acid and analyzed by matrix-assisted laser desorption ionization time-of-flight.
Biochemical Analysis of RIG-I Binding to Nucleic Acid—Native gel assay used recombinant proteins mixed with various nucleic acids in ATPase reaction buffer and separated on non-denaturing PAGE and stained with ethidium bromide UV cross-linking. Nucleic acids were end-labeled using T4 polynucleotide kinase and [γ-32P]ATP. A standard cross-linking reaction used a mixture of recombinant protein (200 ng) with and without competitors in ATPase buffer. Cross-linking was for 2 min at 3600 μJ/cm2. The reaction was separated in a denaturing 4-12% protein gel.
Pulldown assays were performed by binding biotinylated ODN2006 to streptavidin resin overnight in buffer A. A slurry of resin (30 μl) containing bound ∼3 μg of biotinylated 2006 was mixed with 1 μg of recombinant R-gHR and 2 μg of BSA in the presence or absence of equimolar amounts (5 μm) of competitors (poly(I:C) was used at 1 and 2 μg) and incubated at room temperature for 2 h. The unbound fraction was removed by centrifugation, and the pellet was washed with the same buffer as above. The final pellet was resuspended in 15 μl of SDS-PAGE loading buffer and separated on 4-12% denaturing protein gel along with the unbound fraction followed by staining with Coomassie Blue.
Fluorescence Anisotropy Assay—Fluorescein isothiocyanate-labeled RNA for the binding assays were synthesized, purified by high performance liquid chromatography, and quantified by the manufacturer (IDT technologies; Coralville, IA). The probes were diluted to 200 nm in ATPase buffer. For the formation of dsRNA, the probes were heated with a 1:1 molar ratio of the complementary RNA in an 80 °C water bath and then allowed to cool slowly to room temperature. Fluorescence measurements were made at room temperature with a PerkinElmer Life Sciences luminescence spectrometer LS55 using cuvettes with an optical path length of 0.4 cm. Measurements were taken with an integration time of 1 s and a slit width of 5 nm. The excitation and emission wavelengths were 495 and 520 nm, respectively. Anisotropy values were recorded 20 s after each protein addition to allow the sample to reach equilibrium. The total volume of protein added was less than 5% that of the final volume. Represented anisotropy values are the average of 10 measurements. Binding data were analyzed by nonlinear least square fitting using KaleidaGraph software (Synergy Software, Reading, PA). The simple hyperbola equation ΔA = BmaxX/(X + Kd) was used to determine the dissociation constant (Kd). In this equation ΔA is the anisotropy change caused by the ligand binding, Bmax is the maximum anisotropy change, and X is the total concentration of the input protein.
Differential Scanning Fluorometry—Differential scanning fluorometry was performed in an Eppendorf Mastercycler EP Realplex machine. Each sample contains 20-μl solutions of RIG-I (1 μm), SYPRO orange (Molecular probes), and ligand (5 μm) in the ATPase buffer. The 96-well plate containing all of the samples was heated at a rate of 1.0 °C/min in a real-time PCR machine (Eppendorf) from 25 to 95 °C, and the fluorescence intensity was measured with excitation/emission wavelengths of 470/550 nm. TM values were calculated using the software provided by Eppendorf. Each sample was tested in triplicate, and the results were duplicated in at least two independent assays.
Negative-stained Electron Microscopy and Three-dimensional Reconstructions—A JEOL 1200 TEM operated at 100 kV was used for routine negative staining and direct size measurements. Electron micrographs were digitalized using an Epson Projection 3200 scanner at 1200 dots/inch, corresponding to 5.5 Å/pixel at the specimen level. Three-dimensional reconstruction of different proteins reported in this paper were calculated using the EMAN software package (22) run under Linux and following procedures similar to those described by Sun et al. (23). Briefly, particles of the protein of interest (R-HR, 1900 particles; R-HR dimer + dsRNA, 2500 particles; or R-HR dimer + ODN2006, 3300 particles) were selected using the EMAN boxer routine, filtered, and centered. A set of class averages were generated from these centered particles with no assumed symmetry. An initial three-dimensional model was generated from a set of representative class averages and was used for refinement. The reconstructions were iteratively refined until the structure was stable as judged by Fourier shell correlation (FSC). The appropriate molecular mass (R-HR, 80 kDa; R-HR dimers, 167 kDa) was used for the surface-rendering threshold of the three-dimensional structure. Three-dimensional reconstructions were visualized using the University of California, San Francisco Chimera software package (24).
R-HR dimers complexed with 2006 were subjected to EMAN multi-refinement to separate the R-HR monomers present in the gel filtration fraction from the dimers (See “Results” and “Discussions”). The particle set corresponding to dimeric forms was used for further refinement.
RESULTS
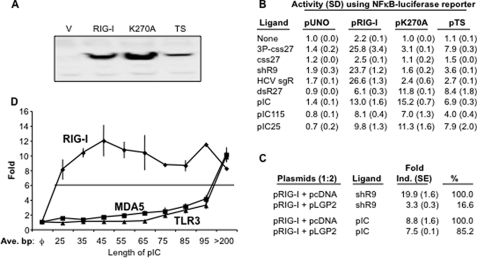
Agonists for RIG-I Activation of Signaling in Cells—Transient transfection of 293T cells was used to test agonists for RIG-I-mediated signaling. To confirm that the signaling observed is due to RIG-I, we used two mutants, K270A and T409A/S411A (referred to as TS). Mutant K270A has a defect in the Walker A motif of the helicase-like domain and was shown to inactivate RIG-I signaling (5). Residues Thr-409 and Ser-411 was suggested to be important for poly(I:C) binding (20). These mutants are both expressed in the cells, as determined by Western blot analysis (Fig. 1A), although mutant TS may be expressed at a reduced level.
FIGURE 1.
Single- and double-stranded ligands have different properties for RIG-I signaling. A, Western blot analysis of RIG-I and mutants. RIG-I protein and the two alanine substitution mutants are expressed in transiently transfected 293T cells. The Western blot was probed with a polyclonal antibody against C-terminal region of RIG-I purchased from Santa Cruz Biotechnology, Inc. V, vector. B, the effects of two RIG-I mutations on response to different ligands. Numbers denote -fold induction that is the ratio of the induced versus uninduced samples assayed. The assay was performed using luciferase reporter regulated by a promoter containing NF-κB binding sites. Each number represents the mean of at least three independent assays, and the value of 1 S.E. is shown in parentheses. Throughout this work the following names are used for the various ligands; 3P-css27 is a 27-nt RNA produced by in vitro transcription, and css27 is the same RNA that lacks 5′-triphosphates. ShR9 is a 60-nt hairpin RNA produced by in vitro transcription (see supplemental Fig. 1). HCV sgR is the hepatitis C virus subgenomic replicon RNA transcribed from linearized plasmid pFK/I389neo/NS3-3′/5.1. DsR27 is a double-stranded RNA made by annealing two single-stranded oligonucleotides of 27-nt each; pIC is poly(I:C) purchased from Invitrogen and has a molecular mass that is in excess of 200 bp with very little fragments below this length. pIC115 and pIC25 are poly(I:C)s of 115 and 25 bp, respectively. C, effects of shR9-induced RIG-I activity in the presence of LGP2. The ratio of RIG-I to LGP2 is 1:2. D, the effects of poly(I:C) length on induction of RIG-I activation of an NF-κB reporter in HEK293T cells. The same series of pIC was used to examine MDA5 and TLR3 signaling and yielded different responses. Each point is the graph shows the mean and S.E. of three independent assays.
