Abstract
The mechanisms by which aniline exposure elicits splenotoxic response, especially the tumorigenic response, are not well-understood. Splenotoxicity of aniline is associated with iron overload and generation of reactive oxygen species (ROS) which can cause oxidative damage to DNA, proteins and lipids (oxidative stress). 8-Hydroxy-2’-deoxyguanosine (8-OHdG) is one of the most abundant oxidative DNA lesions resulting from ROS, and 8-oxoguanine glycosylase 1 (OGG1), a specific DNA glycosylase/lyase enzyme, plays a key role in the removal of 8-OHdG adducts. This study focused on examining DNA damage (8-OHdG) and repair (OGG1) in the spleen in an experimental condition preceding a tumorigenic response. To achieve that, male Sprague-Dawley rats were subchronically exposed to aniline (0.5 mmol/kg/day via drinking water for 30 days), while controls received drinking water only. Aniline treatment led to a significant increase in splenic oxidative DNA damage, manifested as a 2.8-fold increase in 8-OHdG levels. DNA repair activity, measured as OGG1 base excision repair (BER) activity, increased by ~1.3 fold in the nuclear protein extracts (NE) and ~1.2 fold in the mitochondrial protein extracts (ME) of spleens from aniline-treated rats as compared to the controls. Real-time PCR analysis for OGG1 mRNA expression in the spleen revealed a 2-fold increase in expression in aniline-treated rats than the controls. Likewise, OGG1 protein expression in the NEs of spleens from aniline-treated rats was ~1.5 fold higher, whereas in the MEs it was ~1.3 fold higher than the controls. Aniline treatment also led to stronger immunostaining for both 8-OHdG and OGG1 in the spleens, confined to the red pulp areas. It is thus evident from our studies that aniline-induced oxidative stress is associated with increased oxidative DNA damage. The BER pathway was also activated, but not enough to prevent the accumulation of oxidative DNA damage (8-OHdG). Accumulation of mutagenic oxidative DNA lesions in the spleen following exposure to aniline could play a critical role in the tumorigenic process.
Keywords: Aniline, Spleen, Reactive oxygen species, DNA damage, 8-OHdG, OGG1, Base excision repair, Immunochemical localization
Introduction
Aniline, an aromatic amine, is extensively used in chemical and drug industries. Exposure to aniline leads to toxic responses in the spleen, which are characterized by splenomegaly, increased erythropoietic activity, hyperpigmentation, hyperplasia, fibrosis, and a variety of primary sarcomas of the spleen after chronic exposure in rats (Goodman et al., 1984; Weinberger et al., 1985; Bus and Popp, 1987; Khan et al., 1993, 1997, 1999a, 2003, 2006; Pauluhn, 2004). However, the molecular mechanisms by which aniline exerts its toxic effects in the spleen, especially the formation of various types of sarcomas and/or tumorigenesis are not known. Earlier studies have shown that aniline exposure is associated with oxidative stress in the spleen, as evident from the observed increases in lipid peroxidation and protein oxidation in that organ (Khan et al., 1997, 1999a, 2003). An important and well-validated biomarker of oxidative DNA damage is the DNA guanine base oxidation product 8-hydroxy-2’-deoxyguanosine (8-OHdG). This DNA lesion is highly mutagenic causing GC to TA transversions (Thomas et al., 1997) and has been commonly quantified as a steady-state estimate of oxidative stress in tissues. Oxidative DNA damage is one of the most common threats to genomic stability, and is also proposed to play an important role in numerous pathological conditions, including cancer (Kondo, et al., 2000; Caporaso, 2003; Cooke et al., 2003; Gackowski et al., 2005).
The DNA base excision repair (BER) pathway represents a critical step in the maintenance of genome stability. This pathway plays an important role in prevention of disease through the removal of oxidized bases. The 8-OHdG lesion, which represents oxidative DNA damage, is removed from DNA by the BER enzyme 8-oxoguanine glycosylase (OGG1). It has been hypothesized that OGG1 deficiency could be associated with a mutator phenotype and could participate in carcinogenesis (Radicella et al., 1997; Thomas et al., 1997; Blons et al., 1999; Dherin et al., 1999; Lan et al., 2004). Accumulation of 8-OHdG and/or perturbation of OGG1 activity were also reported to be a predictive marker for susceptibility to cancer (Gackowski et al., 2003).
Our previous short-term exposure study showed increased oxidative DNA damage in the spleen of rats following aniline exposure (Wu et al., 2005), which could potentially lead to mutagenic and/or carcinogenic response in the spleen. However, evidence supporting the consequences of increased free radical attack on DNA in the spleen following aniline exposure is still very limited, and the status and extent of splenic DNA damage following a subchronic exposure to aniline are not known. More importantly, no information is available on the status of DNA repair activity (OGG1) in the spleen following aniline insult.
The focus of this study was, therefore, to examine oxidative DNA damage, and its repair dynamics (BER) in both nuclear and mitochondrial protein extracts of rats exposed subchronically to aniline. Because aniline exposure could also affect OGG1 activity by altering its transcriptional and/or its translational activities, mRNA and protein expression of OGG1, and its immunohistochemical localization in the spleen of aniline-treated animals have also been investigated.
Materials and methods
Animal and treatment
Male Sprague-Dawley rats (~200 g), obtained from Harlan (Indianapolis, IN), were housed in wire-bottom cages over absorbent paper with free access to tap water and Purina rat chow. The animals were acclimatized in a controlled-environment animal room (temperature, 22 °C; relative humidity, 50%; photoperiod, 12-h light/dark cycle) for 7 days prior to treatment. The experiments were performed in accordance with the guidelines of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee of University of Texas Medical Branch. The animals were divided into two groups of six rats each. One group of animals was given 0.5 mmol/kg/day of aniline (~97%; Aldrich, Milwaukee, WI) via drinking water (pH of the solution adjusted to ~6.8) (Khan et al, 1993, 1999a, 2006, Wang et al, 2005), whereas the other group received drinking water only and served as controls. After 30 days, the rats were euthanized under nembutal (sodium pentoparbital) anesthesia and the spleens were removed immediately, blotted, weighted and stored at -80 °C until further analysis. A portion of spleen was snap-frozen in liquid nitrogen and stored at -80 °C for RNA isolation. Also, portions of the spleen from control and aniline-treated rats were fixed in 10% neutral buffered formalin for histological processing.
