Abstract
The SNARE complex is a key regulator of vesicular traffic, executing membrane fusion between transport vesicles or organelles and target membranes. A functional SNARE complex consists of four coiled-coil helical bundles, three of which are supplied by Q-SNAREs and another from an R-SNARE. Arabidopsis thaliana VAMP727 is an R-SNARE, with homologs only in seed plants. We have found that VAMP727 colocalizes with SYP22/ VAM3, a Q-SNARE, on a subpopulation of prevacuolar compartments/endosomes closely associated with the vacuolar membrane. Genetic and biochemical analyses, including examination of a synergistic interaction of vamp727 and syp22 mutations, histological examination of protein localization, and coimmunoprecipitation from Arabidopsis lysates indicate that VAMP727 forms a complex with SYP22, VTI11, and SYP51 and that this complex plays a crucial role in vacuolar transport, seed maturation, and vacuole biogenesis. We suggest that the VAMP727 complex mediates the membrane fusion between the prevacuolar compartment and the vacuole and that this process has evolved as an essential step for seed development.
INTRODUCTION
In eukaryotic cells, correct transport of newly synthesized proteins and recycling of preexisting proteins are essential for fundamental cell activities such as maintenance of organelle functions, response to the environment, cellular homeostasis, and intercellular communication. The transport of cargo between donor and acceptor organelles is generally mediated by transport vesicles, which bud from one compartment and discharge cargo at the destination compartment. Specific membrane fusion between transport vesicles and target membranes ensures the accuracy of transport and is mediated by the SNARE (soluble N-ethyl-maleimide sensitive factor attachment protein receptors) complex that assembles into a tight cluster of four coiled-coil helices (Chen and Scheller, 2001; Jahn et al., 2003; Jürgens, 2004). The helical region in the SNARE molecule is called the SNARE motif and is also used as an earmark to classify the SNAREs into four groups, Qa-, Qb-, Qc-, and R-SNAREs. Each of these groups provides one of the four components of a SNARE complex, and only correct combinations of cognate SNAREs generate the functional SNARE complexes that drive specific membrane fusions (Bock et al., 2001; Antonin et al., 2002; Kloepper et al., 2007).
In Arabidopsis thaliana, at least 64 SNARE molecules have been identified (Sanderfoot et al., 2000; Uemura et al., 2004; Sanderfoot, 2007). This number is larger than those of yeast and human, indicating that the endomembrane system in plant cells has a complex organization. The SNAREs in Arabidopsis can also be classified into the Qa-, Qb-, Qc-, and R-groups, each of which is further divided into several subgroups according to sequence similarity (Sanderfoot, 2007). Interestingly, some of these subgroups, such as Syntaxin of plant 1 (SYP1) Qa-SNAREs, have expanded greatly in specific lineages in green plants, suggesting a specific evolution of the plant membrane trafficking system. For example, KNOLLE/SYP111, which is expressed in dividing cells of various developing tissues, mediates vesicle fusion at the cell plate during cytokinesis in Arabidopsis (Lukowitz et al., 1996). Also in Arabidopsis, impairment of plasma membrane–localized SYR1/SYP121/PEN1 results in an increased incidence of pathogen penetration (Collins et al., 2003), and the double mutant of SYP121 and its paralog SYP122 shows dwarfism and necrotic cell death (Assaad et al., 2004). These results suggest that the SYP1 Qa-SNARE subfamily has diverged to fulfill various secretion-related functions in plants.
Plants also seem to have assigned specialized functions to the Q-SNAREs in endocytic and vacuolar systems. Two Arabidopsis Q-SNARE genes, SYP22/VAM3/SGR3 and VTI11, were previously identified as responsible for the sgr3 (for shoot gravitropism 3) and zig (for zig-zag)/sgr4 mutant phenotypes, respectively, which are characterized by defective shoot gravitropic responses (Kato et al., 2002; Yano et al., 2003). Biochemical analysis further demonstrated that SYP22 (Qa) and VTI11 (Qb) constitute a complex that also contains SYP51 (Qc) (Sanderfoot et al., 2001a; Yano et al., 2003). Based on what has been uncovered about the four groups that make up the SNARE complex, this complex should contain another SNARE molecule, probably R-SNARE. Furthermore, the precise function of this complex in membrane trafficking has remained elusive.
While knowledge about the important functions of Q-SNAREs is accumulating, quite limited information is available for R-SNAREs in plants, although unique organization and divergence in sequence suggest that R-SNARE should also have acquired plant-specific functions (Sanderfoot, 2007).
R-SNAREs are classified into two groups, longins and brevins; longins harbor an N-terminal profilin-like fold called the longin domain consisting of 120 to 140 amino acid residues, while brevins lack this domain (Filippini et al., 2001; Gonzalez et al., 2001; Rossi et al., 2004b). One prominent feature in the organization of plant R-SNAREs is that there are no brevins in land plants, including angiosperms, lycophytes, and bryophytes, but that brevins are known to function in multiple steps of membrane traffic in animal cells (Rossi et al., 2004a; Uemura et al., 2005). Instead, plant longins have diverged significantly, especially in land plants. In Arabidopsis, 14 longin-type R-SNAREs (one Sec22-like, two Ykt6-like, and 11 VAMP7-like longins) are encoded in the genome. Among these longins, VAMP727 has a characteristic primary structure; it harbors an insertion of 20 amino acids in the longin domain. This type of VAMP7 member is found only in seed plants (Sanderfoot, 2007), suggesting that VAMP727 functions in physiological events specific to seed plants.
Through the systematic analysis of the subcellular localization of Arabidopsis SNARE molecules using a transient expression method in protoplasts, we have previously identified the seed plant–specific VAMP727 as a unique R-SNARE predominantly localized on endosomes (Uemura et al., 2004). We have also demonstrated that Rab5 homologs of Arabidopsis, ARA7 and RHA1, colocalize with VAMP727 (Ueda et al., 2004), consistently with their endosome localization. This endosomal population, in which these proteins are located, most probably corresponds to the prevacuolar compartment (PVC) in the biosynthetic vacuolar transport pathway because ARA7 and RHA1 have been shown to reside on the multivesicular PVC (Haas et al., 2007). The subcellular localization of VAMP727 implies its role in membrane fusion in the endocytic and/or vacuolar transport pathways, but the precise molecular function of VAMP727 remains undetermined.
In this study, we combined genetics, bioimaging, and biochemistry approaches to explore the molecular and physiological functions of this seed plant–unique R-SNARE as well as its binding partners. With this multidisciplinary approach, we have demonstrated that VAMP727 is the fourth component of the SNARE complex containing SYP22, VTI11, and SYP51, which seems to mediate the fusion between the PVC and the vacuole. We have also demonstrated that this complex is essential for seed maturation. We propose a role of VAMP727 in the evolution of seeds, specifically related to adaptation to life on land.
RESULTS
vamp727 Shows Synthetic Lethality with syp22
We first identified a mutant of VAMP727 with a T-DNA insertion in the fourth intron (GABI_060G05), in which we could not detect expression of VAMP727 by RT-PCR (Figure 1A). In spite of the expected importance of VAMP727 for cellular functions, the vamp727 mutant showed no visibly abnormal phenotype (Figures 1B and 1C). To understand the function of VAMP727, we then obtained double mutants for the vamp727 allele and a variety of mutant alleles for endocytic Rabs and SNAREs. Most of them, including ara7, rha1, ara6, and syp21, still showed no detectable genetic interactions. However, a mutant of a Qa-SNARE, syp22-1, which causes multiple abnormal phenotypes, including serrated and wavy leaves, semidwarfism, and late flowering (Figure 1B), turned out to exhibit a very strong genetic interaction with vamp727.
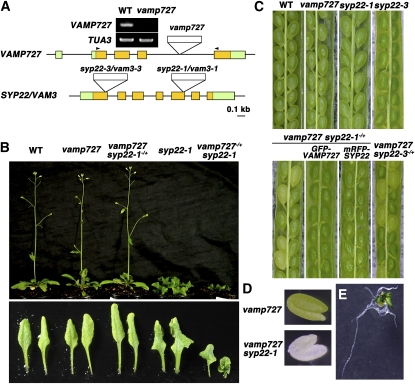
Figure 1.
Synthetic Lethal Interaction between vamp727 and syp22.
(A) Schematic structures of VAMP727 and SYP22 genes and positions of T-DNA insertions. Expression of VAMP727 was not detected in the vamp727 mutant by RT-PCR. Arrowheads indicate the positions of PCR primers.