Different ss- and dsRNAs were tested for their ability to activate RIG-I using the NF-κB promoter-driven luciferase reporter assay (Fig. 1B and supplemental Fig. 1). These include in vitro transcribed 27-nt RNA named 3P-css27, the same RNA lacking 5′-triphosphates (css27), an in vitro transcribed 60-nt hairpin RNA named shRNA9, and in vitro transcribed HCV subgenomic replicon RNA of 9 kilobases (HCV sgR), an unphosphorylated 27-bp dsRNA named dsR27 that has the sequence as css27 in one of its strands, heterogeneous poly(I:C), and poly(I:C) of 115 bp (pIC115) and 25 bp (pIC25).
RIG-I-mediated signaling could be induced by all the agonists tested in Fig. 1B except css27, consistent with reports that ssRNA require a 5′-triphosphate to act as an RIG-I agonist (25, 26). Double-stranded RNAs dsR27, pIC25, pIC115, and poly(I:C) were able to activate RIG-I signaling in the absence of the 5′-triphosphates from 6- to 13-fold (Fig. 1B). The activation is specific to RIG-I as signaling was observed only in cells transfected with RIG-I-expressing plasmid and not in vector-transfected cells (pUNO, Fig. 1B). Furthermore, mutants K270A and TS showed significantly lower signaling with 3P-css27, shR9, and HCV sgR compared with wild type RIG-I (Fig. 1B).
An unexpected observation was that mutants K270A and TS were fully capable of signaling at levels similar to that of wild type RIG-I with ligands dsR27, pIC25, pIC115, and poly(I:C) (Fig. 1B). These results suggest that dsRNA agonists may induce RIG-I-specific signaling independent of the activities conferred by the ATPase domain. The signaling by mutant TS with poly(I:C) was contrary to the report of Takahasi et al. (20), but the substitutions in TS did affect signaling with other ligands. These results show that signaling by RIG-I is very much influenced by the properties of the ligand.
To confirm whether dsRNA and ssRNA induce RIG-I signaling differently, we tested the affect of LGP2 on RIG-I signaling with shR9 and poly(I:C) (Fig. 1C). LGP2 has been proposed to antagonize the signaling by RIG-I through the formation of heterodimeric complexes (18). An alternative model is that LGP2 could limit the amount of ligand that is accessible to RIG-I by binding dsRNA with higher affinity and cooperativity (8). Although LGP2 interfered with shR9-mediated RIG-I signaling, it did not significantly affect poly(I:C)-induced signaling (Fig. 1C). These observations support the hypothesis that ssRNA and dsRNA is recognized differently by RIG-I.
The poly(I:C) used varied from 25 bp to a length that averaged ∼500 bp (Fig. 1B). To determine whether poly(I:C) length could affect signaling, we hydrolyzed commercial preparations of poly(I:C) with sodium hydroxide and separated the products on preparative denaturing gels. Segments of the gel were eluted to obtain different lengths of poly(I:C). The average lengths were determined with an analytical gel stained with toluidine blue (data not shown). These poly(I:C)s were then tested for their ability to induce RIG-I signaling (Fig. 1D). Poly(I:C) of up to 19 bp were unable to activate RIG-I signaling. In addition, heterogeneous poly(I:C)s with average lengths of >200 bp were not a better agonist for RIG-I signaling than ones of ∼45 bp, suggesting that dsRNA length and/or an concentration will affect RIG-I signaling (25). Last, to determine whether the effect of ligand length is specific to RIG-I, the same set of RNAs was tested with the cytoplasmic DEAD-box helicase, MDA5, and the membrane-associated receptor Toll-like receptor 3 (TLR3) (Fig. 1D). For both MDA5 and TLR3, increasing poly(I:C) length increased the signaling (Fig. 1D). These results show that response to dsRNA by RIG-I is distinguishable from that of MDA5 and TLR3. Altogether, results in this section demonstrate that RIG-I can recognize agonists by a complex set of rules, including their length, whether they are single- or double-stranded, or whether they possess 5′-triphosphates.
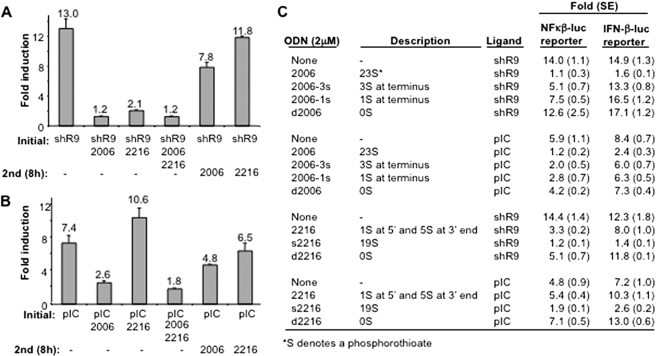
Antagonists of RIG-I Signaling—Modified ODNs with phosphorothioate backbones can bind to the ectodomain of TLR3 and inhibit signaling (26). We wanted to assess whether ODNs such as 2006 can affect RIG-I activity in the cell-based assay (Fig. 2). When 2006 was transfected into 293T cells expressing RIG-I in the absence of an agonist, no significant change in reporter activity was observed (data not shown). However, when ODN2006 (2 μm) was transfected along with the agonist shR9, RIG-I-dependent reporter activity was reduced to less than 10% (Fig. 2A). Comparable inhibition of RIG-I signaling was observed with 2006c that lacks the CpG motif, demonstrating that the inhibition is independent of TLR9 signaling (data not shown). 2216 (containing CpG motifs) as well as 2216c (lacking CpG motifs) had inhibitory effects when they were transfected along with agonist shR9, confirming that the CpG motif is not required for ODNs to act as an antagonist to RIG-I (Fig. 2A and data not shown). Finally, 2006 or 2216 transfected into cells 8 h after shR9 had decreased inhibitory activity, suggesting that they compete with shR9 for recognition by RIG-I (Fig. 2A).
FIGURE 2.
