Abstract
Proteins synthesized in the endoplasmic reticulum (ER) of eukaryotic cells must be folded correctly before translocation out of the ER. Disruption of protein folding results in the induction of genes for ER-resident chaperones, for example, BiP. This phenomenon is known as the ER stress response. We report here that bZIP60, an Arabidopsis thaliana basic leucine zipper (bZIP) transcription factor with a transmembrane domain, is involved in the ER stress response. When compared with wild-type Arabidopsis plants, homozygous bzip60 mutant plants show a markedly weaker induction of many ER stress-responsive genes. The bZIP60 protein resides in the ER membrane under unstressed condition and is cleaved in response to ER stress caused by either tunicamycin or DTT. The N-terminal fragment containing the bZIP domain is then translocated into the nucleus. Cleavage of bZIP60 is independent of the function of Arabidopsis homologs of mammalian S1P and S2P proteases, which mediate the proteolytic cleavage of the mammalian transcription factor ATF6. In Arabidopsis, expression of the bZIP60 gene and cleavage of the bZIP60 protein are observed in anthers in the absence of stress treatment, suggesting that the ER stress response functions in the normal development of active secretory cells.
INTRODUCTION
Secretory proteins are synthesized and folded in the endoplasmic reticulum (ER) of eukaryotic cells. Proper folding of proteins is necessary for transport to their final destinations. When this process is perturbed, unfolded proteins accumulate in the ER, inducing expression of many genes. Among them are genes encoding ER-resident chaperones, such as BiP, and enzymes involved in protein folding, such as protein disulfide isomerase. This phenomenon is conserved among eukaryotic cells and is referred to as the ER stress response or the unfolded protein response (Mori, 2000; Kaufman et al., 2002; Rutkowski and Kaufman, 2004).
There is growing recognition that the ER stress response also plays a role in a wide variety of normal cellular processes (Harding et al., 2001; Reimold et al., 2001; Scheuner et al., 2001; Iwakoshi et al., 2003; Lipson et al., 2006). Mammalian cells specialized for secretion, including plasma cells, pancreatic β-cells, hepatocytes, and osteoblasts, require an intact ER stress response. For instance, an intact ER stress response is required for terminal differentiation of B lymphoid cells to plasma cells, during which the ER compartment expands fivefold to accommodate the large increase in immunoglobulin synthesis (Iwakoshi et al., 2003).
In plants, the ER stress response was initially described in the floury-2 endosperm mutant of maize (Zea mays), which produces an aberrant 24-kD α-zein storage protein with a defective signal peptide processing site (Boston et al., 1991; Fontes et al., 1991). The defective storage protein accumulates as a membrane-anchored protein in the ER and in ER-derived protein bodies. As a result, seeds exhibit the ER stress response, with dramatically elevated levels of BiP and other ER-resident chaperones (Coleman et al., 1995; Gillikin et al., 1997). BiP expression is upregulated during seed development in soybean (Glycine max), rice (Oryza sativa), pumpkin (Cucurbita maxima), and Douglas fir (Pseudotsuga menziesii) when large amounts of seed storage proteins are synthesized, folded, and assembled in the ER (Forward and Misra, 2000; Kalinski et al., 1995; Hatano et al., 1997; Muench et al., 1997). BiP expression is induced under stress conditions, such as salt stress and high sugar levels, and in response to pathogens (Koiwa et al., 2003; Wang et al., 2005; Tajima and Koizumi, 2006). The function of the ER stress response in these cellular processes has yet to be elucidated.
The signal transduction mechanism that triggers the ER stress response has been characterized extensively in yeast and mammalian cells. In mammalian cells, two transcription factors, XBP1 and ATF6, activate ER stress-responsive genes. XBP1 is activated by IRE1, an ER membrane–localized protein kinase/ribonuclease. Upon perception of ER stress, IRE1's ribonuclease domain is activated to catalyze the spliceosome-independent splicing of XBP1 mRNA, which encodes a basic leucine zipper (bZIP) transcription factor (Yoshida et al., 2001a). The splice removes 26 nucleotides from the XBP1 mRNA, resulting in a frame shift that causes production of a nonfunctional protein. The XBP1 protein, which has a transcription-activation domain at the C terminus, is synthesized from the spliced mRNA and enhances target gene expression through cis-elements designated the ER stress response element (ERSE), ERSE-II, or the unfolded protein response element (UPRE) (Yoshida et al., 1998; Wang et al., 2000; Kokame et al., 2001; Shen et al., 2001; Yamamoto et al., 2004).
ATF6 is a transmembrane protein located in the ER membrane and has a bZIP domain on its cytoplasmic side. In response to ER stress, ATF6 protein is transported to the Golgi apparatus, where it is sequentially cleaved by the site-1 and site-2 proteases (S1P and S2P) (Haze et al., 1999; Lee et al., 2002). S1P cleaves ATF6 on the luminal side to generate ATF6 with a shorter luminal domain. The first cleavage by S1P allows S2P to recognize ATF6 and cleave its transmembrane segment. This two-step processing liberates the cytoplasmic fragment containing the bZIP domain from the membrane. It then translocates into the nucleus, where it activates downstream genes through ERSE or ERSE-II elements, cooperating with the NF-Y transcription factor complex (Yoshida et al., 2000, 2001b). Knockouts of both of the two ATF6 genes have been reported to cause embryonic lethality in mice, further suggesting that the ER stress response plays a critical role in normal development (Yamamoto et al., 2007).
The molecular mechanism of the ER stress response is much less well understood in plants than in yeast or mammalian cells. Tunicamycin, an inhibitor of N-glycosylation commonly used to induce ER stress in yeast and mammals, also induces the ER stress response in plants (Koizumi et al., 1999; Iwata and Koizumi, 2005b; Urade, 2007). Although IRE1 homologs have been identified in Arabidopsis thaliana and rice (Koizumi et al., 2001; Okushima et al., 2002), their involvement in the ER stress response has not been analyzed. We previously reported that the Arabidopsis bZIP60 gene is transcriptionally activated by tunicamycin (Iwata and Koizumi, 2005a). bZIP60 is a protein of 295 amino acids that has a bZIP domain and an adjacent putative transmembrane domain (Figure 1A). Expression of bZIP60ΔC, a truncated form of bZIP60 lacking the transmembrane domain (Figure 1A), activated BiP and calnexin (CNX) promoters through ERSE and P-UPRE cis-elements in a transient expression assay, suggesting that bZIP60 functions in a manner analogous to ATF6 during the ER stress response. bZIP28 has also been reported to be involved in the ER stress response (Liu et al., 2007a; Tajima et al., 2008). bZIP28 activates expression of BiP genes in response to ER stress through cis-elements P-UPRE and ERSE, and T-DNA insertion mutants of bZIP28 show reduced induction of all BiP genes.
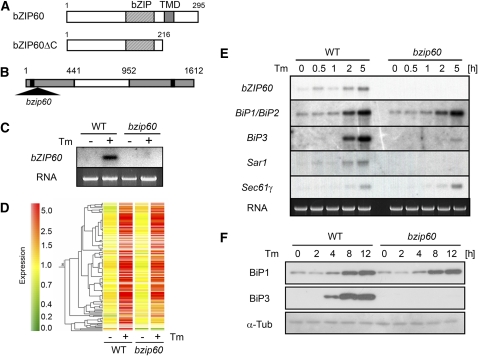
Figure 1.
Characterization of ER Stress-Inducible Genes in Wild-Type Plants and Plants with a T-DNA Insertion in the bZIP60 Gene.
(A) A schematic representation of the bZIP60 protein and the C-terminally truncated form, designated bZIP60ΔC. The locations of the bZIP domain (bZIP) and the transmembrane domain (TMD) are indicated.
(B) A schematic representation of the bZIP60 gene. The T-DNA insertion in the bzip60 mutant is represented by a triangle. Shaded boxes represent exons, and the white box represents an intron. The positions of a start and stop codon are indicated by black boxes.
(C) RNA gel blot analysis of wild-type and bzip60 mutant plants. Total RNA (bottom panel) was extracted from wild-type and bzip60 Arabidopsis leaves floated onto water containing 5 μg/mL tunicamycin (+) or 0.1% DMSO (−; as a solvent control) for 12 h and analyzed by RNA gel blotting (top panel).
(D) Expression of genes activated more than threefold by tunicamycin in wild-type plants compared with wild-type and bzip60 mutant seedlings. RNA was extracted from wild-type and bzip60 seedlings before (−) and after (+) treatment with 5 μg/mL tunicamycin and used for microarray analysis. Each horizontal bar represents a single gene. Green indicates a relatively low level of expression, and red indicates a relatively high level of expression of a given mRNA.
(E) RNA gel blot analysis of several genes in the wild type and bzip60 mutant treated with tunicamycin. Total RNA was extracted from wild-type and bzip60 mutant seedlings treated with 5 μg/mL tunicamycin for the indicated times, and 5 μg of total RNA was loaded into each lane.
(F) Immunoblot analysis of changes in BiP protein levels in wild-type and bzip60 mutant seedlings in response to tunicamycin. Wild-type and bzip60 seedlings were treated as in (E). Protein was extracted at the indicated times, and 20 μg of protein for each sample was loaded on an SDS-PAGE gel and analyzed by immunoblotting with anti-BiP1 and BiP3 antibodies as well as an anti-α-tubulin antibody as a loading control.
Here, we present evidence that bZIP60 is both involved in the plant ER stress response and upregulated and activated during normal development in anther cells specialized for secretion. We show that many of the ER stress-responsive genes are less strongly induced in homozygous bzip60 mutant plants than in wild-type plants. We present evidence that the bZIP60 protein resides in the ER membrane in the unstressed condition and that it is cleaved in response to ER stress, allowing the N-terminal fragment containing the bZIP domain to translocate into the nucleus to function as a transcription factor. We report that cleavage of bZIP60 does not depend on the Arabidopsis homologs of the mammalian S1P and S2P proteases that cleave the ATF6 transcription factor during ER stress. Moreover, cleavage of the bZIP60 protein is observed in actively secreting anther cells in unstressed plants, suggesting a possible function for the ER stress response during normal anther development.