(B) Phenotypes of vamp727, syp22-1, and vamp727syp22-1 mutants: 30-d-old wild-type and mutant plants (top panel) and the seventh and eighth leaves from individual plants shown in the top panel (bottom panel) are presented.
(C) Embryonic lethal phenotype of vamp727 syp22 double mutants. Seedpods collected from Arabidopsis plants with the indicated genotypes are shown. The double mutant seeds exhibit yellowish appearance. Embryonic lethality of vamp727 syp22-1 was rescued by GFP-tagged VAMP727 and mRFP-tagged SYP22, indicating that these chimeric proteins are functional.
(D) Sibling embryos from the vamp727−/− syp22-1−/+ plant. The white embryo was verified to be a double homozygous mutant by genotyping.
(E) vamp727 syp22-1 mutant embryo excised before seed dehydration and cultured on MS medium for 45 d, showing that vamp727 syp22-1 plants can produce leaf and root structures.
We could not find any double homozygous mutants (vamp727−/−syp22-1−/−) in progenies derived from the self-pollinated plants with homozygous vamp727 and heterozygous syp22-1 mutations (vamp727−/−syp22-1−/+), even though the parents were indistinguishable from wild-type plants (Figure 1B). The segregation ratio obtained by the PCR-based genotyping (vamp727−/−syp22-1+/+:vamp727−/−syp22-1−/+:vamp727−/−syp22-1−/− = 113: 195: 0) suggested that the double mutant was embryonic lethal but not gametophytic lethal; thus, we examined the premature seeds in the seedpods of vamp727−/−syp22-1−/+ plants. As shown in Figure 1C, approximately one-fourth of the seed population exhibited a yellowish appearance (green:yellow = 545:176), which contained white embryos with an apparently normal shape (Figure 1D). Genotyping revealed that all of these white embryos harbored double homozygous mutations, and a genomic fragment containing the VAMP727 gene rescued this embryonic lethal phenotype (Figure 3). The synthetic lethality with vamp727 was also confirmed for another syp22 allele, syp22-3 (Figures 1A and 1C). We also generated mutant plants with heterozygous vamp727 and homozygous syp22-1 mutations (vamp727−/+syp22-1−/−). In this mutant, phenotypes of syp22, such as dwarfism and serration of leaves, were severely exaggerated (Figure 1B), suggesting that vamp727 and syp22 mutations affected one another synergistically. These genetic interactions between vamp727 and syp22 mutations strongly suggest that VAMP727 and SYP22 function coordinately in the same membrane traffic pathway.
Figure 3.
Duplication of VAMP727 Suppresses the syp22-1 Mutation.
(A) Plants transformed with a construct expressing VAMP727 from its native promoter show suppression of the syp22-1 mutant phenotype. Thirty-day-old Arabidopsis plants with the indicated genotypes (top panel) and the seventh and eighth leaves sampled from individuals in the top panel (bottom panel). Two independent lines of mutants transformed with exogenous VAMP727 are shown.
(B) Duplication of other members of the VAMP7 family does not suppress the syp22-1 mutation. Thirty-three-day-old Arabidopsis plants with the indicated genotypes (top panel) and the seventh and eighth leaves sampled from individuals in the top panel (bottom panel). Two independent lines of mutants transformed with VAMP727, VAMP721, or VAMP713 are shown.
We then examined the embryogenesis of the double mutants in more detail, observing embryos in cleared seedpods from the vamp727−/−syp22-1−/+ mutant plants. As shown in Figure 2, the growth of double mutants was delayed through embryogenesis. However, the morphology of developing embryos did not appear to be aberrant, and they eventually grew into apparently normal mature embryos except for a chlorotic phenotype (Figure 2F). When embryos at this stage were excised and cultured on Murashige and Skoog (MS) medium, the double mutant embryos developed roots with a normal appearance and leaf-like structures (Figure 1E). However, once dehydrated, double mutant seeds shrank and never germinated (Figure 2G). These results indicated that SYP22 and VAMP727 are essential in the final stage of seed maturation or in the rehydration/germination process but not in earlier stages of embryogenesis. The vamp727 syp22-1 double mutant plants cultured on MS medium slowly but continuously generated leaf-like structures and roots for several months; however, these plants did not bolt or flower. Thus, cooperative functions of VAMP727 and SYP22 are not only essential during the late stage of seed maturation but also are important for postembryonic growth.
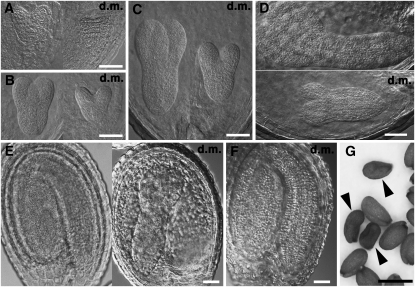
Figure 2.
Embryogenesis of the vamp727 syp22-1 Double Mutant Is Delayed but Eventually Almost Completed.
(A) to (E) The vamp727 syp22-1 double mutant embryos and their wild-type-appearing siblings in the same seedpods collected from the vamp727 syp22-1−/+ plant. d.m., double mutant.
(F) The double mutant embryos seem to almost complete embryogenesis, but it takes another couple of days after wild-type-appearing siblings do so.
(G) The vamp727 syp22-1 double mutant seeds shrink after desiccation. Arrowheads indicate double mutant seeds.
Bars = 50 μm in (A) to (F) and 500 μm in (G).
VAMP727 Is a Multicopy Suppressor of syp22
In the yeast system, overexpression of R-SNAREs frequently suppresses mutations in cognate Q-SNAREs (Sacher et al., 1997; Jantti et al., 2002; Graf et al., 2005). To test whether overexpression of VAMP727 affects the syp22 mutation, we transformed syp22-1 plants with a genomic fragment containing the complete VAMP727 gene. Surprisingly, all macroscopic phenotypes of syp22-1, including wavy serrated leaves and semidwarfism, were suppressed in the transformants (Figure 3A). This suppression activity was specific to VAMP727; duplication of the other VAMP7 members, VAMP713 and VAMP721, which reside on the vacuolar membrane and the plasma membrane, respectively, did not suppress the syp22-1 mutation (Figure 3B). We also generated vamp727 syp22-1 double mutant plants that were complemented by the exogenous VAMP727 gene by transforming vamp727−/− syp22-1−/+ with the genomic sequence of VAMP727. As shown in Figure 3, vamp727 syp22-1 with VAMP727 resulted in phenotypes indistinguishable from syp22-1. Thus, a single exogenous VAMP727 gene could not suppress syp22-1 without endogenous VAMP727. These results demonstrate that duplication of the VAMP727 gene was sufficient to suppress the syp22-1 mutation.
There are several possible mechanisms of syp22-1 suppression by exogenous VAMP727. Three SNARE molecules (two Q-SNAREs and one R-SNARE) can execute membrane fusion in vitro (Chen et al., 2005), and SYP22 might be dispensable in vivo when VAMP727 exists abundantly. In addition, two Q-SNAREs and two R-SNAREs can form a complex in vitro that executes membrane fusion when membrane lipid composition is modified (Fratti et al., 2007). If this is also the case in Arabidopsis, VAMP727 might be able to replace SYP22 when excess VAMP727 is available. However, it is most likely that VAMP727 overexpression increases the incorporating efficiency for other Qa-SNAREs that have functions redundant with SYP22. The SYP21 protein could be a candidate for such a Qa-SNARE because it is also involved in traffic from the PVC (Foresti et al., 2006).
The intimate genetic interaction between VAMP727 and SYP22 strongly suggests that these SNARE molecules function quite proximally in membrane trafficking.
VAMP727 and SYP22 Are Expressed throughout Arabidopsis Plants
The striking and strong genetic interactions described above, the synthetic lethal interaction between vamp727 and syp22 during seed maturation and suppression of syp22-1 by duplicated VAMP727, encouraged us to examine the possibility that VAMP727 and SYP22 function in the same SNARE complex. If they do, these genes must be expressed in the same cells in Arabidopsis. RT-PCR analysis indicated that these genes were ubiquitously expressed in all organs we examined (Uemura et al., 2004), but whether they were expressed in the same tissues was still unclear. To complete a detailed analysis of their expression patterns, we constructed transgenic Arabidopsis expressing translational fusion of VAMP727 or SYP22 with β-glucuronidase (GUS) under control of their own promoters. GUS staining indicated that both genes were expressed abundantly and ubiquitously in all tissues we examined, including embryos at late developmental stages, which are affected most severely in the vamp727 syp22 double mutant (Figure 4). This expression pattern was also confirmed in transgenic Arabidopsis plants expressing fluorescent protein–tagged VAMP727 and SYP22 under regulation of their own promoters, as described below. Thus, VAMP727 and SYP22 are, in fact, expressed in the same tissues and most likely in the same cells.
