Abstract
The multi-component soluble form of methane monooxygenase (sMMO) catalyzes the oxidation of methane through activation of O2 at a non-heme biferrous center in the hydroxylase component, MMOH. Reactivity is limited without binding of the sMMO effector protein, MMOB. Past studies show that mutations of specific MMOB surface residues cause large changes in rates of individual steps in the MMOH reaction cycle. To define the structural and mechanistic bases for these observations, CD, MCD, and VTVH MCD spectroscopies coupled with Ligand Field calculations are used to elucidate changes occurring near and at the MMOH biferrous cluster upon binding of MMOB and the MMOB variants. Perturbations to both the CD and MCD are observed upon binding wild-type MMOB and the MMOB variant that similarly increases O2 reactivity. MMOB variants that do not greatly increase O2 reactivity fail to cause one or both of these changes. LF calculations indicate that reorientation of the terminal glutamate on Fe2 reproduces the spectral perturbations in MCD. Although this structural change allows O2 to bridge the diiron site and shifts the redox active orbitals for good overlap, it is not sufficient for enhanced O2 reactivity of the enzyme. Binding of the T111Y-MMOB variant to MMOH induces the MCD, but not CD changes, and causes only a small increase in reactivity. Thus, both the geometric rearrangement at Fe2 (observed in MCD) coupled with a more global conformational change that may control O2 access (probed by CD), induced by MMOB binding, are critical factors in the reactivity of sMMO.
The soluble form of methane monooxygenase (sMMO) from methanotrophic bacteria catalyzes the NAD(P)H- and O2-dependent oxidation of the extremely stable C–H bond of methane to yield methanol under ambient conditions with the following stoichiometry:(1, 2)
Although methane is the natural substrate of sMMO, the enzyme is also able to catalyze the oxidation of other substrates, including aromatic, saturated, unsaturated, linear, branched, halogenated, and cyclic hydrocarbons, making it potentially useful in biosynthetic and bioremediation processes.(3–6) sMMO from Methylosinus trichosporium (OB3b) is comprised of three components,(7) each necessary for efficient oxidation. The reductase component (MMOR) contains an FAD cofactor and a [2Fe-2S] cluster, and it is responsible for transferring the reducing equivalents from NAD(P)H to MMOH.