RESULTS
Isolation of a T-DNA Insertion Mutant of bZIP60
We isolated a T-DNA insertion mutant of bZIP60 from the SALK collection. The first exon was disrupted by insertion of the T-DNA in this mutant (Figure 1B). A homozygous bzip60 mutant plant was isolated and disruption was confirmed by PCR. Sequencing of the T-DNA flanking region showed that the T-DNA had inserted 16 nucleotides downstream from the ATG start codon. RNA gel blot analysis further showed that bZIP60 transcripts were undetectable in bzip60 plants treated with tunicamycin (Figure 1C). When wild-type and bzip60 plants were grown in soil under normal growth conditions (16-h-light and 8-h-dark cycle at 22°C), there was no apparent difference in plant growth or morphology.
Microarray Analysis of Wild-Type and bzip60 Mutant Seedlings
Our previous observation that bZIP60 lacking its transmembrane and C-terminal domains (bZIP60ΔC, Figure 1A) can activate BiP and CNX promoters (Iwata and Koizumi, 2005a) prompted us to ask whether induction of ER stress-inducible genes is affected in bzip60 mutant plants. We therefore compared the expression profile of genes induced by ER stress in wild-type and bzip60 mutant plants using Agilent Arabidopsis 2 Oligo Microarrays containing 60-mer oligonucleotides for each of 21,500 genes derived from the ATH1 version 3 database of The Institute for Genomic Research. RNA for the microarray analysis was isolated from 10-d-old wild-type and bzip60 tunicamycin-treated and control seedlings.
Of the 21,500 genes represented on the microarray, 19,583 genes showed significant signal intensity, and of these, 129 were activated more than threefold by tunicamycin treatment in wild-type seedlings (Figure 1D, Table 1). The relative changes in gene expression for these 129 genes in wild-type and bzip60 seedling are shown in Supplemental Table 1 online. The numerically dominant component of the tunicamycin response comprises 25 genes encoding ER chaperones and folding enzymes, including BiP, GRP94 (Klein et al., 2006), J protein (Yamamoto et al., 2008), calreticulin (Christensen et al., 2008), calnexin, protein disulfide isomerase (Houston et al., 2005), ERO1, and ROC7, and enzymes involved in modification, such as glycosylation. An additional 21 of the tunicamycin-induced genes code for proteins involved in protein transport into or through the secretory pathway. These include the Sec61 translocon complexes and signal peptidases as well as small GTPases involved in vesicular transport (Sar1 and Arf). Genes coding for proteins of the ER-associated degradation pathway were also induced. These include a Der1-like transmembrane protein for retrotranslocation of unfolded proteins from the ER to the cytosol (Kirst et al., 2005) and proteins related to ubiquitin/proteasome system, such as ubiquitin ligases and F-box proteins. In addition, genes coding for proteins involved in signal transduction, including the transcription factors bZIP60, were also upregulated. An additional 25 genes involved in various other metabolic processes and 34 genes of unknown function were upregulated as well.
Table 1.
Summary of the Microarray Experiment
| Number of Genes | ||
|---|---|---|
| Category | Induced More Than Threefold in the Wild Type | Induction Is Reduced in bzip60 |
| Protein folding | 25 | 15 |
| Secretory pathway | 21 | 10 |
| Protein degradation | 11 | 6 |
| Transcription factor | 13 | 7 |
| Others | 25 | 5 |
| Unknown | 34 | 11 |
| Total | 129 | 54 |
Numbers of genes whose induction is induced more than threefold by tunicamycin treatment in wild-type seedlings for corresponding categories are shown. A number of genes whose induction is significantly reduced in bzip60 mutant seedlings compared with wild-type seedlings (Welch's t test, P < 0.05; see Methods) is shown for each category as well. A list of these genes can be obtained from Supplemental Table 1 online.
Among the 129 genes induced by tunicamycin treatment in wild-type seedlings, 54 showed a significantly lower level of induction in bzip60 than in wild-type seedlings (Welch's t test, P < 0.05). These are identified in boldface type in Supplemental Table 1 online. Among the chaperone genes, the BiP3 gene was clearly less strongly induced in bzip60 than in wild-type seedlings, whereas induction of BiP1 and BiP2 was similar in both. Several genes were subjected to RNA gel blot analysis to confirm the microarray results. These included the three BiP genes, bZIP60, Sar1 (At1g09180), and Sec61γ (At4g24920). As shown in Figure 1E, induction was much less marked for BiP3 and undetectable for Sar1 in bzip60 compared with wild-type seedlings, while induction of others was unaffected, as indicated by the microarray data. The lower level of BiP3 gene induction was reflected at the protein level as well, with little or no increase in BiP3 protein in response to tunicamycin treatment in bzip60 mutant seedlings compared with wild-type seedlings (Figure 1F). By contrast, BiP1 protein accumulated at similar levels in wild-type and bzip60 mutant seedlings treated with tunicamycin, reflecting similar levels of gene induction (Figures 1E and 1F).
The ER stress response is considered to be mediated by the cis-elements designated ERSE and P-UPRE (a combination of ERSE-II and XBP1-BS or UPRE) (Oh et al., 2003). We therefore searched the promoter sequences of genes upregulated by tunicamycin for such motifs (ERSE, ERSE-II, ERSE-L, XBP1-BS, and UPRE; see Methods) and found them in 68 of the 129 upregulated genes (see Supplemental Table 1 online). Among them, ERSE or ERSE-L elements were found in 54 genes, and XBP1-BS or UPRE elements were found in 24 genes. We found that these elements were overrepresented (P = 1.28E-22 for ERSE or ERSE-L elements and P = 7.65E-11 for XBP1-BS or UPRE elements; see Methods), showing that these motifs were enriched among ER stress-responsive genes. Among the 54 genes whose induction was clearly lower in bzip60 mutants than in wild-type seedlings, ERSE or ERSE-L elements were found in 28 genes and XBP1-BS or UPRE elements were found in 12 genes. Thus, only about half of the bZIP60-responsive genes had known ER stress-responsive motifs within 0.5 kb of the transcription start site.
To confirm that the reduced induction of the 54 putative bZIP60-responsive genes was due to disruption of the bZIP60 gene, we performed a complementation experiment by introducing a genomic fragment containing the bZIP60 gene into bzip60 mutant plants and then analyzed the response of the transformed plants to tunicamycin. As shown in Supplemental Figure 1 online, induction of the BiP3 gene was restored by introduction of the genomic fragment containing the bZIP60 gene. This confirms that the reduced induction of ER stress-responsive genes was due to disruption of the bZIP60 gene.
Evidence That bZIP60 Is a Transcription Factor in Vivo
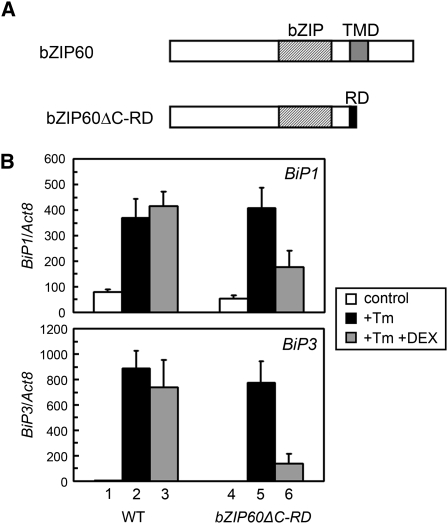
We previously reported that the N-terminal fragment of bZIP60 activates several ER stress-responsive promoters in a protoplast assay (Iwata and Koizumi, 2005a). To provide further evidence that bZIP60 activates ER stress-responsive genes in vivo, we asked whether the minimal repression domain (RD) from SUPERMAN, an Arabidopsis transcriptional repressor regulating flowering time, could override transcription induction by bZIP60. The RD domain consists of the six amino acids DLELRL, and fusion of RD has been reported to convert a transcriptional activator to a strong transcriptional repressor (Hiratsu et al., 2002, 2004). We fused the C terminus of the bZIP60ΔC coding sequence to RD coding sequence to generate the bZIP60ΔC-RD construct and expressed it from a dexamethasone (DEX)-inducible promoter (Figure 2A). We transformed wild-type Arabidopsis plants with the resulting DEX-bZIP60ΔC-RD construct and performed quantitative RT-PCR (qRT-PCR) to investigate the effect of DEX on tunicamycin-induced BiP expression. As shown in Figure 2B, BiP1 and BiP3 could be induced by tunicamycin in wild-type and DEX-bZIP60ΔC-RD plants (columns 1, 2, 4, and 5). Addition of DEX to DEX-bZIP60ΔC-RD seedlings suppressed induction of these genes by tunicamycin (columns 5 and 6) but did not interfere with their tunicamycin induction in wild-type seedlings (columns 2 and 3). Thus, expression of the bZIP60ΔC-RD gene suppressed induction of BiP1 and BiP3 by tunicamycin treatment. Moreover, the inhibitory effect of DEX treatment was greater for the BiP3 gene than for the BiP1 gene.
Figure 2.
Converting bZIP60 to a Repressor Interferes with ER Stress Induction of BiP Genes.
(A) A schematic representation of bZIP60ΔC-RD.
(B) qRT-PCR analysis of BiP transcripts in wild-type and DEX-bZIP60ΔC-RD plants with and without tunicamycin (Tm) and DEX. RNA was extracted from 10-d-old seedlings of wild-type and DEX-bZIP60ΔC-RD plants treated with tunicamycin (5 μg/mLl) alone or a combination of tunicamycin (5 μg/mL) and DEX (10 μM) for 2 h and used for qRT-PCR analysis. DMSO and ethanol were used at a final concentration of 0.1% as solvent controls for tunciamycin and DEX, respectively. The abundance of BiP transcripts was normalized to that of Act8 transcripts. Data represent means with sd of three independent experiments.
