Abstract
The Arabidopsis thaliana resistance gene RPW8 triggers the hypersensitive response (HR) to restrict powdery mildew infection via the salicylic acid–dependent signaling pathway. To further understand how RPW8 signaling is regulated, we have conducted a genetic screen to identify mutations enhancing RPW8-mediated HR-like cell death (designated erh). Here, we report the isolation and characterization of the Arabidopsis erh1 mutant, in which the At2g37940 locus is knocked out by a T-DNA insertion. Loss of function of ERH1 results in salicylic acid accumulation, enhanced transcription of RPW8 and RPW8-dependent spontaneous HR-like cell death in leaf tissues, and reduction in plant stature. Sequence analysis suggests that ERH1 may encode the long-sought Arabidopsis functional homolog of yeast and protozoan inositolphosphorylceramide synthase (IPCS), which converts ceramide to inositolphosphorylceramide. Indeed, ERH1 is able to rescue the yeast aur1 mutant, which lacks the IPCS, and the erh1 mutant plants display reduced (∼53% of wild type) levels of leaf IPCS activity, indicating that ERH1 encodes a plant IPCS. Consistent with its biochemical function, the erh1 mutation causes ceramide accumulation in plants expressing RPW8. These data reinforce the concept that sphingolipid metabolism (specifically, ceramide accumulation) plays an important role in modulating plant programmed cell death associated with defense.
INTRODUCTION
Plants have evolved disease resistance (R) genes to protect themselves from the attack of various pathogens. Characterized plant R genes can be divided into several classes based on their predicted protein structures and functions. The majority of them encode proteins containing a nucleotide binding site and leucine-rich repeat motifs (NB-LRR). These R proteins can be further subdivided into two major subclasses: those having an N-terminal coiled-coil (CC) domain and those containing an N-terminal domain resembling the cytoplasmic signaling domain of the Drosophila Toll and human Interleukin-1 (TIR) transmembrane receptors (Dangl and Jones, 2001).
The Arabidopsis thaliana R gene RPW8 mediates broad-spectrum resistance to powdery mildew (Xiao et al., 2001). The RPW8 locus is not present in Arabidopsis accession Columbia (Col-0), but accession Ms-0 contains two homologous, functional R genes named RPW8.1 and RPW8.2 (Xiao et al., 2001). These two genes are hereafter referred to as RPW8 unless otherwise indicated. RPW8 is unique among the cloned R genes because the deduced RPW8 proteins contain an N-terminal transmembrane domain and one to two CCs and show no significant homology to other proteins, thus belonging to a unique R protein category (Dangl and Jones, 2001; Xiao et al., 2001). RPW8 confers broad-spectrum resistance to powdery mildew isolates from four Golovinomyces species that are capable of infecting hundreds of dicot plant species, including many economically important crops (Xiao et al., 2001). RPW8-mediated resistance is associated with typical plant defense responses similar to those regulated by classic NB-LRR genes, including rapid generation of H2O2 at the fungal penetration sites, induction of pathogenesis-related (PR) genes, and triggering of the hypersensitive response (HR), a form of programmed cell death (PCD) at the site of infection.
The HR is often, although not always, associated with defenses activated by various plant R genes against biotrophic and hemibiotrophic pathogens. HR-like cell death has been used as a phenotypic readout of defense activation in many genetic screens for identifying components that negatively regulate plant defense signaling (Lorrain et al., 2003). Many Arabidopsis mutants with spontaneous HR-like lesions and/or constitutive activation of defense marker genes have been isolated, and some of the responsible genes may indeed act as negative regulators in pathways leading to PCD activation and defense responses (Hammond-Kosack and Parker, 2003; Lorrain et al., 2003). Such studies have implicated many different cellular processes in the regulation of plant PCD pathway(s) that may be associated with plant defenses, including chlorophyll metabolism (Mach et al., 2001; Pruzinska et al., 2003), ion channeling (Clough et al., 2000; Balague et al., 2003; Jurkowski et al., 2004), protein phosphorylation/dephosphorylation (Frye et al., 2001; He et al., 2004), fatty acid homeostasis and modification (Kachroo et al., 2001, 2004), callose synthesis (Nishimura et al., 2003), superoxide production (Kliebenstein et al., 1999; Epple et al., 2003), lipid metabolism (Brodersen et al., 2002; Liang et al., 2003; Lorrain et al., 2004; Tang et al., 2005), and autophagy (Liu et al., 2005).
To understand RPW8 signaling, we examined the genetic interaction between RPW8-induced cell death and previously characterized defense response mutants and thereby established that salicylic acid (SA), and the SA pathway components ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), PHYTOALEXIN-DEFICIENT4 (PAD4), EDS5, and NONEXPRESSOR OF PR1, are required for RPW8-dependent HR and resistance (Xiao et al., 2003, 2005). These SA pathway components are not only activated by the TIR-NB-LRR R proteins for specific resistance but are also required for a basal level of resistance that operates even in susceptible hosts (Falk et al., 1999; Jirage et al., 1999; Nawrath et al., 2002). We also observed that the scale of the RPW8-triggered H2O2 accumulation and HR resulting from powdery mildew challenge was influenced by the genetic background and environmental conditions, with a varying number of cells involved in the HR at the fungal invasion site (Xiao et al., 1997, 2003). Furthermore, plants overexpressing RPW8 from its native promoters exhibit spontaneous HR-like cell death (SHL) in the absence of any pathogens via a SA-dependent feedback amplification circuit (Xiao et al., 2003) that appears to be negatively regulated by EDR1, which encodes a mitogen-activated protein kinase kinase kinase implicated in defense signaling (Frye et al., 2001; Xiao et al., 2005). Hence, it is conceivable that the RPW8-triggered accumulation of H2O2 and HR is a double-edged sword that must be under tight control of both positive and negative regulation.
In order to further dissect the RPW8 signaling pathway, we have conducted a genetic screen using T-DNA tagging for mutations that enhance RPW8-mediated HR-like cell death (designated erh). Here, we report the isolation and characterization of erh1, in which an Arabidopsis homolog (At2g37940) of animal sphingomyelin synthase (SMS) and protozoan inositolphosphorylceramide synthase (IPCS) is knocked out. We provide evidence that ERH1 encodes an active plant IPCS and that loss of function of ERH1 results in higher levels of ceramides in plants expressing RPW8. Ceramides are lipid molecules composed of a long-chain base and an amide-linked acyl chain. They serve as precursors to form more complex sphingolipids by the addition of various sugar residues or phosphate-containing head groups to the ceramide moiety (Dunn et al., 2004). In yeast, protozoans, and plants, IPCS catalyzes the transfer of an inositol phosphate head group from phosphatidylinositol to ceramide, producing inositolphosphorylceramide (IPC) (Nagiec et al., 1997; Bromley et al., 2003; Denny et al., 2006). Ceramide derivatives not only provide structural integrity to eukaryotic cell membranes but also are believed to play important roles as second messengers in diverse cellular and physiological processes, such as cell differentiation and apoptosis in animals (Lei et al., 2007; Phan et al., 2007; Sanchez et al., 2007). Evidence is accumulating to suggest similar functions for sphingolipids in plants. For example, ceramide accumulation due to loss of function of ACCELERATED CELL DEATH5 (ACD5), encoding a ceramide kinase, results in PCD in Arabidopsis (Liang et al., 2003). Results from this study provide evidence that sphingolipid (ceramide) metabolism plays an important regulatory role in plant PCD associated with RPW8-mediated broad-spectrum disease resistance.
RESULTS
Isolation and Phenotypic Characterization of erh1
Arabidopsis accession Col-0 lacks RPW8 and is susceptible to powdery mildew (Xiao et al., 2001; Orgil et al., 2007). We selected a Col-0 line containing RPW8 as a transgene (ST8) for mutagenesis. ST8 (the eighth generation of line S6 reported in Figure 1D of Xiao et al., 2003) contains two copies of RPW8 under control of the native promoters and exhibits an intermediate level of SHL (Xiao et al., 2003). Thus, this line could be used to screen for mutations suppressing or enhancing RPW8-mediated SHL. In the screen for erh mutations (see Methods), we identified >20 putative erh mutants belonging to at least five complementation groups.
The first characterized erh mutant (erh1/ST8) was from a complementation group that contained two mutant lines from the same seed pool, and these two mutants were found to be derived from the same T1 individual by sequencing the genomic–T-DNA junctions. The erh1/ST8 mutant was backcrossed to ST8. All F1 plants had the same phenotype as ST8, indicating that the mutation was recessive. A total of 259 F2 plants grown in soil were scored for SHL and the presence of the T-DNA (reported by basta herbicide resistance). Of these, 26.6% (69 of 259) had the SHL phenotype seen in erh1/ST8, and these individuals were also all resistant to basta. These results suggested that a single T-DNA insertion in erh1/ST8 caused the enhanced SHL phenotype. A backcross (two generations)–purified erh1/ST8 mutant line was used for further characterization.
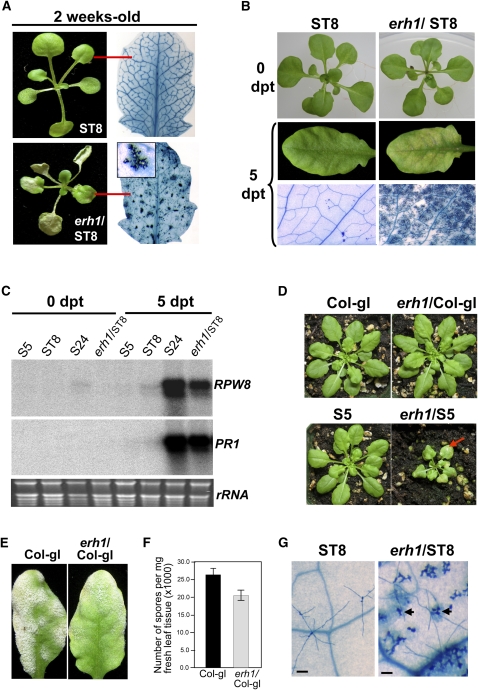
One week after seed germination in soil in short-day conditions (8 h of light, 16 h of dark; see Methods for plant growth conditions), very small lesions characteristic of SHL started to appear in erh1/ST8 cotyledons and true leaves and soon propagated to form big lesions that eventually engulfed the whole cotyledon/true leaf in a few days (Figure 1A). A close examination of the trypan blue–stained leaves showed that cell death first occurred in mesophyll cells around vascular tissues and then spread to neighboring cells, forming clusters of dead cells, which appeared as confined lesions to the naked eye (Figure 1A). Even though massive SHL often kills most of the growing leaves, erh1/ST8 plants survived long enough to set seeds. However, the size of mutant plants was ∼1/10th that of the ST8 parental line at 5 weeks old. Interestingly, like RPW8 overexpression–triggered SHL (Xiao et al., 2003), cell death in erh1/ST8 was also suppressed in plants grown on Murashige and Skoog (MS) agar medium (Figure 1B). However, when 6-week-old erh1/ST8 plants were transplanted from MS agar medium to autoclaved soil, massive SHL developed in mature and young leaves of erh1/ST8 plants in 5 to 7 d (Figure 1B). This response was comparable to the SHL seen in RPW8 transgenic line S24, which contains at least four copies of RPW8 and displays lethal SHL if grown in soil (Xiao et al., 2003). No visible cell death was observed in plants of the parental line ST8 at 10 d after transplanting (Figure 1B).
