Abstract
Plant–parasitic cyst nematodes secrete a complex of cell wall–digesting enzymes, which aid in root penetration and migration. The soybean cyst nematode Heterodera glycines also produces a cellulose binding protein (Hg CBP) secretory protein. To determine the function of CBP, an orthologous cDNA clone (Hs CBP) was isolated from the sugar beet cyst nematode Heterodera schachtii, which is able to infect Arabidopsis thaliana. CBP is expressed only in the early phases of feeding cell formation and not during the migratory phase. Transgenic Arabidopsis expressing Hs CBP developed longer roots and exhibited enhanced susceptibility to H. schachtii. A yeast two-hybrid screen identified Arabidopsis pectin methylesterase protein 3 (PME3) as strongly and specifically interacting with Hs CBP. Transgenic plants overexpressing PME3 also produced longer roots and exhibited increased susceptibility to H. schachtii, while a pme3 knockout mutant showed opposite phenotypes. Moreover, CBP overexpression increases PME3 activity in planta. Localization studies support the mode of action of PME3 as a cell wall–modifying enzyme. Expression of CBP in the pme3 knockout mutant revealed that PME3 is required but not the sole mechanism for CBP overexpression phenotype. These data indicate that CBP directly interacts with PME3 thereby activating and potentially targeting this enzyme to aid cyst nematode parasitism.
INTRODUCTION
Cyst nematodes, Heterodera and Globodera spp, are sedentary parasites of plant roots in many economically important cropping systems where they cause severe yield loss (Barker and Koenning, 1998; Wrather et al., 2001). The soybean cyst nematode, Heterodera glycines, is regarded as the most serious pathogen problem in soybean production worldwide, and the sugar beet cyst nematode (Heterodera schachtii) also is a devastating pathogen to many plant species in addition to sugar beets. To sustain their subsequent sedentary parasitic stages, cyst nematodes induce the formation of elaborate feeding sites, termed syncytia, whose etiology includes substantial cell wall reorganization and dissolution in addition to dramatic cytoplasmic and nuclear changes (Williamson and Hussey, 1996). The parasitic feats of cyst nematodes during penetration, migration, syncytium formation and maintenance, and feeding are aided by proteinaceous stylet secretions, which are the expression products of specific parasitism genes in nematode esophageal gland cells (Davis et al., 2004, 2008). Parasitism proteins are synthesized as preproteins with N-terminal signal peptides targeting them to the gland cell endoplasmic reticulum and the secretory pathway (Qin et al., 2000; Wang et al., 2001). Nematode parasitism proteins are developmentally produced and released through the stylet into plant tissues and cells to promote the parasitic interaction. While two subventral gland cells are most active during the early stages of parasitism (i.e., root penetration and migration) and early events of syncytium formation, the single dorsal gland cell becomes most active in the later stages of syncytium formation, maintenance, as well as during feeding.
A large number of H. glycines parasitism genes were identified by different approaches, most recently the microaspiration of esophageal gland cell cytoplasm followed by the construction and mining of gland cell cDNA libraries (Gao et al., 2001, 2003; Wang et al., 2001; de Boer et al., 2002). Among the subventral gland-produced parasitism proteins of H. glycines was a short protein with high similarity to the cellulose binding domain (CBD) of previously identified H. glycines endoglucanases (Smant et al., 1998). Additionally, actual binding to cellulose was documented experimentally; therefore, this protein was termed cellulose binding protein, or Hg CBP (Gao et al., 2004). Cyst nematode endoglucanases produced in the subventral glands and secreted during the migratory phase are thought to be key parasitic tools to breach root cell walls (Smant et al., 1998; de Boer et al., 1999; Wang et al., 1999; Goellner et al., 2001). Cyst nematode endoglucanase expression sharply decreases as syncytia are initiated (de Boer et al., 1999; Goellner et al., 2000); subsequently, plant endoglucanase expression is upregulated within the developing feeding cells (Goellner et al., 2001).
The plant cell wall provides essential mechanical strength and rigidity and also acts as a physical barrier against pathogens. The primary cell wall is a network of crystalline cellulose microfibrils embedded in a matrix of hemicelluloses and pectins. Cellulose is a particularly difficult polymer to degrade, as it is insoluble and is present as hydrogen-bonded crystalline fibers (Doi and Kosugi, 2004). Microorganisms have evolved the ability to break down plant cell walls, which involves the production and secretion of a large number of cellulase enzymes that can act synergistically (Collmer and Keen, 1986). In the case of aerobic fungi and bacteria, the majority of these cellulases have a specific modular structure consisting of a catalytic domain, responsible for hydrolysis, and a CBD responsible for attachment of the enzymes to the insoluble cellulose substrate.
CBD-containing proteins that have no apparent hydrolytic activity but exhibit cellulose binding activity were first isolated from the cellulolytic bacterium Clostridium cellulovorans (Shoseyov and Doi, 1990). This CBP exhibited a relatively high affinity for cellulose and was found to be essential for the degradation of crystalline cellulose (Goldstein et al., 1993). It is thought that CBD determines the efficiency of degradation of insoluble cellulose by concentrating cellulase catalytic domains on the surface of the insoluble cellulose substrate (Tomme et al., 1998; Carrard et al., 2000).
More than 200 CBDs, either as CBPs alone or as part of catalytic proteins, have been identified from different organisms and classified into different families according to their amino acid sequence similarities in the CAZY database (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/cbm.html). CBDs also differ in their binding affinity and substrate specificity (Carrard et al., 2000; McCartney et al., 2006). In plant-parasitic nematodes, the first CBP gene to be identified was Mi CBP-1 from the root-knot nematode Meloidogyne incognita (Ding et al., 1998). Mi CBP-1 encodes a signal peptide, a C-terminal CBD, and a 71–amino acid N-terminal region with unknown function. These latter two domains are joined by a linker peptide. Mi CBP-1 was found to bind to cellulose and plant cell walls but lacked cellulase activity. Strong evidence for secretion and involvement of Mi CBP-1 in pathogenesis was provided by the detection of this protein in stylet secretions. Hg CBP from H. glycines was the second such cDNA from a nematode (Gao et al., 2003, 2004) and the first CBP identified in nematodes consisting only of a signal peptide and a CBD. Like Mi CBP-1, recombinant Hg CBP protein had no hydrolytic activity on carboxymethyl-cellulose but was able to bind to cellulose in an in vitro assay. Similarly, the potato cyst nematode (Globodera rostochiensis) secrets a noncellulase CBD-containing protein. However, another region of this protein was similar to plant expansins, and cell wall–softening activity could be shown experimentally (Qin et al., 2004). Hg CBP does not have similarity to expansins, and its functional roles in plant–nematode interactions still remain elusive. However, the developmental expression patterns of Hg CBP during different nematode life stages suggest a role in pathogenesis most likely during the establishment and maintenance of the nematode feeding sites (Gao et al., 2004).
Very limited data regarding the effects of CBPs on the living plant cell are available. It has been shown that a recombinant bacterial CBD enhanced elongation of different plant cells in vitro (Shpigel et al., 1998). Similarly, a bacterial CBD from C. cellulovorans was also found to modulate plant growth in transgenic poplar plants leading to a significant increase in biomass production in selected clones when compared with the wild type (Shoseyov et al., 2001). Although these data indicate that CBDs can modulate plant growth, the mechanisms by which these unique CBDs affect plant development or susceptibility to plant-parasitic nematodes still remain obscure. In this study, we used an integrative approach to investigate the function of CBP during cyst nematode parasitism. Interestingly, we discovered an unexpected mode of action of cyst nematode CBPs that entails a direct interaction with a plant endogenous pectin methyl esterase protein.
RESULTS
Identification of the Sugar Beet Cyst Nematode CBP-1 cDNA
To make use of the extensive resources and technical advantages offered by the model plant Arabidopsis thaliana, which is a host to the sugar beet cyst nematode H. schachtii (Sijmons et al., 1991), we isolated the H. schachtii homolog of H. glycines CBP using a homology-based PCR approach. Identical to the Hg CBP (Gao et al., 2004), the H. schachtii CBP cDNA (Hs CBP) contained an open reading frame of 396 bases encoding a 132–amino acid protein with an N-terminal signal peptide for secretion and a CBD of 107 amino acids. The H. schachtii protein had 94% identity and 97% similarity to the CBP of H. glycines (see Supplemental Figure 1 online).
Localization of CBP Transcripts
One of the hallmark characteristics of parasitism genes is their exclusive expression in the secretory esophageal gland cells. To provide evidence that CBP is expressed uniquely in the esophageal gland cells, the tissue localization of CBP mRNA was analyzed in different parasitic life stages of H. schachtii using in situ mRNA hybridization. The digoxigenin-labeled antisense cDNA probes of CBP hybridized exclusively with transcripts accumulating within the subventral esophageal gland cells (see Supplemental Figure 2A online). No signal was detected when the control sense cDNA probes were used. The Hg CBP was also shown to hybridize exclusively to the subventral esophageal gland cells in H. glycines (Gao et al., 2003).