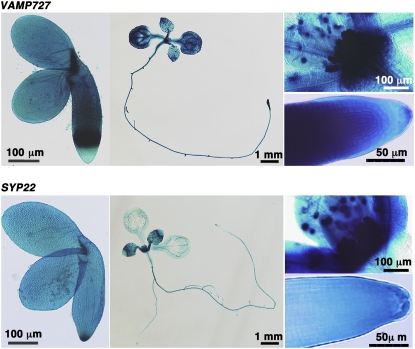
Figure 4.
Promoter-Reporter Assay of VAMP727 and SYP22.
cDNA for GUS was connected in frame to the coding region of VAMP727 or SYP22 and introduced into Arabidopsis. Both proteins were expressed throughout Arabidopsis plants, including maturing embryos (left), 7-d-old seedlings (center), and apices of shoots (right, top) and roots (right, bottom) of 7-d-old seedlings.
VAMP727 and SYP22 Colocalize on a Subpopulation of PVCs Closely Associated with the Vacuolar Membrane
Among the 14 R-SNAREs encoded in the Arabidopsis genome, VAMP727 was the only one localized almost exclusively on endosomes in protoplasts (Uemura et al., 2004). To verify this localization in intact plant cells, we generated transgenic Arabidopsis expressing green fluorescent protein (GFP)–tagged VAMP727. To avoid possible mislocalization due to overexpression or ectopic expression, we constructed a translational fusion between the genomic VAMP727 sequence and GFP and introduced it into the vamp727 mutant. This chimeric gene was functional, rescuing the embryonic lethality of the vamp727 syp22-1 mutant (Figure 1C). As shown in Figure 5A, GFP-VAMP727 localized predominantly on mobile punctate organelles in the cytoplasm in root tip cells, and vacuolar membrane localization was also observed in some cells with a high expression level (see Supplemental Figure 1C online). Consistent with the previous result in protoplasts (Ueda et al., 2004; Uemura et al., 2004), these organelles were labeled by a tracer of endocytosis, FM4-64, 15 to 20 min after uptake (Figure 5A), indicating that VAMP727 was indeed on endocytic organelles in Arabidopsis plants. We then examined whether VAMP727 colocalizes with other known endosomal proteins in plants. To avoid overexpression, endosomal proteins were tagged with fluorescent proteins and expressed under regulation of the authentic regulatory elements (5′- and 3′-flanking sequences and introns). The VAMP727 partially colocalized with the multivesicular endosomal proteins RHA1 and SYP21, which are a conventional RAB5 homolog and a Qa-SNARE, respectively (Figures 5B and 5C). By contrast, VAMP727 rarely localized on the SYP43-positive trans-Golgi network (Figure 5D), which was recently also proposed to function as an early endosome in plant cells (Dettmer et al., 2006; Chow et al., 2008). Thus, VAMP727 is an R-SNARE mainly residing on the multivesicular endosomes/PVCs.
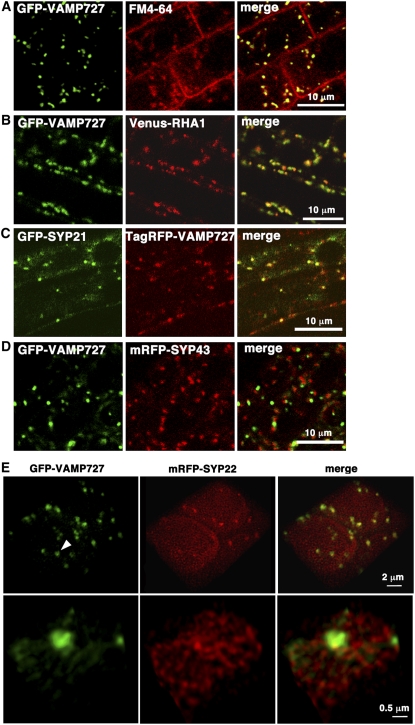
Figure 5.
Subcellular Localization of VAMP727 and SYP22.
(A) GFP-VAMP727 localized on punctate organelles in root cells, which were also labeled by the endocytic tracer FM4-64 20 min after uptake.
(B) GFP-VAMP727 colocalizes with Venus-RHA1, a RAB5 homolog on the multivesicular endosomes/PVC.
(C) TagRFP-VAMP727 colocalizes with GFP-SYP21, a Qa-SNARE on the PVC.
(D) GFP-VAMP727 and a Qa-SNARE on the TGN, mRFP-SYP43, are on distinct organelles.
(E) Three-dimensional images showing colocalization of VAMP727 and SYP22 at subdomains between endosomes and vacuoles. GFP-VAMP727 and mRFP-SYP22 were coexpressed under regulation of their own regulatory elements in Arabidopsis plants. A series of confocal images were deconvolved and reconstructed to generate three-dimensional images. Bottom panels are higher-magnification images of an endosome/PVC pointed by the arrowhead, which seems to be tethered to the vacuolar membrane.
To examine the colocalization between VAMP727 and SYP22, we also constructed transgenic Arabidopsis expressing monomeric red fluorescent protein (mRFP)-SYP22. As with GFP-VAMP727, the open reading frame sequence for mRFP was inserted into the genomic fragment containing the SYP22 gene, which was then introduced into the syp22 mutant. mRFP-SYP22 was also functional because it rescued the embryonic lethality of the vamp727 syp22-1 double mutant (Figure 1C). We crossed vamp727 syp22-1 mutants expressing GFP-VAMP727 and mRFP-SYP22 and observed leaf primordia of 5-d-old seedlings of F2 plants using a high-performance confocal laser scanning microscope we recently developed (Nakano, 2002; Matsuura-Tokita et al., 2006). In combination with the deconvolution technique, it allows multicolor observation with extremely high spatial resolution. As shown in Figure 5E, GFP-VAMP727 was localized on punctate organelles in the cytoplasm. On the other hand, mRFP-SYP22 was localized on the vacuolar membrane and small punctate dots, many of which seemed to be associated with the vacuolar membrane. Consistent with the intimate genetic interactions between VAMP727 and SYP22, these dots colocalized with subdomains of GFP-VAMP727–positive endosomes (Figure 5E; see Supplemental Figure 1 online). Magnified observation further demonstrated that SYP22 and VAMP727 colocalized at an endosomal subdomain, where the endosome appeared to be associated with the vacuolar membrane (Figure 5E; see Supplemental Figure 1A and Supplemental Movie 1 online). We also constructed the transgenic Arabidopsis expressing SYP22 and VAMP727 tagged with a different combination of fluorescent proteins, in which SYP22 was also found to colocalize with VAMP727 on the PVC (see Supplemental Figure 1 online). These observations strongly suggest that SYP22 and VAMP727 are components of the same SNARE complex, which could mediate membrane fusion at the PVC.
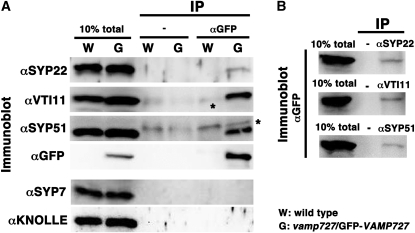
VAMP727 Is Coimmunoprecipitated with SYP22, SYP51, and VTI11
It has been proposed that SYP22, a Qa-SNARE, functions by forming a complex with Qb- VTI11 and Qc-SYP51 (Sanderfoot et al., 2001a; Yano et al., 2003). Our genetic and colocalization data strongly suggested that VAMP727, an R-SNARE, is the fourth component of this SNARE complex. To demonstrate it more clearly, we decided to examine direct interaction between VAMP727 and the above three Q-SNAREs in Arabidopsis cells by coimmunoprecipitation. For this experiment, we used the vamp727 mutant expressing GFP-VAMP727 under control of the authentic regulatory elements, including its promoter and introns. As described above, the chimeric gene coding for GFP-VAMP727 was functional because it complemented the vamp727 mutation (Figure 1C). The lysate prepared from a plant of this line was used for the immunoprecipitation with the anti-GFP monoclonal antibody, and the immunoprecipitates were subjected to immunoblotting using the antibodies indicated in Figure 6A and Supplemental Figure 2 online. SYP22, VTI11, and SYP51 were all coimmunoprecipitated by the anti-GFP antibody, clearly indicating that these four molecules are in the same complex. This interaction was specific because KNOLLE and SYP7, a Qa-SNARE functioning in cell plate formation and a Qc-SNARE on the plasma membrane (Lukowitz et al., 1996), respectively, were not coprecipitated in this experiment (Figure 6A). Conversely, anti-SYP22, anti-VTI11, and anti-SYP51 antibodies all coimmunoprecipitated GFP-VAMP727 (Figure 6B; see Supplemental Figure 2B online). Thus, VAMP727 is indeed the fourth component of the SNARE complex containing SYP22, VTI11, and SYP51.