Modified ssDNAs are potent antagonists for RIG-I signaling. A, effects of modified ODN on RIG-I signaling induced by shR9. This experiment also incorporates a timing of addition, and ODN(s) was either transfected into the cells at the same time as the agonist shR9 or 8 h after shR9 transfection. The mean of the -fold induction is shown as a number above the bars, and the range for one S.E. is shown by the lines above the bar. B, the inhibitory effects of ODNs with poly(I:C) (pIC) as the agonist can be different from that of cells induced by shR9. Note that 2216 can slightly enhance the stimulatory effect of pIC. This result has been reproduced in more than five independent assays. As in panel A, the timing of the introduction of the ligands is shown below the graph. C, the phosphorothioate backbone of the ODNs is a major determinant for inhibitory activity of ODNs. Role of phosphorothioate backbone is analyzed using NF-κB and IFN-β reporters. The number and locations of the phosphorothioates in the ODNs are listed under the descriptions.
Given that RIG-I signaling can depend on the agonist used, we examined whether ODNs can affect RIG-I activation by poly(I:C) (Fig. 2B). Although 2006 inhibited poly(I:C)-induced RIG-I signaling, 2216 lost its inhibitory activity. In fact, 2216 showed a modest increase in RIG-I signaling in the presence of poly(I:C). The addition of both the inhibitory 2006 and the non-inhibitory 2216 together with poly(I:C) resulted in inhibition of signaling, demonstrating that the inhibitory effect of 2006 is dominant. Again, treatment with the 2006 8 h after poly(I:C) resulted in only a modest inhibition. Interestingly, 2216 did not induce RIG-I when it was transfected in 8 h after pIC (Fig. 2B).
Phosphorthioates Are Critical for ODNs to Act as Antagonists—The differential effects of 2006 and 2216 on RIG-I signaling could be due to their sequence and/or the number of backbone modifications. Phosphorothioates can increase DNA stability in cells (27). 2006 contains entirely phosphorothioates (a total of 23, 23S), whereas 2216 has six (1 between the two 5′-most two bases and 5 at the 3′-terminus). To examine the roles of the phosphorothioates, we made three derivatives of 2006 without changing the base sequence: 1) d2006, which has all phosphodiesters, 2) 2006-3s, which has three phosphorothioates at each terminus, and 3) 2006-1s, which has one phosphorothioate at each terminus. In addition, we made two derivatives of 2216 without changing the base sequence: 1) s2216, which has all backbones groups replaced with phosphorothioates, and 2) d2216, which contains only phosphodiesters.
The inhibitory activity exhibited by 2006 was lost with d2006 in the presence of agonist shR9 (Fig. 2C). Furthermore, with NF-κB-luciferase reporter assay, 2006-3s and 2006-1s exhibited an inhibitory activity intermediate between 2006 and d2006, demonstrating that the number of phosphorothioates is correlated with antagonist activity of ODNs. With 2216, the same correlation between the numbers of phosphorothioates was observed; s2216 (19S) was more inhibitory to RIG-I signaling in comparison to 2216 (6S) and d2216 (0S). Interestingly, with reporters driven by the IFN-β promoter elements, 2006-3s and 2006-1s, did not show inhibition of RIG-I signaling as observed with NF-κB-luciferase reporter. However, 2216 derivatives showed a similar decrease in RIG-I signaling with both reporters except that d2216 did not inhibit IFN-β reporter (Fig. 2C).
With poly(I:C) as agonist, reducing the number of phophorothioates in the oligonucleotide decreased the antagonistic effect. Interestingly, whereas 2216 (6S) increased signaling by poly(I:C), whereas d2216 (without phosphorothioates) further enhanced the agonist activity of poly(I:C). Interestingly, both NF-κB and IFN-β reporters showed a similar trend in inhibiting RIG-I signaling when poly(I:C) was the agonist, although the level of inhibition was less with the IFN-β luciferase. These results show that ssDNAs can be the activator as well as inhibitor of RIG-I signaling depending on the backbone modification and the DNA sequence.
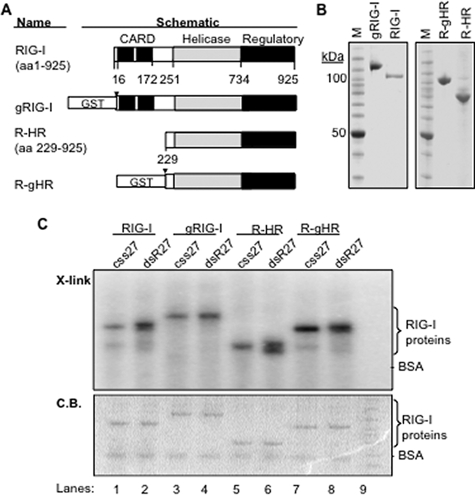
Ligand Binding by RIG-I—The differing effects of agonists and antagonists could be due to different affinities for RIG-I binding. Alternatively, they could have similar affinities but induce RIG-I to form different conformations that could affect activation of signal transduction. Consistent with the second model, Takahasi et al. (20) have observed that single and long double-stranded ligands resulted in different proteolytic fragments from RIG-I. To examine the mechanism of action of the agonist and antagonists further, we used baculovirus to produce several versions of the recombinant RIG-I proteins. The schematics of the constructs are shown in Fig. 3A.
FIGURE 3.
Ligand binding by recombinant RIG-I protein and its derivatives. A, schematics of recombinant proteins expressed by baculovirus-infected insect cells. Two constructs expressed are full-length RIG-I and RIG-I with CARD domain deleted (R-HR). These constructs are expressed with and with out the GST tag that could be removed by treatment with thrombin. The site of cleavage is denoted with a black triangle. The constructs with GST are depicted with prefix g. The three domains of the RIG-I protein are shown in different colors, with key residue numbers shown. aa, amino acids. B, SDS-PAGE of the purified recombinant proteins. Each gel image shows the GST-tagged protein and the version in which the GST was removed and then purified away. M, molecular mass standards. C, results of UV cross-linking assay to examine ligand binding by the four versions of RIG-I protein. The top panel contains an image from an autoradiogram depicting the complexes of the RIG-I with the ligands named above the gel image. The bottom image contains the same gel that was stained with Coomassie Blue (C.B.) to reveal the location of RIG-I and its derivatives along with the internal negative control, BSA.
Full-length RIG-I protein tended to aggregate soon after elution from second purification column, but the addition of a GST tag to the N terminus of RIG-I in a protein named gRIG-I improved solubility. We also generated R-HR and R-gHR, an untagged and GST-tagged version of RIG-I without CARD domains but containing helicase and regulatory domains (Fig. 3A). In both proteins, the GST could be removed by thrombin cleavage, and the resultant purified proteins are named RIG-I and R-HR. The GST-tagged and GST-removed proteins were all purified to greater than 90% purity (Fig. 3B).
