Abstract
Root symbioses with arbuscular mycorrhizal fungi and rhizobial bacteria share a common signaling pathway in legumes. Among the common symbiosis genes are CASTOR and POLLUX, the twin homologous genes in Lotus japonicus that encode putative ion channel proteins. Here, we show that the orthologs of CASTOR and POLLUX are ubiquitously present and highly conserved in both legumes and nonlegumes. Using rice (Oryza sativa) as a study system, we employ reverse genetic tools (knockout mutants and RNA interference) to demonstrate that Os-CASTOR and Os-POLLUX are indispensable for mycorrhizal symbiosis in rice. Furthermore, a cross-species complementation test indicates that Os-POLLUX can restore nodulation, but not rhizobial infection, to a Medicago truncatula dmi1 mutant.
Many terrestrial plants can grow under nutrient-limiting conditions by forming mutually beneficial root symbioses with soil microbes. These underground symbiotic networks contribute significantly to the functionality and sustainability of agricultural and natural ecosystems. The most widespread symbiosis is arbuscular mycorrhiza (AM), the “fungus root” formed between the vast majority of vascular flowering plants and biotrophic fungi belonging to the phylum Glomeromycota (Smith and Read, 1997). The AM symbiosis originated more than 400 million years ago and possibly played a key role in helping the first plants to colonize land (Remy et al., 1994; Redecker et al., 2000; Heckman et al., 2001). Initiation of the AM symbiosis is mediated by signal exchanges between the two symbiotic partners. The host plant releases strigolactones, known as “branching factors,” into the rhizosphere that induce hyphal branching, thereby increasing the chance of AM fungi contacting the root (Buee et al., 2000; Akiyama et al., 2005; Akiyama and Hayashi, 2006; Besserer et al., 2006). The fungal partner subsequently produces yet unknown diffusible signals, termed “Myc factors,” to trigger the host responses (Kosuta et al., 2003; Navazio et al., 2007). During the process, AM fungi enter the plant root and form highly ramified, tree-like fungal structures, called arbuscules, within the inner cortical cells (Harrison, 1997; Smith and Read, 1997). AM fungi also develop extensively branched hyphae outside the plant root. The extraradical hyphae function to expand the rhizosphere, thereby enhancing access to mineral nutrients, particularly inorganic phosphate, from the soil, whereas the intraradical hyphae (arbuscules) serve as an intracellular microbe-plant interface where the soil nutrients move to the plant and plant photosynthates flow to the fungus (Harrison, 1997, 2005). In addition to the supply of mineral nutrients to the plant, AM symbioses also improve plant health through enhanced tolerance to biotic and abiotic stresses (Ruiz-Lozano, 2003; Liu et al., 2007; Pozo and Azcón-Aguilar, 2007).
In addition to the AM symbiosis, certain members of the Eurosid I angiosperms can enter into a root symbiosis with soil bacteria (Soltis et al., 1995). The symbiosis culminates in the formation of the root nodule, an optimal microenvironment for bacteria to fix atmospheric nitrogen and for nutrient exchange between the symbiotic partners. The root nodule symbiosis takes place in two major forms, involving either the gram-negative bacteria of the family Rhizobiaceae, collectively called rhizobia, or the gram-positive Frankia bacteria that are filamentous actinomycetes (Pawlowski and Bisseling, 1996). The majority of leguminous plants as well as a nonleguminous plant, Parasponia, can interact with rhizobia, whereas a diverse group of nonleguminous plants, so called actinorhizal plants, are able to enter into an interaction with Frankia bacteria. The legume-rhizobia symbiosis is best characterized at the cellular and molecular levels. The interaction is set in motion by an intimate communication between the host and rhizobia (Long, 1996; Spaink, 2000). The legume roots secrete into the rhizosphere (iso)flavonoids that attract the rhizobia to the root and trigger the production of a bacterial signal known as Nod factor. Nod factors are chitin-like lipochitooligosaccharides that are essential for activating the host symbiotic responses, including an early ion influx, calcium spiking, root hair deformation and curling, transcriptional reprogramming of the host symbiotic genes, and cortical cell divisions, that ultimately result in the formation of the rhizobium-infected root nodules (for review, see Oldroyd and Downie, 2004, 2006, 2008; Stacey et al., 2006; Jones et al., 2007).
Although the AM and rhizobial symbioses are morphologically distinct, the two are mechanistically related in legumes. A number of legume genes that are required for nodulation also are essential for the AM interaction (Kistner et al., 2005). Moreover, a subset of the host genes that are induced during legume-rhizobia symbiosis also is up-regulated during AM symbioses (Albrecht et al., 1999; Kistner and Parniske, 2002). The overlap of the two symbiotic pathways has led to the hypothesis that the evolutionarily younger legume root nodule symbiosis may have evolved from the more ancient AM symbiosis (LaRue and Weeden, 1994; Gianinazzi-Pearson, 1996; Zhu et al., 2006).
In recent years, the development of genetic and genomic tools for the two model legumes Medicago truncatula and Lotus japonicus has greatly facilitated the cloning of genes required for root symbioses (Stacey et al., 2006). Analysis of these genes has begun to reveal the Nod factor and mycorrhizal signaling pathways. The Nod factors are likely perceived directly by the receptor-like kinases, such as Lj-NFR1/Mt-LYK3 and Lj-NFR5/Mt-NFP, that contain peptidoglycan-binding LysM domains in the extracellular region (Limpens et al., 2003a; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Smit et al., 2007). Downstream of the Nod factor receptors are a set of proteins that play a dual role in AM and nodulation symbioses. These proteins include the Leu-rich-repeat receptor kinase Mt-DMI2/Ms-NORK/Lj-SYMRK/Ps-SYM19 (Endre et al., 2002; Stracke et al., 2002), the ion channel proteins Lj-CASTOR and Lj-POLLUX/Mt-DMI1/Ps-SYM8 (Ane et al., 2004; Imaizumi-Anraku et al., 2005; Edwards et al., 2007), the two nucleoporins Lj-NUP85 and Lj-NUP133 (Kanamori et al., 2006; Saito et al., 2007), the Ca2+/calmodulin-dependent protein kinase Mt-DMI3/Lj-CCaMK/Ps-SYM9 (Levy et al., 2004; Mitra et al., 2004; Tirichine et al., 2006), and the DMI3-interacting protein Mt-IPD3/Lj-CYCLOPS (Messinese et al., 2007; Chen et al., 2008; Yano et al., 2008). Except for Mt-DMI3 and Mt-IPD3, all of these common symbiosis components act upstream of the Nod factor-induced calcium spiking (Ehrhardt et al., 1996; Oldroyd and Downie, 2004, 2006). Mt-DMI3 and Mt-IPD3 function downstream of calcium spiking and are presumably responsible for decoding the calcium spiking signal (Oldroyd and Downie, 2006), which subsequently activates the nodulation-specific transcription factors, such as Mt-NSP1, Mt-NSP2, Mt-ERN1, and Lj-NIN, leading to transcriptional reprogramming of the host symbiotic genes (Schauser et al., 1999; Kalo et al., 2005; Smit et al., 2005; Middleton et al., 2007). In contrast to the Nod factor signaling, the mycorrhiza-specific signaling components beyond the common symbiosis pathway are largely unknown.