Extraction of genomic DNA and DNA digestion for 8-OHdG assay
Genomic DNA was extracted from spleen tissues by using a DNA extraction kit (Easy-DNA Kit, Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. The purity of DNA preparations was assessed by A260/A280 ratio. One hundred μl (~100 μg) of individual genomic DNA samples were digested with nuclease P1 (20 μg nuclease P1 dissolved in 20 mM sodium acetate buffer, pH 4.8) by incubating at 37 °C for 30 min followed by treatment with alkaline phosphatase (AP) (1.3 U or 100 μg AP in 1 M Tris-HCl, pH 7.4) and incubation at 37 °C for 1 hr (Wu et al., 2005).
Determination of 8-OHdG in DNA digests
8-OHdG in DNA digests was quantitated using an enzyme-linked immunosorbent assay (ELISA) kit (Tsai et al., 2001; Wu et al., 2005), essentially as described by the manufacturer (BIOXYTECH 8-OHdG-EIA kit, Oxis, Portland, OR). Briefly, 50 μl of the standard or DNA digests from control and aniline-treated rats (in duplicate) were added to ELISA plate wells precoated with 8-OHdG, followed by addition of 50 μl 8-OHdG specific antibody (monoclonal) and incubated at 37 °C for 1 hr with gentle mixing. After incubation, the plate was washed thoroughly with the wash buffer and 100 μl of the secondary antibody (goat IgG conjugated to horseradish peroxidase) was added and incubated for an additional hr at 37 °C. After washing with wash buffer, 100 μl chromogen substrate (3’,3’,5’,5’- tetramethylbenzidine) was added to each well. The plate was then incubated at room temperature in the dark for 15 min with continuous shaking followed by addition of 100 μl stop solution (1 M H3PO4) to terminate the reaction. The absorbance was measured at 450 nm after 3 min.
Preparation of nuclear extracts (NEs) for OGG1 activity and Western blotting
The nuclear protein extracts (NEs) were prepared according to the method of Ma et al. (Ma et al., 2004), with minor modifications. Spleen tissues (from control and aniline-treated rats) were cut into smaller pieces, homogenized briefly with a loose glass pestle in cold hypotonic buffer [10 mM HEPES-KOH, 10 mM KCl, 100 μM EDTA, 100 μM EGTA, 1 mM DTT, 0.5 mM PMSF, 2 μg/ml pepstatin, and a complete protease inhibitor cocktail (Roche, Germany)], and incubated on ice for 20 min. Tissues were then homogenized with a tight pestle and centrifuged at 800 g for 4 min to obtain nuclear pellets. Pellets were gently washed two times with homogenizing buffer. Nuclear proteins were extracted in a high salt buffer (20 mM HEPES-KOH, 410 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 2 μg/ml pepstatin, and protease inhibitor cocktail) by incubating for 45 min on ice with reverse mixing at intervals of 10 min. The NEs were cleared by centrifugation (16,000 g, 10 min) and adjusted to 15% with glycerol and stored at -80 °C until further analysis.
Preparation of mitochondrial extracts (MEs) for OGG1 and Western blotting
The mitochondrial protein extracts (MEs) were prepared according to the method of Souza-Pinto et al. (Souza-Pinto et al., 2001), with minor modifications. Spleen tissues (from control and aniline-treated rats) were cut into smaller pieces, homogenized briefly with a loose glass pestle in cold 1X MSHE buffer (10 mM HEPES-KOH buffer, pH 7.4, 210 mM mannitol, 70 mM sucrose, 150 μM spermine, 100 μM EGTA, 750 μM spermidine, 5 mM DTT, 1 mM EDTA and protease inhibitor). The homogenate was then centrifuged at 500 g for 8 min to remove nuclear pellets. The supernatant were then centrifuged at 10,000 g for 8 min to obtain crude mitochondrial pellets. Pellets were gently resuspended in 1X MSHE buffer and centrifuged on 50% percoll gradient at 50,000 g for 60 min at 4 °C. The mitochondrial pellets were gently washed once with homogenizing buffer and mitochondrial proteins were extracted in a high salt buffer (20 mM HEPES-KOH, pH 7.4, 305 mM KCl, 5 mM DTT, 1 mM EDTA, 0.1% Triton X-100, and 5% glycerol) by incubating for 30 min on ice. MEs were cleared by centrifugation (15,000 g, 20 min) and adjusted to 15% with glycerol and stored at -80 °C until further analysis. Protein concentration was determined using Bio-Rad detergent compatible (DC) protein assay reagents (Bio-Rad Laboratories, Inc., Hercules, CA).
Oligonucleotide 5’ end-labeling for BER assay
Oligonucleotide containing a single 8-oxoG lesion (sequence shown below) was obtained from Trevigen (Gaithersburg, MD). Oligonucleotides (5.5 pmol) were end-labeled using 5 μCi of [γ-32P]ATP (3000 Ci/mmol, Perkin-Elmer Life & Analytical Sciences, Boston, MA) and T4 polynucleotide kinase (New England BioLabs, Ipswich, MA) and then passed through a G-25 spin column (GE Healthcare, Piscataway, NJ), for purifying the radiolabeled oligonucleotide, and annealed with 1.5-2 fold complementary oligonucleotide by gradual cooling to room temperature.
8-OxoG (O): C Oligonucleotide substrate sequence
*5’-GAA CTA GTG OAT CCC CCG GGC TGC-3’
3’-CTT GAT CAC CTA GGG GGC CCG ACG-5’
OGG1 activity assay
Glycosylase activities were measured in vitro in the NEs or MEs with synthetic end-labeled double-stranded oligonucleotide substrates containing 8-oxoG:C adducts (targeted by OGG1). Glycosylase assays were done as described earlier (Hill et al., 2001; Englander and Ma, 2006) with slight modifications. Briefly, the reactions were done in 20 μl with 40 μg NEs or MEs, 1 nM end-labeled substrate, and reaction buffer (10 mM HEPES-KOH, pH 7.6, 50 mM KCl, 1.0 mM DTT, 1.5% glycerol, 2.25 mM EDTA) and final NaCl concentration adjusted to 90 mM. Incubation was done at 37 °C for indicated times and was terminated with with 5X alkaline loading buffer (0.5M NaOH, 97% formamide, 10 mM EDTA, pH 8, 0.025% bromophenol blue, 0.025% xylene cyanol) and heated at 95 °C for 4 min. Positive control reactions were assembled using recombinant human OGG1 enzyme from Trevigen. Reaction mixtures were resolved on 15% polyacrylamide-7 M urea gels in Tris-borate buffer (89 mM Tis-HCl, 89 mM H3BO3, 2 mM EDTA, pH ~8.3) at 16 mA for 150 min, and products were visualized by autoradiography and quantified on Phosphorimager (GE Healthcare). Phosphorimager values, representing percent of substrate cleavage within the linear range of each reaction, were converted into cleavage-product amounts. Values from 6 individual rats and 3 cleavage assays per extract were averaged and plotted as means ± SD.