To determine whether the recombinant proteins were active, we tested their ability to bind RNA. Unphosphorylated 27-nt ssRNA (css27 and dsR27) were end-labeled and used as probe for the UV cross-linking assay followed by denaturing PAGE (Fig. 3C). RIG-I, R-HR, gRIG-I and R-gHR cross-linked to both ssRNA and dsRNA with one phosphate at the 5′ end (used to radiolabel the RNA), demonstrating that a 5′-triphosphate is not required for ssRNA binding by RIG-I. The presence of a single phosphate on css27 was insufficient to induce RIG-I signaling in cells (data not shown). In these assays BSA served as an internal negative control, and it was not cross-linked to the RNAs to significant levels. This suggests that GST-tagged and untagged versions of RIG-I and R-HR could interact with RNAs in a similar fashion and that the CARD domain is not required for RNA recognition.
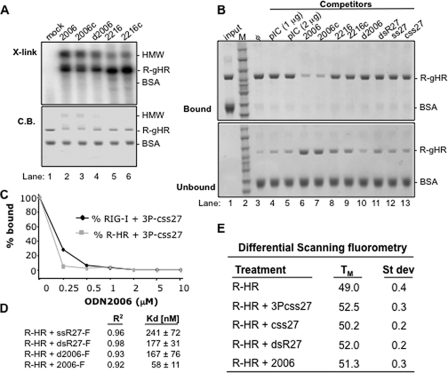
Agonist and Antagonist Binding to RIG-1—Because some ODNs could antagonize RIG-I-dependent signaling, we examined their binding to R-gHR (Fig. 4A). R-gHR was cross-linked to the ODNs specifically, whereas BSA did not. d2006 also cross-linked well to R-gHR even though it lacked phosphorothioate bonds. The cross-linking reaction revealed a prominent higher molecular weight band that could be a dimer of RIG-I (Fig. 4A). This putative dimer is more abundant in the presence of 2006 and 2006c, the two best antagonists we examined. Coomassie Blue staining of the gel containing the R-gHR protein revealed that R-gHR is also present in this position of the gel (Fig. 4A).
FIGURE 4.
Properties of antagonist binding to RIG-I or its derivatives. A, results from a UV cross-linking assay demonstrating that purified R-gHR can be cross-linked to ODNs. The top panel is an autoradiogram of the cross-linked products in a SDS-PAGE. The bottom panel is the SDS-PAGE stained to reveal the locations of the proteins used in the reaction. BSA is used as an internal negative control in all of the reactions. HMW denotes an oligomeric form of RIG-I that is preferentially detected with strong antagonists. B, results of a pulldown assay examining competition of different ligands with R-gHR. The assay uses biotinylated 2006 bound to streptavidin resin, which can specifically bind R-gHR, but not the internal specificity control, BSA. The binding assay was performed in the presence of various competing ligands shown on the top of the gel. C, a competition assay examining UV cross-linking to a internally labeled 5′-triphosphorylated RNA in the presence of increasing concentrations of antagonist 2006. D, fluorescent anisotropy to determine affinity for ligand. Fluorescein isothiocyanate-labeled ligands were used to measure Kd values using R-HR protein, and the R2 values show the fit of the data to the binding isotherm. E, results of differential scanning fluorometry used to measure the melting point (TM) of RIG-I in the presence of different ligands. Results shown are from three independent experiments.
To confirm that ODNs can interact with R-gHR, we examined how different ligands could compete for binding of 2006 to R-gHR. The assay uses a streptavidin resin coupled to 2006 containing a 3′ biotin to generate 2006 resin. After the 2006 resin was incubated with R-gHR and BSA in the presence of different competitors, the bound and unbound materials were collected and analyzed by SDS-PAGE. In the absence of competitors the 2006-resin selectively pulled R-gHR into the bound fraction (Fig. 4B, compare lanes 1 and 3). Poly(I:C) at 1 and 2 μg or 5 μm dsR27 and ssRNAs were inefficient in disrupting R-gHR binding to the 2006-resin (Fig. 4B, lanes 4, 5, and 11-13) when compared with the same concentrations of 2006 and 2006c (lanes 6 and 7). These results suggest that RIG-I preferentially bind strong antagonists (2006 and 2006c) than RNA agonists. Finally, poor antagonists 2216, 2216c, d2006 (2006 without phosphorothioate backbone), and ssRNAs were weaker competitors.
To further confirm the preferential binding of strong antagonists relative to ssRNAs, we performed a UV cross-linking analysis using labeled 3P-css27 as the probe. Binding to RIG-I and R-HR was assessed in the presence of increasing amounts of 2006 (Fig. 4C). 2006 reduced 3P-css27 binding to both RIG-I and R-HR in a concentration-dependent manner. When present at equal concentrations to 3P-css27, 2006 reduced binding of 3P-css27 to less than 30%.
To quantitatively examine the interaction between R-HR and RNA, a fluorescence anisotropy assay was used. Binding isotherms to fluorescein isothiocyanate-labeled css27, dsR27, d2006, and 2006 were generated and fitted into simple hyperbola to derive the Kd values (Fig. 4D). Both ssR27 and dsR27 as well as d2006 showed similar Kd values, 241, 177, and 167 nm, respectively. However, 2006 had a binding constant of 58 nm, consistent with above observations that this antagonist is preferentially bound to RIG-I than the RNA agonists.
The preferential binding of the antagonists begs the question, How do they disrupt signaling? One possibility is that agonists and antagonist induce different conformations of the protein. To compare the complexes formed, we first used differential scanning fluorometry (Fig. 4E). This assay uses the dye SYPRO orange that fluoresces brighter in the presence of hydrophobic environment, such as denatured proteins (28). If RIG-I exists in a different conformation in the presence of agonists and antagonists, the change in fluorescence as a function of temperature might be different. We will use the midpoint of the change in fluorescence as the apparent TM of the complexes. Using a real time PCR machine to regulate the temperature of the reaction, multiple independent samples of R-HR incubated with a 5 m excess of 3P-css27, css27, dsR27, 2006, and SYPRO orange were assayed in a 96-well plate. None of the ligands induced a change in SYPRO orange fluorescence in the absence of protein (data not shown). R-HR had a TM of 49.0 °C in the absence of any ligand (Fig. 4E). In the presence of agonists 3Pcss27 and dsR27, the TM increased to 52.5 and 52 °C, respectively. This suggests that the binding of agonist increased the stability of RIG-I to thermal denaturation. However, in the presence of css27, which can bind R-gHR but cannot activate signaling in cells, the TM was 50.2 °C. With 2006, the TM was 51.3 °C. All values were highly reproducible in three independent sets of assays with multiple independent samples per set (Fig. 4E). These results suggest that RIG-I may form multiple distinguishable complexes with different ligands.