Intriguingly, all cloned legume symbiosis genes, including both the common symbiosis genes and genes only required for rhizobial symbiosis, have orthologs in nonlegumes (Zhu et al., 2006). This finding offers an opportunity to address the evolution of root symbioses in plants by characterizing ortholog functionality across the legume and nonlegume boundary. For example, the Lj-NFR1 ortholog in Arabidopsis (Arabidopsis thaliana; At1g21630) has been shown to be essential for chitin signaling, indicating an evolutionary relationship between chitin and Nod factor perception (Zhu et al., 2006; Miya et al., 2007; Wan et al., 2008). As part of our effort to address the function of nonlegume orthologs of legume genes required for root symbioses, we seek to determine whether the common symbiosis genes in legumes also are required for mycorrhizal symbioses in nonlegumes using rice (Oryza sativa) as a study model. We as well as others have demonstrated that Os-DMI3, the rice ortholog of Mt-DMI3/Lj-CCaMK/Ps-SYM9, is required for AM symbiosis in rice and able to complement a M. truncatula dmi3 mutant (Godfroy et al., 2006; Chen et al., 2007). Two recent studies revealed that the Mt-DMI2/Ms-NORK/Lj-SYMRK/Ps-SYM19 orthologs from the two actinorhizal plants, Casuarina glauca and Datisca glomerata, are essential for root symbioses with both AM fungi and Frankia bacteria (Gherbi et al., 2008; Markmann et al., 2008). Here, we extend these studies to include Os-CASTOR and Os-POLLUX, the rice orthologs of the twin common symbiosis genes Lj-CASTOR and Lj-POLLUX. We demonstrate that the rice orthologs of CASTOR and POLLUX, namely Os-CASTOR and Os-POLLUX, are indispensable for mycorrhizal symbiosis in rice and that Os-POLLUX can restore nodulation, but not rhizobial infection, to a M. truncatula dmi1 mutant.
RESULTS AND DISCUSSION
Antiquity and Evolution of the CASTOR and POLLUX Homologs in Plants
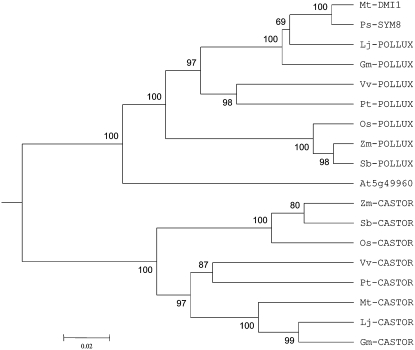
CASTOR and POLLUX are two homologous genes encoding putative ion channels that are components of the common symbiosis pathway in L. japonicus (Imaizumi-Anraku et al., 2005). The twin genes possibly evolved from an ancient gene duplication event that dated before the monocot-dicot divergence (Zhu et al., 2006; Fig. 1). Under this evolutionary scenario, the duplicated gene copies have degenerated to perform complementary rather than redundant functions. It was speculated that CASTOR and POLLUX, analogous to many other ion channels, may act as heteromultimeric complexes (Jiang et al., 2002; Imaizumi-Anraku et al., 2005). The fact that CASTOR and POLLUX homologs also are present in Physcomitrella patens (moss), a basal lineage of land plants that can establish root symbioses with AM fungi (Ane et al., 2004), suggests that the progenitor of CASTOR and POLLUX is ancestral to land plants.
Figure 1.
Phylogenetic tree (unrooted) of CASTOR and POLLUX homologs in M. truncatula (Mt), L. japonicus (Lj), soybean (Gm), poplar (Pt), grapevine (Vv), Arabidopsis (At), rice (Os), sorghum (Sb), and maize (Zm). The tree was based on the C-terminal approximately 650 amino acids of the proteins. Sequence alignments were performed using ClustalX (Thompson et al., 1997) and manually curated. The tree was constructed by MEGA3.1 (Kumar et al., 2004), using the UPGMA method. Numbers below the branches represent the percentages of 1,000 bootstrap replications supporting the particular nodes.
The CASTOR and POLLUX orthologs are ubiquitously present in nearly all examined plant taxa for which sequence information is currently available, including M. truncatula, soybean (Glycine max), poplar (Populus trichocarpa), grapevine (Vitis vinifera), rice, sorghum (Sorghum bicolor), and maize (Zea mays; Supplemental Sequence Data S1). The between-species orthologous relationship of the CASTOR-POLLUX homologs can be readily inferred based on phylogenetic analysis (Fig. 1) and their microsyntenic genomic position (Zhu et al., 2006). The only exception is Arabidopsis, which contains the ortholog of POLLUX (At5g49960) but lacks the ortholog of CASTOR (Ane et al., 2004; Zhu et al., 2006). Notably, Arabidopsis also lacks the orthologs of Mt-DMI2, Mt-DMI3, and Mt-IPD3 (Levy et al., 2004; Zhu et al., 2005, 2006; Messinese et al., 2007). Such gene deletions in Arabidopsis (and likely the lineage leading to the Brassiceae) offer an explanation for the inability of the Brassica plants to form root symbioses with mycorrhizal fungi (Ocampo et al., 1980; Glenn et al., 1985). As shown in Figure 1, the phylogenetic position of At-POLLUX (At5g49960) is not congruent with the species tree, suggesting that purifying selection on At5g49960 may have been relaxed due to a lack of ability of Arabidopsis to establish AM symbiosis.