RNA isolation and real-time PCR
RNA isolation
Total RNA was isolated from spleen tissues using RiboPure kit (Ambion, Austin, TX) as per manufacturer’s instructions. To eliminate contaminating genomic DNA, RNA preparation was treated with RNase free DNase I (DNA-free kit, Ambion, Austin, TX). The total RNA concentration was determined by measuring the absorbance at 260 nm. RNA integrity was verified electrophoretically by ethidium bromide staining and by measuring A260/A280 ratio.
Real-time PCR: SYBR Green detection and Data analysis
The real-time PCR was performed essentially as described earlier (Wang et al., 2005, 2008). Briefly, first-strand cDNA was prepared from isolated RNA by using SuperScript III First-Strand Synthesis Kit (Invitrogen, Carisbad, CA) described earlier (Wang et al., 2005). Quantitative real-time PCR employing a two-step cycling protocol (denaturation and annealing/extension) was carried out using Smart Cycler System as per manufacturer’s instructions (Cepheid, Sunnyvale, CA). The sequence of the forward and the reverse primers of OGG1 for real-time PCR were 5’-CAA CAT TGC TCG CAT CAC TGG-3’ and 5’-ATG GCT TTA GCA CTG GCA CAT ACA-3’, respectively. For each cDNA sample, parallel reactions were performed in triplicate for the detection of 18 S and OGG1. The reaction samples in a final volume of 25 μl contained 2 μl of cDNA templates, 2 μl primer pair, 12.5 μl iQ SYBR Green Supermix and 8.5 μl water. Amplification conditions were identical for all reactions: 95 °C for 2 min for template denaturation and hot start prior to PCR cycling. A typical cycling protocol consisted of three stages: 5 s at 95 °C for denaturation, 30 s at 65 °C for annealing, 30 s at 72 °C for extension, and an additional 6 s hold for fluorescent signal acquisition. To avoid the non-specific signal from primer-dimers, the fluorescence signal was detected 2 °C below the melting temperature (Tm) of individual amplicon and above the Tm of the primer-dimers (Simpson et al., 2000; Rajeevan et al., 2001). A total of 45 cycles were performed for the studies.
Quantitation of PCR was done using the comparative CT method as described in User Bulletin No. 2 of Applied Biosystems (Foster City, CA), and reported as fold difference relative to the calibrator cDNA (QuantumRNA Universal 18 S Standards, Ambion). The fold change in OGG1 cDNA (target gene) relative to the 18 S endogenous control was determined by:
Western blotting for OGG1 in NEs and MEs
Protein extracts (60 μg NEs and 100 μg MEs, respectively) were denatured by heating at 95 °C for 5 min and separated by 12% sodium dodecyl sulfate-polyacylamide gel electrophoresis (SDS-PAGE, Invitrogen, Carlsbad, CA). The separated proteins were transferred onto polyvinylidene difluoride (PVDF) microporous membrane (Millipore Corporation, Billerica, MA) using a transfer buffer (25 mM Tris-HCl, 190 mM glycine, pH 8.4, 10% methanol). The membrane was then blocked with 10% non-fat dry milk and incubated overnight at 4 °C with the rabbit anti-OGG1 antibody (Novus Biologicals, Littleton, CO) at the final concentration of 1.8 μg/ml (1:600 dilution). The membrane was washed three times with PBS-Tween 20 buffer. After incubation with the secondary antibody (anti-rabbit IgG-HRP), the membranes were washed and developed using a chemiluminescence detection kit (ECL, GE Healthcare). As a control, after stripping each membrane with stripping buffer (Boston BioProducts, Worcester, MA), the membrane was reprobed with anti-actin antibody (Sigma, Saint Louis, MO) and developed as above.
Localization of 8-OHdG and OGG1
Paraffin sections were cut and deparaffinized in an oven at 55 °C for 1 hr and treated with xylene and various concentrations of ethanol, and finally rehydrated with water (Khan et al., 2002). The slides were incubated with sodium citrate buffer at 95 °C for 20 min for antigen retrieval and subsequently incubated with various reagents (0.3% H2O2, 10% serum) for blocking the non-specific binding sites, which included quenching of endogenous peroxidase activity with 3% H2O2 in water for 15 min, 5% normal serum (Sigma, St. Louis, MO) for 30 min, and avidin and biotin block solutions (Vector Laboratories, Burlingame, CA) for 15 min each. The sections were then incubated with primary antibodies [anti-8-OHdG (monoclonal, Oxis Int’l Inc., Foster, CA) and anti-OGG1 antibodies, 1:40 and 1:60, respectively] overnight at 4 °C. Immunoreactivity was detected by the ABC Method (Vectastain Elite ABC Kit, Vector Laboratories, Burlingame, CA) with color development using 3,3’-diaminobenzidine (DAB). Mayer’s Hematoxylin was then added as a counterstain for 1 min. The negative controls were immunostained as above, but with goat or horse serum instead of the anti-8-OHdG or OGG1 antibody, respectively. The histological evaluations for staining were done under OLYMPUS BX51 Microscope (Leads Instruments, Inc., Irving, TX).
Statistical Analysis
All data are expressed as means ± SD of six animals in each group. Comparison between the groups was made by P value determination using student’s two-tailed t-test (GraphPad InStat 3 software, La Jolla, CA). A P value of <0.05 was considered to be statistically significant.
Results
Effect of aniline exposure on oxidative DNA damage (8-OHdG formation)
In this study, we used ELISA for determining 8-OHdG because it is highly sensitive, simple, less time-consuming and more economical than other methods (Wu et al., 2005). The 8-OHdG levels in the splenic DNA (with/without aniline treatment) are shown in Fig. 1. The mean 8-OHdG level in the control spleens was 2.69 ± 0.38 ng/mg DNA, whereas in the aniline-treated spleens it was 7.62 ± 0.55 ng/mg DNA. Thus, aniline treatment was associated with a 2.8-fold (p<0.05) increase in 8-OHdG levels in the spleen (Fig. 1).
Fig.1.