Effect of bZIP60 Overexpression on Gene Expression
Because induction of a number of ER stress-responsive genes is much weaker in bzip60 seedlings than in wild-type seedlings, it appeared likely that overexpression of bZIP60 would enhance induction of these genes. Therefore, we generated Arabidopsis suspension culture cells constitutively overexpressing bZIP60 from a cauliflower mosaic virus 35S promoter (bZIP60-OX) and used qRT-PCR to investigate transcriptional induction of ER chaperone genes in response to ER stress. We confirmed that tunicamycin induces the bZIP60 gene in wild-type cells, and we established that the bZIP60 gene is constitutively expressed at a high level in bZIP60-OX cells (Figure 3A). Overexpression of bZIP60 in bZIP60-OX cells was also confirmed at the protein level (see below; Figure 4A). Thus, both the gene and the protein are overexpressed in bZIP60-OX cells. However, BiP1 and BiP3 transcripts were detected at similar levels in unstressed wild-type and bZIP60-OX cells (Figure 3B). These results demonstrate that overexpression of bZIP60 gene does not increase the abundance of BiP transcripts.
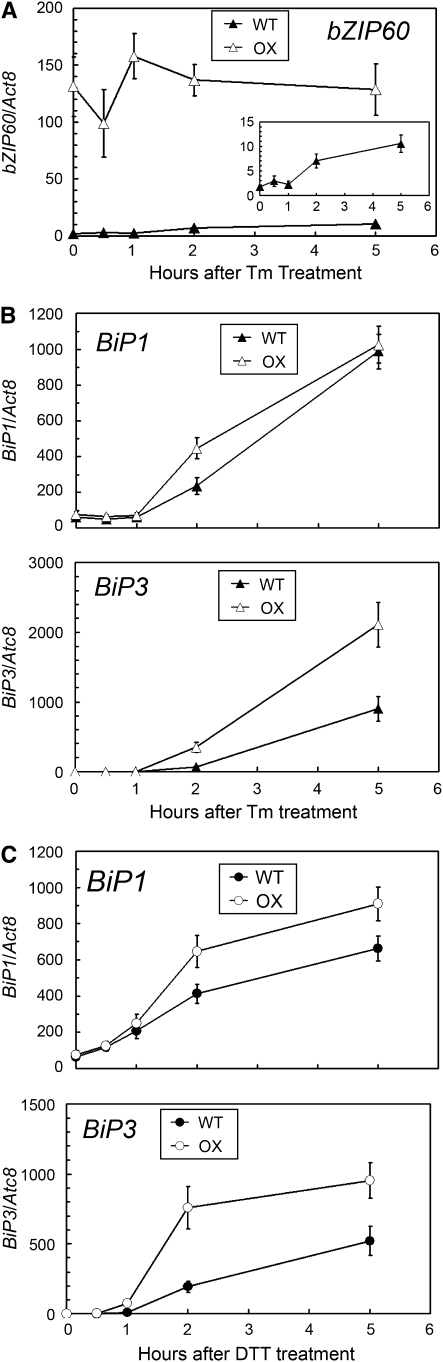
Figure 3.
An Overexpressed bZIP60 Gene Requires Tunicamycin to Affect Stress Gene Expression.
(A) qRT-PCR analysis of bZIP60 transcripts in wild-type and bZIP60-OX cells in response to tunicamycin. RNA was extracted from wild-type and bZIP60-OX Arabidopsis suspension cells treated with 5 μg/mL tunicamycin at each time point and subjected to qRT-PCR analysis. The abundance of bZIP60 transcripts was normalized to that of Act8 transcripts. Inset: An enlarged view of the result obtained with wild-type cells. Data represent means with sd of three independent experiments.
(B) qRT-PCR analysis of BiP transcripts in wild-type and bZIP60-OX in response to tunicamycin. RNA was extracted as in (A). The abundance of BiP transcripts was normalized to that of Act8 transcripts. Data represent means with sd of three independent experiments.
(C) qRT-PCR analysis of BiP transcripts in the wild type and bZIP60-OX in response to DTT. qRT-PCR was performed as in (B), except that the ER stress treatment was 2 mM DTT instead of tunicamycin.
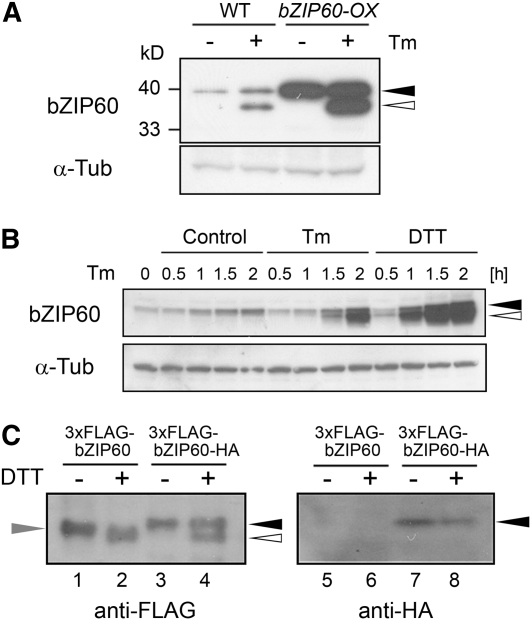
Figure 4.
Proteolytic Processing of bZIP60 in Response to ER Stress.
(A) Detection of bZIP60 protein. Total protein was extracted from wild-type and bZIP60-OX cells treated with 5 μg/mL tunicamycin (+) or 0.1% DMSO (−) for 2 h, and 20 μg of protein was subjected to immunoblot analysis using an anti-bZIP60 antibody as well as an anti-α-tubulin antibody as a loading control. Closed and open triangles in (A) and (B) indicate the positions of the full-length and faster-migrating putative cleaved forms of bZIP60, respectively. Positions of protein marker bands were indicated at the left side.
(B) Time-course analysis of bZIP60 protein by ER stress. Arabidopsis suspension cells were treated with 5 μg/mL tunicamycin (Tm), 2 mM DTT, or 0.1% DMSO (Control) at the indicated times, and total protein extracts (20 μg) were gel-fractionated and subjected to immunoblot analysis using an anti-bZIP60 antibody as well as an anti-α-tubulin antibody as a loading control.
(C) Identification of cleaved bZIP60. Arabidopsis suspension cells expressing either bZIP60 tagged with 3xFLAG at the N terminus (3xFLAG-bZIP60) or bZIP60 tagged with 3xFLAG and HA at the N and C termini, respectively (3xFLAG-bZIP60-HA) were treated with 2 mM DTT for 1 h (+) or maintained as controls (−). Total protein was extracted, and 20 μg of protein was gel-fractionated and subjected to immunoblot analysis using anti-FLAG and anti-HA antibodies. The positions of bZIP60 tagged with both 3xFLAG and HA, and bZIP60 tagged with only 3xFLAG are indicated by closed and gray triangles, and the position of the cleaved form of bZIP60, tagged with 3xFLAG at the N terminus, is indicated by the open triangle.
The abundance of both BiP1 and BiP3 transcripts increased in response to ER stress in both wild-type and bZIP60-OX cells (Figures 3B and 3C). Both BiP1 and BiP3 transcripts were induced in both wild-type and bZIP60-OX cells upon treatment with either tunicamycin (Figure 3B) or DTT, a reducing agent that inhibits disulfide bond formation and induces ER stress (Figure 3C). Thus, despite overexpression of the bZIP60 gene and bZIP60 protein, ER stress-responsive target genes still require the ER stressor for induction. These observations imply the existence of a posttranslational activation step.
Consistent with the greater abundance of bZIP60 protein in bZIP60-OX cells than in wild-type cells, both BiP1 and BiP3 genes were more strongly induced by both tunicamycin and DTT in the bZIP60-OX cells than in the wild-type cells (Figures 3B and 3C). This implies that the overproduced protein contributes to the ER stress response, once activated. The BiP3 gene was more strongly induced by both tunicamycin and DTT treatment in bZIP60-OX cells than in wild-type cells than the BiP1 gene (Figures 3B and 3C). This finding is consistent with the earlier observations that bZIP60 has a more marked effect on the transcription of the BiP3 gene than the BiP1 gene.
Proteolysis and Subcellular Localization of bZIP60 Protein in Response to ER Stress
The results of the foregoing experiments, combined with our earlier report that the N-terminal fragment of bZIP60 lacking the transmembrane domain shows nuclear localization (Iwata and Koizumi, 2005a), suggest that bZIP60 is activated by proteolytic cleavage in or near the transmembrane domain, freeing the N-terminal fragment to translocate to the nucleus to function as a transcriptional activator. To test this hypothesis directly, we used an antibody that recognizes the bZIP60ΔC protein (see Methods and Figure 1A) to investigate the effect of ER stress on the size and intracellular localization of the bZIP60 protein.
Arabidopsis suspension cells were treated with tunicamycin and subjected to immunoblot analysis. As shown in Figure 4A, a single band corresponding to full-length bZIP60 was detected in unstressed cells and a new, faster-migrating band expected for the cleaved form was detected in wild-type cells treated with tunicamycin (Figure 4A, left lanes). Similar results were obtained with bZIP60-OX cells that overexpress bZIP60 (Figure 4A, right lanes). A temporal analysis of bZIP60 protein during tunicamycin treatment revealed that the putative cleaved bZIP60 form first becomes detectable at 1.5 h of tunicamycin treatment (Figure 4B), consistent with the observation that the abundance of BiP1/BiP2 and BiP3 transcripts increases at 2 h after the onset of tunicamycin treatment (Figure 3B).
To determine whether the faster-migrating band is the N-terminal fragment of the bZIP60 protein, rather than a modified form of the intact protein, we incorporated different epitope tags at the N and C termini of bZIP60 and followed their intracellular fate after DTT treatment. A 3xFLAG epitope was added to the N terminus, and an HA epitope was added to the C terminus of the bZIP60 coding sequence. Cauliflower mosaic virus 35S promoter–driven constructs coding for just the FLAG-tagged protein or one tagged with both FLAG and HA epitopes were transformed into wild-type Arabidopsis cells. As shown in Figure 4C, only the slower-migrating band was detected with anti-FLAG antibody in unstressed cells, while either just the faster-migrating band or both were detected with anti-FLAG antibody in DTT-treated cells (Figure 4C, lanes 1 to 4). Neither band was detectable with anti-HA antibody in cells expressing only the FLAG-tagged bZIP60 construct (Figure 4C, lanes 5 and 6), while only the more slowly migrating band was detectable with anti-HA antibody in cells expressing a construct tagged with both FLAG and HA epitopes (Figure 4C, lanes 7 and 8). This shows that the C terminus of bZIP60 is missing from the faster-migrating species, providing direct evidence that the faster-migrating band is a cleaved form of bZIP60 lacking the C terminus.