The CASTOR and POLLUX homologs are highly conserved over the C termini of approximately 650 amino acids, with a sequence identity ranging from 69% to 98% depending on the phylogenetic distances between species (Table I). As shown in Table I, the level of sequence identity between orthologs (80%−98%) is always higher than that between within-species paralogs (72%−76%). Moreover, multiple sequence alignments of the conserved regions revealed numerous amino acid residues that could discriminate between CASTOR and POLLUX orthologs (Supplemental Fig. S1), further supporting the bifurcation of the two orthologous groups in phylogenetic analysis (Fig. 1). In contrast to the C-terminal region, the N terminus of these proteins appears to evolve more rapidly. Within each of the CASTOR and POLLUX orthologous groups, the N-terminal sequences are conserved only between closely related species but highly diverged between distantly related species. Visual and in silico analysis using the SIMPLE algorithm (http://www.biochem.ucl.ac.uk/bsm/SIMPLE/) revealed that the N-terminal regions of CASTOR and POLLUX proteins are rich in simple sequence repeats encoded by simple sequence repeats at the DNA level (data not shown), which may have contributed to the fast-evolution feature of this region (Hancock and Simon, 2005).
Table I.
Percentage amino acid sequence identity between CASTOR and POLLUX homologs
Percentages were calculated based on the C-terminal 650 amino acids of the proteins via the National Center for Biotechnology Information BLAST2 program. Boldface entries indicate percentage identities between orthologs; italic entries indicate percentage identities between within-species paralogs.
| Ps-SYM8 | Lj-POLLUX | Gm-POLLUX | Pt-POLLUX | Vv-POLLUX | Os-POLLUX | Zm-POLLUX | Sb-POLLUX | At5g49960 | Mt-CASTOR | Lj-CASTOR | Gm-CASTOR | Pt-CASTOR | Vv-CASTOR | Os-CASTOR | Zm-CASTOR | Sb-CASTOR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mt-DMI1 | 98 | 94 | 94 | 87 | 86 | 83 | 84 | 84 | 80 | 72 | 75 | 75 | 74 | 74 | 74 | 73 | 74 |
| Ps-SYM8 | 94 | 93 | 87 | 87 | 83 | 84 | 84 | 80 | 72 | 75 | 75 | 74 | 75 | 74 | 73 | 74 | |
| Lj-POLLUX | 93 | 87 | 86 | 83 | 84 | 84 | 81 | 74 | 75 | 76 | 74 | 74 | 74 | 72 | 73 | ||
| Gm-POLLUX | 89 | 87 | 84 | 84 | 84 | 81 | 73 | 75 | 76 | 74 | 75 | 74 | 73 | 74 | |||
| Pt-POLLUX | 90 | 84 | 84 | 85 | 83 | 74 | 76 | 77 | 75 | 75 | 75 | 74 | 75 | ||||
| Vv-POLLUX | 84 | 85 | 85 | 83 | 74 | 76 | 77 | 76 | 74 | 76 | 75 | 75 | |||||
| Os-POLLUX | 96 | 96 | 80 | 73 | 75 | 75 | 74 | 74 | 74 | 75 | 75 | ||||||
| Zm-POLLUX | 98 | 80 | 73 | 75 | 76 | 75 | 74 | 75 | 75 | 76 | |||||||
| Sb-POLLUX | 81 | 73 | 75 | 76 | 74 | 75 | 75 | 76 | 75 | ||||||||
| At5g49960 | 70 | 71 | 72 | 71 | 70 | 70 | 69 | 70 | |||||||||
| Mt-CASTOR | 90 | 92 | 84 | 85 | 83 | 80 | 82 | ||||||||||
| Lj-CASTOR | 95 | 87 | 86 | 85 | 82 | 84 | |||||||||||
| Gm-CASTOR | 88 | 87 | 85 | 82 | 85 | ||||||||||||
| Pt-CASTOR | 88 | 85 | 83 | 84 | |||||||||||||
| Vv-CASTOR | 84 | 82 | 85 | ||||||||||||||
| Os-CASTOR | 91 | 94 | |||||||||||||||
| Zm-CASTOR | 98 |
Characterization of Os-CASTOR and Os-POLLUX in Rice
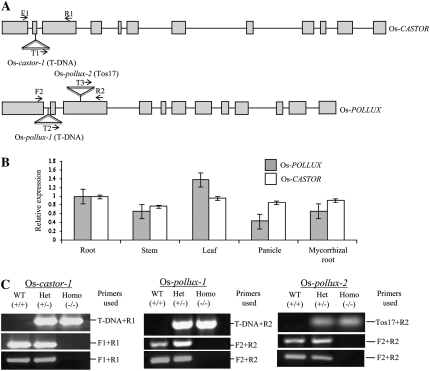
We have selected rice as a model system to assess the function of nonlegume orthologs of the legume symbiosis genes because rice is a mycorrhizal plant with a completely sequenced genome and available resources for high-throughput reverse genetic analysis (Jeong et al., 2006; Chen et al., 2007; Miyao et al., 2007). Os-CASTOR and Os-POLLUX were identified as Os03g62650 and Os01g64980, respectively, in the rice genome (Nipponbare) based on The Institute for Genomic Research Rice Genome Annotation (http://www.tigr.org/tdb/e2k1/osa1/; Zhu et al., 2006). Alignment of full-length cDNAs with the genomic sequences revealed a gene structure of 12 exons for both genes (Fig. 2A), which is conserved with their legume and nonlegume counterparts listed in Table I (Ane et al., 2004; Imaizumi-Anraku et al., 2005). Both Os-CASTOR and Os-POLLUX also produce transcript variants that result from alternative splicing (data not shown). Quantitative reverse transcription (qRT)-PCR analysis indicated that Os-CASTOR and Os-POLLUX are expressed in all tissues tested, including roots, leaves, stems, and panicles, and the expression levels in the root were not enhanced by mycorrhizal colonization (Fig. 2B). This observation was further supported by the analysis of the Rice Gene Index database (http://compbio.dfci.harvard.edu/tgi), from which the cognate expressed sequences of Os-CASTOR (TC285740) and Os-POLLUX (TC285508 and TC334749) were derived from cDNA libraries of various plant tissues. The expression pattern of Os-CASTOR and Os-POLLUX appears to be similar to that observed for Lj-CASTOR and Lj-POLLUX in L. japonicus (Imaizumi-Anraku et al., 2005) but in contrast to that of Mt-DMI1 (orthologous to Lj-POLLUX), which is predominantly expressed in M. truncatula roots (Ane et al., 2004).
Figure 2.