Figure 1.
Isolation and Characterization of erh1.
(A) Soil-grown seedlings of the parental line (ST8) and the erh1/ST8 mutant. A single representative leaf (indicated by red lines) stained with trypan blue is shown beside each seedling. The inset shows a cluster of dead cells at five times magnification.
(B) erh1-enhanced cell death is suppressed in plants grown on MS agar medium. Plants were grown in MS agar medium for 4 weeks and then transplanted into autoclaved soil. Photographs were taken at 0 or 5 d posttransplanting (dpt).
(C) RPW8 expression is enhanced by erh1. Four-week-old plants of the indicated genotypes were transplanted from MS agar medium to autoclaved soil. Total RNA was extracted from fully expanded mature leaves at 0 or 5 dpt, gel-blotted, and probed with a 32P-labeled cDNA mixture of RPW8.1, RPW8.2, and PR1 sequentially. rRNA is shown as a loading control.
(D) erh1-induced cell death is largely RPW8-dependent. The erh1 mutation from erh1/ST8 was introduced into Col-gl and S5 (carrying a single copy of RPW8) by crossing. Photographs were taken at 5 weeks old. Note the SHL (indicated by the red arrow) and reduced stature of erh1/S5.
(E) erh1 in Col-gl results in reduced susceptibility to powdery mildew. Five-week-old, soil-grown plants of Col-gl and erh1/Col-gl were inoculated with G. cichoracearum UCSC1. Representative infected leaves from the indicated genotypes at 10 dpi are shown.
(F) Infected leaves of six plants from each genotype used in (E) were subjected to quantitative measurement of disease susceptibility (see Methods for details) at 10 dpi. Data represent means ± se of three replicate experiments (P < 0.05 based on Student's t test).
(G) erh1/ST8 has more pronounced HR in response to powdery mildew infection. Five-week-old plants were transplanted from MS agar medium to autoclaved soil and maintained at >90% RH. Plants were inoculated with G. cichoracearum UCSC1 immediately after transplanting. At 2 dpi, inoculated leaves were stained with trypan blue. Arrows indicate cell death induced by the fungus. Bars = 100 μm.
The T-DNA–induced mutation in erh1/ST8 was then crossed into Col-gl (Col-0 harboring the glabrous mutation), and an F3 individual that was homozygous for the erh1 mutation and lacked RPW8 was selected for phenotypic examination. Compared with Col-gl, erh1/Col-gl plants were normal in growth and development in the first 6 weeks in short-day conditions (8 h of light; Figure 1D). After 6 weeks, leaves of erh1/Col-gl plants gradually developed a few small chlorotic lesions visible to the naked eye, and trypan blue staining revealed sporadic cell death in erh1/Col-gl leaves (Figure 2B), indicating that erh1 in the absence of RPW8 results in a developmentally regulated mild cell death phenotype. To further confirm if erh1 indeed enhances RPW8-triggered SHL, erh1 was introduced from erh1/Col-gl into line S5, which harbors a single transgene containing RPW8.1 and RPW8.2 under control of their native promoters in Col-gl and does not exhibit SHL (Xiao et al., 2003). As expected, plants of erh1/S5 displayed strong SHL in soil-grown plants at 2 weeks after seed germination and had a reduced stature, reaching ∼30% of the size of the parental S5 line at 5 weeks old (Figure 1D).
Figure 2.
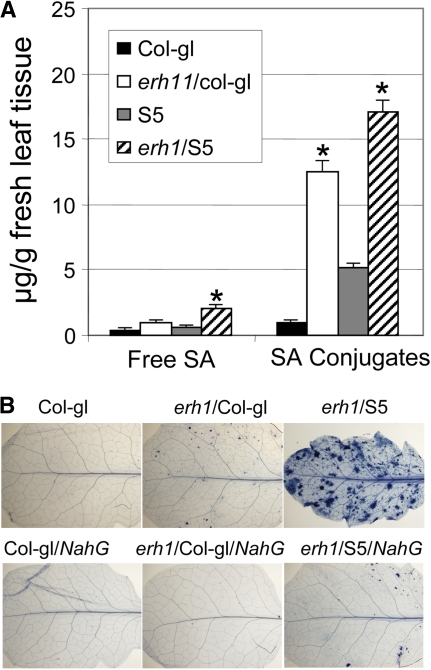
Involvement of SA in the erh1-Mediated Cell Death Phenotype.
(A) The erh1 mutation results in SA accumulation. Leaves of 6-week-old plants of the indicated genotypes were assayed for free and total SA. The amount of conjugated SA is the subtraction of free SA from total SA. Data represent means ± se from three duplicate samples in one experiment. Asterisks indicate that the value of the erh1 mutant is significantly different (P < 0.01) from that of the respective wild-type lines based on Student's t test. This experiment was repeated twice with similar results.
(B) erh1-activated SHL is SA-dependent. erh1 and erh1 plus RPW8 (from line S5) were introduced into the NahG background by crossing, and respective F3 lines homozygous for erh1 or erh1/RPW8 and NahG from the respective crosses were examined for SHL by trypan blue staining at 5 weeks old. [See online article for color version of this figure.]
erh1 Enhances RPW8 Transcription and RPW8-Mediated HR via the SA Signaling Pathway
To determine the mechanism by which erh1 enhances RPW8-mediated cell death, we examined RPW8 transcription and response to pathogens. To see if RPW8 transcription is enhanced by erh1, we checked the expression of RPW8 in the lines S5, ST8, erh1/ST8, and S24 at 0 and 5 d after transplanting from MS agar (SHL-suppressive conditions) to soil (SHL-permissive conditions). As we observed previously, RPW8 expression in S24 plants increased dramatically (Figure 1C) (Xiao et al., 2003), due to the activation of a self-amplification circuit when RPW8 reaches a threshold level under permissive conditions (e.g., soil). Amplification of RPW8 expression correlated with strong induction of PR1 (Figure 1C). We did not observe obvious induction of RPW8 and PR1 in S5 and ST8 plants after transplanting; however, we found that RPW8 and PR1 were highly induced in erh1/ST8 plants at 5 d after transplanting (Figure 1C). These results indicated that the erh1 mutation enhances transcriptional amplification of RPW8, which in turn results in PR1 expression and SHL.
To see if erh1 alters the response to pathogens, we challenged 5-week-old plants of Col-gl, erh1/Col-gl, S5, and erh1/S5 with the powdery mildew isolate Golovinomyces cichoracearum UCSC1. erh1/Col-gl showed slightly reduced fungal mass on the leaf surface compared with wild-type Col-gl (Figure 1E). Quantification of susceptibility (see Methods) at 10 d postinoculation (dpi) showed that the number of spores per milligram of fresh leaf tissue in erh1/Col-gl was 76.5% of that in the wild type (Figure 1F), indicating that erh1/Col-gl is slightly, but significantly (P < 0.05), less susceptible than Col-gl. It was also noted that erh1/Col-gl leaves turned slightly pale at later development stages or after fungal infection.
Under regular infection conditions (Xiao et al., 2005), both S5 and erh1/S5 were resistant to G. cichoracearum UCSC1, making it difficult to assess if erh1 enhances RPW8-mediated response to the pathogen. It has been shown that high humidity (∼90%) attenuates RPW8-transcription and RPW8-triggered resistance to G. cichoracearum UCSC1 (Xiao et al., 2003). We thus grew S5, erh1/S5, ST8, and erh1/ST8 plants in MS agar medium for 5 weeks and then transplanted them into autoclaved soil and continued to grow them at 90% RH. Immediately after transplanting, plants were then inoculated with G. cichoracearum UCSC1. Two days later, inoculated leaves were stained with trypan blue to visualize the cell death and the fungal infection structures. We found that RPW8-mediated HR in response to G. cichoracearum UCSC1 was attenuated in S5 and ST8 under these conditions (Figure 1G), consistent with our previous observation (Xiao et al., 2003). By contrast, in inoculated leaves of erh1/S5 and erh1/ST8, fungal penetration in most cases was found to be associated with host cell death (HR), even though cell death not apparently associated with the fungus was also seen (Figure 1G). No or very little SHL was observed in uninoculated leaves of erh1/S5 and erh1/ST8 over the same time period. This observation indicated that the erh1 mutation in the RPW8 background triggered a more sensitive HR in response to powdery mildew infection, similar to previous observations for edr1 in the RPW8 background (Xiao et al., 2005).
We also tested erh1/Col-gl and Col-gl with the virulent bacterial pathogen Pseudomonas syringae pv maculicola strain ES4326 and the congenic avirulent strain carrying avrRpm1, which elicits RPM1 (a CC-NB-LRR)–dependent HR and resistance (Grant et al., 1995). We did not find any significant difference in bacterial growth between Col-gl and erh1/Col-gl (see Supplemental Figure 1 online), suggesting that ERH1 does not substantially contribute to defense against P. syringae.
To further investigate how the erh1 mutation enhances RPW8-mediated cell death and the defense response, we measured the levels of SA in leaves of 6-week-old plants of Col-gl, erh1/Col-gl, S5, and erh1/S5. As shown in Figure 2A, compared with Col-gl, erh1/Col-gl had higher levels of free SA (∼3×) and SA conjugates (∼12.5×). Similarly, erh1/S5 had higher SA levels (3× for both free SA and SA conjugates) compared with S5. Apparently, the erh1 mutation resulted in increased SA accumulation independent of RPW8. Even though S5 accumulated 1.6 times the free SA and 5.2 times the conjugated SA compared with Col-gl, erh1 in S5 resulted in a further accumulation of free SA and conjugated SA, with levels reaching 5.4 and 14.0 times those in Col-gl, respectively. These data indicate that the erh1 mutation results in increased SA accumulation, possibly due to increased synthesis or decreased turnover, which in turn facilitates the SA-dependent feedback amplification of RPW8 transcription, leading to SHL or HR and resistance.