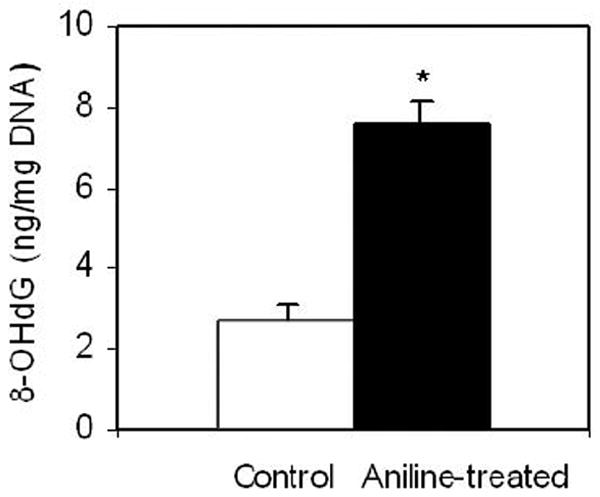
8-OHdG levels in the spleens of control and aniline-treated rats. DNA was extracted from the spleens of control and aniline-treated rats, and 8-OHdG in the DNA digests was quantitated using an ELISA kit. Values are means ± SD of six rats in each group. * p < 0.05 in comparison to controls.
Effect of aniline exposure on DNA repair activity (BER) of the spleen
In mammalian cells, oxidative DNA damage is mostly repaired by the BER pathway. To assess the extent to which BER might be affected by oxidative stress induced by subchronic exposure to aniline, we measured BER activities of NEs and MEs (Figs. 2 and 3, respectively). The assays were conducted with end-labeled oligonucleotides (*8-oxoG:C) which are targeted by OGG1. The cleavage activities were calculated from the cleavage yields generated in the linear range of each reaction. BER activities of NEs and MEs were determined by quantifying radioactivity generated over time in cleavage products of end-labeled oligonucleotides. The pattern of excision activities in NEs (Fig. 2) at 15, 30, 60, and 120 min was 0.33, 0.32, 0.27, and 0.20 fmol/μg protein/h, respectively, for control animals (mean ± SD:0.28 ± 0.04), whereas for aniline-treated animals it was 0.44, 0.45, 0.36, and 0.24 fmol/μg protein/h, respectively (mean ± SD:0.37 ± 0.03). The pattern of excision activities in MEs (Fig. 3) at 22.5, 45, 90, and 180 min was 0.18, 0.15, 0.11, and 0.07 fmol/μg protein/h, respectively, for control animals (mean ± SD:0.13 ± 0.01), whereas for aniline-treated animals it was 0.20, 0.19, 0.14, and 0.09 fmol/μg protein/h, respectively (mean ± SD:0.15 ± 0.02). Our data, thus, show a 1.32-fold (p<0.05) and a 1.15-fold (p<0.05) increase in OGG1 BER activities in the splenic NEs and MEs of aniline-treated rats, suggesting increased capacity to excise oxidized guanines from 8-oxoG:C containing substrates, i.e., OGG1-like activity.
Fig. 2.
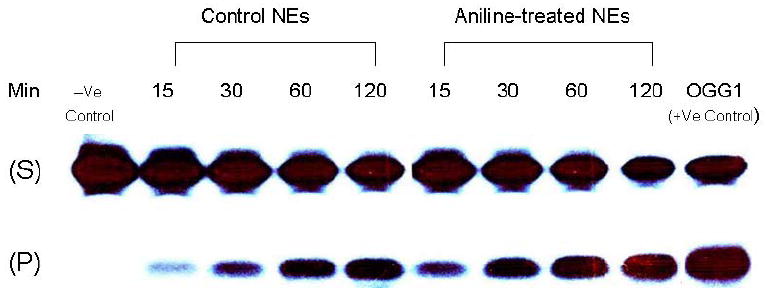
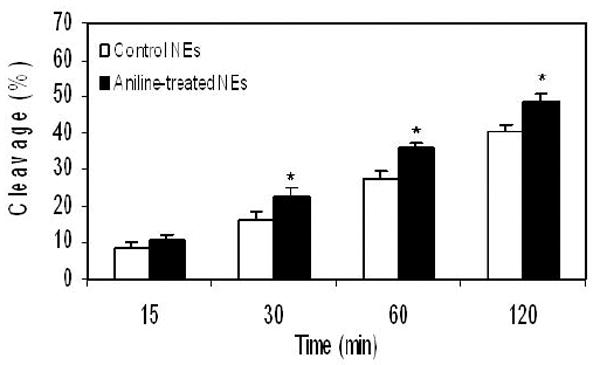
BER activity in the splenic NEs of control and aniline-treated rats. The assay was conducted with end-labeled oligonucleotides (8-oxoG:C) which are targeted by OGG1 (see Methods for details). (A) Autoradiogram of incision products (P) generated over time by cleavage of end-labeled double-stranded oligonucleotides carrying the *8-oxoG:C adducts (S). Negative control (without NEs) and positive control (recombinant OGG1) are included in external lanes. (B) Values from Phosphorimager quantitation of products, generated with NEs from 6 rats per group and 3 cleavage assays per extract, were averaged and plotted as means ± SD. * Indicates that the cleavage is significantly different from controls for each respective time point (p < 0.05).
Fig. 3.
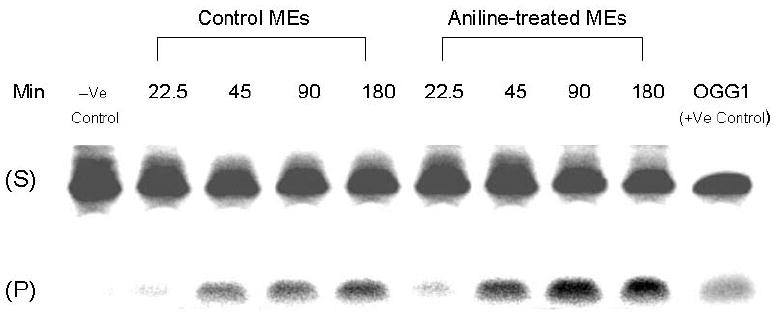
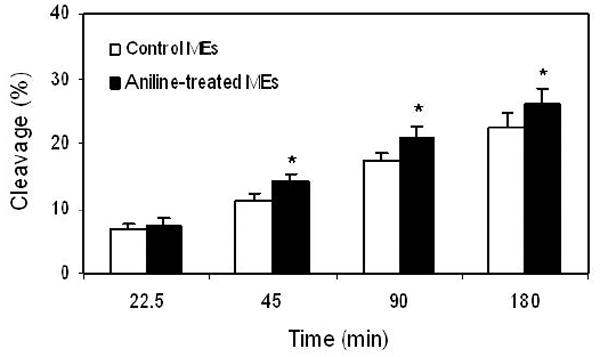
BER activity in the MEs of spleens from control and aniline-treated rats. The assay was conducted with end-labeled oligonucleotides (8-oxoG:C) which are targeted by OGG1 (see Methods for details). (A) Autoradiogram of incision products (P) generated over time by cleavage of end-labeled double-stranded oligonucleotides carrying the *8-oxoG:C adducts (S). Negative control (without MEs) and positive control (recombinant OGG1) are included in external lanes. (B) Values from Phosphorimager quantitation of products, generated with MEs from 6 rats per group and 3 cleavage assays per extract, were averaged and plotted as means ± SD. * Indicates that the cleavage is significantly different from controls for each respective time point (p < 0.05).
mRNA expression of OGG1
To determine the impact of aniline exposure on the expression of OGG1 in the spleen, OGG1 mRNA levels were analyzed by real-time PCR analysis and levels are shown in Fig. 4. As shown in the figure, aniline exposure resulted in a 2-fold increase in OGG1 mRNA expression compared to controls.