Isolation and characterization of Tos17/T-DNA insertion mutants of Os-CASTOR and Os-POLLUX. A, Gene structures of Os-CASTOR and Os-POLLUX and the Tos17/T-DNA insertion sites. The exons and introns are indicated by boxes and lines, respectively. Insertion sites of Tos17/T-DNA are indicated. B, Os-CASTOR and Os-POLLUX expression levels in roots, stems, leaves, panicles, and mycorrhizal roots. Relative transcript abundance was determined by qRT-PCR and normalized against Os-ubiquitin1. Error bars represent sd values from three independent biological replications. C, Identification of homozygous (–/–) insertion mutants by PCR. Top, Identification of positive insertion plants (+/– or –/–) by PCR using a pair of Tos17/T-DNA- and gene-specific primers. Middle, PCR analysis to distinguish between homozygous (–/–) and heterozygous (+/–) mutant plants using a primer pair flanking the Tos17/T-DNA insertion site that allowed the amplification of only the wild-type (WT) allele under given PCR conditions. Bottom, RT-PCR analysis of Os-CASTOR and Os-POLLUX expression in the wild-type and mutant plants. The primer positions are indicated in A.
The regular transcripts of Os-CASTOR and Os-POLLUX encode predicted proteins of 893 and 943 amino acids, respectively, with a domain structure identical to that of their legume orthologs (Ane et al., 2004; Imaizumi-Anraku et al., 2005). Both proteins possess four transmembrane helices, a central region homologous to the RCK (for regulator of conductance of K+) domain of bacterial calcium-gated potassium channels, and several motifs, such as the filter, the pore helix, and the hinge, that are characteristic of the structurally characterized MthK channel from Methanobacterium thermoautotrophicum (Jiang et al., 2002; Fig. 3). Over the C-terminal 650 amino acids, starting from a site between the second and third transmembrane helices, Os-CASTOR and Os-POLLUX are more than 91% identical to their maize and sorghum orthologs and 83% to 85% identical to their legume counterparts (Table I).
Figure 3.
Sequences and domain structures of Os-CASTOR and Os-POLLUX. The characteristic motifs and domains are underlined, including the four transmembrane (TM) helices and the filter, the pore helix, the hinge, and the RCK (for regulation of conductance of K+) domain.
Isolation of Os-castor and Os-pollux Mutants in Rice
We searched the rice mutant databases for putative Tos17 and T-DNA insertion lines to be used for functional analysis of Os-CASTOR and Os-POLLUX in root symbioses (Jeong et al., 2006; Miyao et al., 2007). We identified one T-DNA insertion allele for Os-CASTOR in the genetic background of the japonica rice cv Hawyoung (line no. C04353), hereafter referred to as Os-castor-1 (Jeon et al., 2000; Jeong et al., 2002). In the Os-castor-1 mutant, the T-DNA was inserted into the second 72-bp exon (Fig. 2A). For Os-POLLUX, we obtained two mutant alleles, named Os-pollux-1 and Os-pollux-2 (Fig. 2A). The Os-pollux-1 mutant was a T-DNA insertion line in the genetic background of cv Dongjin (line no. B02432), in which the T-DNA was inserted into the first intron. Os-pollux-2 was a Tos17 insertion line in the genotype Nipponbare derived from tissue culture (line no. NC6423). In Os-pollux-2, the retrotransposon Tos17 was inserted into the third exon. For all three mutants, the expression of Os-CASTOR or Os-POLLUX was disrupted based on RT-PCR analyses (Fig. 2C, bottom).
From progeny of each primary mutant line, positive T-DNA/Tos17 insertion plants were identified by PCR analysis using a pair of T-DNA/Tos17- and gene-specific primers (Fig. 2C, top). A second round of PCR analysis followed to discriminate between homozygous mutant (–/–) and heterozygous (+/–) plants using a primer pair flanking the T-DNA/Tos17 insertion sites that enabled the amplification of only the wild-type alleles under given PCR conditions (Fig. 2C, middle). Since T-DNA and Tos17 mutant lines may comprise multiple insertion sites, the wild-type plants segregated from the progeny of the heterozygous mutant lines were used as additional controls for the experiments described below.
Os-castor and Os-pollux Mutant Plants Are Defective in Mycorrhizal Symbiosis
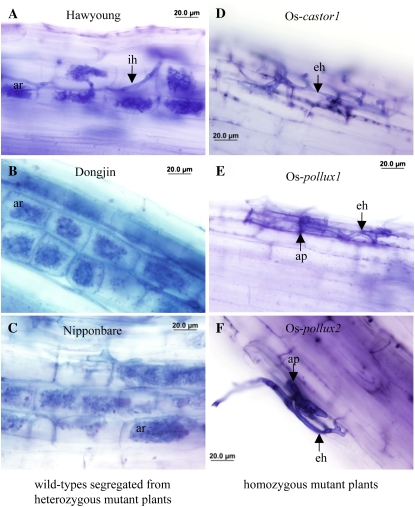
To test whether Os-CASTOR and Os-POLLUX are required for AM symbiosis in rice, we inoculated the mutant and wild-type rice roots with the fungus Glomus intraradices. At 35 d after inoculation, wild-type plants were densely colonized by G. intraradices, exhibiting the range of symbiotic structures typical of a functional symbiosis, including intercellular and intracellular hyphae, vesicles, and arbuscules. In each of the 60 wild-type plants from the genotypes Nipponbare, Hawyoung, and Dongjin, approximately 60% to 85% of the total root length was colonized. Similar levels of colonization also were observed for wild-type plants segregated from heterozygous mutant plants (Fig. 4). In contrast, intracellular fungal structures, including vesicles and arbuscules, were not observed on roots of 60 Os-castor-1, 72 Os-pollux-1, and 60 Os-pollux-2 homozygous mutant plants. For homozygous mutant plants, extraradical hyphae and appressoria were frequently observed on the root surface (Fig. 4), but the fungus was unable to penetrate the roots beyond the epidermis. These observations indicate that the knockout of Os-CASTOR and Os-POLLUX has completely abolished the ability of the AM fungus to enter the plant root. The defective phenotypes were similar to those reported for the castor (i.e. Lj-sym4, Lj-sym22, and Lj-sym71; Bonfante et al., 2000; Senoo et al., 2000) and pollux (i.e. Lj-sym23 and Lj-sym86; Kistner et al., 2005) mutants in L. japonicus, the dmi1 mutants in M. truncatula (Catoira et al., 2000), and the sym8 mutant in pea (Pisum sativum; Balaji et al., 1994). For the weak alleles of the L. japonicus castor and pollux mutants, the fungus was occasionally able to penetrate the cortical cells and form arbuscules (Senoo et al., 2000; Kistner et al., 2005), but this leaky phenotype was not observed for the knockout lines of rice.