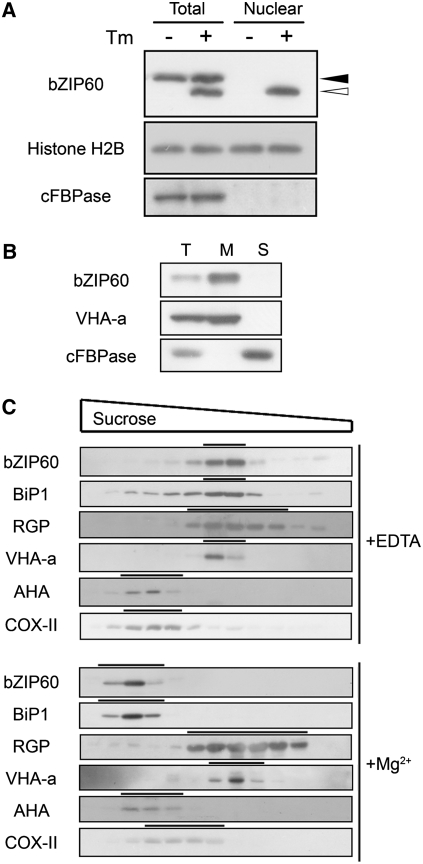
To investigate the intracellular location of the cleaved bZIP60 fragment, nuclear proteins were isolated and subjected to immunoblot analysis. Consistent with previous observations, only full-length bZIP60 was detected in total extracts of unstressed cells, while both the full-length and cleaved forms were detected in extracts of tunicamycin-treated cells (Figure 5A). Neither form was detected in nuclei of untreated cells, while only the cleaved form was detected in the nuclei of cells treated with tunicamycin (Figure 5A). This observation supports the hypothesis that proteolytic cleavage of bZIP60 in response to ER stress liberates the cytosolic N-terminal fragment to translocate into the nucleus.
Figure 5.
Translocation of bZIP60 from the ER to the Nucleus in Response to ER Stress.
(A) Nuclear localization of the cleaved form of bZIP60. Arabidopsis suspension cells were treated with 5 μg/mL tunicamycin (+) or 0.1% DMSO (−) for 2 h, and total protein and nuclear extracts were gel-fractionated and subjected to immunoblot analysis using anti-bZIP60 antibody. Anti-Histone H2B and anti-cFBPase antibodies were used as nuclear and cytosolic markers, respectively. Closed and open triangles indicate the positions of the full-length and cleaved forms of bZIP60, respectively.
(B) Membrane localization of full-length bZIP60 protein. Total protein extract (Total) from untreated Arabidopsis suspension cells was ultracentrifuged at 100,000g for 1 h to obtain pelleted microsomal membrane fraction (M) and soluble fraction (S). Each fraction was analyzed by SDS-PAGE and immunoblotting using anti-bZIP60 antibody. Anti-VHA-a and anti-cFBPase antibodies were used as markers for membrane and soluble fractions, respectively.
(C) Sucrose gradient centrifugation. A microsomal fraction from untreated Arabidopsis suspension cells was fractionated through a 15 to 55% sucrose gradient. Aliquots of fractions were analyzed by SDS-PAGE and immunoblotting using an anti-bZIP60 antibody as well as antibodies for various subcellular marker proteins; anti-BiP1 (ER), anti-RGP (Golgi), anti-VHA-a (vacuolar membrane), anti-AHA (plasma membrane), and anti-COX-II (mitochondria).
To investigate the intracellular localization of full-length bZIP60, we first determined whether bZIP60 associates with the membranes. We prepared the microsomal and soluble fractions from untreated Arabidopsis cells by ultracentrifugation and subjected them to immunoblot analysis. As shown in Figure 5B, bZIP60 protein was detected in the microsomal fraction, indicating membrane localization. A microsomal preparation from unstressed cells was further fractionated on a sucrose density gradient, followed by immunoblot analysis for bZIP60 and various subcellular marker antibodies. We used two kinds of buffers throughout this analysis, one containing EDTA and the other containing MgCl2. Mg2+ keeps the ER membrane–ribosome interaction intact so that ER-localized proteins sediment more rapidly in a buffer containing MgCl2 than in buffer containing EDTA. As shown in Figure 5C, bZIP60 cofractionates with BiP1 in both buffers. bZIP60 sedimented more rapidly in the Mg2+-containing buffer than in the buffer containing EDTA, as did the ER marker BiP1. The sedimentation rates of the other subcellular markers used were unaffected by the presence of Mg2+ (Figure 5C). The cofractionation of bZIP60 with the ER marker BiP1 establishes its presence in the ER. The foregoing experiments show that the bZIP60 protein resides in the ER membrane under unstressed condition and that it is cleaved and translocates to the nucleus in ER-stressed cells.
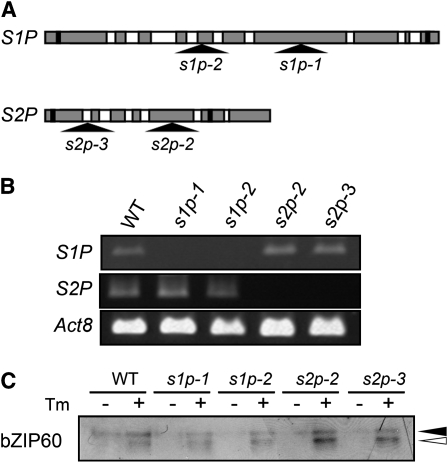
Golgi-localized proteases S1P and S2P are known to mediate the proteolytic cleavage of the mammalian membrane-bound transcription factor ATF6 (Ye et al., 2000). A survey of the genomic database revealed that Arabidopsis has one gene each with significant homology to mammalian S1P and S2P genes. The S1P homolog is encoded by At5g19660, which has been identified previously (Liu et al., 2007b), and S2P is encoded by At4g20310. To determine whether these proteases cleave bZIP60, T-DNA insertion mutants of S1P and S2P were isolated from the SALK and GABI-Kat collections (Figure 6A). Plants homozygous for each of two independent insertion alleles of each gene were isolated and confirmed to be null mutations by RT-PCR (Figure 6B). Seedlings of these mutants were treated with tunicamycin and subjected to immunoblot analysis. As shown in Figure 6C, bZIP60 cleavage was observed in both s1p and both s2p mutants as well as in wild-type plants. Thus, neither the Arabidopsis S1P nor the S2P homolog appears to be involved in bZIP60 cleavage. Since both S1P and S2P must cleave mammalian ATF6 in order for it to be released to translocate to the nucleus, we conclude that bZIP60 cleavage is either performed by different proteases or by a different mechanism than that reported in mammalian cells.
Figure 6.
S1P and S2P Proteases Are Not Responsible for Cleavage of bZIP60.
(A) A schematic representation of the S1P and S2P genes. Shaded boxes represent exons, and white boxes represent introns. The positions of start and stop codons are indicated by black boxes. The locations of the T-DNA insertions in s1p and s2p mutants are shown by triangles.
(B) RT-PCR analysis of s1p and s2p mutants. RNA was extracted from 10-d-old seedlings of wild-type, s1p, and s2p mutants and used for RT-PCR analysis. Act8 was analyzed as a control.
(C) Effect of disruption of S1P and S2P genes on bZIP60 cleavage. Ten-day-old seedlings of wild-type, s1p, and s2p mutants were treated with 5 μg/mL tunicamycin (+) or 0.1% DMSO (−) for 1.5 h, and total protein extracts (20 μg) were fractionated by SDS-PAGE, followed by immunoblot analysis using anti-bZIP60 antibody. Closed and open triangles indicate the positions of the full-length and cleaved forms of bZIP60, respectively.
Activation of bZIP60 without Stress Treatment
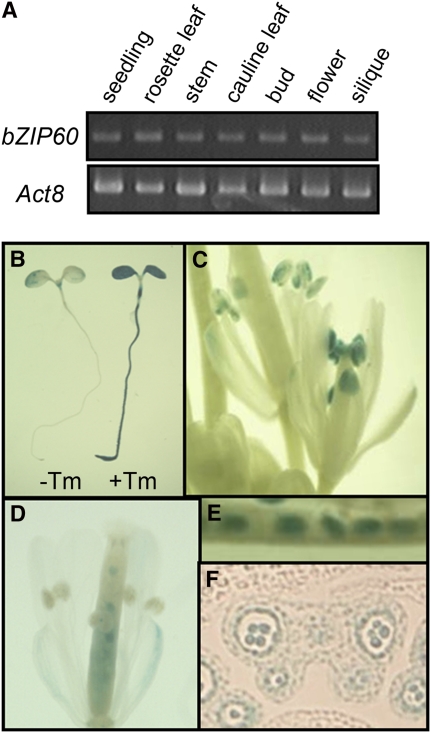
To gain insight into the physiological function of the ER stress response, we analyzed untreated organs and tissues for the presence of the cleaved and uncleaved forms of bZIP60. We first performed RT-PCR to detect expression of the bZIP60 gene. As shown in Figure 7A, the bZIP60 transcripts were detected at similar levels in all tissues examined. Transgenic plants carrying a chimeric gene consisting of the bZIP60 promoter (∼1.2 kb) and a β-glucuronidase (GUS) gene were used to further investigate the expression pattern of the bZIP60 promoter. When seedlings were treated with tunicamycin, GUS staining was obviously enhanced (Figure 7B), indicating that the promoter contains a sequence necessary for the ER stress response. Under normal growth condition, marked GUS staining was observed in anthers and in immature seeds (Figures 7C to 7E). We observed GUS staining of both pollen grains and tapetal cells in sectioned anthers (Figure 7F).
Figure 7.
Tissue-Specific Expression of the bZIP60 Gene and Promoter.
(A) RT-PCR analysis of bZIP60 expression in various tissues. RNA was extracted from the indicated tissues and used for RT-PCR analysis. Act8 was analyzed as a control.