To obtain further supporting evidence for this hypothesis, we individually introduced NahG and pad4-1 into erh1/Col-gl and erh1/S5 by crossing. It is known that NahG (a bacterial gene encoding a SA hydrolase) and pad4-1 (a loss-of-function mutation of PAD4) suppress SA accumulation. We found that both NahG and pad4-1 largely abolished erh1-triggered mild SHL in Col-gl or stronger SHL in S5 (Figure 2B), indicating that SA accumulation indeed contributes to erh1-triggered cell death.
Cloning of ERH1
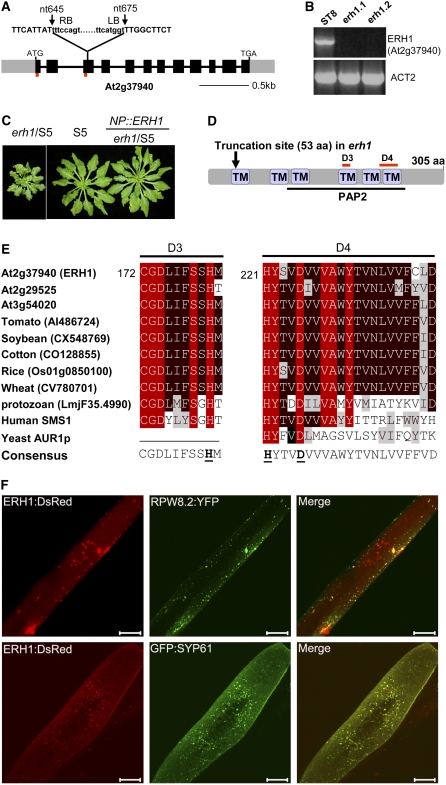
We used inverse PCR (Does et al., 1991) to locate the insertion position of the T-DNA in erh1/ST8, and the results were confirmed by PCR with gene-specific primers (see Methods). Sequencing of the PCR products revealed that erh1/ST8 had a T-DNA insertion in the third exon of At2g37940 (Figure 3A), an expressed gene with unknown function. RT-PCR showed that At2g37940 expression was completely knocked out in erh1/ST8 (Figure 3B). Thus, At2g37940 is the ERH1 candidate gene.
Figure 3.
Cloning of ERH1 and Subcellular Localization of the Protein.
(A) Schematic gene structure of ERH1 (At2g37940) and the position of the T-DNA insertion. Two short red bars indicate the positions of the primers used for RT-PCR in (B).
(B) RT-PCR with the parental line and two erh1/ST8 siblings for ERH1 and ACT2.
(C) Genetic complementation of erh1 with ERH1 under the control of the native promoter (NP).
(D) Predicted protein structure of ERH1. PAP2, phosphatidic acid phosphatase–related2; TM, transmembrane. D3 and D4 are two conserved domains found in functional homologs.
(E) Alignment of two conserved motifs, D3 and D4, from predicted plant ERH1-like protein sequences, a kinetoplastid (L. major) IPCS, human SMS1, and yeast (Saccharomyces cerevisiae) AUR1p. Identical residues are highlighted in red, and conservative residues are highlighted in black or gray. Underlined in the consensus are three residues that form a catalytic triad required for enzymatic function (Huitema et al., 2004).
(F) Subcellular localization of ERH1. Plasmid DNA of 35S:ERH1-DsRed was mixed in equal amounts with that of NP:RPW8.2-YFP or 35S:GFP-SYP61 (a cis-Golgi marker) or other constructs (see Supplemental Figure 4C online) and introduced into Col-0 leaf epidermal cells by bombardment (see Methods). Expression of the fluorescence-tagged proteins was imaged with a laser confocal microscope at 24 h after bombardment. Bars = 10 μm.
Next, we cloned the genomic sequence of At2g37940 and placed it under the control of the native promoter (∼1.8 kb upstream of the ATG start codon) or the 35S promoter and introduced the corresponding DNA constructs into erh1/S5 (Figure 3C). Expression of At2g37940 from the native or the 35S promoter fully complemented the erh1-mediated SHL phenotype. We also made an RNA interference (RNAi) construct (cDNA of At2g37940 in RNAi vector pJawohl8; Feys et al., 2005) to target At2g37940 and introduced it into Col-gl and ST8. We found that 29% (18 of 62) and 34% (20 of 58) of transgenic lines displayed phenotypes similar to those seen in erh1/Col-gl and erh1/ST8, respectively. These genetic data clearly indicated that At2g37940 is indeed ERH1.
ERH1 Shows Homology to Protozoan IPCS
Based on the annotation in the Arabidopsis database (http://www.Arabidopsis.org), ERH1 (At2g37940) is predicted to contain 12 exons and 11 introns, with a coding sequence of 918 bp (Figure 3A). We sequenced the cDNA product from Col-0 and confirmed the predicted exon–intron structure. ERH1 is predicted to encode a protein of 305 amino acids, with a molecular mass of 35 kD. The protein contains six transmembrane domains, which are predicted by TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) and ConPred II (http://bioinfo.si.hirosaki-u.ac.jp/∼ConPred2/) (Figure 3D), and has a weak phosphatidic acid phosphatase–related2 domain. ERH1 has two homologs in the Arabidopsis genome, At2g29525 and At3g54020, which respectively share 64 and 86% sequence identity with ERH1 at the protein level. At2g29525 and At3g54020 were thus designated ERH1-like1 (ERHL1) and ERHL2, respectively (see Supplemental Figure 2A online). A BLAST search identified numerous EST clones from various plant species showing 60 to 80% identity at the amino acid level to ERH1, suggesting that ERH1 and its homologs are highly conserved in plants.
The highest similarity to ERH1 identified outside the plant kingdom is to animal SMS (Huitema et al., 2004). For example, ERH1 shows 27% sequence identity and 44% similarity over a 148–amino acid stretch to human SMS1 (NP_671512; Hs SMS1). Similarity was also detected between ERH1 and the IPCS (LmjF35.4990; Lm IPCS) identified in a kinetoplastid protozoan (Leishmania major) (Denny et al., 2006). Although the amino acid sequence similarity in both cases is not high, the overall protein structures of ERH1, Hs SMS1, and Lm IPCS are very similar: all three are predicted to contain six transmembrane domains and a domain showing weak similarity to phosphatidic acid phosphatase–related2. Moreover, ERH1 and Lm IPCS show higher sequence similarity (∼30 to 45% identity at the amino acid level) in the two domains designated D3 and D4 (Figures 3D and 3E), which are conserved between animal SMSs and the lipid phosphate phosphatase family (Waggoner et al., 1999). A BLAST search with the Hs SMS1 and Lm IPCS protein sequences also identified ERH1 and its two Arabidopsis homologs as the most similar sequences in the Arabidopsis genome. Collectively, these data suggest that ERH1 may be a functional homolog of protozoan IPCSs or animal SMSs.
SMSs in animals transfer the phosphorylcholine moiety from phosphatidylcholine onto the primary hydroxyl of ceramide, producing sphingomyelin and diacylglycerol (Ullman and Radin, 1974; Voelker and Kennedy, 1982; Marggraf and Kanfer, 1984; Huitema et al., 2004; Tafesse et al., 2007), whereas IPCSs in yeast and protozoa catalyze the production of inositolphosphorylceramide and diacylglycerol from ceramide and phosphatidylinositol (Nagiec et al., 1997; Denny et al., 2006) (see Supplemental Figure 2B online). We reasoned that ERH1 may be a functional homolog of Lm IPCS and yeast AUR1p, since plants have been known to synthesize phosphoinositol-containing sphingolipids and sphingomyelin has not been detected in yeast and plants (Carter et al., 1958; Kaul and Lester, 1975; Markham et al., 2006).
ERH1 Encodes an Active Plant IPCS
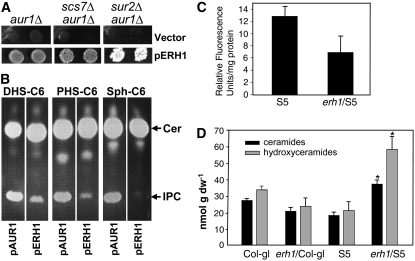
To test if ERH1 indeed encodes an IPCS, we first asked if expression of ERH1 in Saccharomyces cerevisiae could substitute for AUR1, an essential gene that encodes the sole IPCS (AUR1p) in yeast. The coding sequence of ERH1 was cloned into the yeast vector pADH1-LEU2. This recombinant plasmid (pERH1) was introduced into the yeast aur1Δ mutant that contained the yeast AUR1 gene in the pRS316-AUR1 plasmid. This plasmid rescues the lethal phenotype of aur1Δ and is URA3+-marked, enabling counterselection on fluoroorotic acid (FOA). Following transformation with pERH1, but not the pADH1 empty vector, the aur1Δ mutant was able to grow on medium containing FOA (Figure 4A), indicating that it was able to lose the pRS316-AUR1 plasmid. This result demonstrates that ERH1 can substitute for AUR1p. We also found that expression of ERH1 could substitute for AUR1p in the aur1Δ/scs7Δ mutant, which lacks the ceramide-associated fatty acyl chain C2-hydroxylase (Haak et al., 1997) (Figure 4A), indicating that, like AUR1p, ERH1 can use non-C2-hydroxy ceramides as a substrate. Furthermore, we found that ERH1 could substitute for AUR1p in the aur1Δ/sur2Δ mutant, which lacks the sphinganine (d18:0, dihydrosphingosine; DHS) C4-hydroxylase that catalyzes the formation of the major yeast long-chain base phytosphingosine (PHS; t18:0). These data indicate that ERH1 has a broad substrate specificity independent of the hydroxylation status of the ceramide (Figure 4A).
Figure 4.
ERH1 Encodes an IPCS.
(A) Complementation of the yeast aur1 mutation by ERH1. ERH1 was cloned in the pADH-LEU2 plasmid. The parental and the recombinant plasmids were introduced into three yeast strains lacking AUR1p alone (aur1Δ), AUR1p and ceramide-associated fatty acyl chain C2-hydroxylase (aur1Δ/scs7Δ), or AUR1p along with sphinganine hydroxylase (aur1Δ/sur2Δ). The respective yeast cells were grown on SD medium (−Leu) containing FOA.
(B) Yeast cells expressing ERH1 possess IPCS activities. Microsomes were prepared from aur1Δ yeast cells harboring pAUR1 or pERH1 and assayed for IPCS activities using the indicated ceramide substrates. Note that IPCS activities are reflected by the production of IPC (bottom bands) from the ceramide substrate (strong top bands). Cer, ceramide; Sph, sphingosine.