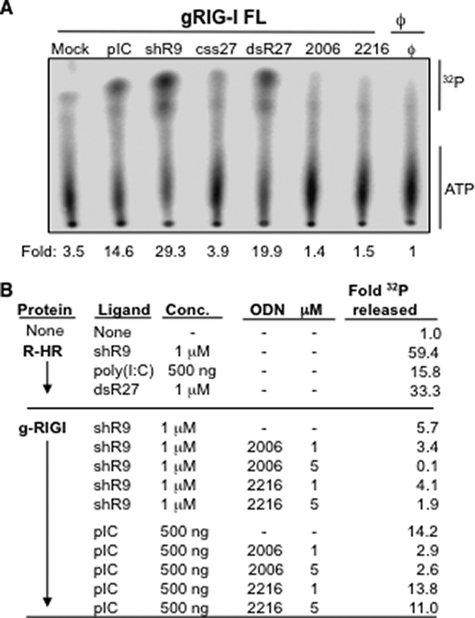
Effects of Agonists and Antagonists on RIG-I ATPase Activity—We examined whether binding to agonist and antagonist would affect the RIG-I ATPase activities. To establish that our proteins had ATPase activity, we incubated the purified gRIG-I protein with [γ-32P]ATP in the absence and presence of ligand (Fig. 5). gRIG-I exhibited 3.5-fold higher ATP hydrolysis compared with a reaction without protein (Fig. 5A). However, in the presence of poly(I:C), shR9, or dsR27, gRIG-I ATPase activity increased to 14.6-, 29.3-, and 19.9-fold. These results show that a ligand is needed to induce RIG-I ATPase activity and are consistent with the observations of Cui et al. (19). Unphosphorylated css27 was a poor inducer of ATPase activity (Fig. 5A). R-HR also had ligand-induced ATPase activity, indicating that the presence of the GST tag or the absence of the CARD domain did not grossly affect RIG-I ATPase activity (Fig. 5B).
FIGURE 5.
Effects of agonists and antagonists on RIG-I ATPase activity. A, a demonstration that the ATPase activity of the GST-tagged version of RIG-I is stimulated by RNA agonists and that antagonist ODNs do not stimulate ATPase activity. B, further characterization of ATPase activity. The top portion of the results shows that R-HR retains ligand-induced ATPase activity despite lacking the CARD domain. The bottom portion shows that antagonist oligonucleotide 2006 can inhibit the ATPase activity of RIG-I. 2216 inhibited only shR9-dependent ATPase activity but not poly(I:C)-dependent ATPase activity.
Next, we examined whether antagonists 2006 and 2216 will affect RIG-I ATPase activity. Both were unable to induce ATPase activity in the absence of RNA ligand (Fig. 5A). However, 2006 present at a 1:1 ratio with agonist shR9 reduced RIG-I ATPase activity to ∼60%. A 5:1 ratio of 2006 to shR9 reduced ATPase activity to background. More modest reduction in shR9-induced RIG-I ATPase activity was observed with 2216 (Fig. 5B). At a 5:1 ratio of 2216 to shR9, a ∼66% reduction was observed (Fig. 5B). These results are consistent with those from cell-based assays that showed 2216 to be a poorer antagonist in comparison to 2006.
Because RIG-I recognized poly(I:C) with distinct properties when compared with shR9 in the cell-based assays (Fig. 1), we tested how antagonists will affect poly(I:C)-induced RIG-I ATPase activity. Poly(I:C)-dependent ATPase activity of gRIG-I was inhibited by 2006 but not by 2216 even at a 5:1 ratio of 2216 to poly(I:C) (Fig. 5B). Remarkably, these results with the purified protein mirror the effects we observed with the antagonists in the cell-based assays (Fig. 2). Furthermore, the results of the ATPase assay indicate that the functional consequence of the agonist and antagonist binding are quite different despite evidence that both bind to RIG-I.
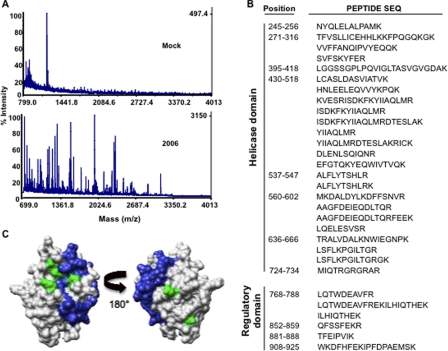
Preliminary Mapping of Antagonist Binding by RIG-I—We wanted to map where antagonist bound to RIG-I using a reversible cross-linking peptide fingerprinting assay. A similar assay was previously used to map template recognition by the hepatitis C virus polymerase (29) and also substrate recognition by the severe acute respiratory syndrome coronavirus endoribonuclease (30). Briefly, 2006 was synthesized with a 3′ amine to allow cross-linking to NHS-succimidyl ester-modified resin (26). R-HR cross-linked to 2006 on the NHS resin was digested with trypsin, and the resin was extensively washed in an attempt to remove peptides that are not covalently linked. Peptides that remained were then heated to 70 °C for 3 h to reverse the cross-linking and then concentrated for mass spectrometry (Fig. 6A). Thirty peptides within 1 dalton of the mass expected from a theoretical trypsin digestion were identified. Multiple overlapping peptides were observed that indicate missed cleavages by trypsin, which could have resulted from the cross-linking, providing confidence to the correct assignment of the peptides (Fig. 6B and supplemental Table 1).
FIGURE 6.
Analysis of the domains in RIG-I that contacts the antagonist 2006. A, a sample of the mass spectra generated by the analysis of the peptides that can be cross-linked to a ligand. The top spectrum shows the background for a mock reaction, whereas the bottom panel shows the spectrum where 2006 was present in the reversible cross-linking reaction. B, a summary of the peptides identified from the reactions with 2006. C, location of the peptides found in the RIG-I regulatory domain that were cross-linked to 2006. The green colors show the location of residues His-830, Ile-875, and Lys-888 that were previously identified to interact with dsRNA by Cui et al. (19).
Twenty-four of the 30 RIG-I-derived peptides mapped to the helicase region and 6 to the C-terminal RD of RIG-I, which approximately corresponds to the difference in mass between the helicase and RD and suggests equal binding of 2006 to the two domains (Fig. 6B). Takahasi et al. (20) and Cui et al. (19) have determined the structure of the RD. When the locations of the cross-linked peptides were highlighted on the x-ray structure (PDB code 2QFB), they mapped to a region near the positively charged groove where three residues His-830, Ile-875, and Lys-888 (shown in green in Fig. 6C) shown to be important for RIG-I activity was observed (19). These data are consistent with our results showing that the antagonists could compete with agonist binding. Furthermore, because the antagonist 2006 could interfere with both ssRNA and dsRNA-induced RIG-I activation of signaling, 2006 can bind to both the helicase and RD. Takahasi et al. (20) had suggested that that the helicase domain contacts poly(I:C).