Figure 4.
Os-castor and Os-pollux are defective in AM symbiosis. A to C, Roots of wild-type plants segregated from heterozygous mutants formed arbuscules upon inoculation with G. intraradices. D to F, Roots of homozygous mutants failed to form AM symbiosis, despite the presence of fungal hyphae on the root surface. Photographs were taken from roots at 5 weeks after inoculation with G. intraradices. Mycorrhizal colonization was assessed by trypan blue staining according to the procedures described by Koske and Gemma (1989). Stained roots were examined using a light microscope (Olympus BX40F-3), and images were captured by a microscope digital camera system (Olympus DP71). ap, Appressorium; ar, arbuscule; eh, extraradical hypha; ih, intraradical hypha.
Transcriptional profiling has revealed a number of host genes that were expressed exclusively in the root colonized by AM fungi (Harrison et al., 2002; Liu et al., 2003; Guimil et al., 2005; Kistner et al., 2005). Thus, the expression of these AM-specific genes can serve as a molecular marker for the occurrence of functional symbiotic interaction between the two symbionts. In addition to the phenotypic analysis at the cytological level, we analyzed the expression of Os-PT11, a rice mycorrhiza-specific phosphate transporter (Paszkowski et al., 2002), in roots of the wild type and mutants under inoculated and noninoculated conditions. The results showed that Os-PT11 was expressed only in the wild-type roots inoculated with G. intraradices and not in the mutant roots (Fig. 5A).
Figure 5.
Expression of the rice AM-specific phosphate transporter Os-PT11 in the mutant and/or RNAi lines of Os-CASTOR and Os-POLLUX under inoculated and noninoculated conditions. A, Expression of Os-PT11 in the wild-type (WT) and mutant roots under inoculated and noninoculated conditions. WT* indicates wild-type plants segregated from a corresponding heterozygous mutant plant. B, Expression of Os-PT11 in the two RNAi lines, Os-CASTORi-1 and Os-CASTORi-2, during AM symbiosis. Numbers in parentheses indicate cycle numbers of the RT-PCR. The rice Actin gene was used as a control.
Since only a single mutant allele of Os-CASTOR was available for this study, we generated stable transgenic silencing lines (Nipponbare) by RNA interference (RNAi) to provide further evidence for the requirement of Os-CASTOR in AM symbiosis. The RNAi silencing cassette consists of a 400-bp inverted-repeat sequence from the third exon of Os-CASTOR. We selected two independent transgenic RNAi knockdown lines, designated Os-CASTORi-1 and Os-CASTORi-2, for further analysis. qRT-PCR analysis revealed a 58% to 72% reduction of Os-CASTOR mRNA levels in the transgenic T1 plants. Remarkably, symbiotic development in the transgenic RNAi roots was significantly impaired. While all wild-type plants segregated from the T1 progeny were normally colonized by the AM fungus, less than 2% of the root segments of the transgenic plants contained arbuscules (two of 163 from 21 Os-CASTORi-1 plants and one of 169 from 13 OsCASTORi-2 plants), despite the frequent presence of extraradical hyphae and appressoria on the root epidermis (data not shown). Again, the observed defective phenotypes were further supported by the differential expression of Os-PT11 between RNAi and wild-type plants (Fig. 5B). Taken together, cytological and molecular evidence strongly indicates that Os-CASTOR and Os-POLLUX are required for the establishment of AM symbiosis in rice.
Os-POLLUX Can Restore Nodulation, But Not Rhizobial Infection, to a M. truncatula dmi1 Mutant
To determine whether the nonlegume CASTOR/POLLUX orthologs possess an equivalent function to their legume counterparts, we introduced two Os-POLLUX full-length cDNAs (AK067564 and AK073102), under the control of the 35S promoter, into the M. truncatula dmi1-1 mutant (allele C71; Catoira et al., 2000; Ane et al., 2004) using Agrobacterium rhizogenes-mediated hairy root transformation (Boisson-Dernier et al., 2001). AK067564 and AK073162, differing at their 3′ ends due to alternative splicing, encode predicted proteins of 943 and 965 amino acids, respectively, of which the 943-amino acid version is canonical based on sequence alignments with other POLLUX orthologs. Nevertheless, the first 941 amino acids are shared between the two isoforms. It is noteworthy that both cDNA clones provided by the Rice Genome Resource Center in Japan contain a point mutation likely resulting from the RT process: there was an insertion of T at position 2,136 of AK067564 and an A-to-T transversion at position 593 of AK073012. Both errors were corrected for the transformation experiments described below. We did not observe any obvious differences resulting from using the two constructs, suggesting that the two protein isoforms are equally functional.
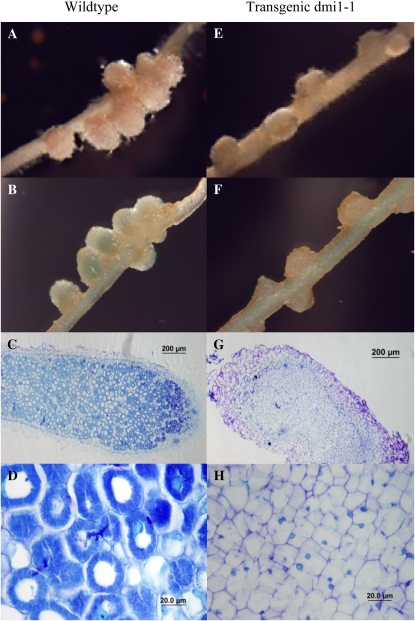
Upon inoculation with Sinorhizobium meliloti, the M. truncatula dmi1-1 mutant fails to exhibit root hair curling, infection thread formation, cortical cell division, and nodule development (Catoira et al., 2000). To determine whether Os-POLLUX was able to complement the nonnodulation phenotype of the dmi1-1 mutant, we inoculated the composite transgenic plants with S. meliloti strain 2011 carrying the lacZ reporter gene in plasmid pXLGD4 (Catoira et al., 2000). Two weeks after inoculation, nodule-like organs were observed on roots of 36 of 59 dmi1-1 plants transformed with Os-POLLUX. However, unlike wild-type plants, which form long, pink, cylindrical nodules, the transgenic mutant roots produce small, white, round nodules that are similar to the phenotypes of the nitrogen fixation mutants of M. truncatula (Starker et al., 2006). GUS staining and microscopy of sectioned nodules revealed that the small white nodules were devoid of bacteria and no infection threads were observed; thus, these nodules were nonfunctional (Fig. 6). The lack of nitrogen-fixing ability of these white nodules also was reflected by the poor growth and chlorotic leaves of the composite transgenic plants under nitrogen-starving conditions. Although Os-POLLUX is required for AM symbiosis in rice, the 35S:Os-POLLUX failed to complement the M. truncatula dmi1-1 mutant for AM symbiosis (data not shown).