(C) In vitro assays for plant IPCS activity. Leaf lysates were prepared from 6-week-old plants and subjected to IPCS assays using dodecanoyl-NBD-d-erythro-DHS as a substrate. IPCS activity was measured using relative fluorescence units. Data are means ± se (P < 0.015 based on Student's t test of the values of the two genotypes; n = 4).
(D) erh1 results in ceramide accumulation in plants expressing RPW8. Sphingolipids were extracted from leaves of 8-week-old plants of the indicated genotypes and measured essentially as described previously (Markham and Jaworski, 2007). Values are means ± se (n = 5). Asterisks indicate significance at P < 0.01 compared with S5 based on Student's t test. dw, dry weight.
To gain direct biochemical evidence that ERH1 is an IPCS, we prepared microsomes from the aur1Δ mutant yeast harboring a plasmid expressing yeast AUR1 (pAUR1) or ERH1 (pERH1) and analyzed the IPCS activity using PHS-, DHS-, and sphingosine-containing 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)) (NBD) C6-ceramides. As shown in Figure 4B, microsomes from ERH1-expressing yeast cells transferred inositolphosphate to ceramides containing either DHS or PHS, albeit at a lower efficiency compared with that from AUR1p-expresssing cells. However, the IPCS activity of the ERH1-containing microsomes toward sphingosine-containing ceramide was very weak, as indicated by the barely detectable band of the IPC product (Figure 4B).
To confirm that ERH1 is an active plant IPCS, total protein extracts were prepared from leaves of 6- to 8-week-old S5 and erh1/S5 plants (see Methods). The extracts were then coincubated with soy phosphatidylinositol and dodecanoyl-NBD-d-erythro-DHS as substrates, and the IPCS activities were measured by quantification of the fluorescent IPC product. We chose dodecanoyl-NBD-d-erythro-DHS as a substrate because NBD-ceramide having DHS as the long-chain base appeared to be a better substrate than NBD-ceramide having sphingosine in our yeast assays (Figure 4B). As shown in Figure 4C, compared with S5 (100%), erh1/S5 showed reduced IPCS activity (53.3%), indicating that ERH1 accounts for about half of the total IPCS activity in mature leaf tissue. The partial reduction in the IPCS activities in erh1/S5 is not unexpected, since the Arabidopsis genome contains two ERH1 homologs (see Supplemental Figure 2A online).
Given the biochemical function of ERH1 as a plant IPCS, one would predict that loss of function of ERH1 may result in the accumulation of ceramides and/or reduced levels of IPC or more complex glycosylated IPCs. To test this, we measured total sphingolipids from leaves of 6-week-old plants of Col-gl, erh1/Col-gl, S5, and erh1/S5. Comparative analyses of the sphingolipid profiles revealed that the total amounts of ceramides and hydroxyceramides in erh1/S5 were significantly higher (P < 0.01) than those in the other three genotypes (Figure 4D) and that there were no significant differences in other sphingolipids (see Supplemental Figures 3A to 3E online). Significant accumulation of ceramides in erh1/S5 but not in erh1/Col-gl correlated with higher levels of SA and much more spontaneous cell death in the former and thus implies a synergistic relationship between RPW8 expression, SA and ceramide accumulation, and PCD.
ERH1 Is Largely Localized to the trans-Golgi Network
To test if ERH1 is also expressed in other organs and tissues apart from leaves, we checked the expression pattern of ERH1 by RT-PCR and found that it was expressed in all organs and tissues examined (see Supplemental Figure 4A online). Ceramide is synthesized at the endoplasmic reticulum and translocated to the Golgi compartment for conversion to more complex sphingolipids (Hanada et al., 2003). Consistent with this, yeast AUR1p and human SMS1 are found to be located to the Golgi apparatus (Levine et al., 2000; Tafesse et al., 2007). We thus asked if ERH1 also resides in the Golgi compartment. For this, we made a fusion construct under the control of the 35S promoter such that ERH1 is fused with yellow fluorescent protein (YFP) at its C terminus. We first introduced the construct into erh1/S5 and found that it rescued the SHL phenotype, indicating that the fusion protein is functional. However, confocal examination of the leaves of erh1/S5 or Col-0 plants transgenic for 35S:ERH1-YFP could not detect obvious fluorescence, implying that the protein expression level is very low. We thus took a transient expression approach to determine its likely subcellular localization. Transient overexpression of 35S:ERH1-YFP in onion (Allium cepa) epidermal cells gave a punctate distribution of YFP fluorescence (see Supplemental Figure 4B online), suggesting a likely endomembrane localization. To find the specific cellular compartment(s) in which ERH1 is localized, we made the 35S:ERH1-DsRed construct and bombarded leaves of Col-0 with the plasmid DNA of either 35S:ERH1-YFP or 35S:ERH1-DsRed, with plasmids containing 35S:mCherry-AtWAK2-HEDL (Nelson et al., 2007), 35S:ERD2-GFP (daSilva et al., 2004), and 35S:GFP-SYP42 and 35S:GFP-SYP61 (Sanderfoot et al., 2001; Uemura et al., 2004) producing marker proteins for the endoplasmic reticulum, the Golgi bodies, and the trans-Golgi network, respectively. ERH1:DsRed did not show obvious colocalization with mCherry:AtWAK2:HEDL or ERD2:GFP; however, it did colocalize with GFP:SYP42 and GFP:SYP61, indicating that ERH1 is localized to the trans-Golgi network (Figure 3F; see Supplemental Figure 4C online). We also biolistically introduced plasmids expressing RPW8.2:YFP and ERH1:DsRed into Col-0 epidermal cells. Interestingly, ERH1 and RPW8.2 showed overlapping localization (Figure 3F).
ERH1 Is Upregulated in the RPW8 Overexpression Background and during HR
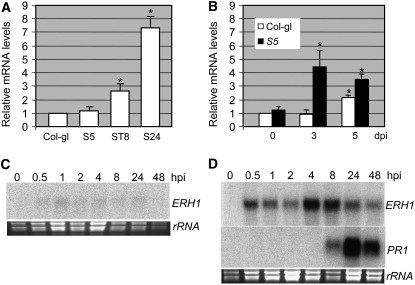
As ERH1 has been defined as a putative negative regulator of RPW8 signaling (Figure 1), we reasoned that ERH1 may be expressed at higher levels in the RPW8 background in order to curb RPW8-triggered HR or SHL. To test this idea, we measured the ERH1 mRNA levels in leaves of 6-week-old soil-grown Col-gl, S5, and ST8 plants and in leaves of S24 plants grown on MS agar medium (no cell death) for 5 weeks and then transplanted into soil for 5 d (cell death initiated). We found that the ERH1 mRNA levels in ST8 and S24 were 2.7 and 7.3 times that in Col-gl, while there was no significant difference between S5 and Col-gl (Figure 5A). This pattern of ERH1 expression correlates with the higher copy number of RPW8 in lines S5, ST8, and S24 (one, two, and four, respectively) and the degree of RPW8-trggered SHL phenotypes (none, mild, and severe, respectively), suggesting that the function of ERH1 is mechanistically associated with RPW8-mediated cell death.
Figure 5.
Induction of ERH1.
(A) RPW8 overexpression leads to enhanced ERH1 expression. Expression levels of ERH1 in 6-week-old plants (except S24, which was grown in MS agar for 5 weeks and transplanted to soil for 5 d) were measured by quantitative RT-PCR using TaqMan chemistry. The relative mRNA levels in RPW8-expressing lines were calculated relative to that of Col-gl (1.0). Data represent means ± se from three replicate experiments. Asterisks indicate significance at P < 0.01 compared with Col-gl based on Student's t test.
(B) ERH1 is induced to higher levels by powdery mildew. Six-week-old plants were inoculated with G. cichoracearum UCSC1. cDNA was synthesized from mRNA extracted from leaf tissues of the indicated genotypes at 0, 3, or 5 dpi and used for transcript quantification by real-time RT-PCR. The relative mRNA levels were calculated relative to that of Col-gl at 0 dpi (1.0). Data represent means ± se from three replicate experiments. Asterisks indicate significance at P < 0.01 compared with Col-gl based on Student's t test.
(C) and (D) ERH1 is induced by avirulent P. syringae. Total RNA was extracted from leaves of 6-week-old Col-0 plants infiltrated with the virulent P. syringae pv maculicola ES4326 strain (OD600 = 0.0002) (C) or the avirulent strain carrying AvrRpm1 (D) at the indicated time points, gel-blotted, and probed with 32P-labeled ERH1 or PR1 cDNA. hpi, hours postinoculation.
To examine if expression of ERH1 correlates with defense signaling, we asked whether exogenous SA application can induce ERH1. We sprayed Col-0 plants with 0.5 mM SA or water and measured the ERH1 mRNA levels by quantitative RT-PCR at 24 and 48 h after treatment. There was no significant change (<1.5 fold) before and after SA or water treatment (see Supplemental Figure 5 online). To see if ERH1 expression correlates with HR activation, we measured the ERH1 mRNA levels in Col-0 and S5 prior to inoculation and at 3 or 5 dpi with powdery mildew. As shown in Figure 5B, we found that ERH1 was slightly but significantly induced in Col-gl by powdery mildew inoculation at 5 dpi (2.2×). By contrast, ERH1 was more strongly (4.4×) induced at 3 dpi in S5, and the level dropped slightly (3.5×) at 5 dpi compared with Col-gl or S5 without infection. The higher ERH1 expression at 3 dpi in S5 coincided with the onset of HR activated by RPW8 in response to fungal invasion (Xiao et al., 2005).
Next, we examined ERH1 expression in response to inoculation with a virulent or an avirulent bacterial pathogen. Six-week-old Col-0 wild-type plants were inoculated with the virulent P. syringae pv maculicola strain ES4326 or the congenic avirulent strain carrying avrRpm1, which elicits RPM1-dependent HR and resistance (Grant et al., 1995). ERH1 was expressed at a very low level in unchallenged leaves and was barely induced by inoculation of the virulent strain (Figure 5C). By contrast, ERH1 was rapidly induced by the avirulent strain as early as 0.5 h after inoculation before the onset of HR. The induction of ERH1 peaked at 4 to 8 h after inoculation before the defense marker gene PR1 was highly induced (Figure 5D).