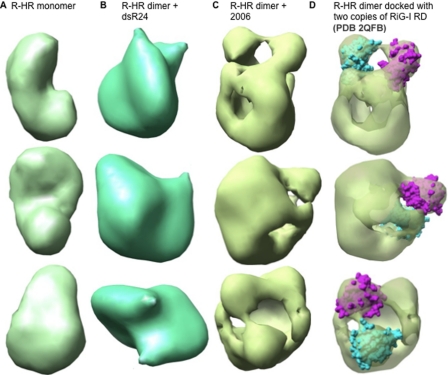
Electron Microscopy and Single Molecule Reconstruction—Thus far we have defined the functional consequence of RIG-I binding to agonists and antagonists. In addition, the differential scanning fluorometry result indicates that agonists and antagonists can result in RIG-I forming distinguishable complexes. To better visualize the conformations, we purified dimers of R-HR in complex with an agonist and an antagonist using gel filtration columns and reconstructed their three-dimensional electron density using negatively stained single particles (Fig. 7 and supplemental Fig. 2). R-HR was used in this analysis because it can bind to agonists and antagonists (Fig. 3C, lanes 4 and 6), and it exhibits ligand-dependent ATPase activity (Fig. 5B). We have also observed that R-HR could act as a dominant negative inhibitor of RIG-I (data not shown). In a related work, we were able to reconstruct that the RIG-I molecule can exist in an open and closed conformation that correlates to the suppression of ligand-independent signaling.3
FIGURE 7.
Reconstruction of monomers of the R-HR domain of RIG-I and R-HR in complex with the agonist dsR24 or the antagonist 2006. The R-HR molecule reconstructed was a monomer, whereas the ligand-bound forms were clearly dimers. A, R-HR monomer. B, R-HR dimer in complex with dsR24. C, R-HR dimer in complex with 2006. D, same as C but with crystal structure of RIG-I regulatory domains docked. Two molecules of RIG-I regulatory domain (shown in cyan and purple) are docked on each R-HR molecule of the dimer. More data for the building of these molecular models are shown in supplemental Fig. 4.
To help identify the RIG-I subunits, we first reconstructed the R-HR monomers (Fig. 7A). About 1900 particles were selected, and the reconstructed structure was resolved to 34 Å (supplemental Fig. 3). The R-HR structure has resemblance to the overall shape of the LGP2 monomer (8).
Next we reconstructed R-HR in complex with dsR24 and 2006 complex (Fig. 7, B and C). Consistent with the gel filtration result (supplemental Fig. 2), the particles appeared as dimers in the electron micrographs, although some monomers and larger structures were also observed (supplemental Fig. 3). More than 6000 particles were subjected to Multi-refine using the EMAN software package to separate the minority of the monomers from the dimers, and the dimers were reconstructed to be 29 and 27 Å for the complexes with dsRNA (to be named the dsRNA dimer) and with 2006 (to be named the 2006 dimer), respectively (supplemental Fig. 3).
Both the dsRNA-bound dimer and the 2006-bound dimer appear to be arranged as head-to-head dimers that are wrapped around a central axis. However, the overall structures of the two dimers were quite different. The two subunits of R-HR dimer complexed with dsRNA were more closely wrapped around the central axis, indicative of additional interaction between the protein subunits. In contrast, the dimer complexed to 2006 has significant space within the complex and occupies a significantly larger diameter than the more compact dsRNA dimer (supplemental Fig. 4). The monomer within the complex resembles the more open structure of RIG-I that we observed under high salt.3 We further tried to dock the RIG-I regulatory domain into the 2006 dimer (cyan and purple structures in Fig. 7D). Notably, two regions that have density that are compatible in volume with the regulatory domain of RIG-I appear to contact each other in the 2006 dimer (Fig. 7, C and D). Overall, electron microscopy reconstruction reveals a dramatic different conformation between the agonist- and antagonist-bound RIG-I dimer.
DISCUSSION
RIG-I has an important role in detecting the infection of RNA viruses including those that cause hepatitis C and influenza. We attempted to better understand how RIG-I could recognize agonists and antagonists and found a complex set of features that can influence signaling.
RIG-I Interaction with Agonists—A remarkable property of RIG-I is that it can interact with and respond to a range of RNA ligands. Single-stranded RNAs require a 5′-triphosphate to activate RIG-I signaling, consistent with the reports in the literature (31, 32). Recently it was shown that the C-terminal regulatory domain of the RIG-I interacts with triphosphates of the RNA and induces dimerization (19, 20). Our reconstructions of R-HR with dsRNA as well as 2006 generated structures that were potential dimers (Fig. 7). Taken together these results suggest that helicase motif did not interfere with dimer formation and probably aids in the dimerization process.
We note that ssRNAs lacking a 5′-triphosphate can be bound by RIG-I in vitro but cannot activate signal transduction (Fig. 3C). In contrast, dsRNAs do not require 5′-triphosphates for binding or to induce signaling. The length of the dsRNA is an important criterion, as dsRNA of 19 bp or less was not capable of inducing RIG-I signaling. Average lengths of ∼25 bp or longer are required for robust signaling (Fig. 1D). Notably, MDA5 and TLR3 activated signal transduction better with dsRNAs of lengths in excess of 100 bp (Fig. 1D and Refs. 25, 33, and 34); thus, these three receptors that recognize RNAs can respond to different dsRNAs.
The complex effects of the ligands may be due to RIG-I having at least two ligand binding domains. The one in the regulatory domain has been demonstrated to bind RNAs with triphosphates, and the helicase domain has been implicated to contact dsRNA such as poly(I:C) (19, 20). The hairpin RNA shR9 has features of both dsRNA and a ssRNA with triphosphate and, thus, could allow some insight into the preference for triphosphorylated RNA versus dsRNA (supplemental Fig. 1). ShR9 does not have blunt ends like dsR27 (supplemental Fig. 1) and behaves more similarly to the less structured 3Pcss27 than poly(I:C) or dsR27 in the cell-based assay (Fig. 1B). Furthermore, mutations K270A and T409A/S411A affected signaling by shR9 in a manner more similar to that of 3P-cssR27. Also, it was shown that 3′ overhangs (as in shR9) and impairs the ability of dsRNA to induce RIG-I signaling (35). These results suggest that the recognition of the triphosphate by the RIG-I RD has more of an impact than recognition of the double-stranded portion of the RNA.
We observed that both gRIG-I and R-HR have ATPase activity that can be activated by triphosphorylated ssRNA or dsRNAs lacking triphosphates. These results suggest that the CARD domain is not required for interaction with RNA or for ATPase activity, although we cannot rule out some regulatory effects of the CARD domain on the ATPase activity of the helicase domain. However, given that both poly(I:C) and heteropolymeric dsRNA dsR27 can induce RIG-I mediated signaling, it is likely that activation of ATPase activity of the helicase domain is not specific for the sequence of the dsRNA.