Developmental Expression Pattern of CBP
The expression profile of a given parasitism gene will identify the most likely time of activity of the corresponding parasitism protein. Therefore, we assessed the CBP expression profile using real-time RT-PCR through the six H. schachtii developmental stages of eggs, preparasitic second-stage juvenile (pre-J2), parasitic J2 (par-J2), third-stage juvenile (J3), fourth-stage juvenile (J4), and adult females. CBP mRNA steadily increased from eggs to J3 and then rapidly declined after the J3 stage (see Supplemental Figure 2B online). This expression pattern is very similar to that previously observed in H. glycines (Gao et al., 2004; Elling et al., 2007a) and supports a role in pathogenicity as a secreted protein. The expression peak at the J3 stage again suggests a role during the early phases of syncytium formation.
Transgenic Arabidopsis Plants Expressing CBP Are Altered in Morphology and Susceptibility to H. schachtii
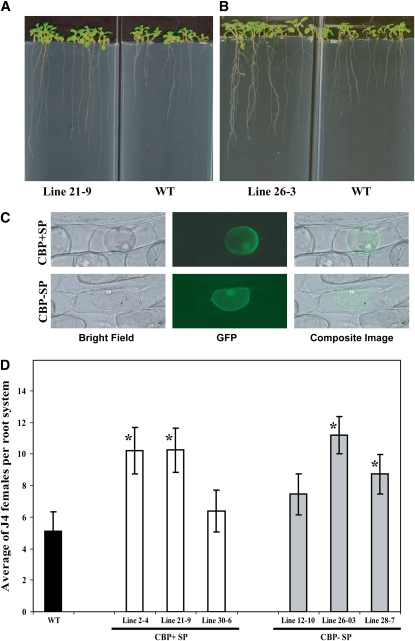
It has been shown that CBDs can affect plant growth and development in vitro and in planta (Shpigel et al., 1998; Shoseyov et al., 2001). Therefore, it was particularly interesting to see if CBP overexpression produces similar phenotypes. In addition, if CBP has a function in cell wall modification, one can reasonably postulate that ectopic expression in Arabidopsis should alter cell wall–associated phenotypes. To investigate this, we transformed Arabidopsis with gene constructs containing the 35S cauliflower mosaic virus (CaMV) promoter driving the Hs CBP cDNA either with the native signal peptide (SP+CBP) or without (SP−CBP). Presence or absence of the signal peptide was thought to potentially be helpful in discerning if the site of CBP action is in the apoplast or the symplast of the host plant, respectively. For each of the two constructs, multiple independent homozygous T3 lines were identified. Lines 2-4, 21-9, and 30-6, containing CBP with signal peptide, and lines 12-10, 26-3, and 28-7, containing CBP without signal peptide, were found to express CBP at high levels as quantified by real-time RT-PCR and were used in further studies. Seeds from these six homozygous lines were planted on nutrient medium, and root length was measured 9 d after planting. Interestingly, expression of SP+CBP as well as SP−CBP produced plants with significantly increased root lengths (Figures 1A and 1B) ranging from 21.68 mm ± 0.91 mm to 33.09 mm ± 2.03 mm in the SP+CBP transgenic lines and 26.67 mm ± 1.57 mm to 33.88 mm ± 0.82 mm in the SP−CBP transgenic lines compared with the wild-type control (16.71 mm ± 0.97 mm), while no statistically significant differences were detected between lines with and without the signal peptide coding sequence. Also, no significant differences were observed in the aboveground parts of any CBP transgenic lines when compared with the wild type. These dramatic changes in morphology document that the nematode-produced CBP has a biological function in planta. Also, the in planta expression phenotype changes are reminiscent of those seen by others investigating bacterial CBDs in planta, suggesting a conserved mode of action of both nematode and bacterial molecules, although bacterial CBDs also influenced shoot phenotypes (Shpigel et al., 1998).
Figure 1.
Characterization of Hs CBP.
(A) and (B) Expression of Hs CBP in Arabidopsis increases root length. Homozygous T3 lines expressing either Hs CBP with (A) or without (B) signal peptide exhibited significantly longer root systems ranging from 21.68 mm ± 0.91 mm to 33.09 mm ± 2.03 mm in the transgenic lines expressing CBP with signal peptide and 26.67 mm ± 1.57 mm to 33.88 mm ± 0.82 mm in the transgenic lines expressing CBP without signal peptide compared with wild-type C24 (16.71 mm ± 0.97 mm) at 10 d after planting as determined by unadjusted paired t tests (P < 0.01). This observation was true with all six homozygous T3 lines tested.
(C) Subcellular localization of Hs CBP. Hs CBP cDNA with or without signal peptide-coding sequence was fused to the GFP and GUS reporter genes and expressed in onion epidermal cells. After plasmolysis, GFP fluorescence was retained inside the protoplast in both cases.
(D) Transgenic Arabidopsis plants expressing Hs CBP showed enhanced susceptibility to H. schachti. Homozygous T3 lines expressing either Hs CBP with (lines 2-4, 21-9, and 30-6) or without (lines 12-10, 26-3, and 28-7) the native signal peptide were planted on modified Knop's medium, and 2-week-old seedlings were inoculated with ∼250 surface-sterilized J2 H. schachtii nematodes. Two weeks after inoculation, the number of J4 female nematodes per root system was determined. Data are presented as the mean ± se. Mean values significantly different from the wild type are denoted by an asterisk as determined by unadjusted paired t tests (P < 0.05). Identical results were obtained from at least two independent experiments.
Since the signal peptide had no effect on plant phenotype, it can be postulated that, most likely, the nematode signal peptide does not properly work in planta and that the observed plant phenotypes are due to a cytoplasmic accumulation of CBP. To test this hypothesis, we fused the 3′-ends of both SP+CBP and SP−CBP cDNAs to the green fluorescent protein (GFP) and β-glucuronidase (GUS) reporter genes (SP+CBP:GFP:GUS and SP−CBP:GFP:GUS) under the control of double CaMV 35S promoters. Both constructs were delivered into onion epidermal cells by biolistic bombardment. After 24 h of incubation following bombardment, the onion epidermal cells were treated with 1 M sucrose solution to separate cytoplasm from cell walls (plasmolysis). For both constructs, GFP signals were localized only within transformed plant cells rather than in cell walls (Figure 1C), suggesting that the native nematode signal peptide in fact did not function properly in planta and that the observed CBP overexpression phenotypes are due to events that are initiated in the cytoplasm.
Secretion of CBP by cyst nematodes into host plants should facilitate parasitism. Therefore, we determined the susceptibility of the same six CBP-expressing transgenic Arabidopsis lines described above in nematode infection assays in which the numbers of female nematodes were determined and used to quantify plant susceptibility. Four out of the six transgenic lines expressing CBP cDNAs were significantly more susceptible to H. schachtii than the wild type (Figure 1D). The remaining two lines also showed increases in susceptibility; however, they were statistically nonsignificant. It is important to mention that there was a clear relationship between CBP expression level and the degree of susceptibility. The two lines that showed the lowest susceptibility among all test lines were those with the lowest CBP expression levels. In other words, these lines required between 1.6 and 3.57 additional PCR cycles to reach the transgene detection threshold (see Supplemental Table 1 online). These results establish that CBP can function in planta in a manner conducive to successful cyst nematode parasitism.
To evaluate whether this elevated level of susceptibility was a specific result of CBP functioning as a cyst nematode parasitism protein or merely was a function of the increased root area available for penetrating nematodes, two lines (21-9 and 26-3) showing the most significant increase in root length were inoculated with H. schachtii J2 along with the wild-type control, and 4 d after inoculation the total number of penetrated nematodes was counted. Data from two independent experiments revealed no statistically significant differences between the tested lines and the wild-type control (see Supplemental Figure 3 online). These data indicate that the increased susceptibility to H. schachtii is not due to elevated penetration rates conditioned by enlarged roots but rather is due to postpenetration effects of the H. schachtii CBP parasitism protein.
Similarly, all six transgenic Arabidopsis lines expressing CBP cDNAs were inoculated with the root knot nematode M. incognita, which uses a different penetration and in planta migration strategy and induces different feeding cells than cyst nematodes. Although slightly elevated, no statistically significant effect of CBP on plant susceptibility to M. incognita was observed (see Supplemental Figure 4 online), indicating that CBP functions specifically to modulate parasitic success of cyst nematodes.