Combined, these results indicated that ERH1 is specifically associated with RPW8-triggered SHL and defense-associated HR.
erh1 and acd5 Activate Cell Death Additively or Synergistically
It was reported that acd5, a loss-of-function mutation in ACD5 encoding a ceramide kinase, results in ceramide accumulation and developmentally regulated cell death (Liang et al., 2003). To test if ceramide accumulation is indeed associated with SHL and enhanced defense responses in erh1/S5 plants, we first inoculated 5-week-old acd5 plants before development of cell death lesions with G. cichoracearum UCSC1 and found that, compared with Col-0, acd5 had clear enhanced resistance to the pathogen (Figure 6A), which is more obvious than that seen in erh1/Col-gl (Figure 1E).
Figure 6.
Additive/Synergistic Effects of acd5 and erh1 on Cell Death.
(A) acd5 has enhanced resistance to powdery mildew. Five-week-old plants were inoculated with G. cichoracearum UCSC1, and disease phenotypes at 8 dpi were shown by two representative leaves for each genotype. Powdery mildew infection can be seen as white powdery coating on Col-0 leaves.
(B) Six-week-old, short-day-grown plants of the indicated genotypes. Note the leaf cell death and reduced plant stature of acd5/S5.
(C) Four-week-old, short-day-grown plants of the indicated genotypes. Note the leaf cell death and reduced plant stature of erh1/acd5.
(D) Two-week-old, short-day-grown plants of the indicated genotypes. Note the collapsed cotyledons and stunted growth of erh1/acd5/S5.
(E) Silencing a rice ERH1 homolog leads to cell death and reduced plant stature. Two-and-one-half-months-old wild-type Nipponbare rice and one representative rice line transgenic for an RNAi construct targeting a rice ERH1 homolog (Os01g0850100) are shown. Arrows indicate dead leaves.
(F) RT-PCR analysis of the indicated genes from four rice transgenic lines along with the parental wild type.
Next, we introduced acd5 into the S5 background to see if it could enhance RPW8-triggered SHL. Compared with acd5, which did not develop cell death until ∼6 weeks old under our growth conditions (8 h of light, ∼80% RH), acd5/S5 developed necrotic cell death lesions at ∼4 weeks old and had slightly reduced plant stature (Figure 6B). Cell death of acd5/S5 also became more severe at later developmental stages compared with that of acd5.
We then made the erh1/acd5 double mutant and found that, compared with the single mutants, the double mutant plants developed earlier (at ∼4 weeks old) and more severe necrotic lesions (Figure 6C).
Finally, we introduced these two mutations into S5 by crossing and found that the erh1/acd5/S5 plants developed very strong cell death on cotyledons and true leaves at 1 week after seed germination in soil (Figure 6D) but managed to set seeds when grown in MS agar and then in soil despite greatly reduced plant stature.
These results suggest that there is an apparent additive or synergistic effect between acd5- and erh1-mediated cell death in leaf tissues and that ceramide accumulation in erh1 may account for accelerated cell death activation in the RPW8 background.
Downregulation of the Rice Homolog of ERH1 Leads to SHL and Reduced Plant Stature
To see if the function of ERH1-like genes is conserved in crop species, we generated rice (Oryza sativa) transgenic lines carrying an RNAi construct targeting the probable ortholog of rice ERH1 (Os01g0850100; designated Os ERH1). Among four transgenic lines analyzed, two showed obvious SHL and reduced plant stature (∼70% of the wild-type level) (Figure 6E). RT-PCR showed that only these two lines (lines 46 and 51) with SHL had reduced mRNA levels of Os ERH1 (Figure 6F). This result suggests a likely conserved function in the control of PCD for ERH1-like genes across plant species.
DISCUSSION
By taking a genetic approach, we have identified At2g37940 (ERH1) as a potential negative regulator of RPW8-mediated cell death associated with defense activation. Sequence homology analysis suggested that ERH1 may be the long-sought functional homolog of protozoan and yeast IPCSs that convert ceramide to IPC. Indeed, we found that ERH1 could complement the yeast IPCS (aur1) mutation and that expression of ERH1 in aur1Δ yeast cells and loss of function of ERH1 in Arabidopsis plants accounted for the complemented and reduced IPCS activities in the respective organisms (Figures 4B and 4C). Thus, our work has led to the identification of a plant IPCS gene and provided evidence for a critical role of sphingolipid metabolism in the regulation of plant PCD associated with pathogen resistance.
Sphingolipids are known to function in all eukaryotic cells as membrane structural components and as bioactive molecules involved in signal transduction and cell regulation (Dunn et al., 2004; Lynch and Dunn, 2004). Numerous reports have shown that spingolipids such as ceramide play essential regulatory roles in the induction of animal apoptosis (Lei et al., 2007; Phan et al., 2007; Sanchez et al., 2007). The aberrant cell death in each of the two Arabidopsis mutants (acd11 and acd5) was most likely caused by perturbation of sphingolipid metabolism, which is supported by the in vitro enzymatic activities of ACD11 and ACD5 as a sphingosine transfer protein and a ceramide kinase, respectively (Brodersen et al., 2002; Liang et al., 2003). These parallel emerging pictures for the role of sphingolipids in cell death regulation, from both the plant and animal kingdoms, suggest that there may be a common, fundamental mechanism of cell fate regulation by sphingolipids. This speculation is supported by the findings that mycotoxins such as fumonisin and AAL (Alternaria alternate f. sp lycopersici) toxin, which disrupt sphingolipid metabolism by inhibiting sphinganine N-acyltransferase, promote cell death in both animals (Wang et al., 1996b; Desai et al., 2002) and plants (Wang et al., 1996a).
Given that ERH1 is homologous to animal SMSs that convert ceramide to spingomyelin, the predominant sphingolipid in mammals (Hakomori, 1983), and that inhibition of SMS activities induces apoptosis (Meng et al., 2004; Ding et al., 2008), our finding that erh1 enhances RPW8-dependent cell death and resistance to powdery mildew in Arabidopsis further augments the notion that there may be a common mechanism in cell death regulation or execution in both plants and animals.
However, sphingomyelin has not been detected in plants (Dunn et al., 2004), posing a question regarding what biochemical function ERH1 may have in plants. Like plants and yeast, the kinetoplastid protozoa synthesize IPC rather than sphingomyelin. By utilizing bioinformatic and functional genetic approaches, Denny et al. (2006) isolated a functional ortholog of AUR1 (LmjF35.4990) in the kinetoplastids (L. major) and demonstrated that it possessed IPCS-like activity when expressed in mammalian cells. Interestingly, like ERH1, the protozoan IPCS shows higher homology to animal SMSs than to yeast AUR1p. Thus, it was not a surprise to us that our genetic and biochemical assays demonstrated that ERH1 indeed encodes an active plant IPCS (Figures 4A to 4C). It is worth noting that the sequence motif H-[YFWH]-X2-D-[VLI]-X2-[GA]-X3-[GSTA], which is shared by previously characterized lipid phosphate phosphatase family members and AUR1p homologs in fungi (Waggoner et al., 1999), is also conserved in animal SMSs, Lm IPCS, and ERH1 (defined as the D4 domain in Huitema et al., 2004) (Figure 3E), and the His and Asp residues (underlined) that are assumed to be required for catalytic activity (Neuwald, 1997) are absolutely conserved in all of these homologs (Figure 3E).
The identification of ERH1 as a plant IPCS supports the notion that biochemically there exists a divergence at an early step of ceramide metabolism among kingdoms: while ceramide is converted to sphingomyelin in animals, it is converted to IPC in fungi, kinetoplastids, and plants (see Supplemental Figure 2B online) (Denny and Smith, 2004). However, because both the plant (e.g., ERH1) and the protozoan (e.g., Lm IPCS) functional homologs of yeast AUR1p show higher homology to animal SMSs than to yeast AUR1p, it seems likely that the dichotomic split with regard to sphingolipid biosynthesis might have occurred more than once. Phylogenetic analysis of animal SMSs and yeast, protozoan, and plant IPCSs suggests that at least three evolutionary splits might have occurred concerning the initial step of ceramide metabolism (see Supplemental Figure 2C and Supplemental Data Set 1 online).
Based on the biochemical function of ERH1 (i.e., as a plant IPCS), loss of function of ERH1 is expected to cause two direct consequences: accumulation of ceramide and reduction of IPC or its more readily detectable glycosylated derivatives (GIPCs). Our sphingolipid profiling results showed that there were significant increases (P < 0.01) of ceramide and hydroxylceramide levels in erh1/S5 compared with those in S5. Interestingly, the increase in ceramides appeared to be as t18:0 (PHS)-containing ceramide species, of which a significant amount was t18:0 c16:0 (palmitic acid) (Figure 4D). No significant differences were detected in the levels of GIPCs, glucosylceramides, sphingoid long-chain bases, and their phosphorylated derivatives (see Supplemental Figures 3A to 3E online). It is possible that levels of GIPCs may have been maintained in erh1/S5 through some compensatory mechanisms, although, given the higher amounts of GIPCs present in plants, any depletion in GIPC levels may have been too small to detect. Elevated levels of ceramides and hydroxyceramides in erh1/S5 are consistent with the hypothesis that ceramide plays an important role in the activation of RPW8-dependent cell death. Additional evidence supporting this came from our genetic and phenotypic analyses of acd5 in combination with erh1 and RPW8. We found that acd5 plants, which have elevated levels of ceramides (Liang et al., 2003), showed enhanced resistance to powdery mildew and also exhibited exacerbated cell death in the S5 (RPW8) background (Figures 6A and 6B). Moreover, the acd5/erh1 double mutant developed an earlier and much stronger cell death phenotype in both the Col-0 and S5 backgrounds (Figures 6C and 6D). It was recently reported that loss of function of EDR2 causes enhanced resistance to powdery mildew in Arabidopsis and that the inhibition of fungal growth coincides with pathogen-induced host cell death (Tang et al., 2005; Vorwerk et al., 2007). EDR2 encodes a novel protein that contains a pleckstrin homology domain and a steroidogenic acute regulatory protein–related lipid-transfer domain, both found in the human ceramide transport protein CERT (Hanada et al., 2003). It is thus possible that edr2-mediated enhanced powdery mildew resistance may be attributed to perturbation of ceramide metabolism.
Cell death in erh1/S5 appears to be influenced by environmental conditions. Similar to the observation we made previously with S24 (Xiao et al., 2003), SHL was suppressed in erh1/S5 plants grown on MS agar plates, even though other growth conditions (i.e., light, temperature, and RH) were similar or identical. How MS agar medium conditions inhibit SHL development remains to be determined. Cell death in erh1/S5 also appears to be developmentally regulated. SHL became visible in true leaves of 2-week-old seedlings of erh1/S5, and the plant stature was permanently reduced. By contrast, acd5/S5 plants appeared to have normal growth and development in the first 4 weeks, reaching a similar rosette size as S5, and then developed big necrotic lesions that in some cases propagated to engulf the whole leaf. Despite an earlier occurrence and a more severe phenotype, the pattern of cell death in acd5/S5 is similar to that observed in acd5/Col-0 (Greenberg et al., 2000) but different from that in erh1/S5 (Figures 1 and 6). These observations suggested that ERH1 and ACD5 may fulfill different but essential biological functions in different or perhaps overlapping developmental stages.