(B) to (F) GUS histochemical staining of bZIP60 promoter:GUS fusions in transgenic plants.
(B) Five-day-old seedlings were treated with 5 μg/mL tunicamycin (+Tm) or 0.1% DMSO (−Tm; as a solvent control) for 10 h and subjected to histochemical staining for GUS activity.
(C) to (F) GUS histochemical staining without stress treatment.
(C) Flowers.
(D) A flower 1 d after fertilization
(E) Immature seeds.
(F) A section of a bud with GUS expression in pollen grains and tapetal cells.
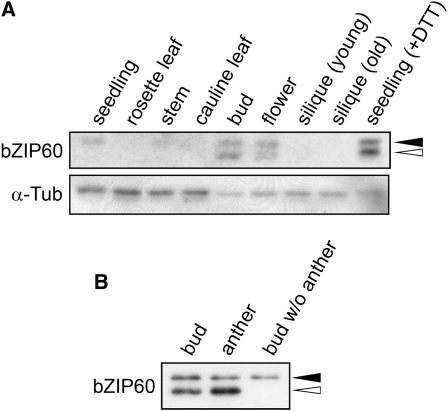
We used immunoblot analysis to detect endogenous bZIP60 without stress treatment. As shown in Figure 8A, cleaved bZIP60 was detected in buds and flowers. Immunoblot analysis after dissection of buds showed that the cleaved bZIP60 was in anthers (Figure 8B). The tapetum is a highly secretory tissue that produces pollen surface proteins, and pollen grains are poised for the high secretory activity that supports rapid pollen tube growth (Hepler et al., 2001; Hsieh and Huang, 2007). The observation that these tissues contain the transcriptionally active N-terminal fragment of bZIP60 suggests that the genes activated by this transcription factor are upregulated in cells with high secretory activity.
Figure 8.
Detection of bZIP60 Protein without Stress Treatment.
(A) Total protein extracts (10 μg) from each tissue were fractionated by SDS-PAGE, followed by immunoblot analysis using anti-bZIP60 antibody as well as an anti-α-tubulin antibody as a loading control. A protein extract from seedlings treated with 2 mM DTT for 1 h was loaded as a control to indicate the full-length and cleaved forms of bZIP60. Closed and open triangles indicate the positions of the full-length and cleaved forms of bZIP60, respectively.
(B) Detection of bZIP60 protein within buds. Anthers and the other parts were separated from five buds, proteins were extracted, fractionated by SDS-PAGE, and subjected to immunoblot analysis using anti-bZIP60 antibody. Closed and open triangles indicate the positions of the full-length and cleaved forms of bZIP60, respectively.
DISCUSSION
We have presented evidence that bZIP60 is a posttranslationally activated transcription factor involved in the Arabidopsis ER stress response. The results of microarray analysis of wild-type plants identified 129 genes that showed increases in transcript abundance of threefold or more in response to tunicamycin treatment. A majority of the proteins encoded by the induced genes are involved in protein folding, transport, secretion, or degradation. These observations are consistent with those reported in earlier studies using an 8000-gene microarray (Martinez and Chrispeels, 2003; Noh et al., 2003) and a fluid microarray (Kamauchi et al., 2005). ERSE and UPRE cis-elements were identified in the 500-bp upstream sequences of 68 of the 129 upregulated genes. Although these characteristic cis-elements were not identified in the other genes, it does not mean they do not contain such elements since they may be located outside of the 500-bp region examined.
We were unable to establish a simple correlation between the expression profile, currently known ER stress-responsive cis-elements, and the bZIP60 transcription factor. Thus, although 54 of the 129 genes observed to be upregulated by ER stress in wild-type plants showed a reduced transcriptional response to ER stress in homozygous bzip60 mutant plants, not all of these contain either an ERSE or UPRE element within 0.5 kb of the transcription start site. Furthermore, not all genes that contain an ERSE or UPRE element showed a reduced transcriptional response to ER stress in bzip60 mutant seedlings. However, it is clear that bZIP60 is at least one of the transcription factors that mediate gene activation in the ER stress response, since induction of many ER stress-responsive genes, including BiP3 and Sar1B is substantially reduced in bzip60 mutant plants lacking the bZIP60 protein. It appears likely that bZIP60 is the main regulator of BiP3 gene expression but makes only a minor contribution to BiP1/BiP2 regulation. This inference is supported by the observation that expression of the transcriptional repressor bZIP60ΔC-RD, derived by adding a repressor domain to the C-terminal sequence of bZIP60, more strongly interfered with tunicamycin induction of BiP3 than that of BiP1. It is also supported by the observation that overexpression of bZIP60 enhances induction of the BiP3 gene more than that of the BiP1 gene.
Our observations also imply that induction of the ER stress response is complex in Arabidopsis, since many genes, including BiP1/BiP2, are still inducible in bzip60 mutant plants. Even the transcriptional induction of BiP3, although significantly reduced in bzip60 mutants, was not completely abolished. It was recently reported that bZIP28 is also involved in induction of BiP genes during the Arabidopsis ER stress response (Liu et al., 2007a). Thus, it appears that bZIP60 and bZIP28 perform complementary and partially overlapping regulatory functions in the Arabidopsis ER stress response. IRE1 homologs are candidate molecules mediating the Arabidopsis ER stress response in view of their roles in the ER stress responses of other organisms. However, plant homologs of IRE1 have not, to date, been shown to function in the plant ER stress response. Other possible candidates are the transcription factors highly induced by tunicamycin treatment identified by our microarray analysis (see Supplemental Table 1 online).
We have presented evidence that the bZIP60 protein resides in the ER membrane under unstressed condition and is cleaved in response to ER stress, translocating into the nucleus to function as a transcription factor. This is consistent with the observation that overexpressing the bZIP60 gene increased the abundance of the full-length bZIP60 protein but did not activate BiP genes in the absence of ER stress. We observed a further increase, rather than a decrease, in the abundance of full-length bZIP60 protein following ER stress, although the cleaved form of bZIP60 is derived from the full-length form. Similar observations have been reported for other membrane-bound transcription factors, such as the mammalian ER stress transducer BBF2H7 (Kondo et al., 2007) and the Arabidopsis transcription factor bZIP17 activated in response to salt stress (Liu et al., 2007b). The most likely explanation is that the full-length form of bZIP60 turns over during unstressed conditions and that both the full-length and cleaved forms are stabilized under ER stress conditions.
The mechanism by which a membrane-bound transcription factor is activated by proteolytic cleavage is called regulated intramembrane proteolysis (RIP), and we have presented evidence here that bZIP60 is activated by RIP. Indeed, the transmembrane domains of bZIP60 and its orthologs contain a Pro residue (see Supplemental Figure 2 online), which has been considered to unwind the α-helix to be efficiently cleaved by RIP proteases (Akiyama et al., 2004), as do other RIP-regulated transcription factors, including ATF6. However, the RIP mechanism by which bZIP60 is activated in response to ER stress in Arabidopsis differs from that known to be involved in activation of ATF6. In the case of ATF6, the Golgi localization signal in the luminal domain of ATF6 is masked by BiP binding, and dissociation of BiP in response to ER stress allows translocation of ATF6 to the Golgi, where it is cleaved by S1P and S2P proteases (Shen et al., 2002). Recent studies have identified OASIS and CREBH as additional bZIP transcription factors with a transmembrane domain involved in the mammalian ER stress response, both of which are targets for S1P and S2P cleavage (Kondo et al., 2005; Zhang et al., 2006).
We showed here that proteolytic cleavage of bZIP60 is not affected in T-DNA insertion mutants of the genes coding for the Arabidopsis S1P and S2P homologs. This observation is consistent with the observation that bZIP60 does not contain the consensus sequence for S1P cleavage, RxxL or RxL. Also, the luminal region, which is thought to function as a sensor for ER stress, is much longer in ATF6 and other bZIPs than in bZIP60. The fact that bZIP60 has a relatively short luminal domain raises the possibility that bZIP60 itself does not function as an ER stress sensor but that the sensor is a bZIP60-interacting protein or the protease that cleaves bZIP60. In the case of another RIP-regulated transcription factor, the mammalian sterol regulatory element binding protein (SREBP), accessory proteins SCAP and INSIG act as sterol sensors and regulate transport of SREBP to Golgi, where it is cleaved by S1P and S2P proteases in a similar manner as ATF6 (Yang et al., 2002; Radhakrishnan et al., 2007; Sun et al., 2007). Such accessory proteins might function to regulate bZIP60 activation. Taken together with the fact that bZIP60 homologs occur in several plant species, but not in yeast or mammals (see Supplemental Figure 2 online), we conclude that bZIP60 is a plant-specific RIP-regulated bZIP transcription factor involved in the ER stress response. Further analysis is required to clarify the mechanisms of signal perception and proteolytic cleavage.
RIP has been recognized as a versatile key mechanism in bacterial and mammalian cells (Weihofen and Martoglio, 2003; Wolfe and Kopan, 2004), and evidence for RIP's importance is also emerging in plants (Schutze et al., 2008; Seo et al., 2008). Regulation of cell division has been reported to be controlled by the RIP-regulated transmembrane transcription factor NTM1 (Kim et al., 2006), which belongs to a plant-specific NAC (NAM/ATAF1,2/CUC2) transcription factor family. It was also reported that bZIP17 is activated by S1P-mediated RIP in response to salt stress (Liu et al., 2007b). Moreover, Arabidopsis homologs of rhomboids and signal peptide peptidases, additional families of intramembrane-cleaving proteases, have recently been identified (Kanaoka et al., 2005; Tamura et al., 2008), although substrates of these proteases are unknown. These reports bear the implication that RIP is an important regulatory mechanism in a variety of cellular processes in plants, as it is in animals.
In mammalian cells, the ER stress response is also known to be required for the differentiation of secretory cells. For example, the ER stress response is activated during differentiation of B cells to plasma cells, during which there is a dramatic expansion of the ER to accommodate the increase in immunoglobulin synthesis (Iwakoshi et al., 2003). There is also evidence that components of the ER stress response are required for the secretory functions of pancreatic β cells (Harding et al., 2001; Lipson et al., 2006). Although the role of the plant ER stress response during development remains unknown, we have observed that bZIP60 protein is cleaved in anthers and that the bZIP60 gene is strongly expressed in pollen and tapetal cells.