(7, 8) The cofactor-free, monomeric component B (MMOB) has a regulatory role in catalysis, increasing the turnover number by 150 fold and the rate of initial reaction with O2 by a factor of 1000.(7, 9, 10) The hydroxylase component (MMOH) exists as a dimer of three subunits with an (αβγ)2 structure and contains a binuclear non-heme iron active site that is essential for O2 activation and subsequent methane oxidation.(7, 11, 12)
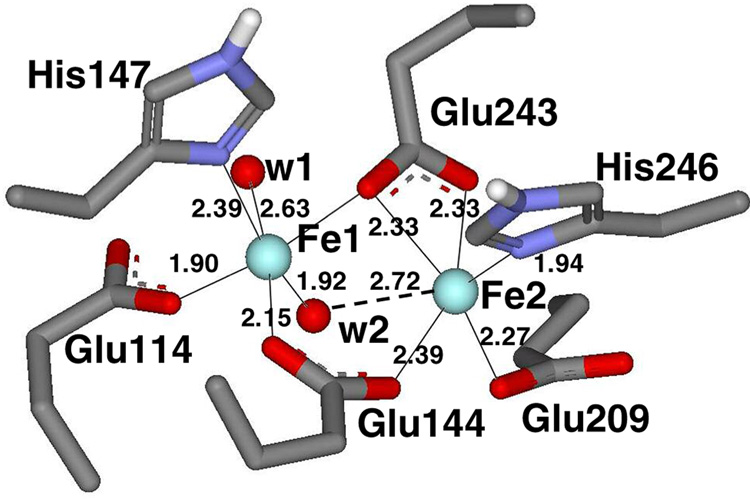
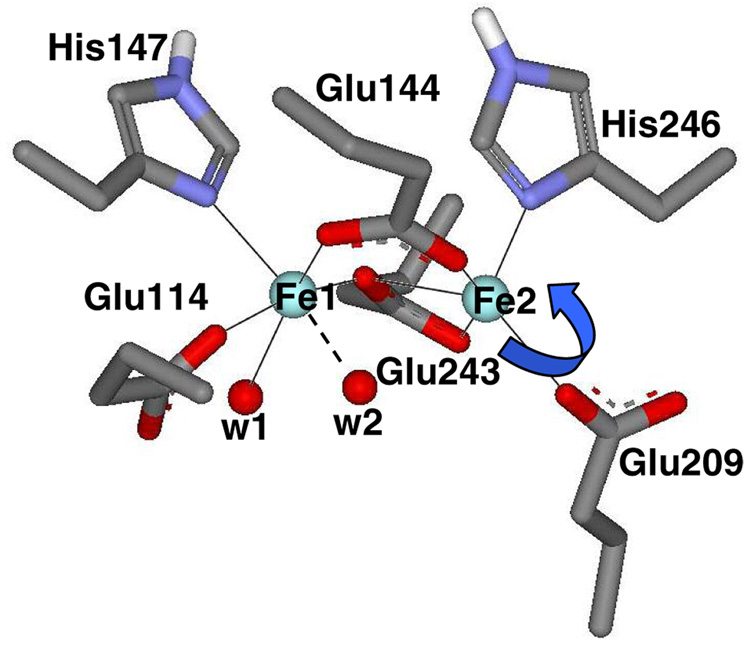
Crystal structures of MMOH in both oxidized, [FeIIIFeIII], and reduced, [FeIIFeII], forms from M. capsulatus (Bath) have been reported,(13–16) while the oxidized structure has been obtained for the enzyme from M. trichosporium OB3b.(12) The overall structures of the two enzymes are very similar, with the binuclear iron cluster located within the α subunit, ~12 Å below the protein surface without direct access to bulk solvent.(12, 13) In the vicinity of the binuclear iron cluster, the amino acid residues are identical or highly conserved, and the structures are essentially superimposable. The various structures of reduced M. capsulatus (Bath) MMOH show some degree of structural flexibility and variability in the number of coordinated water molecules in the active site. In the structure of the chemically reduced enzyme (Figure 1), Glu144 and Glu243 bridge the metal ions in µ-1,3 and µ-1,1 modes, respectively (Glu243 also binds to Fe2 in a bidentate fashion).(14) His246 and monodentate Glu209 complete the Fe2 coordination sphere. Fe1 is also coordinated by His147, monodentate-Glu114, and at least one terminal water molecule, although alternative structures of reduced MMOH suggest that W1 may be more tightly bound than W2 or that W2 may bridge the diiron site.(13–16) CD/MCD spectroscopic studies of MMOH support five-coordinate geometries for both metal ions.(17, 18) In the crystal structures of oxidized MMOH both iron atoms are six-coordinate and triply bridged by two hydroxo or bridged by one hydroxo and one water molecule in addition to the bridging µ-1,3 carboxylate from Glu144.(12, 14)
Figure 1.
Representation of the crystallographically determined (structure generated using PDB file 1MTY) reduced binuclear iron active site of sMMO from Methylococcus capsulatus (Bath). Bonding distances (Å) are shown. CD/MCD, LF, and DFT calculations indicate that W2 is unbound, so that each Fe is 5-coordinate.