Figure 6.
VAMP727 Is Coimmunoprecipitated with SYP22, VTI11, and SYP51.
(A) Plant extracts prepared from wild-type Arabidopsis (W) and transgenic Arabidopsis expressing GFP-tagged VAMP727 (G) were subjected to immunoprecipitation with the anti-GFP monoclonal antibody, which was followed by immunoblotting using indicated antibodies. Precipitates prepared without anti-GFP antibody (−) were loaded as negative controls. SYP22, VTI11, and SYP51 were coimmunoprecipitated with GFP-VAMP727, whereas SYP7 and KNOLLE were not. The asterisks indicate nonspecific bands derived from protein G used for binding immunocomplex.
(B) SYP22, VTI11, and SYP51 were immunoprecipitated from the lysate prepared from Arabidopsis expressing GFP-tagged VAMP727, and coprecipitation of GFP-VAMP727 was confirmed by immunoblotting with the anti-GFP antibody. Precipitates prepared without anti-GFP antibody (−) were loaded as negative controls.
For confirmation of the in vivo interaction between VAMP727 and SYP22, we also examined whether fluorescent resonance energy transfer (FRET) occurs between CFP-VAMP727 and Venus-SYP22 expressed in protoplasts prepared from cultured Arabidopsis cells. As shown in Supplemental Figure 2C online, we observed FRET between these molecules on the PVC associated with the vacuole. This result further demonstrated that VAMP727 and SYP22 form a complex on the PVC in Arabidopsis cells.
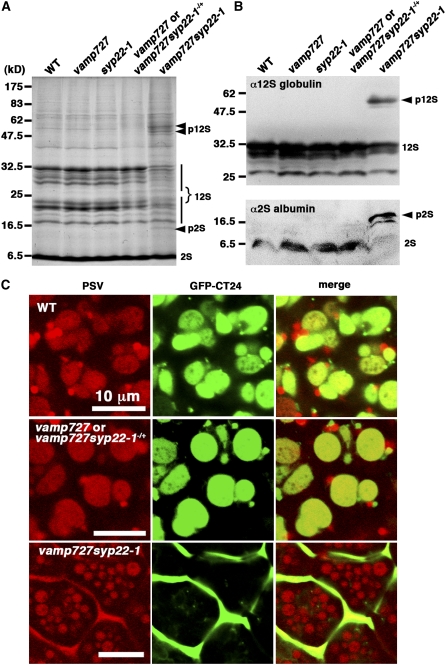
The VAMP727/SYP22 Complex Is Essential for Biogenesis of and Transport to Protein Storage Vacuoles
As described above, the SNARE complex composed of VAMP727, SYP22, VTI11, and SYP51 most likely functions in the membrane fusion between the PVC and the vacuole. If so, impaired function of this complex would cause defects in traffic to the vacuole. To verify this hypothesis, we examined processing of storage proteins in premature embryos. In vamp727 and syp22 mutants, we could not find any difference in the processing of 12S globulins and 2S albumins (Figures 7A and 7B). By contrast, in the embryos of the vamp727 syp22-1 double mutant, the precursors of storage proteins with higher molecular mass accumulated in the double mutant embryos, while normally processed 12S globulins and 2S albumins were also observed in the double mutant (Figure 7A). The immunoblot of 12S globulins and 2S albumins also demonstrated the partial defect in processing of storage proteins (Figure 7B). These results indicated that traffic of storage proteins to the vacuole is partially impaired by the double mutation of vamp727 syp22.
Figure 7.
VAMP727 and SYP22 Play Crucial Roles in Transport of Vacuolar Proteins to Protein Storage Vacuoles.
(A) and (B) vamp727 syp22-1 is defective in the processing of seed storage proteins. Total protein (50 μg/lane) prepared from maturing seeds of wild-type and mutant Arabidopsis were subjected to SDS-PAGE. The precursor protein of 12S globulin (p12S) and 2S albumin (p2S) with higher molecular mass accumulated in vamp727 syp22-1.
(A) Coomassie blue staining showing total protein, with positions for mature and unprocessed globulins indicated.
(B) Immunoblots with anti-12S globulin (top) and 2S albumin (bottom).
(C) GFP-CT24 is mis-secreted outside cells. The GFP-CT24 is secreted to intercellular spaces in the vamp727 syp22-1 double mutants, while it accumulates in protein storage vacuoles in wild-type and wild-type-appearing siblings (vamp727 or vamp727syp22-1−/+) of the double mutants.
We also examined the traffic of an artificial cargo, SP-GFP-CT24, which was successfully used to study the traffic to protein storage vacuoles (PSVs) in Arabidopsis (Nishizawa et al., 2003; Fuji et al., 2007). SP-GFP-CT24 consists of a signal peptide, GFP, and C-terminal 24 amino acids of α′ subunit of β-conglycinin that is sufficient for sorting to PSVs in wild-type Arabidopsis embryos (Nishizawa et al., 2003; Fuji et al., 2007). In wild-type-looking siblings (vamp727 or vamp727syp22-1−/+) excised from seedpods of vamp727 syp22-1−/+ plants, GFP fluorescence was predominantly observed in PSVs, recognized by their autofluorescence, as is the case in the wild type (Figure 7C). The GFP also labeled nonautofluorescent compartments, which probably represent carrier organelles to PSVs. By contrast, in the embryos of the vamp727 syp22-1 double mutant, GFP-CT24 was missecreted to extracellular spaces (Figure 7C). A similar phonotype has been reported for vsr1, which is mutated in the vacuolar sorting receptor required for transport of storage proteins to PSVs (Fuji et al., 2007). These data suggest that the vacuolar transport pathway is impaired in the vamp727 syp22-1 double mutant, while the secretory pathway does not seem to be affected.
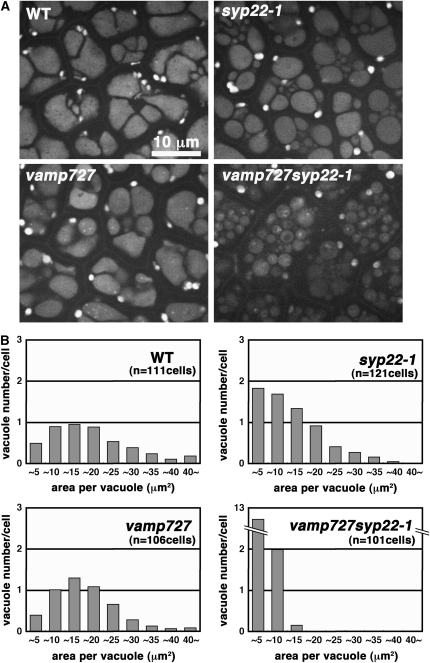
In addition to the mistargeting of storage proteins, significant alteration of PSV morphology was observed in the vamp727 syp22-1 double mutant (Figure 7C). We then examined effects of mutations on the morphology of PSVs in more detail. Mutation in VAMP727 caused a slight but significant reduction of the average section area per vacuole (mean ± sd: 16.6 ± 10.4 μm2 [n = 512 PSVs in 111 cells] in the wild type versus 15.3 ± 8.5 μm2 [n = 532 PSVs in 106 cells] in vamp727; P < 0.05, t test). In the syp22-1 mutant embryo, small PSVs were frequently observed in addition to the PSVs with normal size (Figures 8A and 8B), resulting in a smaller average size of vacuole (10.8 ± 8.2 μm2; n = 805 PSVs in 121 cells). The double mutant of vamp727 syp22-1 exhibited a much more severe defect; most of the PSVs were fragmented and average vacuole size was much reduced (2.9 ± 2.1 μm2; n = 1501 PSVs in 101 cells) compared with PSVs in wild-type or single mutants (Figures 8A and 8B). Thus, vamp727 and syp22-1 mutations affect PSV biogenesis in a synergistic manner, which is consistent with the results of genetic analysis (Figure 1).
Figure 8.