HR cell death is often, although not always, associated with resistance against biotrophic pathogens such as powdery mildew and, to a lesser degree, with resistance against hemibiotrophic pathogens such as P. syringae. It is conceivable that HR cell death must be tightly regulated in plants to avoid unnecessary host cell death at the site of pathogen infection. Therefore, ERH1 may function to curb RPW8-dependent cell death. Consistent with this idea, we found that ERH1 induction occurred before or coincided with the onset of HR cell death, and there was no apparent or only a slight induction of ERH1 during compatible interactions with bacterial and fungal pathogens (Figures 5B and 5C). Mining of gene expression data in the National Center for Biotechnology Information database also found that ERH1 was induced to higher levels by powdery mildew (Nishimura et al., 2003; Stein et al., 2006) and by necrosis- and ethylene-inducing peptide extracted from Fusarium oxysporum (Bae et al., 2006). It remains unclear what role increased expression of ERH1 has during pathogen infection, but it presumably serves to increase GIPC biosynthesis for a defense-related function.
How erh1- and acd5-caused ceramide accumulation enhances RPW8-dependent cell death is not clear. In animals, ceramide accumulation has been known to induce apoptosis via multiple pathways (Pettus et al., 2002). Given that RPW8-triggered SHL, HR, and resistance involve an SA-dependent feedback amplification circuit, one likely scenario is that disruption of ERH1 or ACD5 somehow results in SA accumulation, which further strengthens RPW8 signaling, resulting in cell death. Indeed, the erh1 mutation in Col-gl and S5 resulted in 3 to 12 times increases of free SA and SA conjugates (Figure 2A), supporting this speculation. Similarly, the SA level in acd5 mutant plants also had an approximately fivefold increase compared with the wild type (Greenberg et al., 2000). Furthermore, depleting SA by the expression of NahG or blocking the SA signaling pathway by pad4-1 (in erh1; Figure 2B) and eds5-1 (in acd5; Greenberg et al., 2000) abolished the cell death phenotype caused by erh1 and acd5, respectively. These observations collectively suggest that normal sphingolipid metabolism is probably important for SA homeostasis required for cell survival and that ceramide accumulation may cause overproduction and accumulation of SA via an unknown mechanism, leading to PCD associated with defense signaling. Because erh1/S5 but not erh1/Col-gl showed accumulation of ceramides, we reason that RPW8's function must be somehow affected by erh1, which results in further SA accumulation in plants expressing RPW8, leading to ceramide accumulation, thus explaining erh1-triggered, RPW8-dependent cell death.
In spite of cell death and the activation of defense-related genes (i.e., PR1), acd5 did not show resistance to a virulent bacterial pathogen, P. syringae, leading to the conclusion that SA-dependent cell death and defense gene expression are uncoupled in acd5 and that the cell death in acd5 may mimic disease-caused cell death in plants (Greenberg et al., 2000). In this study, we also found that erh1/Col-gl was as susceptible as the wild type to a virulent P. syringae strain. However, both erh1/Col-gl and acd5/Col-0 had enhanced resistance to powdery mildew. This pattern of defense is consistent with our earlier observation that RPW8-activated defense signaling does not contribute to resistance to P. syringae; instead, RPW8 overexpression results in slightly enhanced susceptibility to this bacterial pathogen (Wang et al., 2007). Combined, these results suggest that subtle differences exist in the efficacy of SA signaling and HR cell death in resistance against biotrophic pathogens (such as powdery mildew) and hemibiotrophic pathogens (such as P. syringae) and that cell death and SA signaling activated by acd5 and erh1 appear to be effective only against biotrophic pathogens. One likely explanation for this phenomenon is that biotrophic pathogens such as powdery mildew strictly require living cells to establish infection and complete their life cycle, and cell death caused by ceramide accumulation may pose a physical barrier for the pathogen to grow and thrive. However, for hemibiotrophic bacterial pathogens such as P. syringae, once they are delivered into host tissues by injection, their multiplication is much faster and may not be affected by cell death caused by ceramide accumulation, even if it is accompanied by the activation of SA signaling.
There are two SMS homologs in the human genome. SMS1 functions in the Golgi, while SMS2 works at the plasma membrane (Tafesse et al., 2007). ERH1 is mainly localized to the trans-Golgi network (Figure 3F). The subcellular localization and function of the two Arabidopsis homologs of ERH1 remain to be determined. Functional redundancy between ERH1 and its homologs is likely, which could explain why erh1-triggered cell death is weaker in the Col-gl background compared with that caused by loss of function of the single-copy ceramide kinase gene (Liang et al., 2003). Construction and characterization of double and triple mutants for ERH1 and its homologs should clarify these questions.
There is indirect evidence for the involvement of ERH1-homologous genes (based on EST clones) in pathogen defense in other plant species. For example, a rice ERH1 homolog (Os05g0287800) was found to be induced by an avirulent pathogen (N1141 strain of Acidovorax avenae) (Fujiwara et al., 2004), and a soybean (Glycine max) ERH1 homolog (CD411105) was slightly induced by SA and Phytophthora sojae, race 1 (Tian et al., 2004). In this study, we have demonstrated that a rice ERH1 homolog (Os01g0850100) also appeared to function as a negative regulator of PCD in rice (Figure 6E), providing genetic evidence for likely functional conservation of ERH1-like genes across plant species. This functional conservation may provide a potential new avenue for enhancing disease resistance of crop species against biotrophic pathogens by targeted downregulation of the corresponding ERH1-like genes using pathogen-inducible promoters.
METHODS
T-DNA Mutagenesis and Mutant Characterization
The Arabidopsis thaliana Col-gl line ST8 (the eighth generation of line S6 reported in Xiao et al., 2003), harboring two copies of a transgene each containing both RPW8.1 and RPW8.2 under the control of their native promoters, developed an intermediate level of confined SHL readily visible to the naked eye at ∼5 weeks after seed germination in short days (8 h of light, 16 h of dark). Plants of the eighth generation from this line were transformed with the binary vector pSLJ755I5 (Jones et al., 1992) that contains the basta herbicide resistance gene for selection of transformants. Approximately 20,000 independent transgenic T1 plants were obtained, and the seeds were collected in 40 pools (∼500 T1 lines per pool). We screened ∼80,000 T2 plants (∼2000 for each pool) and identified >20 mutants belonging to at least five complementation groups with accelerated SHL in 2 to 3 weeks after seed germination. The T-DNA in erh1/ST8 was located by inverse PCR. Specifically, the genomic DNA of the mutant was digested with AseI and then self-ligated using T4 DNA ligase (New England Biolabs). PCR amplification of the T-DNA–genomic DNA junction was done using the primers 5′-CGGGCCTAACTTTTGGTGTGATGA-3′ and 5′-CCCCCCATCGTAGGTGAAGGTGGA-3′, designed based on the right T-DNA border of pSLJ755I5. The specific PCR product was sequenced, and the T-DNA junctions in At2g37940 (ERH1) were determined by PCR with gene-specific primers and primers based on the left or right border of the vector.
Unless otherwise indicated, seeds were sown in Sunshine Mix 1 (Maryland Plant and Supplies) and cold-treated (4°C for 2 d), and seedlings were kept under 22°C, 75% RH, short-day (8 h of light at ∼125 μmol·m−2·s−1, 16 h of dark) conditions for 5 weeks before pathogen inoculation or other treatments. The method for plant growth in MS agar medium was essentially the same as reported (Xiao et al., 2003). Briefly, surface-sterilized seeds were placed on MS agar plates (100 × 20 mm). After cold treatment (4°C for 2 d), the plates were placed in a growth chamber with the same growth conditions mentioned above except that the RH was slightly higher (85 to 90%) on the plates.
Introduction of NahG, pad4-1, or acd5 into erh1/Col-gl or erh1/S5 was done by crossing. Primers for genotyping NahG and pad4-1 were described by Xiao et al. (2005). To make the erh1/acd5 double mutant, erh1/Col-gl was crossed with acd5/Col-0, and F2 individuals homozygous for erh1/acd5 were identified by PCR for the T-DNA inserted in ERH1 and sequencing for the point mutation in ACD5 (Liang et al., 2003).
Pathogen Strains, Inoculation, and Phenotyping
Powdery mildew isolate Golovinomyces cichoracearum UCSC1 was maintained on live eds1-2 or pad4-1 plants. Inoculation, visual scoring, and quantification of disease susceptibility were done as described previously (Xiao et al., 2005). For bacterial infection, fully expanded leaves were infiltrated with a suspension (OD600 = 0.0002) of Pseudomonas syringae pv maculicola ES4326 wild-type strain or the avirulent strain carrying avrRpm1 in 10 mM MgCl2. Quantification of bacterial growth was according to Wang et al. (2007).