To further explore why CBP-expressing Arabidopsis lines are more susceptible to H. schachtii, we measured the size of syncytia formed in transgenic and wild-type Arabidopsis lines. For this purpose, 10 single-nematode syncytia were randomly selected for size measurement from lines 21-9 and 26-3 10 d after inoculation. No statistically significant differences were observed between the size of syncytia from transgenic lines expressing CBP (0.249 mm2 ± 0.01 mm2) and the wild-type control (0.231 mm2 ± 0.018 mm2). These results indicate that the mode of action responsible for increased susceptibility of CBP-expressing Arabidopsis lines does not involve an enlargement of syncytia per se.
CBP Interacts with Arabidopsis Pectin Methylesterase 3
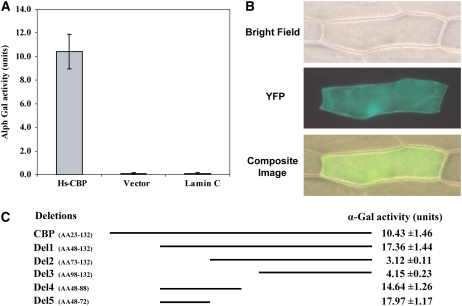
Judging from the fact that CBP lacks an obvious catalytic domain, it is plausible that interactions with other proteins are required for CBP function. We therefore set out to identify host proteins that interact with CBP and used a yeast two-hybrid (Y2H) approach to screen three Arabidopsis Y2H prey libraries, which we generated from roots at 3, 7, and 10 d after H. schachtii inoculation. These libraries, consequently, contained cDNA clones from parasitic H. schachtii at the different time points after infection along with the cDNAs from Arabidopsis plants at various stages of nematode parasitism. After screening of ∼15.62 × 106 yeast colonies from the three prey libraries in roughly equal proportions, we identified 90 clones of candidate interactor (prey) proteins. Prey plasmids were rescued from these yeast colonies, and DNA sequencing revealed that these clones coded for nine independent Arabidopsis proteins. The specificity of these putative interactions was further scrutinized as described in Methods, and only one protein consistently and reproducibly gave a specific interaction with Hs CBP (see Supplemental Figure 5A online). This protein was Arabidopsis pectin methylesterase 3 (PME3; At3g14310). The positive interaction between Hs CBP and PME3 was further confirmed by α-Gal quantitative assays (Figure 2A). To provide additional confirmatory evidence of this interaction, bimolecular fluorescent complementation (BiFC) assays were performed. Hs CBP and PME3 without signal peptides were fused to N-terminal and C-terminal halves of yellow fluorescent protein (YFP), respectively, and coexpressed in onion epidermal cells. The interaction between CBP and PME3 reconstituted the activity of YFP in the cytoplasm of transformed cells (Figure 2B). No YFP fluorescence was obtained when the YFP fragment constructs containing CBP or PME3 were bombarded alone or in combination with empty vectors or an unrelated nematode gene.
Figure 2.
Hs CBP Reproducibly Interacts with PME3.
(A) α-Gal quantitative assays of the CBP/PME3 interaction. Yeast strain AH109 was cotransformed with the prey plasmid in combination with either the CBP bait vector, pGBKT7-lam (expressing lamin C as a GAL-4 DNA-BD fusion), or the empty pGBKT7 bait vector and plated on SD/-Leu/-Trp. Three days after culture, 10 separate colonies per combination were picked to quantify the interaction using α-Gal activity. Activity was seen only in yeast cells containing the PME3 prey plasmid and the CBP bait vector. The experiment was repeated four times with identical results.
(B) BiFC visualization of the CBP/PME3 interaction. Onion epidermal cells were cobombarded with constructs expressing the nEYFP-CBP and cEYFP-PME3 and bright-field, YFP and overlay of bright field, and YFP images were taken 20 h after bombardment.
(C) Mapping the CBP domain involved in the interaction with PME3. Yeast strain AH109 cotransformed with prey plasmid and the different bait constructs indicated in the scheme were streaked on SD/-Leu/-Trp, and 3 d after culture, 10 separate colonies per construct were picked to quantify α-galactosidase activity. The assays were repeated three times with identical results.
Because the Arabidopsis genome contains at least 66 PME-related genes, we tested whether other PME proteins can bind to CBP in Y2H assays as well. In these experiments, PME2 (At1g53830), which shares the highest sequence similarity with PME3 among all PME family members, was cloned with and without signal peptide in the prey vector. Also, PME1 (At1g53840), a nonsecretory protein, was cloned in the prey vector. The potential interactions between these two PME proteins and CBP were tested after cotransformation of prey and bait plasmids into yeast. No interaction between CBP and these preys was detected, indicating that the interaction between CBP and PME3 is highly specific (see Supplemental Figure 5B online).
To identify the region of amino acids of CBP required for the interaction with PME3, a series of deletions of the CBP sequence in the bait vector was generated and the interaction was examined by α-Gal quantitative assay after cotransformation of bait and prey plasmids into the requisite yeast strain. N-terminal Hs CBP amino acid deletions Δ1-72 and Δ1-97 showed only weak interaction with PME3 (Figure 2C). However, the shorter N-terminal amino acid deletion Δ1-47 gave rise to an increased interaction when compared with the complete CBP sequence. This finding indicates that the first 47 amino acids of CBP are not critical for the interaction and that the next 25 amino acids are necessary for binding to PME3. To test this possibility, a bait plasmid containing only amino acids 48 to 88 was generated. This bait construct was found to bind to the prey and gave rise to strong α-Gal activity. Finally, a bait construct containing only amino acids 48 through 72 gave rise to the strongest interaction (Figure 2C), indicating that these 25 amino acid residues are necessary and sufficient for recognition and binding of PME3 in the Y2H system.
Subcellular Localization of PME3 and Temporal Expression during H. schachtii Infection
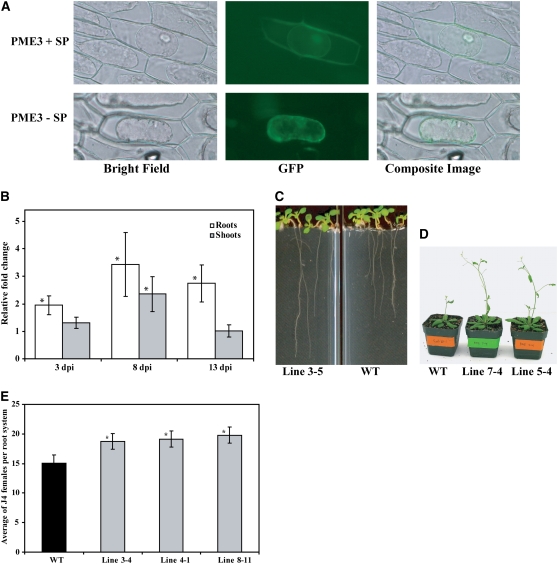
To investigate the subcellular localization of PME3, the 3′-end of the PME3 coding sequence (with or without the native signal peptide coding region) was cloned before the start codon of a joined GFP-GUS coding sequence construct, thus giving rise to a PME3:GFP:GUS translational fusion protein that is big enough to prevent passive diffusion among cell compartments. This gene construct was expressed in onion epidermal cells. After plasmolysis, the GFP signal was detected only in the cytoplasm of the onion cells bombarded with the fusion construct containing PME3 without the signal peptide. However, when the PME3 construct with signal peptide was fused to GFP, the reporter protein was detected both in the cell wall and the cytoplasm (Figure 3A). These results corroborate our Y2H-predicted CBP-PME3 in planta interaction since both proteins can be found within the plant cell cytoplasm.
Figure 3.
Characterization of Arabidopsis PME3.
(A) Subcellular localization of PME3. PME3 cDNA with or without signal peptide-coding sequence was fused to GFP and GUS reporter proteins and expressed in onion epidermal cells. After plasmolysis, GFP signal was detected both in the cell wall and the cytoplasm of the transformed cells expressing whole PME3 (top panel). By contrast, in onion cells expressing PME3 without signal peptide, the GFP signal was detected only in the cytoplasm (bottom panel).
(B) Upregulation of PME3 in response to H. schachtii infection. The mRNA expression level of PME3 was measured by quantitative real-time RT-PCR in wild-type (C24) root and shoot tissues. Infected and noninfected tissues were collected at 3, 8, and 13 d after inoculation (dpi). The fold-change values were calculated using the 2−ΔΔCT method and represent changes of mRNA abundance in infected tissues relative to noninfected controls. Data are the average of three independent biological experiments, each consisting of four technical replicates. Mean values significantly different from 1.0 (no change) are indicated by an asterisk as determined by paired t tests (P < 0.01).
(C) and (D) Transgenic Arabidopsis plants overexpressing PME3 are altered in morphology. Homozygous T3 lines overexpressing PME3 displayed longer roots (C) and taller shoots (D) than the wild type (Col-0).