Fig. 4.
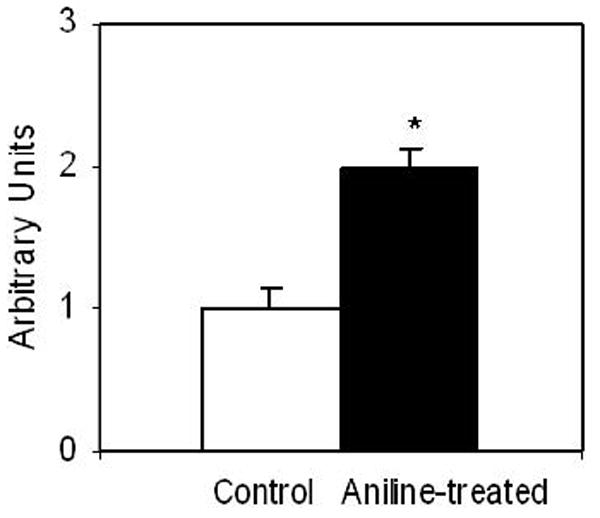
Real-time PCR analysis of OGG1 gene expression in the spleens of control and aniline-treated rats. Total RNA was extracted from spleen tissues, real-time PCR was performed, and the fold change in mRNA expression was determined. Values are means ± SD. *p< 0.05.
OGG1 protein expression in the NEs and MEs
To investigate whether gene expression levels are associated with actual protein levels, OGG1 protein expression in the spleens was also determined by Western blotting. As evident from Fig. 5, the OGG1 protein in the NEs and MEs of aniline-treated rats were 1.5- and 1.3-fold higher, respectively, than the controls (p<0.01).
Fig. 5.
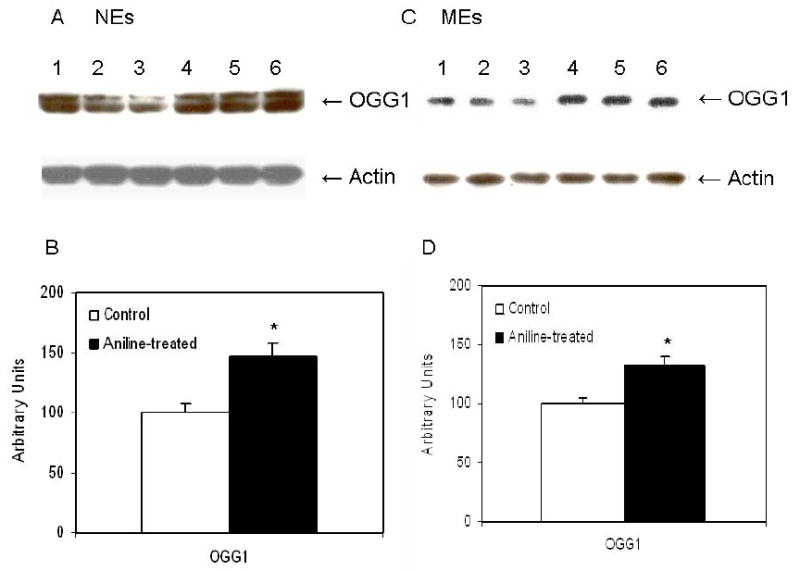
Western blot detection of OGG1 in the NEs (A) and MEs (C) from control and aniline-treated rats. Lanes 1-3: controls; lanes 4-6: aniline-treated. (B, D) Densitometric analyses of OGG1 bands from control and aniline-treated rats. The densitometric analysis of the protein bands was done using Eagle Eye II software. Data from aniline-treated spleen samples are presented in comparison to untreated controls, which were set at 100. Values are means ± SD of three determinations. *p< 0.05.
Immunohistochemical assessment of 8-OHdG and OGG1 in the spleen
Immunohistochemical studies for 8-OHdG and OGG1 were also conducted to demonstrate their expression and cellular localization in the spleen of experimental animals (Figs. 6 and 7). While control spleens showed sparse immunostaining for both 8-OHdG and OGG1, significantly increased immunostaining for both 8-OHdG and OGG1 was evident in the spleens of aniline-treated rats. The immunoreactivity for both 8-OHdG and OGG1 appeared predominantly in the red pulp areas of the spleen.
Fig. 6.
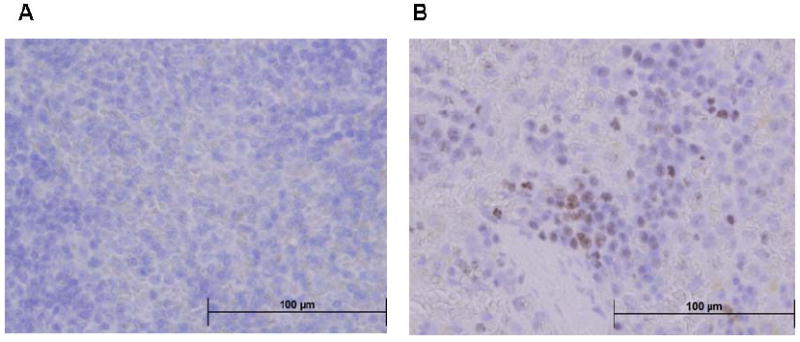
8-OHdG immunohistochemistry in the spleens of control and aniline-treated rats. A: control spleen; B: aniline-treated spleen. Controls showed sparse immunoreactivity for 8-OHdG, whereas aniline-treated spleens showed strong immunoreactivity for 8-OHdG confined to the red pulp areas of the spleen (see Methods for details of immunohistochemistry).
Fig. 7.