Several intermediates formed during MMO-catalyzed reactions have also been characterized through a variety of kinetic and spectroscopic techniques.(19–24) In the first step of catalysis the diferric active site of MMOH is reduced to its diferrous form. Upon addition of O2, diferrous intermediate O is formed, where oxygen is bound to the enzyme but not yet to the binuclear iron site. Binding of O2 to the binuclear site leads to the formation of peroxo intermediates P* and P. The latter peroxo species decays to a high-valent diamagnetic intermediate, Q, that contains a bis-µ-oxo bridged [FeIVFeIV] cluster. Intermediate Q then reacts with the substrate, cleaving a C–H bond and inserting oxygen to form the product complex T. Product is subsequently released from the now diferric binuclear iron cluster to complete the cycle.(25, 26)
The binding of MMOB to MMOH causes changes in the rates of specific steps and the structural characteristics of specific intermediates within this catalytic cycle.(10, 27) In addition to the great increase in the O2 binding rate to form intermediate P mentioned above,(10, 19) the redox potential for reduction of the diferric to diferrous form of the MMOH binuclear iron active site is shifted by −132 mV.(28–30) Conformational changes in the vicinity of the active site apparently occur, causing an alteration of the regiospecificity for hydroxylation of more complex substrates.(31) Finally, changes in the electronic structure of the binuclear cluster in several oxidation states of MMOH are apparent from spectroscopic studies.(7, 18, 28)
Our previous CD/MCD studies revealed that both iron atoms in the reduced MMOH-MMOB complex remain five-coordinate and ferromagnetically coupled, however, the ligand field of one of the two is altered (tentatively assigned to Fe2 in Figure 1).(17, 18) Other studies have suggested that MMOB may also affect O2 and CH4 entry into the active site of MMOH.(32–34) Nevertheless, it is not yet known what specific changes allow MMOB to cause such profound effects on MMOH catalysis, in particular its ability to bind and activate O2.
The NMR solution structures of MMOB were recently determined for the proteins from both M. trichosporium OB3b and M. capsulatus (Bath).(32, 35, 36) Accompanying relaxation measurement in the presence of MMOH allowed the interacting residues of MMOB to be tentatively identified. Site-directed mutagenesis studies showed that several of these residues greatly affected the rates of specific steps in the reaction cycle.(27, 37) A tentative model for the docked structure of the MMOH-MMOB complex(32) was constructed using results from: (i) saturation recovery EPR using spin-labeled MMOB and the oxidized MMOH diiron cluster that identified the docking surface of MMOH,(38) (ii) EPR experiments that measured the immobilization and accessibility of spin-labels placed specifically in MMOB that identified the interaction surface of MMOB,(33) and (iii) cross-linking experiments that provided information on the alignment of the two components.(32) This model shows that MMOB binds above the diiron cluster in MMOH, on the surface formed by the helices that include the ligands to Fe2 (Figure 1, residues Glu209, His246 and Glu243). Recently, the crystal structure of the complex of phenol hydroxylase with its regulatory protein has been reported and shows a similar docking orientation.(39) The MMOH-MMOB docking model shows that MMOB residues Asn107, Ser109, Ser110 and Thr111 are in the center of the protein-protein interface. Mutation of these residues, individually and in groups to produce MMOB variants, significantly affects the rates of reactions in which O2 binds, substrates react with compound Q, and products are released.(27, 34, 37)
The focus of this study is to understand geometric and electronic structural changes in the active site associated with MMOB binding through spectroscopic studies on the wild type (wt) and selected MMOB variants. Each of the three MMOB variants studied, S109A/T111A, N107A/S110A and T111Y, have different effects on the catalytic cycle. The double variant S109A/T111A elicits a very similar MMOH turnover number for CH4 oxidation to that observed when using wt-MMOB, but the turnover number with larger substrates like furan or nitrobenzene is increased slightly (1.2–1.7-fold).(37) More importantly, binding of this MMOB variant enables O2 reactivity similar to that of wt-MMOB (the rate of O2 reactivity measured by P formation is >200 s−1 in both cases). In contrast, binding of the MMOB-N107A/S110A variant leads to a two to three-fold decrease in the NADH consumption rate in the presence of substrates, intermediate P is not observed (indirect measurements suggest the rate of P formation is ~2 s−1), and the maximum amount of Q formed is significantly decreased.(37) This variant also alters the regiospecificity of nitrobenzene hydroxylation, resulting in a different product distribution. It was proposed that this mutation leads to a structural change in the MMOH active site of the MMOH-MMOB complex that allows for a different substrate orientation as compared to the MMOH-wt-MMOB complex.(37) The T111Y variant was shown to have a lower affinity for MMOH (MMOB:MMOH = 10:1 for maximum turnover) and drastically reduces the rate of P formation (3.4 s−1).(37) However, while this mutation leads to a decrease in MMOH reactivity with CH4, the rate constants for the reaction with larger substrates were greatly increased. It has been suggested that MMOB-T111Y may facilitate organic substrate accessibility to the active site of MMOH.