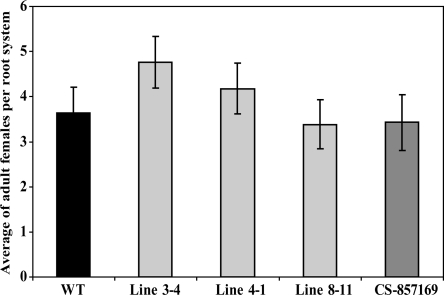
(E) Transgenic Arabidopsis plants overexpressing PME3 revealed increased susceptibility to H. schachtii. Homozygous T3 lines overexpressing PME3 (lines 3-4, 4-1, and 8-11) were planted on modified Knop's medium, and 2-week-old seedlings were inoculated with ∼250 surface-sterilized J2 H. schachtii nematodes. Two weeks after inoculation, the number of J4 female nematodes per root system was determined. Data are presented as the mean ± se. Mean values significantly different from the wild type (Col-0) as determined by unadjusted paired t tests (P < 0.05) are denoted by an asterisk. Identical results were obtained from at least two independent experiments.
If indeed PME3 has a function during cyst nematode parasitism, it is conceivable that PME3 mRNA abundance may change during cyst nematode infection, potentially as a result of the protein–protein interaction or other mechanisms. PME3 mRNA was quantified in triplicate quantitative real-time RT-PCR assays using gene-specific primers designed to discriminate between different members of the PME gene family (see Methods). Two-week-old wild-type Arabidopsis seedlings were inoculated with H. schachtii, and root and shoot tissues were harvested from treated and control plants at 3, 8, and 14 d post inoculation for RNA extraction. Data from three independent experiments revealed that PME3 mRNA was upregulated in H. schachtii–inoculated roots at all time points when compared with noninfected roots. The strongest induction was observed at the 8-d time point (Figure 3B). Interestingly, PME3 mRNA abundance was much weaker in shoots than in roots, and significant upregulation was only seen at the 8-d time point (Figure 3B). Quantifying the gene expression level of PME3 in root versus shoot tissues of 2- to 3-week-old uninfected Arabidopsis plants revealed a >58-fold higher PME3 expression in roots when compared with shoots.
PME3 Expression Levels Correlate with Arabidopsis Susceptibility
To provide additional information about the involvement of PME3 in the plant response to H. schachtii, transgenic Arabidopsis lines overexpressing the full-length PME3 cDNA with the signal peptide and under the control of 35S CaMV promoter were generated and phenotypically investigated. Independent T3 homozygous lines developed longer roots and taller shoots than Col-0 wild-type plants (Figures 3C and 3D). When transgenic lines were assayed for nematode susceptibility, all three T3 homozygous lines tested (3-4, 4-1, and 8-11) supported significantly higher numbers of adult females than wild-type plants (Figure 3E). These findings indicate that, much like CBP, PME3 activity is conducive to H. schachtii parasitism.
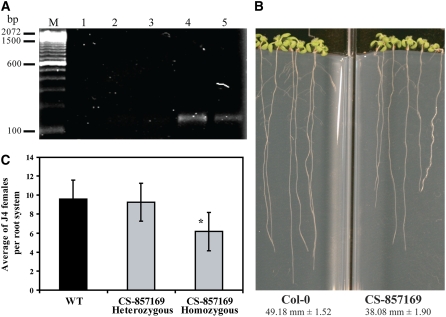
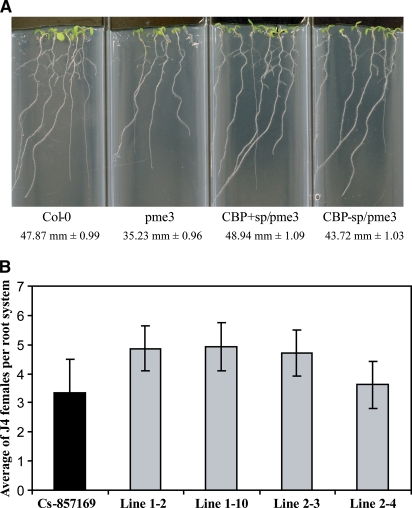
To further explore PME3 function, we screened a T-DNA insertion mutant database at The Arabidopsis Information Resource (Alonso et al., 2003) and identified a T-DNA–tagged PME3 insertional mutant (CS857169). Sequence analysis of this mutant allele revealed that the T-DNA insertion is located in the likely promoter at 305 nucleotides upstream of the start codon. Quantitative real-time RT-PCR analysis of PME3 mRNA abundance in homozygous mutant plants using gene-specific primers failed to detect any transcript even after high numbers of PCR amplification cycles. By contrast, specific amplifications were detected in both heterozygous and wild-type plants (Figure 4A). To address whether the knockout mutation of PME3 affected root length, because we observed larger roots in overexpression lines, we examined 15-d-old seedlings grown on nutrient medium. We determined that the root length of mutant plants was significantly shorter (38.08 mm ± 1.90 mm) than that of the wild type (49.18 mm ± 1.52 mm) (Figure 4B), which is in line with the overexpression phenotype described above. Most importantly, we also determined the susceptibility to H. schachtii of pme3 knockout mutants in three independent experiments. Compared with the wild type, mutant plants were significantly less susceptible to H. schachtii (Figure 4C).
Figure 4.
The pme3 Knockout Mutant Is Altered in Root Length and Susceptibility to H. schachtii.
(A) PME3 mRNA accumulation in the pme3 mutant. PME3 mRNA level was determined by quantitative real-time RT-PCR using gene-specific primers. The real-time RT-PCR products were resolved on syber safe-stained 2% agarose gel. No PCR products were detected after 40 cycles of amplification of cDNA from homozygous mutant plants (lanes 1 to 3), whereas specific amplifications were detected after amplification of cDNA from either heterozygous (lane 4) or wild-type (Col-0) plants (lane 5). Molecular weight marker is a 100 bp ladder (Invitrogen).
(B) The pme3 knockout mutant develops shorter roots than the wild-type (Col-0). Homozygous plants were planted on modified Knop's medium with the wild type (Col-0), and root lengths were measured 15 d after planting. Root length values are averages of at least 30 plants ± se. Differences between pme3 and the wild type were statistically significant as determined by unadjusted paired t tests (P < 0.01).
(C) The pme3 knockout mutant is less susceptible to H. schachtii than the wild type (Col-0). The pme3 knockout mutant (homozygous and heterozygous) and wild-type (Col-0) plants were planted on modified Knop's medium, and 2-week-old seedlings were inoculated with ∼250 surface-sterilized J2 H. schachtii. Two weeks after inoculation, the number of J4 female nematodes per root system was counted. Data are presented as the mean ± se. Mean values significantly different from that of the wild type as determined by unadjusted paired t tests (P < 0.05) are denoted by an asterisk. Identical results were obtained from at least four independent experiments.
As was performed for CBP-expressing Arabidopsis lines, we assayed PME3-overexpressing lines and the knockout mutant for susceptibility to M. incognita. These assays did not reveal altered susceptibility phenotypes of these lines (Figure 5), documenting that the PME3 effects, much like CBP, are specific for the cyst nematode interaction.
Figure 5.
Root-Knot Nematode Susceptibility Is Not Altered in PME3-Overexpressing Lines and the pme3 Knockout Mutant.
Homozygous T3 lines overexpressing PME3 (lines 3-4, 4-1, and 8-11) or the pme3 knockout mutant (CS-857169) as well as wild-type (Col-0) plants were planted on modified Knop's medium, and 2-week-old seedlings were inoculated with ∼250 surface-sterilized J2 M. incognita. Four weeks after inoculation, the numbers of adult egg-laying female nematodes per root system were counted. No statistically significant differences between the tested lines and the wild-type control were detected. Data are presented as the mean ± se. Identical results were obtained from two independent experiments.
To provide additional evidence for the direct connection between CBP and PME3, we measured PME activity in the transgenic plants expressing CBP (line 2-4), the pme3 knockout mutant, and the PME3 overexpression plants (line 3-4) under noninfected conditions. As expected, the pme knockout mutant showed an ∼15% decrease in PME activity, while PME3 overexpression plants exhibited a dramatic increase of PME activity of almost 260% (Table 1). Interestingly, a small but statistically significant increase of PME activity of ∼8% was detected in the CBP-expressing plants compared with the wild-type control (Table 1). These findings support our finding of an effect of CBP on PME function and suggest that CBP interaction with PME3 increases this protein's activity.
Table 1.
PME Activity in Transgenic Plants Overexpressing Hs CBP or PME3 and in the pme3 Knockout Mutant
| Line | PME Activity (nmol H+/min/mg protein) | % | P Value |
|---|---|---|---|
| C24 | 341.3 ± 3.6 | 100 | |
| CBP overexpressing line (2-4) | 367.7 ± 3.9 | 108 | 0.01900 |
| Col-0 | 371.8 ± 1.9 | 100 | |
| pme3 knockout mutant | 317.1 ± 6.0 | 85 | 0.04900 |
| PME3 overexpressing line (3-4) | 964.0 ± 7.6 | 259 | 0.00001 |
PME activity was measured in roots of 5-d-old plants. Data are presented as the mean ± se. Mean values significantly different from the wild-type controls were determined by paired t tests (P < 0.05).