Figure 6.
Complementation of the M. truncatula dmi1-1 mutant (C71) with Os-POLLUX using A. rhizogenes-mediated hairy root transformation (Boisson-Dernier et al., 2001). A full-length cDNA clone of Os-POLLUX (AK067564) was cloned into a binary vector modified from pHellsgate8 driven by the 35S promoter (Helliwell et al., 2002). The binary vector was introduced into the A. rhizogenes strain, ARqua1, and transformed into the roots of the dmi1-1 mutant. A to D, Wild type. E to H, The dmi1-1 mutant transformed with 35S:Os-POLLUX. A and E, Nodule phenotypes of wild-type and transgenic dmi1-1 roots, respectively. B and F, GUS staining of nodules shown in A and E, respectively. C and G, Toluidine blue staining of sectioned nodules from wild-type and transgenic roots, respectively. D and H, Close-ups of the sectioned nodules.
Our observations are similar to those recently reported for the L. japonicus pollux-3 mutant complemented by 35S:Os-POLLUX (Banba et al., 2008). It was also reported that Os-DMI2 (or Os-SYMRK), a “reduced-length” version of the Mt-DMI2/Lj-SYMRK ortholog, restored nodule organogenesis of an Lj-sym10 mutant but failed to support the formation of infection threads, despite the fact that the “full-length” versions from several other nonlegume orthologs can complement both processes (Markmann et al., 2008). These observations are consistent with the finding that the nodule organogenesis and bacterial infection can be uncoupled (Gleason et al., 2006; Tirichine et al., 2006, 2007; Murray et al., 2007). Taken together, these data suggest that (1) in addition to early steps of nodule initiation, the common symbiosis genes also are involved in the control of the infection process and (2) the infection process in nodule development appears to be under more stringent genetic control than the nodule organogenesis. It has been shown that knockdown mutants of the DMI2 orthologs maintained the ability to form nodules but failed to form symbiosomes in the legume nodules (Capoen et al., 2005; Limpens et al., 2005). This taxonomy-specific functionality could be due to adaptation through changes in gene regulation and expression or sequence diversification between leguminous and nonleguminous plants.
CONCLUSION
Root symbioses with AM fungi and nitrogen-fixing bacteria share common signaling components, suggesting that the nitrogen-fixing root nodule symbioses have evolved from the ancient AM symbiosis (Kistner and Parniske, 2002). This hypothesis is further supported by the fact that all of the legume common symbiosis genes are present in nonlegumes that have the ability to establish the AM symbiosis (Zhu et al., 2006). However, the function of these nonlegume orthologs needs to be addressed, in order to gain insight into the evolution of the root symbioses in plants. Thus far, five of the seven known common symbiosis genes have been shown to be required for AM symbiosis in nonlegumes (Chen et al., 2007, 2008; Banba et al., 2008; Gherbi et al., 2008; Markmann et al., 2008). We speculate that all common symbiosis genes in legumes would be required for the AM symbiosis in nonlegumes. Interestingly, those genes required only for rhizobial symbiosis but not essential for the AM symbiosis in legumes are also present in nonlegumes. Elucidation of the function of nonlegume orthologs of nodulation-specific genes will provide further insights into the evolution of root symbioses in land plants.
MATERIALS AND METHODS
Rice and Medicago truncatula Mutants
The rice (Oryza sativa) Tos17 insertion line (NC6423) in the Nipponbare background was provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences in Japan. The rice T-DNA insertion mutants, C04353 and B02432, in the genetic backgrounds of Hawyoung and Dongjin, respectively, were provided by the Pohang University of Science and Technology in Korea. The Medicago truncatula dmi1-1 mutant (C71) was obtained from Dr. Douglas Cook's laboratory at the University of California, Davis.
Isolation of Homozygous Os-castor and Os-pollux Mutant Lines
Seeds of the Tos17/T-DNA insertion lines from the providers were from the progeny of primary transgenic or tissue culture-derived plants. To isolate homozygous mutants, we carried out two rounds of PCR analyses. The first round of PCR was to identify plants with Tos17/T-DNA insertion using the Tos17/T-DNA-specific and Os-CASTOR/Os-POLLUX-specific primer pairs. The second round of PCR was conducted to identify homozygous mutant plants using the primer pairs flanking the putative Tos17/T-DNA insertion sites. The positions of these primers are indicated in Figure 2A. Primer sequences are as follow: F1, 5′-CGATGGTCAGGGATGGTATC-3′; R1, 5′-CGTGTGGCTTTGCTCTATGA-3′; F2, 5′-CGATTTGATCTCTCCCCGTA-3′; R2, 5′-GCTGACAACATAAAGCGCAA-3′; T1, 5′-ACGCTGAACTTGTGGCCGTT-3′; T2, 5′-CCACAGTTTTCGCGATCCAGACTG-3′; T3, 5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′.
Binary Vector Construction and Rice Transformation
For the generation of RNAi knockdown lines of Os-CASTOR, a 400-bp inverted-repeat sequence from the third exon of Os-CASTOR was cloned into the Gateway-enabled pSTARGATE vector (provided by CSIRO Plant Industry in Australia). The expression of the inverted-repeat sequence in pSTARGATE was driven by a ubiquitin promoter. The construct was introduced into Agrobacterium tumefaciens strain EHA105 and transformed to Nipponbare as described by Hiei et al. (1994). Primary transgenic plants (T0) were selfed to obtain the T1 generation for phenotypic analysis of AM symbiosis.