VAMP727 and SYP22 Play Important Roles in Biogenesis of Protein Storage Vacuoles.
(A) Morphology of protein storage vacuoles in cotyledons of wild-type and mutant embryos. Autofluorescence from protein storage vacuoles excited by an Ar+ laser at 488 nm was observed.
(B) Histograms representing a distribution of size and number of vacuoles within a single cell. Area of each vacuole was measured in cells of wild-type and mutant embryos. Note that only small vacuoles are observed in the vamp727 syp22-1 double mutant, while wild-type and single mutant cells contain large vacuoles.
These data indicate that the SNARE complex consisting of VAMP727, SYP22, VTI11, and SYP51 plays crucial roles in PSV biogenesis and transport of storage proteins through the regulation of membrane fusion in the vacuolar transport pathway.
Ultrastructural Analysis of the vamp727 syp22-1 Double Mutants
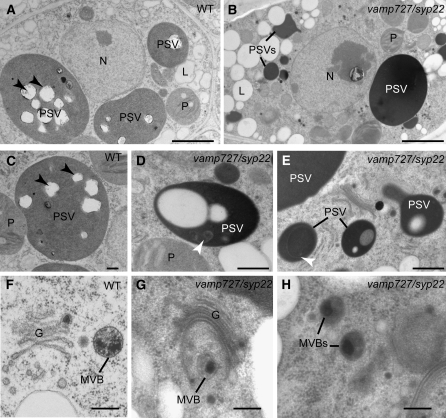
To elucidate the effects of the vamp727 syp22-1 double mutation at the ultrastructural level, we used transmission electron microscopy to observe developing seeds from seedpods collected from vamp727 syp22-1−/+ mutant plants. The double mutant embryos with a slightly chlorotic appearance and their wild-type-appearing siblings (vamp727 or vamp727 syp22-1−/+) at bent cotyledon stage were processed by high-pressure freezing and freeze substitution to preserve organelle morphology and antigenicity. As observed under a light microscope, small vacuole-like compartments ranging from 0.5 to 5 μm in diameter were frequently observed in the vamp727 syp22-1 double mutant embryos (Figures 9B, 9D, and 9E).
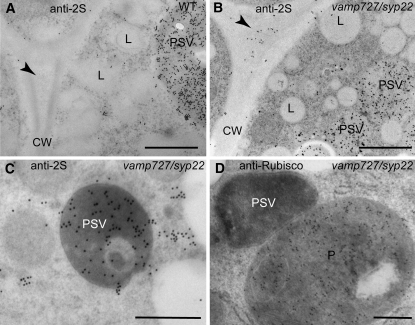
Figure 9.
Structural Analysis of High-Pressure Frozen/Freeze-Substituted Embryos.
vamp727 syp22-1 mutant and wild-type-looking embryos at the bent cotyledon stage were taken from the same siliques growing in vamp727 syp22-1−/+ plants. L, lipid body; N, nucleus; P, plastid. Bars = 2 μm in (A) and (B), 500 nm in (C) to (E), and 200 nm in (F) to (H).
(A) and (B) General overview of embryo cells in embryos with a wild-type appearance (A) and the vamp727 syp22 double mutant (B) cotyledons. The arrowheads indicate the position of the phytin globoid crystals, which often detach from the thin sections.
(C) to (E) PSVs in embryos with a wild type appearance (C) and double mutant ([D] and [E]) cotyledons. Black arrowheads indicate the position of the phytin globoids. Note the smaller size of vacuoles in the vamp727 syp22 double mutant, their electron-dense contents, and the presence of luminal membranes (white arrowheads).
(F) to (H) Golgi stacks (G) and multivesicular bodies (MVBs) in embryos with a wild-type appearance (F) and double mutant ([G] and [H]) cotyledons. The multivesicular bodies in the double mutant are indistinguishable from the wild-type multivesicular bodies in terms of general architecture and size.
Some of these organelles showed electron-dense luminal contents and internal membranes (Figures 9D and 9E). To determine the nature of these compartments, we examined the localization of a vacuolar storage protein 2S albumin and ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) in the vamp727 syp22-1 double mutant embryos and their wild-type-appearing siblings by immunoelectron microscopy. The anti-2S albumin antibodies labeled the lumen of the electron-dense compartments (Figures 10B and 10C), but the anti-Rubisco antibodies did not (Figure 10D), confirming the vacuolar identity of these organelles. In agreement with accumulation of the unprocessed precursor in the double mutant, the 2S albumins were also detected in intercellular spaces in the vamp727 syp22-1 double mutant (Figure 10B). This result further supports our previous observation that the transport of storage proteins is partially impaired in the double mutant, whereas the secretory pathway does not seem to be markedly affected. Intriguingly, we did not find structural abnormalities in the architecture of Golgi and multivesicular endosomes/PVCc (Figures 9E to 9H). This result suggests that the function of the SNARE complex containing SYP22 and VAMP727 is required after biogenesis of the PVC, which could be consistent with their potential role in membrane fusion between the vacuolar membrane and the PVC.
Figure 10.
Immunolocalization of 2S Albumins and Rubisco in Embryos with a Wild-Type Appearance and Double Mutant Embryos.
(A) to (C) Embryos with a wild-type appearance (A) and vamp727 syp22 double mutant cotyledons ([B] and [C]) were labeled with antibodies against the 2S albumins. The antibodies labeled the lumen of the PSVs in both embryos with a wild-type appearance and double mutant embryos, but it also labeled the extracellular space in the double mutant (arrowheads).
(D) Rubisco antibodies heavily labeled the plastids in the double mutant.
CW, cell wall; L, lipid body. Bars = 1 μm in (A) and (B) and 500 nm in (C) and (D).
In spite of the chlorotic appearance, plastid structure and Rubisco distribution seemed to be quite normal in double mutant embryos (Figures 9B and 10D). Why the vamp727 syp22-1 embryo exhibits chlorosis would be an interesting question to examine in a future study.
DISCUSSION
VAMP727 Is Likely an R-SNARE Mediating Membrane Fusion between the PVC and the Vacuole Together with Q-SNAREs SYP22, SYP51, and VTI11
Recent molecular genetic studies have demonstrated that SNARE molecules play important roles in various plant functions including cytokinesis, gravitropism, pathogenic responses, nodulation, hormonal responses, abiotic stress responses, cell differentiation, gametogenesis, and development (Lukowitz et al., 1996; Leyman et al., 1999; Sanderfoot et al., 2001b; Kato et al., 2002; Zhu et al., 2002; Collins et al., 2003; Nuhse et al., 2003; Surpin et al., 2003; Yano et al., 2003; Ohtomo et al., 2005; Leshem et al., 2006; Ueda et al., 2006). These lines of evidence clearly indicate that SNARE-dependent membrane fusion is an essential process to fulfill higher-order plant functions as well as for basic cellular activity. Despite this importance, however, full identification of components has not been achieved, primarily because of the lack of information on R-SNAREs. The only exception is the PEN1 complex, whose full components were identified recently (Kwon et al., 2008). In this study, we have combined approaches in genetics, bioimaging, and biochemistry to identify the seed plant–specific VAMP727 as the R-SNARE in the SYP22/SYP51/VTI11 SNARE complex.
First, we demonstrated that VAMP727 and SYP22 show intimate genetic interactions; vamp727 and syp22 are synthetic lethal, whereas duplicated VAMP727 suppresses syp22. This kind of genetic interaction has been reported for pairs of cognate SNAREs in yeast (Sacher et al., 1997; Jantti et al., 2002; Graf et al., 2005) but not thus far in multicellular organisms. Our results clearly demonstrate that plants are good experimental systems suitable for such a genetic analysis, offering a powerful approach to unveiling how membrane traffic participates in multicellular organisms.
Second, the high-speed and high-sensitivity confocal laser scanning microscopy system we developed allowed multicolor observation with extremely high resolution in time and space (Nakano, 2002; Matsuura-Tokita et al., 2006). Using this system, we demonstrated that VAMP727 and SYP22 colocalize at subdomains of the PVC, which seem to be associated with the vacuolar membrane. By coimmunoprecipitation from plant lysate and FRET analysis, we further demonstrated that VAMP727 forms a complex with SYP22, SYP51, and VTI11 in plant cells. Considering these results together with the impaired PSV biogenesis and the traffic of storage proteins as well as the normal structure of the Golgi and multivesicular bodies in the mutants, we conclude that this SNARE complex mediates membrane fusion between the PVC and the vacuole. The possibility that VAMP727 is also incorporated into other SNARE complexes with different functions is not ruled out either because there are a considerable number of endosomes with VAMP727 that are not associated with vacuoles and bear no SYP22 (see Supplemental Figure 1C online).