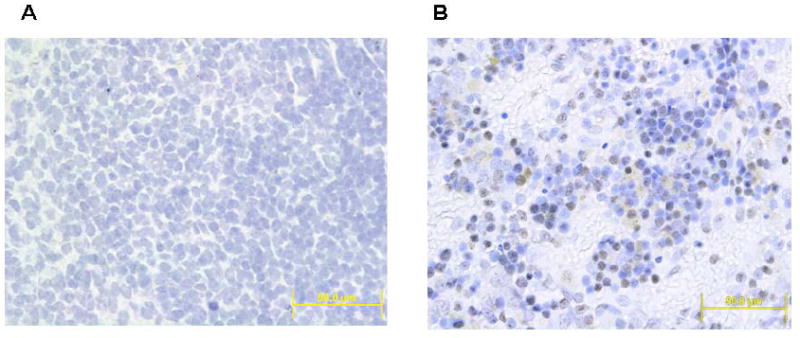
Immunohistochemistry of OGG1 in the spleen of control and aniline-treated rats. A: control spleen; B: aniline-treated spleen. Controls showed scattered immunoreactivity for OGG1, whereas aniline-treated spleens showed strong immunoreactivity for OGG1 confined to the red pulp areas of the spleen (see Methods for details of immunohistochemistry).
Discussion
Previous studies have shown that aniline exposure produces substantial increases, not only in the total iron, but also in the free iron in the spleen (Khan et al., 1993, 1997, 1999b, 2003; Wu et al., 2005). Iron-catalyzed hydroxyl radical formation can generate a multiplicity of oxidative DNA base modifications. Most of these oxidative DNA lesions are known to be mutagenic (McBride et al., 1991; Abalea et al., 1998). Also, DNA damage accumulations could have considerable consequences for the cells in terms of mutagenesis and carcinogenesis (Floyd, 1990; Feig et al., 1994). Persistent DNA damage can result in either arrest or induction of transcription, induction of signal transduction pathways, replication errors, and genomic instability, all of which are known to be associated with carcinogenesis. It is thus not surprising that iron overload is associated with carcinogenesis in animal models and also in human diseases (Bradbear et al., 1985; Toyokuni, 1996). Taking into consideration that aniline exposure leads to iron overload and oxidative stress in the spleen (Khan et al., 1997, 2003), this study focused on determining the status of oxidative DNA damage using 8-OHdG as a biomarker and its repair in the spleen of rats following subchronic exposure to aniline. Data from this study provide evidence that aniline exposure is not only associated with increased formation of 8-OHdG, but is also associated with induction of OGG1 gene expression and OGG1 activity. The OGG1 is a key enzyme in the BER pathway that functions to preferentially excise the highly mutagenic 8-OHdG lesions from DNA (Altieri et al., 2008).
Subchronic aniline exposure in this study led to a 2.8-fold increase in the 8-OHdG levels in the splenic DNA compared to untreated controls. Interestingly, increases in 8-OHdG levels in this study (183%) were much greater in comparison to a previous repeated-dose study conducted in our laboratory (83% after 7 days) (Wu et al., 2005). These data suggest that persistent generation of ROS through subchronic exposure to aniline could lead to higher levels of 8-OHdG in the DNA of the spleens. Our observation of the quantitative changes in 8-OHdG levels was further substantiated by immunohistochemical data, which showed much stronger immunoreactivity for 8-OHdG in the spleens of aniline-treated rats. Increased formation of 8-OHdG as a result of oxidative DNA damage could induce mutations, and is considered to be highly relevant in carcinogenesis (Boiteux and Radicella, 2000; Cheng et al., 1992). Thus, our observation of the increased oxidative DNA damage in the spleen is highly significant, and could lead to serious consequences, if unrepaired.
Repair of the modified bases by their removal from DNA is critical to prevent mutations. BER is the major mechanism for the repair of oxidative DNA base lesions induced by reactive oxygen species (Altieri et al., 2008). One of the key repair enzymes in eukaryotic cells is OGG1, a DNA glycosylase/lyase that removes 8-OHdG and other oxidative guanine adducts from the nuclear and mitochondrial DNA (Boiteux and Radicella, 2000; Bruner et al., 2000; Osterod et al., 2001; Wu et al., 2008). 8-OHdG levels in DNA, therefore, depend upon the balance between its formation and its repair by OGG1 (Hazra et al., 1998; Altieri et al., 2008). The observed increases in our study of the splenic 8-OHdG levels, necessitated the assessment of the extent to which BER might be affected by aniline-induced oxidative stress. This was achieved by measuring OGG1 activity in both nuclear and mitochondrial extracts. The rationale behind studying mitochondrial DNA repair was that mitochondrial DNA contains important genes known to be involved in the oxidative phosphorylation chain, and is highly susceptible to ROS-mediated damage (Zhang et al., 2007). Also, several mitochondrial DNA point mutations and deletions have been linked to various types of cancers (Mandavilli et al., 2002; Ricci et al., 2008). OGG1 activity was significantly higher in both NEs (1.3 fold) and MEs (1.2 fold) of the spleens from aniline-treated rats than the controls, indicating the induction of OGG1 activity in response to increased levels of 8-OHdG. The induction of OGG1 in both NEs and MEs was supported by Western blot data showing 1.5- and 1.3-fold increases in the protein levels of OGG1, and also gene expression data showing a 2-fold increase in splenic OGG1 RNA levels. Taken together, our data thus suggest that aniline induces oxidative DNA lesions, mainly 8-OHdG, that are repaired by OGG1 in the BER pathway.
Since OGG1 is a key BER enzyme, it is not surprising that OGG1 inactivation results in the accumulation of 8-OHdG, and mutations in the OGG1 gene is associated with an increased risk of cancer (Chevillard et al., 1998; Bruner et al., 2000). Expression of both 8-OHdG and OGG1 is also reported to be significantly up-regulated in cancer and experimental studies (Abalea et al., 1998; Chevillard et al., 1998; Hirano et al., 2000). We interpret the induction of OGG1 in our study as a response to an increased level of 8-OHdG, provoked by aniline exposure. However, the oxidative DNA damage induced by aniline is more than the repair capacity, i.e., the repair machinery is overwhelmed and cannot remove all the damaged lesions. Other factors could be involved in determining OGG1 activity in addition to the actual protein levels. For example, it was recently reported that oxidative stress-induced acetylation of OGG1 resulted in significant increases in OGG1 activity and enhanced repair of 8-OHdG (Bhakat et al., 2006).