Hairy Root Transformation of M. truncatula
The dmi1-1 mutant of M. truncatula was transformed with Os-POLLUX using Agrobacterium rhizogenes-mediated hairy root transformation (Boisson-Dernier et al., 2001). Two full-length cDNA clones of Os-POLLUX (AK067564 and AK073162) were cloned into a binary vector modified from pHellsgate8 driven by the 35S promoter (Helliwell et al., 2002). The binary vector was introduced into the A. rhizogenes strain ARqua1 and transformed into the roots of the dmi1-1 mutant. Transformed roots were selected on Färhaeus medium (Färhaeus, 1957) containing 20 mg L−1 kanamycin for 2 weeks at 20°C. The transgenic roots were further confirmed by PCR analysis.
Inoculation of Rice Roots with Glomus intraradices
The AM fungus Glomus intraradices was from Premier Tech Biotechnologies. The inoculation method was as described by Chen et al. (2007). Briefly, the rice plants were grown in 11-cm pots with sterilized Turface covered with 3 cm of sand in a growth chamber with a 13-h-light, 28°C/11-h-dark, 24°C regime. The plants were fertilized twice weekly with half-strength Hoagland solution (Arnon and Hoagland, 1940) supplemented with 100 μm KH2PO4. Roots of 2-week-old rice plants were inoculated by adding 1,000 spores to the sand at 1.5 cm depth. Roots were harvested at 5 weeks after inoculation. A random sample of the root tissues was used for phenotyping analysis, and the remaining tissues were used for RNA isolation.
Mycorrhizal colonization was phenotyped by means of trypan blue staining according to the protocol described by Koske and Gemma (1989). The cleaned roots were first fixed in 50% (v/v) ethanol. The fixed roots were then incubated at 90°C in 10% KOH for approximately 20 min. After rinsing with water, the roots were soaked in 1% HCl at room temperature overnight. The roots were then stained at 90°C for 30 min in an acidic glycerol solution containing 0.1% trypan blue. After destaining in acidic glycerol, the roots were examined using a light microscope (Olympus BX40F-3), and images were captured by a microscope digital camera system (Olympus DP71).
Inoculation of A. rhizogenes-Transformed M. truncatula Roots with Sinorhizobium meliloti
The nodulation assay was conducted as described by Limpens et al. (2003b). Three weeks after transformation, composite plants were transferred to sterile Turface saturated with Färhaeus medium [without Ca(NO3)2] for 3 d at 21°C and 16/8 h of light/dark. Each plant was then inoculated with 1 mL of culture (optical density at 600 nm = 0.1) of Sinorhizobium meliloti strain 2011 carrying the lacZ reporter gene in plasmid pXLGD4 (Catoira et al., 2000). The nodulation was scored 2 weeks after inoculation.
Nodule Sectioning, Staining, and Microscopy
Nodules were stained with X-gal to detect the presence of bacteria. For nodule sectioning, nodules were cut in half longitudinally, placed in FAA solution (100 mL: 45 mL of 95% ethanol, 40 mL of distilled water, 5 mL of glacial acetic acid, and 10 mL of 37% [w/w] formaldehyde) and vacuum infiltrated until they sank. The FAA fixation was followed by several steps of ethanol dehydration (50%, 60%, 70%, 80%, 95%, and two changes of 100% ethanol each for 30 min). The samples were then gradually infiltrated with Hemo-De (20%, 50%, and 75% Hemo-De solutions, each for 30 min, then two changes of 100% Hemo-De each for 1 h). Once hydrated with Hemo-De, the samples were infiltrated with Paraplast Plus by successively adding chips of Paraplast to Hemo-De at 42°C. After removing the Paraplast/Hemo-De solution, melted Paraplast was added and incubated at 60°C for at least 8 h. (This step was repeated for at least six changes of Paraplast.) The samples were then embedded and sectioned. The samples were sectioned with a Leica RM2135 microtome.
For light microscopy, 5-μm-thick sections were dried onto glass slides. The slides went through stepwise de-Paraplast and hydration and were stained with 1% (w/v) toluidine blue in 95% ethanol. Photographs were taken with an Olympus BX40F-3 light microscope, and images were captured by an Olympus DP71 microscope digital camera system.
Analysis of Gene Expression
Total RNA was isolated by the Qiagen Plant RNeasy kit. Two micrograms of RNA was used to perform RT reactions using M-MLV reverse transcriptase (Invitrogen) in a 20-μL reaction mixture. Two microliters of the RT reaction was used as a template in a 20-μL PCR solution. The PCR primers were as follows: Os-Actin, 5′-GCGATAATGGAACTGGTATG-3′ and 5′-CTCCATTTCCTGGTCATAGTC-3′; Os-CASTOR, 5′-CGATGGTCAGGGATGGTATC-3′ and 5′-CGTGTGGCTTTGCTCTATGA-3′; Os-POLLUX, 5′-CGATTTGATCTCTCCCCGTA-3′ and 5′-GCTGACAACATAAAGCGCAA-3′; Os-PT11, 5′-ATGGCTCGACGGACAGTAAG-3′ and 5′-GATCAGCTGGATCATGTACCT-3′. qRT-PCR was performed on the Applied Biosystems StepOne Real-Time PCR System using the SYBR Green I detection kit (Bio-Rad). The Os-ubiquitin gene was selected as a constitutive internal control. PCR primers used for the real-time PCR experiments were as follows: Os-ubiquitin, 5′-TGCACCCTAGGGCTGTCAAC-3′ and 5′-TGACGCTCTAGTTCTTGATCTTCTTC-3′; Os-CASTOR, 5′-CAAGAGGGTGATGAGGTGCTAGTA-3′ and 5′-GGTAACCTCTCATAACCTTGGGTAAT-3′; Os-POLLUX, 5′-CCTCGGATGGAGCGACAA-3′ and 5′-ACGACACCACCACCAATACTCTT-3′.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of the CASTOR and POLLUX homologs.
Supplemental Sequence Data S1. The amino acid sequences of the CASTOR and POLLUX homologs.
Supplementary Material
Acknowledgments
We thank Dr. Douglas Cook for providing seeds of the M. truncatula mutants, the Rice Genome Resource Center for providing the rice Tos17 mutant lines and full-length cDNA clones, and the Pohang University of Science and Technology for providing the rice T-DNA insertion mutants. We also thank Dr. Sharyn Perry for help with nodule sectioning.