Ligand binding by either the RIG-I helicase or regulatory domains can lead to some degree of signal transduction. We base this claim on the observation that mutations in RIG-I such as K270A abolished signal transduction by triphosphorylated ssRNA 3P-css27 and shR9 but retained signaling in the presence of either dsR27 or poly(I:C). Furthermore, although either ssRNA or dsRNA can induce ATPase activity, a mutation in the Walker A motif that is presumed to be defective in ATPase activity did not affect signaling by at least some dsRNA (Fig. 1B). This suggests that ATPase activity is not necessarily a prerequisite for signaling by dsRNA. The T409A/S411A mutation in RIG-I could also signal in the presence of dsRNA but not ssRNA, indicating that dsRNA can bypass some of the requirements for signaling by triphosphorylated ssRNA. Based on in vitro binding analysis, it was reported that T409A/S411A mutation affected poly(I:C) binding (20). However, using a cell-based assay, we observed that this mutant could be induced by poly(I:C). It is possible that the RIG-I-agonist complex undergo different conformational changes upon binding to ss- and dsRNA. Furthermore, LGP2 was unable to inhibit RIG-I activity when poly(I:C) was the agonist. Taken together these results suggest that dsRNA may be able to induce RIG-I signaling by a different mechanism than ssRNA.
RIG-I Interaction with Antagonists—We found that single-stranded oligodeoxynucleotides that are not agonists can modulate RIG-I signaling and the induction of ATPase activity by RNA agonists. Using a reversible cross-linking peptide fingerprinting assay, we found that antagonist 2006 can interact with both the C-terminal regulatory domain as well as the helicase domain (Fig. 6). Inhibition of agonist-dependent ATPase activity by 2006 could be possibly due to its interaction with the ATPase motif. 2006 also interacted with the regulatory domain at the same region that was proposed to bind RNA (19, 20). These data provides a mechanism by which 2006 interferes with RIG-I signaling.
Phosphorothioates in the backbone of these ssDNAs is a key determinant for antagonistic or modulatory activity. Interestingly, signaling by TLR3 can also be antagonized by phosphorothioated ssDNA (26), suggesting that this class of modified oligonucleotides has a propensity to interact with dsRNA binding proteins. An interesting property of the ssDNAs is that the same sequences lacking phosphorothioates or only containing a few phosphorothioates, such as 2216, can act to stimulate RIG-I signaling. This was not observed with TLR3 (26). Also, 2006 modifications showed different levels of inhibition of RIG-I when reporter-driven by NFκB promoter was used but did not have as significant an inhibitory effect on the reporter driven by the IFN-β promoter. This indicates that the correlation of RIG-I inhibition and phosphorothioate bonds is not due to a difference in stability of these antagonists. It is possible that 2006 has pleiotropic effects on innate immunity signaling pathways. We have already documented that 2006 can bind to TLR3 and affect its function in several cell lines. Altogether, these properties of modified ssDNAs suggest that proper design and engineering of the ssDNAs will be needed to regulate signaling by a specific innate immunity pathway.
Comparison to LGP2—Ligand binding by RIG-I has several differences when compared with baculovirus-expressed LGP2 (8). First, purified LGP2 strongly preferred binding to dsRNA than ssRNA. Second, we observed that LGP2 binds dsRNA with cooperativity. In preliminary anisotropy experiments (Fig. 4D), RIG-I binding to ssRNA or dsRNA did not exhibit cooperativity. Based on empirical evidence, we believe that LGP2 will bind dsRNA with higher affinity, but it will also be more discriminating against ssRNA. Last, reconstruction of LGP2 with a dsRNA has a density that could be best explained with a head-to-tail dimer (8), whereas the RIG-I complex to either agonists or antagonists appears to be head-to-head dimers (Fig. 7, B and C). It will be of interest to reconstruct the RIG-I heterodimer with LGP2.
RIG-I Conformational Changes and Signaling—The multitude of responses from RIG-I is complex and likely reflects different RIG-I monomer and oligomer conformations that could lead to different signaling responses. Intramolecular interaction in RIG-I is suggested to sequester the CARD domain and prevent spurious signaling (18). In addition, both the agonists and antagonists appear to induce dimer formation. Differential scanning fluorometry is a quick and sensitive method to monitor protein unfolding. The thermal unfolding of protein can be measured by using the environmentally sensitive dye SYPRO orange whose fluorescence increases with increasing exposure of hydrophobic portions of proteins. This methodology was used to determine binding of compounds to β-amyloid precursor protein-cleaving enzyme 1 (BACE1) (36). Binding of inhibitors to BACE1 protein was considered significant when the change in TM was as low as 0.4 °C (2 times the error). We used a similar methodology to analyze the interaction of RIG-I with its agonists and antagonist (Fig. 4E). Use of differential scanning fluorometry revealed that RIG-I in complex with dsRNA resulted in a structure more stable to thermal denaturation than RIG-I in complex with 2006. The change in the TM observed RNA agonist and 2006 ranged from 2.3 to 3.5 °C, significantly above experimental error from multiple trials in our analyses (Fig. 4E). Interestingly, single molecule reconstruction revealed that the RIG-I dimer complexed to dsRNA was a tighter complex that had more extensive surface interactions than the more open complex with 2006, perhaps accounting for the differences in the TM values of the proteins in complex with agonists and antagonists in solution (Fig. 4E).
The 2006 dimer complex contains four distinct sets of contacts, suggesting that the interaction of the antagonists induces additional interactions. Additional protein-protein contacts were also observed with the Toll protein dimer in complex with the ligand Spatzel (37). It is likely that changes in the oligomer structures of innate immunity receptors will hold the key to downstream signaling.
The resolution of the reconstruction prevents firm assignment of domains, but several features could help reduce the possibilities. First, the reconstructions were performed with R-HR, which lacked the CARD domain that is not needed for ligand binding. R-HR is somewhat better than the full-length RIG-I in binding ligands. Second, in a separate report,3 we captured full-length RIG-I in distinct open and closed conformations in a manner dependent on the salt condition. These structures allowed a reasonable assignment of the RD and CARD by manual docking. A comparable structure is also seen in the 2006 dimer (Fig. 7D). We note with interest that the two putative regulatory domains appear to contact each other in the complex.
Supplementary Material
Acknowledgments
We thank our colleagues at Centocor for helpful discussions concerning the role of innate immunity receptor antagonists.
This work was supported, in whole or in part, by National Institutes of Health Grant 1RO1AI073335 (NIAID; to C. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1-4.