DNA Constructs and Gene Expression
The genomic sequence of ERH1 was amplified by 5′-CACCGGATCCATGACACTTTATATTCGTCGTGAA-3′ and 5′-CCGAATTCACGCGCCATTCATTGTGTT-3′, digested with BamHI and EcoRI, and cloned into the BamHI–EcoRI site of binary vector pSMB under the control of the 35S promoter. An Agrobacterium tumefaciens strain containing 35S:ERH1 in pSMB was used to transform erh1/ST8. Selection of transgenic lines was based on suppression of erh1-mediated strong SHL at 2 weeks old and confirmed by PCR. To express ERH1 from the native promoter, a genomic fragment containing the coding sequence of ERH1 was amplified by 5′-CACCGGATCCATGACACTTTATATTCGTCGTGAA-3′ and 5′-CCGAGCTCACGCGCCATTCATTGTGTT-3′ and cloned into the PstI–BamHI site of pPZP211. Then, an ∼1.8-kb sequence 5′ upstream of the ERH1 start codon was amplified by 5′-CACCCTGCAGACTTGTTTCCTATTTCGGATA-3′ and 5′-CTCGGATCCTTATTATTCTTGTTTGTTGATGATGT-3′ and cloned into the BamHI–SacI site of the pPZP211 plasmid containing the ERH1 coding sequence. The construct was introduced into erh1/S5 and erh1/ST8 by Agrobacterium-mediated transformation (Clough and Bent, 1998). To silence ERH1 in Arabidopsis, ERH1 was amplified by 5′-CACCGGATCCATGACACTTTATATTCGTCGTGAA-3′ and 5′-CCGAATTCACGCGCCATTCATTGTGTT-3′ and cloned to the PENTR/D-TOPO vector (Invitrogen). The fragment was then recombined into the binary Gateway vector pJawohl8 (Feys et al., 2005) using the manufacturer's suggested methods (Invitrogen) and subsequently introduced into Col-0 and ST8 by Agrobacterium-mediated stable transformation. For subcellular localization studies, the ERH1 coding sequence was amplified by 5′-GGGGATCCGATGACACTTTATATTCGTCGTGAA-3′ and 5′-TTGGATCCGCCATTCATTGTGTTATCCGT-3′ and cloned into binary vector pBamEYFP (Wang et al., 2007) such that YFP was translationally fused with ERH1 at the C terminus, resulting in pERH1-YFP. This 35S:ERH1-YFP construct was introduced into Col-0 and erh1/S5 by Agrobacterium-mediated transformation or into onion (Allium cepa) epidermal cells by bombardment. For colocalization analysis, the DsRed coding sequence was amplified with primers 5′-TAGGATCCATGGCCTCCTCCGAGG-3′ and 5′-AAAGATCTCCGCTACAGGAACAGGTGGTG-3′ and used to replace YFP in pERH1-YFP, resulting in pERH1-DsRed. Plasmid DNA for 35S:ERH1-DsRed was cobombarded with plasmid DNA for 35S:mCherry-AtWAK2-HEDL (Nelson et al., 2007), 35S:ERD2-GFP (daSilva et al., 2004), and 35S:GFP-SYP42 or 35S:GFP-SYP61 (Sanderfoot et al., 2001; Uemura et al., 2004) into leaves of Col-0, followed by examination with a laser scanning confocal microscope according to a previous report (Wang et al., 2007). To silence a rice homolog of ERH1 (Os01g0850100) by RNAi, the rice cDNA fragment was amplified by primers 5′-CACCATGGCGGTTTACATCGCTCGGGA-3′ and 5′-TCATGTGCCATTGGGAGTGGCATC-3′, cloned to pENTR/D-TOPO vector (Invitrogen), and then shuttled to the binary vector pANDA under the control of the ubiquitin promoter (Qu et al., 2006). The RNAi construct was introduced into Agrobacterium strain LBA4404, and rice transgenic plants were generated according to a previously published protocol (Qu et al., 2006).
Transcript Analysis
RNA extraction and gel blot analysis were conducted as described previously (Xiao et al., 2005). Quantification of the transcripts of ERH1 by real-time RT-PCR was done according to Xiao et al. (2003). ACT2 was chosen as the normalization standard for TaqMan analysis because of its constitutive expression in nearly all vegetative tissues (An et al., 1996) and its stable expression after pathogen challenge (Feys et al., 2001). cDNA-specific primers were used to measure levels of ACT2 and ERH1 transcripts. The sequences of the primers for ERH1 were 5′-CCGGGACCGACACTTCAG-3′ and 5′-CGGTTTCACTTATGTAGCTTCTCTCTT-3′, and the sequence for the probe was 5′-ATCTTGGCTTCTTTCTTCTTCCGGAGCTTG-3′. PCR was performed on an ABI 7300 sequence detection system (Applied Biosystems). Triplicate cDNA samples were prepared and analyzed for each genotype/treatment.
Yeast Strains, Vectors, and IPCS Assays
For genetic complementation tests with yeast strains lacking the IPCS gene, AUR1, the ERH1 coding sequence was PCR-amplified from an Arabidopsis cDNA library (Paul et al., 2006) and inserted into the SalI site of the yeast expression vector pADH1-LEU2 to generate pERH1. Sequence-confirmed pERH1 was introduced into the yeast aur1Δ mutant harboring the pRS316-AUR1 plasmid, and transformants were selected on medium lacking Leu and then transferred to YPD medium to allow for loss of pRS316-AUR1. Transformants that had lost the URA3+-marked pRS316-AUR1 plasmid were selected on FOA. pERH1 was similarly introduced into the yeast aur1Δ/scs7Δ and aur1Δ/sur2Δ double mutants (Haak et al., 1997) to determine the spectrum of ceramide substrates that could be utilized by ERH1 in yeast. To assay for IPCS activities, microsomes from yeast cells expressing ERH1 were prepared as described previously (Paul et al., 2006) and analyzed for IPCS activity using PHS-, DHS-, and sphingosine-containing NBD C6-ceramides as described previously (Figueiredo et al., 2005).
In Vitro Assay of Plant IPCS Activity
Leaves (∼250 mg) from 6- to 8-week-old plants were homogenated in 4 volumes of buffer (10 mM HEPES, pH 7.8, 2 mM EDTA, 2 mM β-mercaptoethanol, and plant protease inhibitor cocktail [Sigma-Aldrich]) using a Kontes glass tissue grinder. The lysates were centrifuged for 90 s at 13,400g, and the resulting supernatants were used in the enzyme assay immediately or stored at −80°C. Protein concentration of the lysates was determined using the Coomassie (Bradford) protein assay reagent from Pierce. IPCS activities in lysates were assayed using 450 μM soy phosphatidylinositol and 60 μM dodecanoyl-NBD-d-erythro-DHS (Matreya) as substrates, 0.5 mM CHAPS, 10 mM EDTA, 100 mM Tris-HCl (pH 7.5), and 45 μL of leaf lysate (40 to 80 μg of protein) in a total volume of 100 μL. Reactions were performed at 32°C for 40 min in the dark and terminated by the addition of 1.0 mL of methanol. The fluorescent IPC product was recovered by partitioning against tert-butyl methyl ether and water as described previously (Nagiec et al., 1997) and quantified by monitoring fluorescence at excitation and emission wavelengths of 466 and 537 nm, respectively. Assay tubes having all components but terminated with methanol prior to incubation were used as controls.
Other Analysis
Sphingolipids were extracted and measured as described previously (Markham and Jaworski, 2007) except that sphingolipids were separated using a 3.0- × 100-mm, 3.5-μm silica XBD-C18 column (Agilent Technologies). Sequence alignment was done using the AlignX function of the Vector NTI software package (Invitrogen). For SA analysis, leaves of 7-week-old plants grown in short days were used to measure SA levels using a protocol described previously (Vanacker et al., 2001). Three replicates of each genotype were analyzed. This experiment was repeated three times with similar results. Trypan blue staining for cell death was performed according to Xiao et al. (2003). Laser scanning confocal imaging was done as described by Wang et al. (2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At ERH1 (At2g37940); At ERHL1 (At2g29525); At ERHL2 (At3g54020); Os ERH1 (Os01g0850100); AUR1P (NP_012922); Lm IPCS (XP_843601); Hs SMS1 (NP_671512); Hs SMS2 (NP_689834); ACT2 (AT3G18780); and, for EST clones: AI486724 (tomato); CX548769 (soybean); CO128855 (cotton); and CV780701 (wheat).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. erh1/Col-gl Plants Are Not More Resistant to P. syringae.
Supplemental Figure 2. Evolution and Diversification of IPCSs (SMSs).
Supplemental Figure 3. Levels of Sphingolipids in Wild Type and erh1 Mutant Plants.
Supplemental Figure 4. Expression and Subcellular Localization of ERH1.
Supplemental Figure 5. ERH1 Is Not Significantly Induced by SA.
Supplemental Data Set 1. Sequence Alignment of ERH1 and Its Homologs in Arabidopsis, Yeast, Protozoan, and Human.
Supplementary Material
Acknowledgments
We thank Jean Greenberg (University of Chicago) for seeds of acd5, Hua Lu (University of Maryland, Baltimore County) for advice on SA measurement, Robert Dickson (University of Kentucky) for advice on earlier yeast work in the Xiao laboratory, Jane Parker and Dieter Becker (Max Planck Institute for Plant Breeding Research) for the pJawohl8 vector, James Culver (University of Maryland, College Park) for the 35S:ERD2-GFP plasmid, Masa Sato (Kyoto Prefectural University) for the 35S:GFP-SYP42 and 35S:GFP-SYP61 constructs, Undral Orgil for technical support, and Richelle Green and Lori Urban for assistance with mutant screens and maintaining plant growth facilities. This work was supported by the start-up fund from the University of Maryland Biotechnology Institute, by the National Research Initiative of the U.S. Department of Agriculture (Grant 2006-35301-16883 to S.X.), by National Science Foundation 2010 Program funds (Grant MCB-0313466 to T.M.D. and Grant MCB-0312864 to D.V.L.), and by the Howard Hughes Medical Institute program for support of undergraduates (to D.V.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shunyuan Xiao (xiao@umbi.umd.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121. [DOI] [PubMed] [Google Scholar]
- Bae, H., Kim, M.S., Sicher, R.C., Bae, H.J., and Bailey, B.A. (2006). Necrosis- and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiol. 141 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balague, C., Lin, B., Alcon, C., Flottes, G., Malmstrom, S., Kohler, C., Neuhaus, G., Pelletier, G., Gaymard, F., and Roby, D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley, P.E., Li, Y.O., Murphy, S.M., Sumner, C.M., and Lynch, D.V. (2003). Complex sphingolipid synthesis in plants: characterization of inositolphosphorylceramide synthase activity in bean microsomes. Arch. Biochem. Biophys. 417 219–226. [DOI] [PubMed] [Google Scholar]
- Carter, H.E., Celmer, W.D., Galanos, D.S., Gigg, R.H., Lands, W.E.M., Law, J.H., Mueller, K.L., Nakayama, T., Tomizawa, H.H., and Weber, E. (1958). Biochemistry of the sphingolipides. 10. Phytoglycolipide, a complex phytosphingosine-containing lipide from plant seeds. J. Am. Oil Chem. Soc. 35 335–343. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I.C., Lippok, B., Smith, R.K., Jr., and Bent, A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- daSilva, L.L., Snapp, E.L., Denecke, J., Lippincott-Schwartz, J., Hawes, C., and Brandizzi, F. (2004). Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, P.W., Shams-Eldin, H., Price, H.P., Smith, D.F., and Schwarz, R.T. (2006). The protozoan inositol phosphorylceramide synthase: A novel drug target that defines a new class of sphingolipid synthase. J. Biol. Chem. 281 28200–28209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, P.W., and Smith, D.F. (2004). Rafts and sphingolipid biosynthesis in the kinetoplastid parasitic protozoa. Mol. Microbiol. 53 725–733. [DOI] [PubMed] [Google Scholar]
- Desai, K., Sullards, M.C., Allegood, J., Wang, E., Schmelz, E.M., Hartl, M., Humpf, H.U., Liotta, D.C., Peng, Q., and Merrill, A.H., Jr. (2002). Fumonisins and fumonisin analogs as inhibitors of ceramide synthase and inducers of apoptosis. Biochim. Biophys. Acta 1585 188–192. [DOI] [PubMed] [Google Scholar]
- Ding, T., Li, Z., Hailemariam, T., Mukherjee, S., Maxfield, F.R., Wu, M., and Jiang, X.C. (2008). SMS overexpression and knockdown: Impact on cellular sphingomyelin and diacylglycerol metabolism, and cell apoptosis. J. Lipid Res. 49 376–385 [DOI] [PubMed] [Google Scholar]
- Does, M.P., Dekker, B.M., de Groot, M.J., and Offringa, R. (1991). A quick method to estimate the T-DNA copy number in transgenic plants at an early stage after transformation, using inverse PCR. Plant Mol. Biol. 17 151–153. [DOI] [PubMed] [Google Scholar]
- Dunn, T.M., Lynch, D.V., Michaelson, L.V., and Napier, J.A. (2004). A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. (Lond.) 93 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Mack, A.A., Morris, V.R., and Dangl, J.L. (2003). Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. USA 100 6831–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, J.M., Dias, W.B., Mendonca-Previato, L., Previato, J.O., and Heise, N. (2005). Characterization of the inositol phosphorylceramide synthase activity from Trypanosoma cruzi. Biochem. J. 387 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A., Tang, D., and Innes, R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, S., Tanaka, N., Kaneda, T., Takayama, S., Isogai, A., and Che, F.S. (2004). Rice cDNA microarray-based gene expression profiling of the response to flagellin perception in cultured rice cells. Mol. Plant Microbe Interact. 17 986–998. [DOI] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269 843–846. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Silverman, F.P., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak, D., Gable, K., Beeler, T., and Dunn, T. (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272 29704–29710. [DOI] [PubMed] [Google Scholar]
- Hakomori, S. (1983). Chemistry of glycosphingolipids. In Handbook of Lipid Research, Vol. 3, Sphingolipid Biochemistry J.N. Kanfer and S. Hakomori, eds (New York: Plenum Press), pp. 1–164.