Pollen cells have a high secretory capacity to support pollen tube growth after pollination (Hepler et al., 2001). Consistent with this fact, elongated ER stacks have been observed in pollen grains of several plant species (Ciampolini et al., 1988; Weber, 1989; Rodriguez-Garcia and Fernandez, 1990; Polowick and Sawhney, 1993). Tapetal cells secrete large amounts of proteins and lipids to the surface of pollen cells (Hsieh and Huang, 2007). Ultrastructural analyses have shown the presence of extensive layers of ER with dilation in tapetal cells of Arabidopsis and other plant species (Owen and Makaroff, 1995; Fei and Sawhney, 1999; Papini et al., 1999; Zheng et al., 2003). Thus, it appears likely that aspects of the ER stress response are important in the development and function of such secretory cells. It has also been suggested that a high secretory capacity is required for the pathogen response, in which the ability to upregulate the secretory machinery is critical for the efficient production of secreted and vacuole-targeted pathogenesis-related proteins (Wang et al., 2005). Whether the ER stress response mediated by bZIP60 is also involved in the plant's pathogen response will be an interesting topic for future studies.
METHODS
Plant Materials and Growth Conditions
We used Arabidopsis thaliana Col-0 plants and T-DNA insertion mutants in the Col-0 background. Mutants bzip60, s1p-1, and s1p-2 were obtained from the ABRC (Alonso et al., 2003), and s2p-2 and s2p-3 from GABI-Kat (Rosso et al., 2003). The Arabidopsis suspension cell line MM2d was derived from the Landsberg erecta ecotype (Menges and Murray, 2002) and cultured in Murashige and Skoog (MS) (Murashige and Skoog, 1962) medium supplemented with 3% (w/v) sucrose, 200 mg/L KH2PO4, 1 mg/L thiamine hydrochloride, 100 mg/L myoinositol, and 0.2 mg/L 2,4-D at 25°C. A proportion of the cells was transferred to new medium once a week.
Microarray Analysis
Arabidopsis seedlings were grown in half-strength MS medium supplemented with 2% (w/v) sucrose in a 16-h-light/8-h-dark cycle. Total RNA was extracted from 10-d-old seedlings treated with 5 μg/mL tunicamycin (Wako Pure Chemical) for 5 h and from control seedlings not treated with tunicamycin. Total RNA was extracted using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Total RNA samples were processed to cDNA, labeled, and hybridized to Arabidopsis 2 Oligo Microarrays (Agilent Technologies), which were then scanned using an Agilent Technologies Microarray Scanner and processed using Feature Extraction 7.5.1 (Agilent Technologies). Each RNA sample labeled with Cy5 was hybridized competitively with common reference (CR) labeled with Cy3; an equal mixture of each total RNA was used for CR. Hybridization and scanning were performed by Hokkaido System Science. Two independent batches of seedlings were used as the source of RNA for each condition, and the averaged values per condition are shown in Supplemental Table 1 online. Genes whose induction is lower in bzip60 mutant seedlings than in wild-type seedlings were identified by Welch's t test using GeneSpring GX software (Silicon Genetics). A P value cutoff of 0.05 was selected.
We surveyed 500 bp of promoter sequence upstream from the known or predicted transcription start site for the following consensus sequences. Since the first cytosine residue of CCACG of ERSE (CCAAT-N9-CCACG) and ERSE-II (ATTGG-N-CCACG) has been reported to have a minor effect, the consensus sequences for ERSE and ERSE-II used in this search were defined as CCAAT-N10-CACG (for ERSE) and ATTGG-N2-CACG (for ERSE-II). XBP1-BS (GA-TGACGT-GK) and UPRE (TGACGT-GR) sites were used without modification. We also defined an ERSE-like sequence with one mismatch as ERSE-L. To determine whether these cis-elements are overrepresented among the promoters of ER stress-responsive genes, we first determined the frequency of each motif in 500 bp of promoter sequences of all Arabidopsis genes. We then calculated the probability of finding promoter regions having one or more elements in the set of 129 promoter regions of ER stress-responsive genes.
RNA Gel Blot Analysis
Arabidopsis seedlings were grown in half-strength MS medium supplemented with 2% (w/v) sucrose in a 16-h-light/8-h-dark cycle. Total RNA was extracted using the aurintricarboxylic acid method (Gonzalez et al., 1980) from 2-week-old seedlings treated with tunicamycin (5 μg/mL) for the indicated time periods. Five micrograms of RNA was applied per lane and fractionated on a 1.2% agarose gel containing 2% formaldehyde. The RNA was capillary blotted onto a nylon membrane (Hybond N; Amersham Biosciences) in 20× SSC and fixed by UV irradiation. To detect BiP transcripts, ∼200-bp fragments of the 5′ regions of BiP1 and BiP3 were PCR amplified using primers 5′-CAAAAAGAGAGATCGTACGCAAAAG-3′ and 5′-ACTGATCCTAACTTCGTAGCTTCTT-3′ for BiP1 and 5′-ACAAACGAGATCGAAGAAGAGTTCTC-3′ and 5′-ACCGTCCCCAGTTTCTGCTCTTCGC-3′ for BiP3 and used for the labeling reaction. The BiP1 fragment also recognizes BiP2 because the homology between BiP1 and BiP2 is very high. To detect bZIP60, Sar1B, and Sec61γ transcripts, their cDNAs were PCR amplified using primers 5′-ATGGCGGAGGAATTTGGAAGC-3′ and 5′-GAGCTCTCACGCCGCAAGGGTTAAGATTTGG-3′ for bZIP60, 5′-ATGTTTTTATTCGATTGGTTCTATGGAATC-3′ and 5′-CTACTTGATATACTGAGATAGCCA-3′ for Sar1B, and 5′-ATGGACGCCATTGATTCCGTCGTGG-3′ and 5′-CTAAGTAGCACCGACGATGATGTTG-3′ for Sec61γ. These cDNA hybridization probes were labeled with [α-32P]dCTP using a DNA labeling kit (BcaBEST labeling kit; TaKaRa). The membrane was washed with 0.2× SSC, 0.1% SDS at 55°C three times for BiP1 and BiP3 or with 0.2× SSC, 0.1% SDS at 65°C three times for others, and then exposed to x-ray film.
Stable Transformation of Arabidopsis
For complementation of bzip60, a genomic fragment containing ∼2.1 kb of the 5′ sequence upstream from the start codon and 0.6 kb of the 3′ sequence downstream from the stop codon of bZIP60 coding sequence was PCR amplified using primers 5′-CCTCGAGCGTGATGATAATTAGACTAGGAC-3′ and 5′-CGGATCCCCGTAGCTAGTTTCATCGACAAC-3′. The PCR product was digested with XhoI and BamHI and inserted into pBIB-KS and was a gift from Keiji Nakajima (Nara Institute of Science and Technology); pBIB-KS was generated by inserting ori and LacZ DNA fragments of pBluescript II KS (Alting-Mees and Short, 1989) into pBIB (Becker, 1990).
To generate bZIP60-OX, bZIP60 cDNA was amplified using primers 5′-ATGGCGGAGGAATTTGGAAGC-3′ and 5′-GAGCTCTCACGCCGCAAGGGTTAAGATTTGG-3′. The PCR product was digested with BamHI and SacI and used to replace the GUS gene of pBI121 (Clontech Laboratories). To generate DEX-bZIP60ΔC-RD plants, bZIP60ΔC-RD was amplified by PCR using primers 5′-CTCGAGATGGCGGAGGAATTTGGAAGCATA-3′ and 5′-ACTAGTTCACAATCTCAACTCCAAATCAGACTCCTGCTTCGACATCATGG-3′, digested with XhoI and SpeI, and inserted into pTA7002 (Aoyama and Chua, 1997). These constructs were transferred to Agrobacterium tumefaciens C58 by the freeze-thaw method (Holsters et al., 1978). Stable transformation of Arabidopsis plants was performed using the floral dip method (Clough and Bent, 1998). Stable transformation of MM2d cells was performed by coculturing with Agrobacterium (Menges and Murray, 2004).
RT-PCR
Wild-type and DEX-bZIP60ΔC-RD seedlings grown in half-strength MS medium for 10 d were treated with 5 μg/mL tunicamycin, 10 μM DEX (Sigma-Aldrich), or both for 5 h. Wild-type and bZIP60-OX MM2d cells were treated with 5 μg/mL tunicamycin or 2 mM DTT for indicated times. Total RNA was extracted using the RNeasy plant mini kit. RNA (100 ng) was reverse transcribed using the PrimeScript RT reagent kit (TaKaRa) according to the manufacturer's instructions with oligo(dT) primers. Real-time qRT-PCR measurements were performed using a Roche LightCycler 480 (Roche Diagnostics). bZIP60, BiP1, BiP3, and Act8 transcripts were amplified using SYBR Premix Ex Taq (TaKaRa) with primers 5′-CGATGATGCTGTGGCTAAAA-3′ and 5′-TCTCAAGCATTCTCTTTCGAGAT-3′ for bZIP60, 5′-TCAGTCCTGAGGAGATTAGTGCT-3′ and 5′-TGCCTTTGAGCATCATTGAA-3′ for BiP1, 5′-CGAAACGTCTGATTGGAAGAA-3′ and 5′-GGCTTCCCATCTTTGTTCAC-3′ for BiP3, and 5′-TCAGCACTTTCCAGCAGATG-3′ and 5′-ATGCCTGGACCTGCTTCAT-3′ for Act8. Expression values of BiP1 and BiP3 were normalized to those of Act8.
For detecting Act8, S1P, S2P, and bZIP60 transcripts by PCR, cDNAs were amplified for 25 cycles using the primers 5′-CTGTGGACAATGCCTGGACCTGC-3′ for Act8, 5′-GTATCATACACCTCATGGTTTCTTG-3′ and for 28 cycles using the primers 5′-CCAAAAGCCCAAGAACAGCAGGGTCTTC-3′ S1P, 5′-ATGGAAATTTCAGGACGGCGAATGAG-3′ and 5′- TCAGATCACCGACAAAAACAAACGTGCC-3′ for S2P and for 30 cycles using the primers 5′-ATGGCGGAGGAATTTGGAAGC-3′ and 5′-GAGCTCTCACGCCGCAAGGGTTAAGATTTGG-3′ for bZIP60. PCR products were separated by electrophoresis and visualized by ethidium bromide staining.