The fact that the fold-increase in gene expression is not coupled with the same fold-increase in protein expression or OGG1 repair activity could be explained by different mechanisms, including altered mRNA stability and turnover, or translational modifications. For example, it has been reported that over-expression of mammalian BER genes, such as APE, impairs transcriptional regulation of genes (Kaina et al., 2001). The mRNA could decay at several steps during heterogeneous nuclear DNA maturation and pioneer protein synthesis in the nucleus (Byers, 2002). Also, the time frame from transcription to translation may be short to allow the production of new protein. Since the repair activity of OGG1 was somewhat lower than its mRNA or protein expression, it could be generalized that not all the expressed OGG1 proteins have repair activity. Also, increased OGG1 activity may generate an excess of non-instructive repair intermediates to imbalance the repair process, and the imbalanced repair pathways might be mutagenic (Barnes and Lindahl, 2004).
In conclusion, it is evident from our study that aniline-induced oxidative stress in spleen is associated with increases in 8-OHdG levels and induction of 8-OHdG-specific lyase activity. Spleens from aniline-treated rats showed greater OGG1 activity, mRNA and protein levels, and strong OGG1 immunoreactivity, especially in the red pulp areas. Our data suggest that DNA repair in aniline-induced oxidative DNA damage follows BER pathway. Our data also suggest that even though DNA repair pathways were induced, this induction was not enough to prevent the accumulation of DNA oxidation products in genomic DNA. This could represent a critical step in the initiation of a mutagenic and/or carcinogenic response in the spleen. Our study is the first to establish a relationship among aniline exposure, oxidative DNA damage and BER, and may provide a mechanistic explanation for aniline-induced sarcomas of the spleen.
Acknowledgments
This publication was made possible by grant ES06476 from National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIEHS, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abalea V, Cillard J, Dubos MP, Anger JP, Cillard P, Morel I. Iron-induced Oxidative DNA damage and its repair in primary rat hepatocyte culture. Carcinogenesis. 1998;19:1053–1059. doi: 10.1093/carcin/19.6.1053. [DOI] [PubMed] [Google Scholar]
- Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA damage and repair : from molecular mechanisms to health implications. Antioxidants Redox Signaling. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blons H, Radicella JP, Laccourreye O, Brasnu D, Beaune P, Boiteux S, Laurent-Puig P. Frequent allelic loss at chromosome 3p distinct from genetic alterations of the 8- oxoguanine DNA glycosylase 1 gene in head and neck cancer. Mol Carcinog. 1999;26:254–260. [PubMed] [Google Scholar]
- Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- Bradbear RA, bain C, Siskind V, Schofield FD, Webbs S, Azelsen EM, Halliday JW, Bassett ML, Powell LW. Cohort study of internal malignancy in genetic hemochromatosis and other chronic non-alcoholic liver diseases. J Natl Cancer Inst. 1985;75:81–84. [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- Bus JS, Popp JA. Perspectives on the mechanism of action of the splenic toxicity of aniline and structurally related compounds. Food Chem Toxicol. 1987;25:619–626. doi: 10.1016/0278-6915(87)90024-x. [DOI] [PubMed] [Google Scholar]
- Byers PH. Killing the messenger: new insights into nonsense-mediated mRNA. J Clin Invest. 2002;109:3–6. doi: 10.1172/JCI14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso N. The molecular epidemiology of oxidative damage to DNA and cancer. J Natl Cancer Inst. 2003;95:1263–1265. doi: 10.1093/jnci/djg065. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, cause GT and AC substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- Chevillard S, Radicella JP, Levalois C, Lebeau J, Poupon MF, Oudard S, Dutrillaux B, Boiteux S. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene. 1998;16:3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander EW, Ma H. Differential modulation of base excision repair activities during brain ontogeny: Implications for repair of transcribed DNA. Mech Ageing Develop. 2006;127:64–69. doi: 10.1016/j.mad.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Feig DI, Reid TM, Loeb LA. Reactive oxygen species in mutagenesis. Cancer Res. 1994;54:1890S–1894S. [PubMed] [Google Scholar]
- Floyd RA. The role of 8-hydroxyguanine in carcinogenesis. Carcinogenesis. 1990;11:1447–1450. doi: 10.1093/carcin/11.9.1447. [DOI] [PubMed] [Google Scholar]
- Gackowski D, Kowalewski J, Siomek A, Olinski R. Oxidative DNA damage and antioxidant vitamin level: comparison among lung cancer patients, healthy smokers and nonsmokers. Int J Cancer. 2005;114:153–156. doi: 10.1002/ijc.20700. [DOI] [PubMed] [Google Scholar]
- Gackowski D, Speina E, Zielinska M, Kowalewski J, Rozalski R, Siomek A, Paciorek T, Tudek B, Olinski R. Products of oxidative DNA damage and repair as possible biomarkers of susceptibility to lung cancer. Cancer Res. 2003;63:4899–4902. [PubMed] [Google Scholar]
- Goodman DG, Ward JM, Reichardt WD. Splenic fibrosis and sarcomas in F344 rats fed diets containing aniline hydrochloride, p-chloroaniline, azobenzene, o-toluidine hydrochloride, 4,4’-sulfonyldianiline, or DRC Red No. 9. J Natl Cancer Inst. 1984;73:265–273. [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Maidt L, Floyd RA, Mitra S. The presence of two distinct 8- hydroxyguanine repair enzymes in human cells: their porential complementary roles in preventing mutation. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Higashi K, Sakai A, Tsurudome Y, Ootsuyama Y, Kido R, Kasai H. Analyses of oxidative DNA damage and its repair activity in the livers of 3’-methyl-4-dimethylaminoazobenzene-treated rodents. Jpn J Cancer Res. 2000;91:681–685. doi: 10.1111/j.1349-7006.2000.tb00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B, Ochs K, Grösch S, Fritz G, Lips J, Tomicic M, Dunkern T, Christmann M. BER, MGMT, and MMR in defense against alkylation-induced genotoxicity and apoptosis. Prog Nucleic Acid Res Mol Biol. 2001;68:41–54. doi: 10.1016/s0079-6603(01)68088-7. [DOI] [PubMed] [Google Scholar]