Expression of CBP in the pme3 Knockout Mutant
To determine whether CBP affects plant phenotypes exclusively through its interaction with PME3, we expressed CBP in the pme3 knockout mutant, and four nonsegregating T2 lines were phenotypically analyzed. Interestingly, these transgenic plants showed significantly increased root lengths ranging from 43.72 mm ± 1.03 mm to 48.94 mm ± 1.09 mm compared with the pme3 knockout mutant (35.23 mm ± 0.96 mm) (Figure 6A). However, the average increase of 32% was dramatically less than the 64% observed in wild-type plants expressing CBP. When these lines were used in nematode assays to determine plant susceptibility to H. schachtii, we observed a trend of weakly increased plant susceptibility compared with pme3 knockout mutant. However, these changes were statistically not significant (Figure 6B). These data indicate that CBP in fact requires PME3 for the majority of the observed phenotypic effects but that other, minor mechanisms also do exist.
Figure 6.
Overexpression of CBP in the pme3 Knockout Mutant.
pme3 knockout mutant plants expressing CBP with or without signal peptide exhibited significantly longer root systems when compared with the pme3 knockout mutant at 14 d after planting (A) and nonsignificant increases of H. schachtii susceptibility (B) as determined by unadjusted paired t tests (P < 0.05). Root length values are averages of at least 30 plants ± se.
DISCUSSION
Identification of both nematode and host proteins involved in key processes of the compatible plant–nematode interaction will provide insights into the mechanisms of successful parasitism. At the same time, these studies reveal valuable insights into general plant biology as new functions of plant proteins are discovered. The soybean cyst nematode has been shown to produce a unique secretory CBP consisting only of a CBD without a catalytic domain (Gao et al., 2004). Several lines of evidence suggest that CBP is secreted during parasitism. First of all, there is no cellulose within the nematode and CBP genes are not found in any nonparasitic or animal-parasitic nematodes, indicating that the site of action is within the plant tissue. Furthermore, the presence of an N-terminal signal peptide and the absence of transmembrane domain motifs in the protein sequence, together with the CBP gene's exclusive expression in the nematode secretory subventral gland cells, document that CBP is a secreted protein involved in parasitism. If indeed CBP has the important function in pathogenesis as hypothesized, its gene should be expressed during a defined period of parasitic stages. Indeed, we observed high increases of CBP mRNA abundance in the parasitic J2 and an expression peak in the J3 stages, which suggests a role in parasitism during the time period after penetration into the plant root and particularly during early syncytium formation and development. In this period, cell walls are dramatically altered/remodeled during nematode migration as well as during extensive incorporation of root cells into the syncytium. The ability of the recombinant Hg CBP protein to bind to cellulose in in vitro assays (Gao et al., 2004) again supports a role of CBP inside plant tissues where cellulose represents an ubiquitous molecule. Finally, detection of a CBP in stylet secretions of M. incognita J2 (Ding et al., 1998) provides strong evidence that CBPs are secreted proteins with functions in plant tissues during nematode parasitism.
Expressing parasitism genes in plant hosts to assess their effects on plant phenotype and nematode susceptibility provides a meaningful way to functionally characterize parasitism proteins (Wang et al., 2005; Huang et al., 2006). We expressed the Hs CBP cDNA in Arabidopsis, which resulted in increased root length compared with wild-type controls. This observation is in agreement with the previously reported functional role of bacterial CBDs in living plant cells. Shpigel et al. (1998) reported that the CBD of the CBP of C. cellulovorans enhanced elongation of peach pollen tubes and Arabidopsis roots when it was exogenously applied at low concentrations. Recently, transgenic potato plants producing a bacterial CBP were found to be taller and larger than wild-type plants at early stages of development but did not differ significantly at later developmental stages (Safra-Dassa et al., 2006), suggesting that this CBP can function in a stage-dependent manner, potentially correlated with elevated rates of cell division.
The fact that CBP serves an important function during the plant–nematode interaction became evident through the significantly elevated susceptibility of Arabidopsis plants expressing Hs CBP. This result was further confirmed by our finding that the increased root size per se was not the cause of the increase in the number of nematodes penetrating into the roots but that rather a cyst nematode-specific root alteration had taken place that allowed higher parasitic success postpenetration. These data along with our expression analyses indicate that CBP most likely functions not during the penetration phase of infection but rather during early phases of syncytium formation. Interestingly, our data revealed no significant difference in syncytia size between the transgenic CBP-expressing lines and wild-type plants. The functional role of CBP in facilitating nematode parasitism, therefore, could be accelerated feeding site development.
Like CBP, PME3 showed convincing cause to postulate an important role during parasitism. PME3 overexpression lines showed phenotypes similar to CBP overexpression, including a significantly elevated susceptibility to cyst nematodes. By contrast, knocking out PME3 resulted in the exact opposite phenotypes of shorter roots and reduced susceptibility. PME3, therefore, plays a crucial role in cyst nematode parasitism, which also gives additional credence to the conclusion that the interaction with PME3 is of importance for the function of CBP. In addition, the soybean ortholog of PME3 (BE821923) was shown to be upregulated within the actual developing syncytia induced by the soybean cyst nematode H. glycines (Ithal et al., 2007), which provides additional evidence of a role for PME3 in the cyst nematode infection process. Most importantly, a direct functional connection between PME3 and CBP is supported by our finding that transgenic plants expressing Hs CBP exhibited higher PME activity than wild-type controls. The observed 8% increase in PME activity is even more impressive when considering that (1) the employed assay measures all root PME activity, consisting of numerous enzymes and that CBP appears to only interact with PME3; and (2) mutational knockout of PME3 resulted in an activity loss of only 15%. Furthermore, the root preferential expression of PME3 most likely is the cause of the root-specific phenotypes in CBP-expressing Arabidopsis lines and again highlights the relevance of the interaction between both proteins.
Several pieces of evidence have indicated that PMEs contribute to cell growth by regulating the mechanical and chemical properties of plant cell walls via demethylesterification of pectin (Micheli, 2001). Transgenic potato plants overexpressing pectin methylesterase were characterized by relatively rapid elongation at early stages of development (Pilling et al., 2000). The enlarged size of plants overexpressing CBP or PME3 may be attributable to this type of PME action. It has been suggested that PMEs also have a role in resistance to fungal and bacterial pathogens (McMillan et al., 1993; Boudart et al., 1998; Wietholter et al., 2003) by affecting the physicochemical properties of the cell wall to be more accessible to cell wall–digesting enzymes (Lionetti et al., 2007). Interestingly, we did not find significant effects of CBP or PME3 on plant susceptibility to M. incognita. The finding that CBP and PME are specific to cyst nematodes can be explained by the fact that CBP functions during syncytium formation and the fact that root-knot nematodes induce completely different feeding sites, termed giant cells, that are derived through unrelated etiology despite the relatively close phylogenetic relationship between both nematodes.
Due to the new insights provided here, the mechanism by which CBP facilitates nematode parasitism becomes clearer. Taken together, our results show that this parasitism protein functions during syncytium formation through its interaction with PME3. However, it remains unclear where these two proteins interact. Despite our findings that CBP, when produced in planta, is most probably a cytoplasm-localized protein, it appears likely that, ultimately, CBP functions in the cell wall, given the fact that the cell wall is the site of cellulose accumulation and also where PME3 is expected to be localized. In this scenario, CBP would be secreted by the nematode using its stylet into the host plant cytoplasm from where it is hypothesized to be exported into the apoplast and the cell wall. Because we observed PME3 in the cytoplasm as well as the cell wall, interaction with CBP could be in the cytoplasm followed by a potential joint export. Ultimately, further research has to address this remaining question.