This work was supported by the Kentucky Science and Engineering Foundation (grant to H.Z.) and by the U.S. National Science Foundation (grant no. IOS 0640197 to H.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hongyan Zhu (hzhu4@uky.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akiyama K, Hayashi H (2006) Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann Bot (Lond) 97 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Geurts R, Bisseling T (1999) Legume nodulation and mycorrhizae formation: two extremes in host specificity meet. EMBO J 18 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ane JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367 [DOI] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR (1940) Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci 50 463–483 [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Gherardi M, Huguet T, et al (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji B, Ba AM, Larue TA, Tepfer D, Piche Y (1994) Pisum sativum mutants insensitive to nodulation are also insensitive to invasion in vitro by the mycorrhizal fungus, Gigaspora margarita. Plant Sci 102 195–203 [Google Scholar]
- Banba M, Gutjahr C, Miyao A, Hirochika H, Paszkowski U, Kouchi H, Imaizumi-Anraku H (2008) Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol 49 1659–1671 [DOI] [PubMed] [Google Scholar]
- Besserer A, Puech-Pages V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Becard G, Sejalon-Delmas N (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4 e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14 695–700 [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A, Faccio A, Martini I, Schauser L, Stougaard J, Webb J, Parniske M (2000) The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Mol Plant Microbe Interact 13 1109–1120 [DOI] [PubMed] [Google Scholar]
- Buee M, Rossignol M, Jauneau A, Ranjeva R, Becard G (2000) The pre-symbiotic growth of arbuscular mycorrhizal fungi is induced by a branching factor partially purified from plant root exudates. Mol Plant Microbe Interact 13 693–698 [DOI] [PubMed] [Google Scholar]
- Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M (2005) SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci USA 102 10369–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ané JM, Zhu H (2008) OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol 180 311–315 [DOI] [PubMed] [Google Scholar]
- Chen C, Gao M, Liu J, Zhu H (2007) Fungal symbiosis in rice requires an ortholog of a legume common symbiosis gene encoding a Ca2+/calmodulin-dependent protein kinase. Plant Physiol 145 1619–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Heckmann AB, Yousafzai F, Duc G, Downie JA (2007) Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol Plant Microbe Interact 20 1183–1191 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966 [DOI] [PubMed] [Google Scholar]
- Färhaeus G (1957) The infection of white clover root hairs by nodule bacteria studied by a simple slide technique. J Gen Microbiol 16 374–381 [DOI] [PubMed] [Google Scholar]
- Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Peret B, Laplaze L, Franche C, et al (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankiabacteria. Proc Natl Acad Sci USA 105 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V (1996) Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell 8 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Munoz A, Poovaiah BW, Oldroyd GE (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441 1149–1152 [DOI] [PubMed] [Google Scholar]
- Glenn MG, Chew FS, Williams PH (1985) Hyphal penetration of Brassica (Cruciferae) roots by a vesicular-arbuscular mycorrhizal fungus. New Phytol 99 463–472 [Google Scholar]
- Godfroy O, Debelle F, Timmers T, Rosenberg C (2006) A rice calcium- and calmodulin-dependent protein kinase restores nodulation to a legume mutant. Mol Plant Microbe Interact 19 495–501 [DOI] [PubMed] [Google Scholar]
- Guimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JM, Simon M (2005) Simple sequence repeats in proteins and their significance for network evolution. Gene 345 113–118 [DOI] [PubMed] [Google Scholar]
- Harrison MJ (1997) The arbuscular mycorrhizal symbiosis: an underground association. Trends Plant Sci 2 54–56 [Google Scholar]
- Harrison MJ (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59 19–42 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB (2001) Molecular evidence for the early colonization of land by fungi and plants. Science 293 1129–1133 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29 1217–1225 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45 123–132 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417 515–522 [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 17 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, et al (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Parniske M (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7 511–518 [DOI] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koske RE, Gemma JN (1989) A modified procedure for staining roots to detect VA mycorrhizas. Mycol Res 92 486–505 [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Denarie J, Barker DG, Becard G (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- LaRue TA, Weeden NF (1994) The symbiosis genes of the host. In GB Kiss, G Endre, eds, Proceedings of the 1st European Nitrogen Fixation Conference. Officina Press, Szeged, Hungary, pp 147–151
- Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003. a) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2003. b) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55 983–992 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Maldonado-Mendoza I, Lopez-Meyer M, Cheung F, Town CD, Harrison MJ (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50 529–544 [DOI] [PubMed] [Google Scholar]
- Long SR (1996) Rhizobium symbiosis: nod factors in perspective. Plant Cell 8 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Markmann K, Giczey G, Parniske M (2008) Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol 6: e68 [DOI] [PMC free article] [PubMed]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rouge P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20 912–921 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kalo P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, Itoh J, Maekawa M, Murata K, Yatou O, Nagato Y, Hirochika H (2007) A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol 63 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104 [DOI] [PubMed] [Google Scholar]
- Navazio L, Moscatiello R, Genre A, Novero M, Baldan B, Bonfante P, Mariani P (2007) A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol 144 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo JA, Martín J, Hayman DS (1980) Mycorrhizal development in host and non-host plants. I. Mycorrhizal infection in plants grown together. New Phytol 84 27–35 [Google Scholar]
- Oldroyd GE, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59 519–546 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K, Bisseling T (1996) Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell 8 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo MJ, Azcon-Aguilar C (2007) Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 10 393–398 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Redecker D, Kodner R, Graham LE (2000) Glomalean fungi from the Ordovician. Science 289 1920–1921 [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lozano JM (2003) Arbuscular mycorrhizal symbiosis and alleviation of osmotic stress: new perspectives for molecular studies. Mycorrhiza 13 309–317 [DOI] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, et al (2007) NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Senoo K, Solaiman MZ, Kawaguchi M, Imaizumi-Anraku H, Akao S, Tanaka A, Obata H (2000) Isolation of two different phenotypes of mycorrhizal mutants in the model legume plant Lotus japonicus after EMS-treatment. Plant Cell Physiol 41 726–732 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (1997) Mycorrhizal Symbiosis. Academic Press, San Diego
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG (1995) Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA 92 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP (2000) Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol 54 257–288 [DOI] [PubMed] [Google Scholar]
- Stacey G, Libault M, Brechenmacher L, Wan J, May GD (2006) Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol 9 110–121 [DOI] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed]
- Zhu H, Choi HK, Cook DR, Shoemaker RC (2005) Bridging model and crop legumes through comparative genomics. Plant Physiol 137 1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Riely BK, Burns NJ, Ane JM (2006) Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.