- Hammond-Kosack, K.E., and Parker, J.E. (2003). Deciphering plant-pathogen communication: Fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14 177–193. [DOI] [PubMed] [Google Scholar]
- Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., and Nishijima, M. (2003). Molecular machinery for non-vesicular trafficking of ceramide. Nature 426 803–809. [DOI] [PubMed] [Google Scholar]
- He, X., Anderson, J.C., del Pozo, O., Gu, Y.Q., Tang, X., and Martin, G.B. (2004). Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J. 38 563–577. [DOI] [PubMed] [Google Scholar]
- Huitema, K., van den Dikkenberg, J., Brouwers, J.F., and Holthuis, J.C. (2004). Identification of a family of animal sphingomyelin synthases. EMBO J. 23 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1 285–297. [DOI] [PubMed] [Google Scholar]
- Jurkowski, G.I., Smith, R.K., Jr., Yu, I.C., Ham, J.H., Sharma, S.B., Klessig, D.F., Fengler, K.A., and Bent, A.F. (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17 511–520. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., Venugopal, S.C., Lapchyk, L., Falcone, D., Hildebrand, D., and Kachroo, P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 5152–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, K., and Lester, R.L. (1975). Characterization of inositol-containing phosphosphingolipids from tobacco-leaves: Isolation and identification of two novel, major lipids: N-Acetylglucosamidoglucuronidoinositol phosphorylceramide and glucosamidoglucuronidoinositol phosphorylceramide. Plant Physiol. 55 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Dietrich, R.A., Martin, A.C., Last, R.L., and Dangl, J.L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12 1022–1026. [DOI] [PubMed] [Google Scholar]
- Lei, X., Zhang, S., Bohrer, A., Bao, S., Song, H., and Ramanadham, S. (2007). The group VIA calcium-independent phospholipase A2 participates in ER stress-induced INS-1 insulinoma cell apoptosis by promoting ceramide generation via hydrolysis of sphingomyelins by neutral sphingomyelinase. Biochemistry 46 10170–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T.P., Wiggins, C.A., and Munro, S. (2000). Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 11 2267–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H., Yao, N., Song, J.T., Luo, S., Lu, H., and Greenberg, J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Czymmek, K., Tallóczy, Z., Levine, B., and Dinesh-Kumar, S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121 567–577. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., Lin, B., Auriac, M.C., Kroj, T., Saindrenan, P., Nicole, M., Balague, C., and Roby, D. (2004). Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S., Vailleau, F., Balague, C., and Roby, D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8 263–271. [DOI] [PubMed] [Google Scholar]
- Lynch, D.V., and Dunn, T.M. (2004). An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 161 677–702. [DOI] [PubMed] [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marggraf, W.D., and Kanfer, J.N. (1984). The phosphorylcholine acceptor in the phosphatidylcholine:ceramide cholinephosphotransferase reaction. Is the enzyme a transferase or a hydrolase? Biochim. Biophys. Acta 793 346–353. [DOI] [PubMed] [Google Scholar]
- Markham, J.E., and Jaworski, J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21 1304–1314. [DOI] [PubMed] [Google Scholar]
- Markham, J.E., Li, J., Cahoon, E.B., and Jaworski, J.G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281 22684–22694. [DOI] [PubMed] [Google Scholar]
- Meng, A., Luberto, C., Meier, P., Bai, A., Yang, X., Hannun, Y.A., and Zhou, D. (2004). Sphingomyelin synthase as a potential target for D609-induced apoptosis in U937 human monocytic leukemia cells. Exp. Cell Res. 292 385–392. [DOI] [PubMed] [Google Scholar]
- Nagiec, M.M., Nagiec, E.E., Baltisberger, J.A., Wells, G.B., Lester, R.L., and Dickson, R.C. (1997). Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272 9809–9817. [DOI] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51 1126–1136. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F. (1997). An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci. 6 1764–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- Orgil, U., Araki, H., Tangchaiburana, S., Berkey, R., and Xiao, S. (2007). Intraspecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics 176 2317–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S., Gable, K., Beaudoin, F., Cahoon, E., Jaworski, J., Napier, J.A., and Dunn, T.M. (2006). Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J. Biol. Chem. 281 9018–9029. [DOI] [PubMed] [Google Scholar]
- Pettus, B.J., Chalfant, C.E., and Hannun, Y.A. (2002). Ceramide in apoptosis: An overview and current perspectives. Biochim. Biophys. Acta 1585 114–125. [DOI] [PubMed] [Google Scholar]
- Phan, V.H., Herr, D.R., Panton, D., Fyrst, H., Saba, J.D., and Harris, G.L. (2007). Disruption of sphingolipid metabolism elicits apoptosis-associated reproductive defects in Drosophila. Dev. Biol. 309 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska, A., Tanner, G., Anders, I., Roca, M., and Hortensteiner, S. (2003). Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the ACCELERATED CELL DEATH 1 gene. Proc. Natl. Acad. Sci. USA 100 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, S., Liu, G., Zhou, B., Bellizzi, M., Zeng, L., Dai, L., Han, B., and Wang, G.L. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, A.M., Malagarie-Cazenave, S., Olea, N., Vara, D., Chiloeches, A., and Diaz-Laviada, I. (2007). Apoptosis induced by capsaicin in prostate PC-3 cells involves ceramide accumulation, neutral sphingomyelinase, and JNK activation. Apoptosis 12 2013–2024. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Kovaleva, V., Bassham, D.C., and Raikhel, N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, M., Dittgen, J., Sanchez-Rodriguez, C., Hou, B.H., Molina, A., Schulze-Lefert, P., Lipka, V., and Somerville, S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafesse, F.G., Huitema, K., Hermansson, M., van der Poel, S., van den Dikkenberg, J., Uphoff, A., Somerharju, P., and Holthuis, J.C. (2007). Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282 17537–17547. [DOI] [PubMed] [Google Scholar]
- Tang, D., Ade, J., Frye, C.A., and Innes, R.W. (2005). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, A.G., et al (2004). Characterization of soybean genomic features by analysis of its expressed sequence tags. Theor. Appl. Genet. 108 903–913. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Ueda, T., Ohniwa, R.L., Nakano, A., Takeyasu, K., and Sato, M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29 49–65. [DOI] [PubMed] [Google Scholar]
- Ullman, M.D., and Radin, N.S. (1974). The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J. Biol. Chem. 249 1506–1512. [PubMed] [Google Scholar]
- Vanacker, H., Lu, H., Rate, D.N., and Greenberg, J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28 209–216. [DOI] [PubMed] [Google Scholar]
- Voelker, D.R., and Kennedy, E.P. (1982). Cellular and enzymic synthesis of sphingomyelin. Biochemistry 21 2753–2759. [DOI] [PubMed] [Google Scholar]
- Vorwerk, S., Schiff, C., Santamaria, M., Koh, S., Nishimura, M., Vogel, J., Somerville, C., and Somerville, S. (2007). EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol. 7 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner, D.W., Xu, J., Singh, I., Jasinska, R., Zhang, Q.X., and Brindley, D.N. (1999). Structural organization of mammalian lipid phosphate phosphatases: Implications for signal transduction. Biochim. Biophys. Acta 1439 299–316. [DOI] [PubMed] [Google Scholar]
- Wang, H., Li, J., Bostock, R.M., and Gilchrist, D.G. (1996. a). Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Devoto, A., Turner, J.G., and Xiao, S. (2007). Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol. Plant Microbe Interact. 20 966–976. [DOI] [PubMed] [Google Scholar]
- Wang, W., Jones, C., Ciacci-Zanella, J., Holt, T., Gilchrist, D.G., and Dickman, M.B. (1996. b). Fumonisins and Alternaria alternata lycopersici toxins: Sphinganine analog mycotoxins induce apoptosis in monkey kidney cells. Proc. Natl. Acad. Sci. USA 93 3461–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., Brown, S., Patrick, E., Brearley, C., and Turner, J.G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., Calis, O., Patrick, E., Zhang, G., Charoenwattana, P., Muskett, P., Parker, J.E., and Turner, J.G. (2005). The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42 95–110. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M., and Turner, J.G. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291 118–120. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Ellwood, S., Findlay, K., Oliver, R.P., and Turner, J.G. (1997). Characterization of three loci controlling resistance of Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J. 12 757–768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.