Our data further suggest that CBP functions in the targeting (to cellulose-associated pectin) and/or activation of PME3 to reduce the level of methylesterification of pectin in the cell wall. The opposite relationship between PME activity and pectin methylesterification level has been described in the literature (Lionetti et al., 2007). Therefore, it appears most likely that a reduction of cell wall pectin methylesterification through CBP-mediated increased and targeted PME3 activity allows improved access of other cell wall–modifying enzymes to cell wall polymers, thereby accelerating enzymatic activities, which is a requirement for syncytium development. In support, the ability of polygalacturonases and pectate lyases to degrade the pectin main chain has been shown to depend on the activity of PME (Christgau et al., 1996). By contrast, the expected high level of methylesterified pectin in the pme3 knockout mutant would hamper the activity of cell wall–modifying enzymes and consequently reduce their effectiveness resulting in reduced susceptibility, which is what we observed in the nematode susceptibility assays. Expression of CBP in the pme3 knockout mutant resulted in a slight increase of nematode susceptibility of ∼34% compared with the pme3 mutant. However, this percentage is much lower than the 78% observed when CBP was expressed in the wild type, showing that the CBP–PME3 interaction is a required, but not the sole, mechanism for the increased susceptibility observed in CBP-overexpressing lines. Similarly, CBP expression in the pme3 knockout mutant resulted in only a 32% increase of root length compared with the 64% increase when CBP was expressed in the wild type. This is an example of a molecular plant–animal interaction using a bipartite cooperative means of cell wall modification to further the parasitic success of the nematode. In conclusion, reducing the level of methylesterified pectin in the cell walls of presyncytial cells during the early stages of syncytium formation through the activation/targeting of PME3 is the most likely mode of action of CBP in facilitating parasitism.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds were surface-sterilized in 50% bleach for 5 min followed by four rinses in sterile water. Plants were grown under sterile conditions on Murashige and Skoog (MS) medium (PlantMedia) containing 2% sucrose solidified with 0.8% phyto agar (RPI) or in Metro-Mix 200 soil mixture (Sun Gro Horticulture) in a growth chamber (16 h light/8 h dark) at 23°C. Arabidopsis ecotype C24 was used as the wild type for overexpression of Hs CBP, while ecotype Columbia-0 (Col-0) was used for overexpression of PME3. For phenotypic assays of overexpressing lines or knockout mutant lines, the corresponding wild type was used for comparison.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The coding sequences of Hs CBP with or without the nematode signal peptide (SP) sequence were amplified from the full-length cDNA clone (EU328302) with primers CBP(SP) forward (5′-TATAGGATCCATGAATTGGATGCATTATTGTTTAATC-3′) or CBP forward (5′-TATAGGATCCATGTCCACTATTAATTCGGTAACCGTAC-3′) and CBP reverse (5′-ATGATGAGCTCTAGGGCAATTAGCTTAATTGGGTAGG-3′), which contained BamHI or SacI restriction sites (underlined) for directional cloning. PCR amplification was performed using the Expand High Fidelityplus PCR system (Roche) according to the manufacturer's instructions. PCR products were digested by BamHI and SacI, gel purified, and ligated into the binary vector pBI121. Cloned products were verified by sequencing. The coding region of PME3 was amplified from Arabidopsis cDNA as described above using gene-specific primers designed to create the XbaI and XhoI restriction sites in the forward and reverse primers, respectively. The primer sequences used were PME forward 5′-CTCTAGAATGGCACCATCAATGAAAGAAATT-3′ and PME reverse 5′-CCTCGAGTCAAAGACCGAGCGAGAAGGG-3′ (restriction sites are underlined). The amplified products were digested by XbaI and XhoI, gel purified, ligated into XbaI-XhoI restriction sites of the binary vector pBI121, transformed, and confirmed by sequencing. Agrobacterium tumefaciens strain C58 was transformed with the binary plasmids by the freeze-thaw method (An et al., 1988) and used to transform Arabidopsis wild-type C24, Col-0, or pme3 knockout mutant plants as described by Clough and Bent (1998). Transformed T1 plants were screened on MS medium containing 50 mg/L kanamycin to select for transgenic plants. Homozygous T3 seeds were collected from T2 lines after segregation analysis on kanamycin-containing medium.
Root Length Measurements
Seeds were surface sterilized and transferred to Nunc 4 well Polystyrene Rectangular Dishes (Thermo Fisher Scientific) containing MS medium. Plates were incubated in a vertical position in a growth chamber at 23°C under 16-h-light/8-h-dark conditions. At the time of measurement, the root length of at least 10 plants per treatment was measured as the distance between the crown and the tip of the main root in three independent experiments. Statistically significant differences between lines were determined by unadjusted paired t test (P < 0.01).
Nematode Infection Assay
Transgenic Arabidopsis seeds (T3 generation) and wild-type controls (Col-0 and C24) were surface sterilized and planted in a random block design in 12-well Falcon culture plates (BD Biosciences) containing modified Knop's medium (Sijmons et al., 1991) solidified with 0.8% Daishin agar (Brunschwig Chemie). Plants were grown at 24°C under 16-h-light/8-h-dark conditions. Two-week-old seedlings were inoculated with ∼250 surface-sterilized J2 Heterodera schachtii or Meloidogyne incognita nematodes as previously described by Baum et al. (2000). Inoculated plants were maintained under the conditions described above for an additional 2 weeks before the number of H. schachtii J4 females were counted. In the case of M. incognita, the number of J4 females was counted 4 weeks after inoculation.
Nematode Penetration Assay
The penetration rate of H. schachtii second-stage juveniles was studied in lines 21-9 and BV2-26-3 and compared with wild-type Arabidopsis C24. The three lines were planted in a random-block design on modified Knop's medium in 12-well culture plates as described above. At 10 d, each plant was inoculated with 150 surface-sterilized J2 of H. schachtii, and the plates placed back in the incubator for 4 additional days. Then, each well was stained for the presence of penetrating nematodes using a modified version of the protocol from Grundler et al. (1991). In short, nematodes were stained by adding 1 mL of fixation/staining solution (500 mL acetic acid, 500 mL 96% ethanol, and 17 mg acid fuchsin) and incubated at room temperature for 24 h. Each well was then rinsed with water, and 1 mL of destaining solution (600 mL water, 200 mL glycerine, and 200 mL lactic acid) was added. The number of penetrating nematodes in each root system was counted by observing the root systems under bright-field illumination at ×200 using a Zeiss Axiovert 100 microscope. Each plant line was replicated 18 times, and two independent experiments were conducted. Average numbers of penetrating nematodes were calculated, and statistically significant differences were determined in a modified t test using the statistical software package SAS.
Syncytial Measurements
Syncytial measurements were taken 24 d after inoculation of H. schachtii onto the three subject lines. For each line, 10 single-female syncytia were randomly selected and photographed on a Zeiss Stemi SV11 dissecting microscope using a Zeiss AxioCam MRc5 digital camera. The digital files were then processed using Zeiss Axiovision software (release 4.4). For each photographed syncytium, the image was processed with the Axiovision measure tool and calibrated against the correct scaling of the original image from the Stemi SV11. The measurements were aided by the use of a Wacom Intuos 3 9x12 drawing tablet (Wacom Technology). Using the tablet, the syncytial cells were outlined, and total area for the syncytium was calculated as square millimeters. The individual measurements for the syncytium of each line were used to calculate an average syncytial size for each line tested using the statistical software package SAS. Significance of size differences between lines was determined via a modified t test in SAS (P < 0.05).
RNA Isolation and Quantitative Real-Time RT-PCR
Total RNA was extracted from 100 mg frozen ground plant tissues using the RNeasy plant mini kit (Qiagen) or from 50 mg nematode tissues using the Versagene RNA tissue kit (Gentra Systems) following the manufacturer's instructions. DNase treatment of total RNA was performed using DNase I (Invitrogen). Gene-specific primers to Hs CBP, PME3, H. schachtii actin (AY443352), and Arabidopsis actin (AT1G49240) were designed. For real-time RT-PCR in H. schachtii, 10 ng of DNase-treated RNA were used for cDNA synthesis and PCR amplification using the one-step RT-PCR kit (Bio-Rad) according to the manufacturer's protocol. The PCR reactions were run in an I Cycler (Bio-Rad) using the following program: 50°C for 10 min, 95°C for 5 min, and 40 cycles of 95°C for 30 s and 60°C for 30 s. Following PCR amplification, the reactions were subjected to a temperature ramp to create the dissociation curve, determined as changes in fluorescence measurements as a function of temperature, by which the nonspecific products can be detected. The dissociation program was 95°C for 1 min, 55°C for 10 s, followed by a slow ramp from 55°C to 95°C. For real-time RT-PCR in Arabidopsis, first-strand cDNA was synthesized from DNase-treated RNA using the Advantage RT-for-PCR kit (Clontech) according to the manufacturer's instructions. The synthesized cDNAs then were diluted to a concentration equivalent to 10 ng total RNA/μL and used as a template in real-time RT-PCR reactions using the two-step RT-PCR kit (Bio-Rad) according to manufacturer's protocol. In all cases, at least three independent experiments each with three technical replicates of each reaction were performed. Arabidopsis and nematode actin, as constitutively expressed genes, were used as internal controls to normalize gene expression levels. Quantification of the relative changes in gene expression was performed using the 2−ΔΔCT method as described by Livak and Schmittgen (2001).