Generation of Antibodies
bZIP60ΔC protein was used as an antigen for preparation of an antibody recognizing the bZIP60 protein (Figure 1A). The bZIP60ΔC fragment was PCR amplified using primers 5′-CACCATGGCGGAGGAATTTGGAAGCATAG-3′ and 5′-AGACTCCTGCTTCGACATCATGGTAG-3′ and cloned into pBAD102 vector (Invitrogen) according to the manufacturer's instruction. The recombinant bZIP60ΔC fused with thioredoxin and hexahistidine Trx-bZIP60ΔC-His was expressed in Escherichia coli TOP10 cells (Invitrogen), purified by Ni-NTA agarose (Qiagen), and used to immunize rabbits. The resulting serum was passed through a column containing hexahistidine-tagged thioredoxin to remove antibodies recognizing thioredoxin and hexahistidine, and purified using Trx-bZIP60ΔC-His, resulting in an antibody specifically recognizing bZIP60ΔC. Peptides corresponding to 645 to 658 amino acids of BiP1 or 657 to 670 amino acids of BiP3 were used as antigens for preparation of an antibody recognizing BiP1 and BiP3, respectively. Each antibody raised in a rabbit was affinity purified using the peptide antigens.
Protein Extraction and Subcellular Fractionation
For total protein extracts, Arabidopsis seedlings or MM2d cells were treated with 5 μg/mL tunicamycin or 2 mM DTT for the indicated times and homogenized in an extraction buffer (50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 0.05% Tween 20, 1 mM EDTA, 1 mM PMSF, and 5 μg/mL leupeptin). The homogenate was centrifuged at 12,000g for 10 min. Soluble proteins in the supernatant were used as total proteins. The protein concentration was quantified using a Bio-Rad protein assay kit (Bio-Rad) with BSA as a standard. Nuclear protein was extracted from MM2d cells treated with or without 5 μg/mL tunicamycin for 2 h using a plant nuclei isolation/extraction kit (Sigma-Aldrich) according to the manufacturer's instruction. To determine membrane localization of bZIP60, MM2d cells were collected, homogenized in LE buffer (80 mM Tris-HCl, pH 7.5, 12% sucrose, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 5 μg/mL leupeptin), and centrifuged at 100,000g for 1 h. The resulting pellet was resuspended with the same amount of LE buffer as the supernatant and used for immunoblot analysis. For sucrose gradient centrifugation, MM2d cells were collected and homogenized in LE buffer, which contains EDTA, or LM buffer, which contains Mg2+ (80 mM Tris-HCl, pH 7.5, 12% sucrose, 2 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 5 μg/mL leupeptin). The homogenate was centrifuged at 10,000g for 10 min. The supernatant was further centrifuged at 100,000g for 60 min to collect the microsomal fraction. The pellet was resuspended in the same buffer (0.5 mL), subjected to sucrose gradient (15 to 55%, 11.5 mL), centrifugation at 100,000g for 16 h, and then fractionated into 13 fractions.
Immunoblot Analysis
Protein extracts were loaded on an SDS-PAGE gel (Mini Electrophoresis System; Bio-Rad). After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane, and immunoreactive proteins were detected with ECL Plus protein gel blotting detection reagent (Amersham Biosciences) according to the manufacturer's instructions. The antibodies used were anti-BiP1, anti-BiP3, anti-bZIP60, anti-RGP (Dhugga et al., 1997), anti-VHA-a (Kobae et al., 2004), anti-AHA (Kobae et al., 2004), anti-cFBPase (Strand et al., 2000), anti-COX-II (Agrisera), anti-Histone H2B (Abcam), anti-α-tubulin (Santa Cruz), anti-HA 3F10 (Roche), anti-FLAG M2-HRP (Sigma-Aldrich), anti-rabbit IgG-HRP (Bio-Rad), and anti-rat IgG-HRP (Santa Cruz).
Histochemical Staining for GUS Activity
Generation of a transgenic plant carrying the bZIP60 promoter:GUS fusion was previously described (Iwata and Koizumi, 2005a). Seedlings treated with 5 μg/mL tunicamycin for 10 h and untreated seedlings were incubated with GUS staining solution (0.1 M sodium phosphate buffer, 10 mM EDTA, pH 8.0, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6], 1 mM X-glucuronide, and 0.1% Triton X-100) at 37°C for 16 h. Buds, open flowers, and siliques were immersed in ice-cold 90% acetone for 15 min and then stained with GUS staining solution. Tissues were dehydrated by increasing the ethanol concentration gradually from 70% to absolute ethanol and then subjected to microscopy observation. Dehydrated buds were incubated at 60°C in melted Paraplast Plus (Sherwood Medical). After five changes of Paraplast over 72 h, the tissues were embedded in Paraplast Plus blocks, sectioned into 8-μm slices, and observed under a light microscope.
Accession Numbers
Sequence data can be found in the Arabidopsis Genome Initiative database under accession numbers At1g42990 (bZIP60), At5g28540 (BiP1), At5g42020 (BiP2), At1g09080 (BiP3), At1g09180 (Sar1), At4g24920 (Sec61γ), At5g19660 (S1P), At4g20310 (S2P), and At1g49240 (Act8). Identifiers for T-DNA insertion mutants are Salk_050204 (bzip60), Salk_020530 (s1p-1), Salk_111474 (s1p-2), 459C12 (s2p-2), and 816A08 (s2p-3). Microarray data can be found in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under the accession number E-MEXP-1753 “bzip60 vs wild type in response to tunicamycin.”
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of bzip60.
Supplemental Figure 2. Amino Acid Alignment of bZIP60 Homologs.
Supplemental Table 1. Genes Induced More Than Threefold by Tunicamycin Treatment.
Supplementary Material
Acknowledgments
A part of present study was conducted at the Nara Institute of Science and Technology when Y.I. and N.K. were there. We thank Naoko Yamaguchi (Nara Institute of Science and Technology) for technical assistance and Kazufumi Watanabe (Hokkaido System Science) for statistical analysis of the microarray data. We also thank the ABRC and GABI-Kat for providing T-DNA insertion lines, James A.H. Murray (Cambridge University) for MM2d cells, Kanwarpal S. Dhugga (Pioneer Hi-Bred International) for an anti-RGP antibody, Masayoshi Maeshima (Nagoya University) for anti-VHA-a and anti-AHA antibodies, Keiji Nakajima (Nara Institute of Science and Technology) for pBIB-KS, and Nam-Hi Chua (Rockefeller University) for pTA7002. This work was supported by a Grant-in-Aid for Scientific Research (Grants 19039023 and 20380188) to N.K. and National Science Foundation Grant MCB-0447506 to N.V.F.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nozomu Koizumi (nkoizumi@plant.osakafu-u.ac.jp).
Online version contains Web-only data.
References
- Akiyama, Y., Kanehara, K., and Ito, K. (2004). RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 23 4434–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Alting-Mees, M.A., and Short, J.M. (1989). pBluescript II: Gene mapping vectors. Nucleic Acids Res. 17 9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 605–612. [DOI] [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston, R.S., Fontes, E.B., Shank, B.B., and Wrobel, R.L. (1991). Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell 3 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A., Svensson, K., Persson, S., Jung, J., Michalak, M., Widell, S., and Sommarin, M. (2008). Functional characterization of Arabidopsis calreticulin1a: A key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 49 912–924. [DOI] [PubMed] [Google Scholar]
- Ciampolini, F., Moscatelli, A., and Cresti, M. (1988). Ultrastructural features of Aloe ciliaris pollen. 1. Mature grain and its activation in vitro. Sex. Plant Reprod. 1 88–96. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coleman, C.E., Lopes, M.A., Gillikin, J.W., Boston, R.S., and Larkins, B.A. (1995). A defective signal peptide in the maize high-lysine mutant floury 2. Proc. Natl. Acad. Sci. USA 92 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga, K.S., Tiwari, S.C., and Ray, P.M. (1997). A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: Purification, gene cloning, and trans-Golgi localization. Proc. Natl. Acad. Sci. USA 94 7679–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, H. and Sawhney, V.K. (1999). MS32-regulated timing of callose degradation during microsporogenesisin Arabidopsis is associated with the accumulation of stacked rough ER in tapetal cells. Sex. Plant Reprod. 12 188–193. [Google Scholar]
- Fontes, E.B., Shank, B.B., Wrobel, R.L., Moose, S.P., OBrian, G.R., Wurtzel, E.T. and Boston, R.S. (1991). Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell 3 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forward, B.S., and Misra, S. (2000). Characterization and expression of the Douglas-fir luminal binding protein (PmBiP). Planta 212 41–51. [DOI] [PubMed] [Google Scholar]
- Gillikin, J.W., Zhang, F., Coleman, C.E., Bass, H.W., Larkins, B.A., and Boston, R.S. (1997). A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol. 114 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, R.G., Haxo, R.S., and Schleich, T. (1980). Mechanism of action of polymeric aurintricarboxylic acid, a potent inhibitor of protein–nucleic acid interactions. Biochemistry 19 4299–4303. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zeng, H., Zhang, Y., Jungries, R., Chung, P., Plesken, H., Sabatini, D.D., and Ron, D. (2001). Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7 1153–1163. [DOI] [PubMed] [Google Scholar]
- Hatano, K., Shimada, T., Hiraiwa, N., Nishimura, M., and Hara-Nishimura, I. (1997). A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol. 38 344–351. [DOI] [PubMed] [Google Scholar]
- Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Mitsuda, N., Matsui, K., and Ohme-Takagi, M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321 172–178. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Ohta, M., Matsui, K., and Ohme-Takagi, M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514 351–354. [DOI] [PubMed] [Google Scholar]
- Holsters, M., de Waele, D., Depicker, A., Messens, E., van Montagu, M., and Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163 181–187. [DOI] [PubMed] [Google Scholar]
- Houston, N.L., Fan, C., Xiang, J.Q., Schulze, J.M., Jung, R., and Boston, R.S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137 762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, K., and Huang, A.H. (2007). Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi, N.N., Lee, A.H., Vallabhajosyula, P., Otipoby, K.L., Rajewsky, K., and Glimcher, L.H. (2003). Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4 321–329. [DOI] [PubMed] [Google Scholar]