Why Is VAMP727 Conserved Only in Seed Plants?
In comparison of its structure with other members of the VAMP7 group, the most striking characteristic of VAMP727 is the 20-residue insertion in the longin domain. VAMP727 homologs with a similar peculiar insertion are also found in the genomic sequences of rice (Oryza sativa) and poplar (Populus trichocarpa), indicating that homologs of VAMP727 are well conserved among angiosperms. A similar protein with a more divergent sequence in the insertion region can also be found in the EST database of pine tree (Pinus taeda), but there is no close homolog of VAMP727 in moss (Physcomitrella patens) or green algae, such as Chlamydomonas reinhardtii (Sanderfoot, 2007). Thus, VAMP727 appears to be conserved only in seed plants, which implies that VAMP727 plays important roles in functions specialized to seed plants.
Our results in this study are entirely consistent with this hypothesis. The development of the vamp727 syp22 double mutant was arrested at a late step of embryogenesis, suggesting an essential function of the VAMP727 complex in normal seed maturation. The small PSVs or faulty processing of storage proteins are probably not alone responsible for the lethality because many mutants with similar phenotypes have been reported to be viable (Shimada et al., 2003a, 2003b; Li et al., 2006; Shimada et al., 2006; Fuji et al., 2007; Tamura et al., 2007). There should be additional defects in the vamp727 syp22 mutant that severely affect the viability of seeds.
The machinery impaired by the vamp727 syp22 mutations seems to play more important roles in the late stages of seed maturation or rehydration/germination processes than in the early embryonic and postembryonic developmental stages because double mutant embryos can develop roots and leaf-like structures when cultured on the growth medium before dehydration. Thus, the vamp727 syp22 defect is expected to be associated with some seed-specific functions, such as dehydration tolerance, which must have been important in evolving seeds. The lineage with the VAMP727 homolog might be more adapted to land life, which is also supported by the fact that all the seed plants whose genomes have been sequenced so far conserve the VAMP727 homologs.
SYP22 Seems to Function in Multiple Steps in the Vacuolar Transport Pathway
Arabidopsis SYP22 was originally isolated by screening of a cDNA library for genes that complement the yeast vam3 mutation (Sato et al., 1997). Two groups then reported distinct localizations of SYP22, on the vacuolar membrane (Sato et al., 1997; Uemura et al., 2002) and on the PVC (Sanderfoot et al., 1999), suggesting that SYP22 resides on multiple compartments. In this study, we have confirmed that SYP22 is localized on both vacuoles and a part of PVCs, using the mRFP-tagged SYP22, which is fully functional as shown by its ability to complement the syp22 knockout mutant. The PVC-localized population of SYP22 probably mediates the membrane fusion between vacuoles and PVCs, as indicated in this study. On the other hand, a considerable amount of SYP22 is localized on the vacuolar membrane where VAMP727 does not colocalize. Taking also into account that a rather small portion of SYP22 was recovered in the complex with VAMP727 in the coimmunoprecipitation experiment, it is likely that SYP22 also functions in other SNARE complexes on the vacuolar membrane.
In yeast, Vam3p has been demonstrated to mediate homotypic vacuolar fusion by forming a complex with two Q-SNAREs (Vti1p and Vam7p) and one R-SNARE (Nyv1p) (Ungermann et al., 1999; McNew et al., 2000). Vti1p and Vam7p are homologs of VTI11 and SYP5 in Arabidopsis, respectively; thus, plants and yeast seem to share common Q-SNAREs for membrane fusion with vacuolar membranes. On the other hand, there is no obvious ortholog of Nyv1p in Arabidopsis. Among Arabidopsis VAMP7 members with substantial divergence, three of four VAMP71 members, VAMP711, VAMP712, and VAMP713, are demonstrated to be localized to the vacuoles when expressed transiently in protoplasts (Uemura et al., 2004). These R-SNAREs may cooperate with SYP22 to execute the homotypic vacuolar fusion in plant cells, which plays a collaborative role with the VAMP727 complex in the vacuole biogenesis. The proteomic analysis of the vacuolar membrane fraction in fact identified VAMP711 in this fraction with SYP22, SYP51, and VTI11 (Carter et al., 2004), which is also consistent with this idea.
Defects of vamp727 syp22 in the PSV biogenesis and the transport of storage proteins to vacuoles are partial; thus, the SNARE complex containing VAMP727 and SYP22 is important to but not essential for vacuole biogenesis and vacuolar transport. This inference suggests that other SNARE complexes also function in vacuole biogenesis. It is also possible that other proteins, such as SYP21 and VAMP71, replace the functions of SYP22 and VAMP727 when the authentic components are impaired, which should be verified in future studies.
Another interesting question to ask is about the upstream regulation of the SNARE complex identified in this study. In plant cells, Rab5 homologs (ARA7 and RHA1) are localized on the multivesicular PVCs (Haas et al., 2007), and another subgroup, Rab7, is found on the vacuolar membrane (Saito et al., 2002). In yeast, the assembly of the SNARE complex composed of Vam3p, Vti1p, Vam7p, and Nyv1p is accelerated by Ypt7p, the homolog of Rab7, via interaction with the conserved HOPS complex. The HOPS complex is an effector of Ypt7p, and at the same time it acts as a guanine nucleotide exchange factor for Ypt7p, through which the HOPS complex functions as a tethering factor between fusing vacuoles. Recently, a novel endosomal tethering complex sharing four of six components with the HOPS complex, the CORVET complex, was identified (Peplowska et al., 2007). Interestingly, the CORVET complex interacts preferentially with Vps21p, a yeast Rab5 homolog. Because there are putative homologs for the all components of HOPS and CORVET in Arabidopsis (Rojo et al., 2001), another interesting future project will be to examine whether the VAMP727 complex, a seed plant–unique SNARE complex, functions downstream of the conserved tethering complexes HOPS and CORVET.
METHODS
Plant Materials and Plasmids
syp22-1 (SALK_060946) and syp22-3 (SALK_075924) were obtained from ABRC (Alonso et al., 2003), and vamp727 (GABI_060G05) was from GABI-KAT (Rosso et al., 2003). Mutants were backcrossed at least three times with wild-type Arabidopsis thaliana (Columbia).
For complementation and suppression analyses, the 5.5-kb VAMP727 fragment (including 2.0 kb of the 5′- and 1.0 kb of the 3′-flanking sequences), the 5.9-kb SYP22 fragment (including 3.2 kb of the 5′- and 1.2 kb of the 3′-flanking sequences), the 4.4-kb VAMP721 fragment (including 2.0 kb of the 5′- and 0.9 kb of the 3′-flanking sequences), and the 4.3-kb VAMP713 fragment (including 2.3 kb of the 5′- and 0.8 kb of the 3′-flanking sequences) were used. Primer sequences used for amplification of genomic fragments are listed in Supplemental Table 1 online. Translational fusions between cDNA for fluorescent proteins and Arabidopsis genes were generated by the technique of fluorescent tagging of full-length proteins (Tian et al., 2004). The cDNA encoding GFP, Venus, TagRFP, or mRFP was inserted in front of the start codon of each gene: VAMP727 (including 2.0 kb of the 5′- and 1.0 kb of the 3′-flanking sequences), SYP22 (3.2 kb of the 5′- and 1.2 kb of the 3′-flanking sequences), SYP21 (2.8 kb of the 5′- and 1.1 kb of the 3′-flanking sequences), SYP43 (2.0 kb of the 5′- and 0.8 kb of the 3′-flanking sequences), and RHA1 (3.0 kb of the 5′- and 1.3 kb of the 3′-flanking sequences). Amplified genomic or chimeric fragments were subcloned into the binary vector, pGWB1 (a kind gift from T. Nakagawa), which were used for transforming Arabidopsis plants. The same promoter regions were also used for promoter-reporter analysis. The cDNA for GUS was connected in frame to the first exon of SYP22 or the fifth exon of VAMP727. The plasmid containing SP-GFP-CT24 and the transgenic plant expressing SP-GFP-CT24 were kindly provided by S. Utsumi. Transformation of Arabidopsis plants was performed by floral dipping using Agrobacterium tumefaciens (strain GV3101∷pMP90) (Clough and Bent, 1998).