Subcellular Localization
The Hs CBP and PME3 coding sequences with and without signal peptide-encoding regions were amplified using gene-specific primer pairs containing HindIII and XbaI restriction enzyme sites in the forward and reverse primers, respectively. For CBP, the primer sequences used were CBP(SP) forward (5′-TATAAAGCTTATGAATTGGATGCATTATTGTTTAATC-3′) or CBP forward (5′-TATAAAGCTTATGTCCACTATTAATTCGGTAACCGTAC-3′) and CBP reverse (5′-ATGATTCTAGATTTTTTTGCATTGTTTGCTGGTTGGA-3′). For PME3, primer sequences used were PME3(SP) forward (5′-TATAAAGCTTATGGCACCATCAATGAAAGAAATTTTTTC-3′) or PME3 forward (5′-TATAAAGCTTATGATCTCTGCCGGAGCTTCAAAAGCC-3′) and PME3 reverse (5′-ATGATTCTAGATAAGACCGAGCGAGAAGGGGAAACCG-3′). The resulting amplified fragments were cloned into the respective sites in the modified pRJG23 vector (Grebenok et al., 1997) before the start codon of GFP fused into the GUS reporter gene and under the control of double CaMV 35S promoter. All constructs were confirmed by DNA sequencing. These constructs were delivered into onion epidermal cells by biolistic bombardment as described by Elling et al. (2007b). After bombardment, epidermal peels were incubated for 24 h in the dark. Plasmolysis of onion epidermal cells was achieved by soaking the cells in 1 M sucrose solution for 15 min. The subcellular localization of the fused proteins was visualized using a Zeiss Axiovert 100 microscope. The transient transformation experiments were repeated at least three times independently.
Y2H Assays
A Y2H screening was performed as described in the BD Matchmaker Library Construction and Screening Kits (Clontech). The complete coding sequence of CBP was fused to the GAL4 DNA binding domain (BD) of pGBKT7 to generate pGBKT7-CBP and then introduced into Saccharomyces cerevisiae strain Y187 to generate the bait strain. Three Arabidopsis cDNA libraries from roots of ecotype C24 at 3, 7, and 10 d after H. schachtii infection were generated in S. cerevisiae strain AH109, as fusion to the GAL4 activation domain (AD) of pGADT7-Rec2 vector. Screening for interacting proteins and subsequent analyses were performed as described in Clontech protocols.
BiFC Analysis of Hs CBP and PME3
The CBP cDNA without signal peptide was PCR-amplified using forward primer (5′-TATAGAATTCATCCACTATTAATTCGGTAACCGTAC-3′) and reverse primer (5′-ATGATTCTAGATCATTTTTTGCATTGTTTGCTGGTTGGA-3′) containing EcoRI and XbaI restriction sites (underlined), respectively, and cloned into EcoRI-XbaI sites of pSAT4-nEYFP-C1 to generate pSAT4-nEYFP-CBP. Meanwhile, the full-length PME3 cDNA without signal peptide was PCR amplified using forward primer (5′-TATAGAATTCAATCTCTGCCGGAGCTTCAAAAGCC-3′) and reverse primer (5′-ATGATTCTAGATCAAAGACCGAGCGAGAAGGGGAAACCG-3′) containing EcoRI and XbaI restriction sites (underlined), respectively, and cloned into EcoRI-XbaI sites of pSAT4-cEYFP-C1(B) to generate pSAT4-cEYFP-PME3. Both plasmids were confirmed by sequencing. For coexpression, particle bombardment was performed using onion epidermal cells. Gold particles (1.6 μm diameter) (Bio-Rad) were washed with 100% ethanol and coated with 1.5 μg of each DNA using standard procedures. cDNA-coated gold particles were bombarded at 1100 p.s.i. and 9 cm distance using a Biolistic Particle Delivery System PDS-1000/He (Bio-Rad). Bombarded tissues were incubated at 25°C in darkness for ∼24 h before being assayed for YFP activity. The bright-field and fluorescent images were taken using the Zeiss Axiovert 100 microscope with appropriate YFP filter.
PME Activity
The PME-containing protein fractions were extracted from root tissues using a high-salt buffer as described by Ren and Kermode (2000). The root tissues of 5-d-old Arabidopsis plants were collected and fully ground in liquid nitrogen and then homogenized with 200 μL PME extraction buffer (0.1 M citrate acid, 0.2 M Na2HPO4, and 1 M NaCl, pH 5.0). The homogenized materials were incubated on ice for 1 h, during which they were mixed three times at 20-min intervals, and finally centrifuged for 10 min at 14,000 rpm at 4°C. The supernatant was collected and the crude protein content was quantified using a Pierce BCA-200 protein assay kit (Pierce) following the manufacturer's instructions. PME activity was assayed according to the method described by Richard et al. (1994). Two micrograms of protein samples were added to 4 mL of substrate solution containing 0.5% citrus pectin (Sigma-Aldrich), 0.2 M NaCl, and 0.002% methyl red, pH 6.8, for 1 h at 37°C. Pectin de-esterification decreases the pH, thus changing the color from yellow to red. The color change was measured using a spectrophotometer (UV-Vis Spectrophotometer; Cary 50 Bio) as OD at 525 nm. A calibration curve was obtained by adding 5 to 30 μL 0.01 M HCl to 4 mL of substrate solution and measuring the respective OD values at 525 nm. PME activity of different samples (nmol H+/min/mg protein) was obtained according to the calibration curve.
In Situ Hybridization
Specific forward and reverse primers for the Hs CBP cDNA clone were used to synthesize a digoxigenin (DIG)-labeled sense and antisense cDNA probes (Roche) by PCR. In situ hybridizations were performed using mixed parasitic stages of H. schachtii as described by de Boer et al. (1998). Hybridization signals within the nematodes were detected with alkaline phosphatase-conjugated anti-DIG antibody and substrate, and specimens were observed with a Zeiss Axiovert 100 inverted light microscope.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis actin (AT1G49240), H. schachtii actin (AY443352), Hg CBP (AY340946), Hs CBP (EU328302), PME1 (At1g53840), PME2 (At1g53830), and PME3 (At3g14310). The Arabidopsis Information Resource stock number for PME3 insertional mutant is CS857169.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of Hg CBP and Hs CBP.
Supplemental Figure 2. In Situ Hybridization and Developmental Expression Level of Hs CBP
Supplemental Figure 3. Hs CBP Expression in Arabidopsis Does Not Alter Nematode Penetration into Roots.
Supplemental Figure 4. Hs CBP Expression in Arabidopsis Does Not Alter Susceptibility to the Root-Knot Nematode.
Supplemental Figure 5. Hs CBP Specifically Interacts with PME3.
Supplemental Table 1. Quantification of Hs CBP Expression Levels in Transgenic Arabidopsis Lines Using Quantitative Real-Time RT-PCR.
Supplementary Material
Acknowledgments
This is a journal paper of the Iowa Agriculture and Home Economics Station, Ames, IA, supported by Hatch Act and State of Iowa funds. This work was funded by USDA National Research Initiative Competitive Grants Program Award 2005-35604-15434 and by grants from the Iowa Soybean Association. We thank Stanton Gelvin for providing the BiFC vectors and Calvin Tan for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Thomas J. Baum (tbaum@iastate.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- An, G., Ebert, P.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual, S.B. Gelvin, R.A. Schilperoort, and D.P.S. Verma, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), 1–19.