- Khan MF, Gu Y, Alock NW, Boor PJ, Ansari GAS. Oxidative stress in splenotoxicity of aniline. Fundam Appl Toxicol. 1997;35:22–30. doi: 10.1006/faat.1996.2259. [DOI] [PubMed] [Google Scholar]
- Khan MF, Kaphalia BS, Boor PJ, Ansari GAS. Subchronic toxicity of aniline hydrochloride in rats. Arch Environ Contant Toxicol. 1993;24:368–374. doi: 10.1007/BF01128736. [DOI] [PubMed] [Google Scholar]
- Khan MF, Kannan S, Wang J. Activation of transcription factor AP-1 and mitogen-activated protein kinases in aniline-induced splenic toxicity. Toxicol Appl Pharmacol. 2006;210:86–93. doi: 10.1016/j.taap.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Alcock NW, Boor PJ, Ansari GA. Iron exacerbates aniline-associated splenic toxicity. J Toxicol Environ Health. 1999b;57:173–184. doi: 10.1080/009841099157746. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Ansari GAS, Boor PJ. Malondialdehyde-protein adducts in spleens of aniline-treated rats: immunohistochemical detection and localization. J Toxicol Environ Health, Part A. 2003;66:93–102. doi: 10.1080/15287390306464. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Boor PJ, Ansari GAS. Oxidative modification of proteins and lipids in aniline-induced splenic toxicity. Toxicol Sci. 1999a;48:134–140. doi: 10.1093/toxsci/48.1.134. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Tipnis UR, Ansari GA, Boor PJ. Protein adducts of malondialdehyde and 4-hydroxynonenal in livers of iron loaded rats: quantitation and localization. Toxicology. 2002;173:193–201. [PubMed] [Google Scholar]
- Kondo S, Toyokuni S, Tanada T, Hiai H, Onodera H, Kasai H, Immamura M. Overexpression of the hOGG1 gene and high 8-hydroxy-2’-deoxyguanosine (8-OHdG) lyase activity in human colorectal carcinoma: regulation mechanism of the 8-OHdG level in DNA. Clinical Cancer Res. 2000;6:1394–1400. [PubMed] [Google Scholar]
- Lan Q, Mumford JL, Shen M, Demarini DM, Bonner MR, He X, Yeager M, Welch R, Chanock S, Tian L, Chapman RS, Zheng T, Keohavong P, Caporaso N, Rothman N. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25:2177–2181. doi: 10.1093/carcin/bgh240. [DOI] [PubMed] [Google Scholar]
- Ma H, Lee HM, Englander EW. N-terminus of the rat adenine glycosylase MYH affects excision rates and processing of MYH-generated abasic sites. Nucleic Acids Res. 2004;32:4332–4339. doi: 10.1093/nar/gkh758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandavilli BS, Santos JH, Van Houten B. Mitochondrial DNA repair and aging. Mutat Res. 2002;509:127–151. doi: 10.1016/s0027-5107(02)00220-8. [DOI] [PubMed] [Google Scholar]
- McBride TJ, Preston BD, Loeb LA. Mutagenic spectrum resulting from DNA damage by oxygen radicals. Biochemistry. 1991;30:207–213. doi: 10.1021/bi00215a030. [DOI] [PubMed] [Google Scholar]
- Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis. 2001;22:1459–1463. doi: 10.1093/carcin/22.9.1459. [DOI] [PubMed] [Google Scholar]
- Pauluhn J. Subacute inhalation toxicity of aniline in rats: analysis of time-dependence and concentration-dependence of hematotoxic and splenic effects. Toxicol Sci. 2004;81:198–215. doi: 10.1093/toxsci/kfh187. [DOI] [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and Characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeevan MS, Ranamukhararachchi DG, Vernon SD, Unger ER. Use of real time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods. 2001;25:443–4451. doi: 10.1006/meth.2001.1266. [DOI] [PubMed] [Google Scholar]
- Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffe RSW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol. 2008;294:C413–C422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- Simpson DA, Feeney S, Boyle C, Stitt AW. Retinal VEGF mRNA measured by SYBR green I fluorescence: a versatile approach to quantitative PCR. Mol Vision. 2000;6:178–183. [PubMed] [Google Scholar]
- Souza-Pinto NC, Hogue BA, Bohr VA. DNA repair and aging in mouse liver: 8-oxodg glycosylase activity increase in mitochondrial but not in nuclear extracts. Free Radic Biol Med. 2001;30:916–923. doi: 10.1016/s0891-5849(01)00483-x. [DOI] [PubMed] [Google Scholar]
- Thomas D, Scot AD, Barbey R, Padula M, Boiteux S. Inactivation of OGG1 increases the incidence of GC→TA transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol Gen Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- Toyokuni S. Iron-induced carcinogenesis: the role of redox regulation. Free Radic Biol Med. 1996;20:553–566. doi: 10.1016/0891-5849(95)02111-6. [DOI] [PubMed] [Google Scholar]
- Tsai K, Hsu T-G, Hsu K-M, Cheng H, Liu T-Y, Hsu C-F, Kong C-W. Oxidative DNA damage in human peripheral leukocytes induced by massive aerobic exercise. Free Radic Biol Med. 2001;31:1465–1472. doi: 10.1016/s0891-5849(01)00729-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Kannan S, Li H, Khan MF. Cytokine gene expression and activation of NF-kappa B in aniline-induced splenic toxicity. Toxicol Appl Pharmacol. 2005;15:36–44. doi: 10.1016/j.taap.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang G, Ansari GAS, Khan MF. Activation of oxidative stress-responsive signaling pathways in early splenotoxic response of aniline. Toxicol Appl Pharmacol. 2008;230:227–234. doi: 10.1016/j.taap.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger MA, Albert RH, Montgomery SB. Splenotoxicity associated with splenic sarcomas in rats fed high doses of DRC Red No. 9 or aniline hydrochloride. J Natl Cancer Inst. 1985;75:681–690. [PubMed] [Google Scholar]
- Wu M, Zhang Z, Che W. Suppression of a DNA base excision repair gene, hOGG1, increases bleomycin sensitivity of human lung cancer cell line. Toxicol Appl Pharmacol. 2008;228:395–402. doi: 10.1016/j.taap.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Wu X, Kannan S, Ramanujam VM-S, Khan MF. Iron release and oxidative DNA damage in splenic toxicity of aniline. J Toxicol Environ Health, Part A. 2005;68:657–666. doi: 10.1080/15287390590921757. [DOI] [PubMed] [Google Scholar]
- Zhang H, Mizumachi T, Carcel-Trullols J, Li L, Naito A, Spencer HJ, Spring PM, Smoller BR, Watson AJ, Margison GP, Higuchi M, Fan CY. Targeting human 8-oxoguanine DNA glycosylase (hOGG1) to mitochondria enhances cisplatin cytotoxicity in hepatoma cells. Carcinogenesis. 2007;28:1629–1637. doi: 10.1093/carcin/bgm072. [DOI] [PubMed] [Google Scholar]