RT-PCR
Total RNA was extracted from rosette leaves of 14-d-old wild-type and vamp727 mutant plants using RNAqueous-4PCR (Ambion), and cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen). Primer sets used for PCR amplification were VAMP727f (5′-ATGAGTCAAAAGGGTTTGAT-3′) and VAMP727r (5′-TTCGATCTTCTCTCCTCTGT-3′), and TUA3f (5′-GGACAAGCTGGGATCCAGG-3′) and TUA3r (5′-CGTCTCCACCTTCAGCACC-3′). PCR conditions were as follows: 97°C (2 min); 27 cycles of 97°C (30 s), 60°C (30 s), 72°C (1 min), and 72°C (10 min).
Light Microscopy
For labeling with FM4-64, Arabidopsis roots were soaked in MS cell culture medium (Murashige and Skoog, 1962) plus 4 μM FM4-64 on ice for 10 min. Samples were washed twice, incubated for 20 min at 23°C, and then observed under a confocal laser scanning microscope (model LSM510; Carl Zeiss). Plants expressing GFP- and Venus-tagged proteins were observed under the LSM510 equipped with a META device (Carl Zeiss). For plants expressing GFP-VAMP727 and mRFP-SYP22, leaf primordia were observed under a custom-made high-performance microscope system, which has been described elsewhere (Nakano, 2002; Matsuura-Tokita et al., 2006). For three-dimensional imaging, the objective lens was oscillated vertically to the sample plane by a piezo actuator system (Yokogawa Electric). Deconvolution analysis was performed with Volocity software (Improvision). For the observation of PSVs, autofluorescence from embryos excited by an Ar+ laser at 488 nm was observed under a fluorescence microscope (model BX51; Olympus) equipped with a confocal scanner unit (model CSU10; Yokogawa Electric) and a cooled CCD camera (model ORCA-AG; Hamamatsu Photonics). Images were processed with IPLab software (BD Biosciences). For quantitative analysis, size and number of the PSVs were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/).
Electron Microscopy
Arabidopsis embryos at bent cotyledon stage were high-pressure frozen and freeze-substituted in 0.2% uranyl acetate (Electron Microscopy Sciences) plus 0.2% glutaraldehyde (Electron Microscopy Sciences) in acetone at −80°C for 72 h and warmed to −50°C for 24 h. After several acetone rinses, these samples were infiltrated with Lowicryl HM20 (Electron Microscopy Sciences) for 72 h and polymerized at −50°C under UV light for 48 h.
For immunolabeling, sections were mounted on formvar-coated nickel grids and blocked for 20 min with a 5% (w/v) solution of nonfat milk in PBS containing 0.1% Tween 20. The sections were incubated in the primary antibodies (1:10 in PBS-Tween 20) for 1 h, rinsed in PBS containing 0.5% Tween 20, and then transferred to the secondary antibody (anti-rabbit IgG 1:10) conjugated to 15-nm gold particles for 1 h. Controls omitted the primary antibodies.
Antibodies
Anti-SYP51 and anti-SYP7 antibodies were kind gifts from M.H. Sato (Kyoto Prefectural University), and anti-SYP22 antibody was kindly provided by Y. Wada (Osaka University). Anti-KNOLLE antibody was the generous gift of G. Jürgens (University of Tübingen), and anti-12S globulin and anti-2S albumin antibodies used for immunoblotting were supplied by I. Hara-Nishimura (Kyoto University). Anti-GFP monoclonal antibodies were purchased from Abcam and Nacalai Tesque. Anti-2S albumin large chain and anti-Rubisco antibodies used for immunoelectron microscopy were kindly provided by A. Scarafoni (University of Milan) (Scarafoni et al., 2001) and Juan Jose Guiamet (University of La Plata) (Martínez et al., 2008), respectively.
For the anti-VTI11 antibody, the cytoplasmic region of VTI11 (residues 1 to 195) was cloned into the BamHI-HindIII site of an expression vector, pET-32a (+) (Novagen) slightly modified to produce a His-tagged protein at the N terminus. The His-VTI11 fusion protein was expressed in Escherichia coli Rosetta (DE3) pLysS (Novagen). The fusion protein was solubilized with 8 M Urea, purified on a Ni-NTA agarose (Qiagen) column, and then used for immunizing rabbits. Specificity of the antibody was confirmed with a zig-1 mutant.
Immunoprecipitation
Immunoprecipitation from detergent extracts was performed as described previously (Yano et al., 2003) with minor modifications. Root tissues collected from Arabidopsis cultivated in liquid medium for 17 d were homogenized on ice in extraction buffer (50 mM HEPES-KOH, pH 6.5, 10 mM potassium acetate, 100 mM sodium chloride, 5 mM EDTA, and 0.4 M sucrose) supplemented with a protease inhibitor cocktail (Roche). The homogenate was passed through Miracloth (Calbiochem) to remove debris and then centrifuged at 20,000g to yield a membrane pellet. The pellet was resuspended in extraction buffer containing 1% Triton X-100, 5% PreserveX-QML (QBI biosciences) and protease inhibitor cocktail (Roche) and incubated at 4°C for 3 h. Insoluble material was repelleted at 9000g, and the supernatant (total protein extract) was incubated at 4°C for 4 h with protein G-MicroBeads (Miltenyi Biotec), which were preloaded with anti-GFP (Abcam), anti-SYP22, anti-VTI11, or anti-SYP51 antibody. Samples were then applied to microcolumns attached to the magnetic field of the micro-MACS separator (Miltenyi Biotec) and washed five times with extraction buffer plus 1% Triton X-100. Immunoprecipitates were then eluted and subjected to immunoblotting.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At3g54300 (VAMP727), At5g46860 (SYP22), At5g39510 (VTI11), At1g16240 (SYP51), At5g11150 (VAMP713), At1g08560 (KNOLLE/SYP111), At3g09740 (SYP71), At5g16830 (SYP21), At3g05710 (SYP43), and At5g45130 (RHA1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. VAMP727 and SYP22 Colocalize at the Subdomain of the PVC Closely Associated with the Vacuole.
Supplemental Figure 2. VAMP727 Forms a Complex with SYP22, VTI11, and SYP51.
Supplemental Table 1. Primers Sequences Used for the Amplification of Genomic Fragments.
Supplemental Movie 1. VAMP727 and SYP22 Colocalize at the Tethering Site between the PVC and the Vacuole.