- Barker, K.R., and Koenning, S.R. (1998). Development of sustainable systems for nematode management. Annu. Rev. Phytopathol. 36 165–205. [DOI] [PubMed] [Google Scholar]
- Baum, T.J., Wubben II, M.J.E., Hardy, K.A., Su, H., and Rodermel, S.R. (2000). A screen for Arabidopsis thaliana mutants with altered susceptibility to Heterodera schachtii. J. Nematol. 32 166–173. [PMC free article] [PubMed] [Google Scholar]
- Boudart, G., Lafitte, C., Barthe, J.P., Frasez, D., and Esquerré-Tugayé, M.T. (1998). Differential elicitation of defense responses by pectic fragments in bean seedlings. Planta 206 86–94. [Google Scholar]
- Carrard, G., Koivula, A., Söderlund, H., and Béguin, P. (2000). Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 97 10342–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christgau, S., Kofod, L.V., Halkier, T., Anderson, L.N., Hockauf, M., Dorreich, K., Dalboge, H., and Kauppinen, S. (1996). Pectin methyl esterase from Aspergillus aculeatus: Expression cloning in yeast and characterization of the recombinant enzyme. Biochem. J. 319 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Collmer, A., and Keen, N.T. (1986). The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24 383–409. [Google Scholar]
- Davis, E.L., Hussey, R.S., and Baum, T.J. (2004). Getting to the roots of parasitism by nematodes. Trends Parasitol. 20 134–141. [DOI] [PubMed] [Google Scholar]
- Davis, E.L., Hussey, R.S., Mitchum, M.G., and Baum, T.J. (2008). Parasitism proteins in nematode-plant interactions. Curr. Opin. Plant Biol. 11 360–366. [DOI] [PubMed] [Google Scholar]
- de Boer, J.M., McDermott, J.P., Davis, E.L., Hussey, R.S., Popeijus, H., Smant, G., and Baum, T.J. (2002). Cloning of a putative pectate lyase gene expressed in the subventral esophageal glands of Heterodera glycines. J. Nematol. 34 9–11. [PMC free article] [PubMed] [Google Scholar]
- de Boer, J.M., Yan, Y., Bakker, J., Davis, E.L., and Baum, T.J. (1998). In situ hybridization to messenger RNA of Heterodera glycines. J. Nematol. 30 309–312. [PMC free article] [PubMed] [Google Scholar]
- de Boer, J.M., Yan, Y., Wang, X., Smant, G., Hussey, R.S., Davis, E.L., and Baum, T.J. (1999). Developmental expression of secretory β-1,4-endoglucanases in the subventral esophageal glands of Heterodera glycines. Mol. Plant Microbe Interact. 12 663–669. [DOI] [PubMed] [Google Scholar]
- Ding, X., Shields, J., Allen, R., and Hussey, R.S. (1998). A secretory cellulose-binding protein cDNA cloned from the root-knot nematode (Meloidogyne incognita). Mol. Plant Microbe Interact. 11 952–959. [DOI] [PubMed] [Google Scholar]
- Doi, R.H., and Kosugi, A. (2004). Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 7 541–551. [DOI] [PubMed] [Google Scholar]
- Elling, A.A., Davis, E.L., Hussey, R.S., and Baum, T.J. (2007. b). Active uptake of cyst nematode parasitism proteins into the plant cell nucleus. Int. J. Parasitol. 37 1269–1279. [DOI] [PubMed] [Google Scholar]
- Elling, A.A., et al. (2007. a). Divergent evolution of arrested development in the dauer stage of Caenorhabditis elegans and the infective stage of Heterodera glycines. Genome Biol. 8 R211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, B., Allen, R., Davis, E.L., Baum, T.J., and Hussey, R.S. (2004). Molecular characterization and developmental expression of a cellulose-binding protein gene in the soybean cyst nematode Heterodera glycines. Int. J. Parasitol. 34 1377–1383. [DOI] [PubMed] [Google Scholar]
- Gao, B., Allen, R., Maier, T., Davis, E.L., Baum, T.J., and Hussey, R.S. (2001). Identification of putative parasitism genes expressed in the esophageal gland cells of the soybean cyst nematode (Heterodera glycines). Mol. Plant Microbe Interact. 14 1247–1254. [DOI] [PubMed] [Google Scholar]
- Gao, B., Allen, R., Maier, T., Davis, E.L., Baum, T.J., and Hussey, R.S. (2003). The parasitome of the phytonematode Heterodera glycines. Mol. Plant Microbe Interact. 16 720–726. [DOI] [PubMed] [Google Scholar]
- Goellner, M., Smant, G., deBoer, J.M., Baum, T.J., and Davis, E.L. (2000). Isolation of beta-1,4-endoglucanase genes of Globodera tabacum and their expression during parasitism. J. Nematol. 32 154–165. [PMC free article] [PubMed] [Google Scholar]
- Goellner, M., Wang, X., and Davis, E.L. (2001). Endo-beta-1,4-endoglucanase expression in compatible plant nematode interactions. Plant Cell 13 2241–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, M.A., Takagi, M., Hashida, S., Shoseyov, O., Doi, R.H., and Segel, I.H. (1993). Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 175 5762–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok, R.J., Pierson, E., Lambert, G.M., Gong, F.C., Afonso, C.L., Haldeman-Cahill, R., Carrington, J.C., and Galbraith, D.W. (1997). Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11 573–586. [DOI] [PubMed] [Google Scholar]
- Grundler, F., Betka, M., and Wyss, U. (1991). Influence of changes in the nurse cell system (syncytium) on sex determination and development of the cyst nematode Heterodera schachtii: Total amounts of proteins and amino acids. Phytopath. 81 70–74. [Google Scholar]
- Huang, G., Dong, R., Allen, R., Davis, E.L., Baum, T.J., and Hussey, R.S. (2006). A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant Microbe Interact. 19 463–470. [DOI] [PubMed] [Google Scholar]
- Ithal, N., Recknor, J., Nettleton, D., Hearne, L., Maier, T., Baum, T.J., and Mitchum, M.G. (2007). Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol. Plant Microbe Interact. 20 293–305. [DOI] [PubMed] [Google Scholar]
- Lionetti, V., Raiola, A., Camardella, L., Giovane, A., Obel, N., Pauly, M., Favaron, F., Cervone, F., and Bellincampi, D. (2007). Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 143 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- McCartney, L., Blake, A.W., Flint, J., Bolam, D.N., Boraston, A.B., Gilbert, H.J., and Knox, J.P. (2006). Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc. Natl. Acad. Sci. USA 103 4765–4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, G.P., Hedley, D., Fyffe, L., and Pérombelon, M.C.M. (1993). Potato resistance to soft-rot Erwinias is related to cell wall pectin esterification. Physiol. Mol. Plant Pathol. 42 279–289. [Google Scholar]
- Micheli, F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6 414–419. [DOI] [PubMed] [Google Scholar]
- Pilling, J., Willmitzer, L., and Fisahn, J. (2000). Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta 210 391–399. [DOI] [PubMed] [Google Scholar]
- Qin, L., Kudla, U., Roze, E.H., Goverse, A., Popeijus, H., Nieuwland, J., Overmars, H., Jones, J.T., Schots, A., Smant, G., Bakker, J., and Helder, J. (2004). Plant degradation: A nematode expansin acting on plants. Nature 427 30. [DOI] [PubMed] [Google Scholar]
- Qin, L., Overmars, H., Helder, J., Popeijus, H., van der Voort, J.R., Groenink, W., van Koert, P., Schots, A., Bakker, J., and Smant, G. (2000). An efficient cDNA-AFLP-based strategy for the identification of putative pathogenicity factors from the potato cyst nematode Globodera rostochiensis. Mol. Plant Microbe Interact. 13 830–836. [DOI] [PubMed] [Google Scholar]
- Ren, C., and Kermode, A.R. (2000). An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol. 124 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, L., Qin, L.X., Gadal, P., and Goldberg, R. (1994). Molecular cloning and characterization of a putative pectin methylesterase cDNA in Arabidopsis thaliana (L.). FEBS Lett. 355 135–139. [DOI] [PubMed] [Google Scholar]
- Safra-Dassa, L., Shani, Z., Danin, A., Roiz, L., Shoseyov, O., and Wolf, S. (2006). Growth modulation of transgenic potato plants by heterologous expression of bacterial carbohydrate-binding module. Mol. Breed. 17 355–364. [Google Scholar]
- Shoseyov, O., and Doi, R.H. (1990). Essential 170-kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc. Natl. Acad. Sci. USA 87 2192–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov, O., Shani, Z., and Shpigel, E. (2001). Transgenic plants of altered morphology. US patent 6184440.
- Shpigel, E., Roiz, L., Goren, R., and Shoseyov, O. (1998). Bacterial cellulose-binding domain modulates in vitro elongation of different plant cells. Plant Physiol. 117 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons, P.C., Grundler, F.M.W., Von Mende, N., Burrows, P.R., and Wyss, U. (1991). Arabidopsis thaliana as a new model host for plant parasitic nematodes. Plant J. 1 245–254. [Google Scholar]
- Smant, G., et al. (1998). Endogenous cellulases in animals: Isolation of beta-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl. Acad. Sci. USA 95 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomme, P., Boraston, A., McLean, B., Kormos, J., Creagh, A.L., Sturch, K., Gilkes, N.R., Haynes, C.A., Warren, R.A., and Kilburn, D.G. (1998). Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 715 283–296. [DOI] [PubMed] [Google Scholar]
- Wang, X., Meyers, D., Yan, Y., Baum, T., Smant, G., Hussey, R., and Davis, E. (1999). In planta localization of a β-1,4-endoglucanase secreted by Heterodera glycines. Mol. Plant Microbe Interact. 12 64–67. [DOI] [PubMed] [Google Scholar]
- Wang, X., Mitchum, M.G., Gao, B., Li, C., Diab, H., Baum, T.J., Hussey, R.S., and Davis, E.L. (2005). A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol. Plant Pathol. 6 187–191. [DOI] [PubMed] [Google Scholar]
- Wang, X.H., Allen, R., Ding, X.F., Goellner, M., Maier, T., de Boer, J.M., Baum, T.J., Hussey, R.S., and Davis, E.L. (2001). Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Mol. Plant Microbe Interact. 14 536–544. [DOI] [PubMed] [Google Scholar]
- Wietholter, N., Graessner, B., Mierau, M., Mort, A.J., and Moerschbacher, B.M. (2003). Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol. Plant Microbe Interact. 16 945–952. [DOI] [PubMed] [Google Scholar]
- Williamson, V.M., and Hussey, R.S. (1996). Nematode pathogenesis and resistance in plants. Plant Cell 8 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrather, J.A., Stienstra, W.C., and Koenning, S.R. (2001). Soybean disease loss estimates for the United States from 1996 to 1998. Can. J. Plant Pathol. 23 122–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.