- Iwata, Y., and Koizumi, N. (2005. a). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y., and Koizumi, N. (2005. b). Unfolded protein response followed by induction of cell death in cultured tobacco cells treated with tunicamycin. Planta 220 804–807. [DOI] [PubMed] [Google Scholar]
- Kalinski, A., Rowley, D.L., Loer, D.S., Foley, C., Buta, G., and Herman, E.M. (1995). Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta 195 611–621. [DOI] [PubMed] [Google Scholar]
- Kamauchi, S., Nakatani, H., Nakano, C., and Urade, R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272 3461–3476. [DOI] [PubMed] [Google Scholar]
- Kanaoka, M.M., Urban, S., Freeman, M., and Okada, K. (2005). An Arabidopsis Rhomboid homolog is an intramembrane protease in plants. FEBS Lett. 579 5723–5728. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C.Y., and Arnold, S.M. (2002). The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 3 411–421. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Kim, S.G., Park, J.E., Park, H.Y., Lim, M.H., Chua, N.H., and Park, C.M. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst, M.E., Meyer, D.J., Gibbon, B.C., Jung, R., and Boston, R.S. (2005). Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol. 138: 218–231. [DOI] [PMC free article] [PubMed]
- Klein, E.M., Mascheroni, L., Pompa, A., Ragni, L., Weimar, T., Lilley, K.S., Dupree, P., and Vitale, A. (2006). Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J. 48 657–673. [DOI] [PubMed] [Google Scholar]
- Kobae, Y., Uemura, T., Sato, M.H., Ohnishi, M., Mimura, T., Nakagawa, T., and Maeshima, M. (2004). Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 45 1749–1758. [DOI] [PubMed] [Google Scholar]
- Koiwa, H., Li, F., McCully, M.G., Mendoza, I., Koizumi, N., Manabe, Y., Nakagawa, Y., Zhu, J., Rus, A., Pardo, J.M., Bressan, R.A., and Hasegawa, P.M. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N., Martinez, I.M., Kimata, Y., Kohno, K., Sano, H., and Chrispeels, M.J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127 949–962. [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N., Ujino, T., Sano, H., and Chrispeels, M.J. (1999). Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 121 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokame, K., Kato, H., and Miyata, T. (2001). Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J. Biol. Chem. 276 9199–9205. [DOI] [PubMed] [Google Scholar]
- Kondo, S., Murakami, T., Tatsumi, K., Ogata, M., Kanemoto, S., Otori, K., Iseki, K., Wanaka, A., and Imaizumi, K. (2005). OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7 186–194. [DOI] [PubMed] [Google Scholar]
- Kondo, S., Saito, A., Hino, S., Murakami, T., Ogata, M., Kanemoto, S., Nara, S., Yamashita, A., Yoshinaga, K., Hara, H., and Imaizumi, K. (2007). BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol. Cell. Biol. 27 1716–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K., and Kaufman, R.J. (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson, K.L., Fonseca, S.G., Ishigaki, S., Nguyen, L.X., Foss, E., Bortell, R., Rossini, A.A., and Urano, F. (2006). Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4 245–254. [DOI] [PubMed] [Google Scholar]
- Liu, J.X., Srivastava, R., Che, P., and Howell, S.H. (2007. a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X., Srivastava, R., Che, P., and Howell, S.H. (2007. b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2004). Cryopreservation of transformed and wild-type Arabidopsis and tobacco cell suspension cultures. Plant J. 37 635–644. [DOI] [PubMed] [Google Scholar]
- Mori, K. (2000). Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101 451–454. [DOI] [PubMed] [Google Scholar]
- Muench, D.G., Wu, Y., Zhang, Y., Li, X., Boston, R.S., and Okita, T.W. (1997). Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 38 404–412. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Noh, S.J., Kwon, C.S., Oh, D.H., Moon, J.S., and Chung, W.I. (2003). Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311 81–91. [DOI] [PubMed] [Google Scholar]
- Oh, D.H., Kwon, C.S., Sano, H., Chung, W.I., and Koizumi, N. (2003). Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun. 301 225–230. [DOI] [PubMed] [Google Scholar]
- Okushima, Y., Koizumi, N., Yamaguchi, Y., Kimata, Y., Kohno, K., and Sano, H. (2002). Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol. 43 532–539. [DOI] [PubMed] [Google Scholar]
- Owen, H.A., and Makaroff, C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185 7–21. [Google Scholar]
- Papini, A., Mosti, S., and Brighigna, L. (1999). Programmed-cell-death events during tapetum development of angiosperms. Protoplasma 207 213–221. [Google Scholar]
- Polowick, P.L., and Sawhney, V.K. (1993). An ultrastructural study of pollen development in tomato (Lycopersicon esculentum). II. Pollen maturation. Can. J. Bot. 71 1048–1055. [Google Scholar]
- Radhakrishnan, A., Ikeda, Y., Kwon, H.J., Brown, M.S., and Goldstein, J.L. (2007). Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA 104 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold, A.M., Iwakoshi, N.N., Manis, J., Vallabhajosyula, P., Szomolanyi-Tsuda, E., Gravallese, E.M., Friend, D., Grusby, M.J., Alt, F., and Glimcher, L.H. (2001). Plasma cell differentiation requires the transcription factor XBP-1. Nature 412 300–307. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Garcia, M.I. and Fernandez, M.C. (1990). Ultrastructural evidence of endoplasmic reticulum changes during the maturation of the olive pollen grain (Olea europaea L., Oleaceae). Plant Syst. Evol. 171: 221–231.
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Rutkowski, D.T., and Kaufman, R.J. (2004). A trip to the ER: Coping with stress. Trends Cell Biol. 14 20–28. [DOI] [PubMed] [Google Scholar]
- Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R.J. (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7 1165–1176. [DOI] [PubMed] [Google Scholar]
- Schutze, K., Harter, K., and Chaban, C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13 247–255. [DOI] [PubMed] [Google Scholar]
- Seo, P.J., Kim, S.G., and Park, C.M. (2008). Membrane-bound transcription factors in plants. Trends Plant Sci. 13: 550–556. [DOI] [PubMed]
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3 99–111. [DOI] [PubMed] [Google Scholar]
- Shen, X., Ellis, R.E., Lee, K., Liu, C.Y., Yang, K., Solomon, A., Yoshida, H., Morimoto, R., Kurnit, D.M., Mori, K., and Kaufman, R.J. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107 893–903. [DOI] [PubMed] [Google Scholar]
- Strand, A., Zrenner, R., Trevanion, S., Stitt, M., Gustafsson, P., and Gardestrom, P. (2000). Decreased expression of two key enzymes in the sucrose biosynthesis pathway, cytosolic fructose-1,6-bisphosphatase and sucrose phosphate synthase, has remarkably different consequences for photosynthetic carbon metabolism in transgenic Arabidopsis thaliana. Plant J. 23 759–770. [DOI] [PubMed] [Google Scholar]
- Sun, L.P., Seemann, J., Goldstein, J.L., and Brown, M.S. (2007). Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. USA 104 6519–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, H., Iwata, Y., Iwano, M., Takayama, S., and Koizumi, N. (2008). Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 374 242–247. [DOI] [PubMed] [Google Scholar]
- Tajima, H., and Koizumi, N. (2006). Induction of BiP by sugar independent of a cis-element for the unfolded protein response in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 346 926–930. [DOI] [PubMed] [Google Scholar]
- Tamura, T., Asakura, T., Uemura, T., Ueda, T., Terauchi, K., Misaka, T., and Abe, K. (2008). Signal peptide peptidase and its homologs in Arabidopsis thaliana–Plant tissue-specific expression and distinct subcellular localization. FEBS J. 275 34–43. [DOI] [PubMed] [Google Scholar]
- Urade, R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274 1152–1171. [DOI] [PubMed] [Google Scholar]
- Wang, D., Weaver, N.D., Kesarwani, M., and Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R.J., and Prywes, R. (2000). Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275 27013–27020. [DOI] [PubMed] [Google Scholar]
- Weber, M. (1989). Ultrastructural changes in maturing pollen grains of Apium nodiflorum L. (Apiaceae), with special reference of the endoplasmic reticulum. Planta 152 69–76. [Google Scholar]
- Weihofen, A., and Martoglio, B. (2003). Intramembrane-cleaving proteases: Controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 13 71–78. [DOI] [PubMed] [Google Scholar]
- Wolfe, M.S., and Kopan, R. (2004). Intramembrane proteolysis: Theme and variations. Science 305 1119–1123. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K., Sato, T., Matsui, T., Sato, M., Okada, T., Yoshida, H., Harada, A., and Mori, K. (2007). Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 13 365–376. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K., Yoshida, H., Kokame, K., Kaufman, R.J., and Mori, K. (2004). Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. (Tokyo) 136 343–350. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., Maruyama, D., Endo, T., and Nishikawa, S.I. (2008). Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol. 49 1547–1562. [DOI] [PubMed] [Google Scholar]
- Yang, T., Espenshade, P.J., Wright, M.E., Yabe, D., Gong, Y., Aebersold, R., Goldstein, J.L., and Brown, M.S. (2002). Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110 489–500. [DOI] [PubMed] [Google Scholar]
- Ye, J., Rawson, R.B., Komuro, R., Chen, X., Dave, U.P., Prywes, R., Brown, M.S., and Goldstein, J.L. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6 1355–1364. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Haze, K., Yanagi, H., Yura, T., and Mori, K. (1998). Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273 33741–33749. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001. a). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2000). ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2001. b). Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K., Shen, X., Wu, J., Sakaki, K., Saunders, T., Rutkowski, D.T., Back, S.H., and Kaufman, R.J. (2006). Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124 587–599. [DOI] [PubMed] [Google Scholar]
- Zheng, Z., Xia, Q., Dauk, M., Shen, W., Selvaraj, G., and Zou, J. (2003). Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15 1872–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.