Supplementary Material
Acknowledgments
We thank the Salk Institute and the Max Planck Institute for Plant Breeding Research for providing T-DNA insertion mutants of Arabidopsis. We also thank G. Jürgens (Tübingen University), I. Hara-Nishimura (Kyoto University), M.H. Sato (Kyoto Prefectural University), T. Nakagawa (Shimane University), S. Utsumi (Kyoto University), A. Scarafoni (University of Milan), and J.J. Guiamet (University of La Plata) for sharing materials. This work was supported by Grants-in-Aid for Scientific Research and the Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (A.N. and T.U.), a research fellowship for young scientists from the Japan Society for the Promotion of Science (K.E.; 195010), and a U.S. National Science Foundation grant (M.S.O.; MCB-0619736). The development of the high-speed and high-resolution microscopic system was supported by funds from the Ministry of Economy, Trade, and Industry of Japan, from the New Energy and Industrial Technology Development Organization, and from RIKEN.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Takashi Ueda (tueda@biol.s.u-tokyo.ac.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Antonin, W., Fasshauer, D., Becker, S., Jahn, R., and Schneider, T.R. (2002). Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9 107–111. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Qiu, J.L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., Wanner, G., Peck, S.C., Edwards, H., Ramonell, K., Somerville, C.R., and Thordal-Christensen, H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, J.B., Matern, H.T., Peden, A.A., and Scheller, R.H. (2001). A genomic perspective on membrane compartment organization. Nature 409 839–841. [DOI] [PubMed] [Google Scholar]
- Carter, C., Pan, S., Zouhar, J., Avila, E.L., Girke, T., and Raikhel, N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Shin, Y.K., and Bassham, D.C. (2005). YKT6 is a core constituent of membrane fusion machineries at the Arabidopsis trans-Golgi network. J. Mol. Biol. 350 92–101. [DOI] [PubMed] [Google Scholar]
- Chen, Y.A., and Scheller, R.H. (2001). SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2 98–106. [DOI] [PubMed] [Google Scholar]
- Chow, C.M., Neto, H., Foucart, C., and Moore, I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J.L., Huckelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977. [DOI] [PubMed] [Google Scholar]
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y.D., and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini, F., Rossi, V., Galli, T., Budillon, A., D'Urso, M., and D'Esposito, M. (2001). Longins: A new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem. Sci. 26 407–409. [DOI] [PubMed] [Google Scholar]
- Foresti, O., daSilva, L.L., and Denecke, J. (2006). Overexpression of the Arabidopsis syntaxin PEP12/SYP21 inhibits transport from the prevacuolar compartment to the lytic vacuole in vivo. Plant Cell 18 2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti, R.A., Collins, K.M., Hickey, C.M., and Wickner, W. (2007). Stringent 3Q.1R composition of the SNARE 0-layer can be bypassed for fusion by compensatory SNARE mutation or by lipid bilayer modification. J. Biol. Chem. 282 14861–14867. [DOI] [PubMed] [Google Scholar]
- Fuji, K., Shimada, T., Takahashi, H., Tamura, K., Koumoto, Y., Utsumi, S., Nishizawa, K., Maruyama, N., and Hara-Nishimura, I. (2007). Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, L.C., Jr., Weis, W.I., and Scheller, R.H. (2001). A novel snare N-terminal domain revealed by the crystal structure of Sec22b. J. Biol. Chem. 276 24203–24211. [DOI] [PubMed] [Google Scholar]
- Graf, C.T., Riedel, D., Schmitt, H.D., and Jahn, R. (2005). Identification of functionally interacting SNAREs by using complementary substitutions in the conserved '0' layer. Mol. Biol. Cell 16 2263–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, T.J., Sliwinski, M.K., Martinez, D.E., Preuss, M., Ebine, K., Ueda, T., Nielsen, E., Odorizzi, G., and Otegui, M.S. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, R., Lang, T., and Sudhof, T.C. (2003). Membrane fusion. Cell 112 519–533. [DOI] [PubMed] [Google Scholar]
- Jantti, J., Aalto, M.K., Oyen, M., Sundqvist, L., Keranen, S., and Ronne, H. (2002). Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 115 409–420. [DOI] [PubMed] [Google Scholar]
- Jürgens, G. (2004). Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20 481–504. [DOI] [PubMed] [Google Scholar]
- Kato, T., Morita, M.T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M., and Tasaka, M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper, T.H., Kienle, C.N., and Fasshauer, D. (2007). An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451 835–840. [DOI] [PubMed] [Google Scholar]
- Leshem, Y., Melamed-Book, N., Cagnac, O., Ronen, G., Nishri, Y., Solomon, M., Cohen, G., and Levine, A. (2006). Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 103 18008–18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman, B., Geelen, D., Quintero, F.J., and Blatt, M.R. (1999). A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283 537–540. [DOI] [PubMed] [Google Scholar]
- Li, L., Shimada, T., Takahashi, H., Ueda, H., Fukao, Y., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18 3535–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84 61–71. [DOI] [PubMed] [Google Scholar]
- Martínez, D.E., Costa, M.L., Otegui, M.S., and Guiamet, J.J. (2008). Senescence-associated vacuoles of tobacco leaves are involved in the degradation of chloroplast proteins. Plant J. 56: 192–206. [DOI] [PubMed]
- Matsuura-Tokita, K., Takeuchi, M., Ichihara, A., Mikuriya, K., and Nakano, A. (2006). Live imaging of yeast Golgi cisternal maturation. Nature 441 1007–1010. [DOI] [PubMed] [Google Scholar]
- McNew, J.A., Parlati, F., Fukuda, R., Johnston, R.J., Paz, K., Paumet, F., Sollner, T.H., and Rothman, J.E. (2000). Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 407 153–159. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nakano, A. (2002). Spinning-disk confocal microscopy—A cutting-edge tool for imaging of membrane traffic. Cell Struct. Funct. 27 349–355. [DOI] [PubMed] [Google Scholar]
- Nishizawa, K., Maruyama, N., Satoh, R., Fuchikami, Y., Higasa, T., and Utsumi, S. (2003). A C-terminal sequence of soybean beta-conglycinin alpha' subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 34 647–659. [DOI] [PubMed] [Google Scholar]
- Nuhse, T.S., Boller, T., and Peck, S.C. (2003). A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J. Biol. Chem. 278 45248–45254. [DOI] [PubMed] [Google Scholar]
- Ohtomo, I., Ueda, H., Shimada, T., Nishiyama, C., Komoto, Y., Hara-Nishimura, I., and Takahashi, T. (2005). Identification of an allele of VAM3/SYP22 that confers a semi-dwarf phenotype in Arabidopsis thaliana. Plant Cell Physiol. 46 1358–1365. [DOI] [PubMed] [Google Scholar]
- Peplowska, K., Markgraf, D.F., Ostrowicz, C.W., Bange, G., and Ungermann, C. (2007). The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell 12 739–750. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Gillmor, C.S., Kovaleva, V., Somerville, C.R., and Raikhel, N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1 303–310. [DOI] [PubMed] [Google Scholar]
- Rossi, V., Picco, R., Vacca, M., D'Esposito, M., D'Urso, M., Galli, T., and Filippini, F. (2004. a). VAMP subfamilies identified by specific R-SNARE motifs. Biol. Cell 96 251–256. [DOI] [PubMed] [Google Scholar]
- Rossi, V., Banfield, D.K., Vacca, M., Dietrich, L.E., Ungermann, C., D'Esposito, M., Galli, T., and Filippini, F. (2004. b). Longins and their longin domains: Regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 29 682–688. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sacher, M., Stone, S., and Ferro-Novick, S. (1997). The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J. Biol. Chem. 272 17134–17138. [DOI] [PubMed] [Google Scholar]
- Saito, C., Ueda, T., Abe, H., Wada, Y., Kuroiwa, T., Hisada, A., Furuya, M., and Nakano, A. (2002). A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 29 245–255. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A. (2007). Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144 6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Assaad, F.F., and Raikhel, N.V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Kovaleva, V., Bassham, D.C., and Raikhel, N.V. (2001. a). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Kovaleva, V., Zheng, H., and Raikhel, N.V. (1999). The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 121 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Pilgrim, M., Adam, L., and Raikhel, N.V. (2001. b). Disruption of individual members of Arabidopsis syntaxin gene families indicates each has essential functions. Plant Cell 13 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M.H., Nakamura, N., Ohsumi, Y., Kouchi, H., Kondo, M., Hara-Nishimura, I., Nishimura, M., and Wada, Y. (1997). The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 272 24530–24535. [DOI] [PubMed] [Google Scholar]
- Scarafoni, A., Carzaniga, R., Harris, N., and Croy, R.R. (2001). Manipulation of the napin primary structure alters its packaging and deposition in transgenic tobacco (Nicotiana tabacum L.) seeds. Plant Mol. Biol. 46 727–739. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Fuji, K., Tamura, K., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2003. a). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100 16095–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T., Koumoto, Y., Li, L., Yamazaki, M., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47 1187–1194. [DOI] [PubMed] [Google Scholar]
- Shimada, T., et al. (2003. b). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278 32292–32299. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Zheng, H., Morita, M.T., Saito, C., Avila, E., Blakeslee, J.J., Bandyopadhyay, A., Kovaleva, V., Carter, D., Murphy, A., Tasaka, M., and Raikhel, N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Takahashi, H., Kunieda, T., Fuji, K., Shimada, T., and Hara-Nishimura, I. (2007). Arabidopsis KAM2/GRV2 is required for proper endosome formation and functions in vacuolar sorting and determination of the embryo growth axis. Plant Cell 19 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H., Nishiyama, C., Shimada, T., Koumoto, Y., Hayashi, Y., Kondo, M., Takahashi, T., Ohtomo, I., Nishimura, M., and Hara-Nishimura, I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47 164–175. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Uemura, T., Sato, M.H., and Nakano, A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40 783–789. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Sato, M.H., and Takeyasu, K. (2005). The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett. 579 2842–2846. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Ueda, T., Ohniwa, R.L., Nakano, A., Takeyasu, K., and Sato, M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29 49–65. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Yoshimura, S.H., Takeyasu, K., and Sato, M.H. (2002). Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes Cells 7 743–753. [DOI] [PubMed] [Google Scholar]
- Ungermann, C., von Mollard, G.F., Jensen, O.N., Margolis, N., Stevens, T.H., and Wickner, W. (1999). Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J. Cell Biol. 145 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, D., Sato, M., Saito, C., Sato, M.H., Morita, M.T., and Tasaka, M. (2003). A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. USA 100 8589–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Gong, Z., Zhang, C., Song, C.P., Damsz, B., Inan, G., Koiwa, H., Zhu, J.K., Hasegawa, P.M., and Bressan, R.A. (2002). OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14 3009–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.