Footnotes
The abbreviations used are: ssRNA, single-stranded RNA; dsRNA, double-stranded RNA; RIG-I, retinoic acid-inducible protein I; CARD, caspase recruitment domains; ODN, oligodeoxynucleotide; TLR3, Toll-like receptor 3; RD, regulatory domain; GST, glutathione S-transferase; BSA, bovine serum albumin; nt, nucleotide; IFN, interferon; HCV, hepatitis C virus.
C. T. Ranjith-Kumar, D. Srisathiyanarayanan, A. Murali, X. Li, W. Dong, M. Sherman, P. Li, and C. C. Kao, manuscript in preparation.
References
- 1.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783-801 [DOI] [PubMed] [Google Scholar]
- 2.Gaspari, A. A. (2006) J. Am. Acad. Dermatol. 54 Suppl. 2, 67-80 [Google Scholar]
- 3.Kobayashi, T., Takaesu, G., and Yoshimura, A. (2006) Nat. Immunol. 7 123-124 [DOI] [PubMed] [Google Scholar]
- 4.Underhill, D. M. (2004) Curr. Opin. Immunol. 16 483-487 [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730-737 [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama, M., and Fujita, T. (2007) J. Biol. Chem. 282 15315-15318 [DOI] [PubMed] [Google Scholar]
- 7.Saito, T., and Gale, M., Jr. (2007) Curr. Opin. Immunol. 19 17-23 [DOI] [PubMed] [Google Scholar]
- 8.Murali, A., Li, X., Ranjith-Kumar, C. T., Bhardwaj, K., Holzenburg, A., Li, P., and Kao, C. C. (2008) J. Biol. Chem. 283 15825-15833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101-105 [DOI] [PubMed] [Google Scholar]
- 10.Sumpter, R., Jr., Loo, Y. M., Foy, E., Li, K., Yoneyama, M., Fujita, T., Lemon, S. M., and Gale, M., Jr. (2005) J. Virol. 79 2689-2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, G., Zhong, J., and Chisari, F. V. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8499-8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foy, E., Li, K., Sumpter, R., Jr., Loo, Y. M., Johnson, C. L., Wang, C., Fish, P. M., Yoneyama, M., Fujita, T., Lemon, S. M., and Gale, M., Jr. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2986-2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981-988 [DOI] [PubMed] [Google Scholar]
- 14.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, J. (2005) Nature 437 1167-1172 [DOI] [PubMed] [Google Scholar]
- 15.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669-682 [DOI] [PubMed] [Google Scholar]
- 16.Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005) Mol. Cell 19 727-740 [DOI] [PubMed] [Google Scholar]
- 17.Loo, Y. M., Owen, D. M., Li, K., Erickson, A. K., Johnson, C. L., Fish, P. M., Carney, D. S., Wang, T., Ishida, H., Yoneyama, M., Fujita, T., Saito, T., Lee, W. M., Hagedorn, C. H., Lau, D. T., Weinman, S. A., Lemon, S. M., and Gale, M., Jr. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6001-6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito, T., Hirai, R., Loo, Y. M., Owen, D., Johnson, C. L., Sinha, S. C., Akira, S., Fujita, T., and Gale, M., Jr. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 582-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui, S., Eisenacher, K., Kirchhofer, A., Brzozka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K. K., Krug, A., and Hopfner, K. P. (2008) Mol. Cell 29 169-179 [DOI] [PubMed] [Google Scholar]
- 20.Takahasi, K., Yoneyama, M., Nishihori, T., Hirai, R., Kumeta, H., Narita, R., Gale, M., Jr., Inagaki, F., and Fujita, T. (2008) Mol. Cell 29 428-440 [DOI] [PubMed] [Google Scholar]
- 21.Guarino, L. A., Bhardwaj, K., Dong, W., Sun, J., Holzenburg, A., and Kao, C. (2005) J. Mol. Biol. 353 1106-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludtke, S. J., Baldwin, P. R., and Chiu, W. (1999) J. Struct. Biol. 128 82-97 [DOI] [PubMed] [Google Scholar]
- 23.Sun, J., Duffy, K. E., Ranjith-Kumar, C. T., Xiong, J., Lamb, R. J., Santos, J., Masarapu, H., Cunningham, M., Holzenburg, A., Sarisky, R. T., Mbow, M. L., and Kao, C. (2006) J. Biol. Chem. 281 11144-11151 [DOI] [PubMed] [Google Scholar]
- 24.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comput. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 25.Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., Hiiragi, A., Dermody, T. S., Fujita, T., and Akira, S. (2008) J. Exp. Med. 205 1601-1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjith-Kumar, C. T., Duffy, K. E., Jordan, J. L., Eaton-Bassiri, A., Vaughan, R., Hoose, S. A., Lamb, R. J., Sarisky, R. T., and Kao, C. C. (2008) Mol. Cell. Biol. 28 4507-4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinozuka, K., Morita, T., and Sawai, H. (1991) Nucleic Acids Symp. Ser. 25 101-102 [PubMed] [Google Scholar]
- 28.Niesen, F. H., Berglund, H., and Vedadi, M. (2007) Nat. Protoc. 2 2212-2221 [DOI] [PubMed] [Google Scholar]
- 29.Kim, Y. C., Russell, W. K., Ranjith-Kumar, C. T., Thomson, M., Russell, D. H., and Kao, C. C. (2005) J. Biol. Chem. 280 38011-38019 [DOI] [PubMed] [Google Scholar]
- 30.Bhardwaj, K., Palaninathan, S., Alcantara, J. M., Yi, L. L., Guarino, L., Sacchettini, J. C., and Kao, C. C. (2008) J. Biol. Chem. 283 3655-3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994-997 [DOI] [PubMed] [Google Scholar]
- 32.Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., and Reis e Sousa, C. (2006) Science 314 997-1001 [DOI] [PubMed] [Google Scholar]
- 33.Leonard, J. N., Ghirlando, R., Askins, J., Bell, J. K., Margulies, D. H., Davies, D. R., and Segal, D. M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 258-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, L., Botos, I., Wang, Y., Leonard, J. N., Shiloach, J., Segal, D. M., and Davies, D. R. (2008) Science 320 379-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marques, J. T., Devosse, T., Wang, D., Zamanian-Daryoush, M., Serbinowski, P., Hartmann, R., Fujita, T., Behlke, M. A., and Williams, B. R. (2006) Nat. Biotechnol. 24 559-565 [DOI] [PubMed] [Google Scholar]
- 36.Lo, M. C., Aulabaugh, A., Jin, G., Cowling, R., Bard, J., Malamas, M., and Ellestad, G. (2004) Anal. Biochem. 332 153-159 [DOI] [PubMed] [Google Scholar]
- 37.Gangloff, M., Murali, A., Xiong, J., Arnot, C. J., Weber, A. N., Sandercock, A. M., Robinson, C. V., Sarisky, R., Holzenburg, A., Kao, C., and Gay, N. J. (2008) J. Biol. Chem. 283 14629-14635 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.