Here, the electronic and geometric structural changes induced in the MMOH active site by binding MMOB and its variants are correlated to corresponding kinetic data to develop an understanding of how this small regulatory component affects the reactivity of MMO. The experimental CD/MCD results and LF calculations are compared with the crystal structures of reduced MMOH to identify possible structural changes associated with wt-MMOB binding and constraints for optimal O2 reactivity.
Experimental Procedures
All commercial reagents were of the highest grade available and were used as obtained without further purification: deuterium oxide (99.9 atom % D; Aldrich), d3-glycerol (98 atom % D; Cambridge Isotope Laboratories), enzyme grade 3-(N-morpholino)propanesulfonic acid (Sigma), sodium hydrosulfite (Fisher), and methyl viologen (Sigma). d3-glycerol was degassed by heating under vacuum overnight followed by approximately 10–15 freeze-pump-thaw cycles at 10−3 Torr.
Soluble MMOH from M. trichosporium OB3b was purified and characterized as described previously.(7) Recombinant wt-MMOB and its variant forms were expressed, purified, and characterized as reported earlier.(27, 35, 37) Protein concentrations were determined by measuring the absorbance at 280 nm, using ε280 = 539 M−1 cm−1 for MMOH, ε280 = 20.8 M−1 cm−1 for wt-MMOB, S109A/T111A, and N107A/S110A variants, and ε280 = 22.3 M−1 cm−1 for the T111Y variant. All specific activity assays were conducted as described previously.(7)
Pure oxidized MMOH and all MMOB variants were exchanged into D2O-containing buffer (200 mM MOPS/5% glycerol, pD = 7.4) and concentrated to ~2 mM and ~10 mM and final volumes of ≤1 mL, respectively by ultrafiltration using 50 mL Amicon ultrafiltration device and YM-30/YM-5 membranes. Concentrated protein solutions were then transferred to conical reaction vials kept on ice (MMOH containing ~100 µM redox mediator methyl viologen, and MMOB variants) and degassed by gentle flushing with O2-scrubbed argon gas on the Schlenk line for at least 30 min at 4 °C. Solid sodium hydrosulfite was made anaerobic by pumping at 10−3 Torr for 30 min. All subsequent protein manipulations were carried out under strictly anaerobic conditions in an argon-filled Vacuum Atmospheres (Hawthorne, CA) glovebox (<1 ppm O2). Once the protein samples were deoxygenated on a Schlenk line at 4 °C, oxidized MMOH was reduced in the anaerobic glove box by addition of excess sodium dithionite (sodium hydrosulfite) from a D2O stock solution (~10–50-fold excess of dithionite relative to MMOH). The reduced MMOH was allowed to equilibrate for approximately 15 minutes on a cold plate in the glove box. Approximately 10-fold excess of dithionite was also added from a D2O stock solution to wt-MMOB and other MMOB variants. Samples of the reduced MMOH-MMOB complexes were prepared by the addition of ~4-fold molar excess of anaerobic MMOB to reduced MMOH. The only exception was MMOB-T111Y variant which has a decreased affinity for MMOH and the final T111Y:MMOH stoichiometry was 10:1 for all CD/MCD experiments.
Samples for CD measurements were transferred to an anaerobic cuvette. To ensure homogeneous glass formation for low temperature MCD experiments, concentrated MMOH-MMOB samples were diluted slowly with degassed d3-glycerol (58–65% v/v). The MCD cell was assembled under argon with a neoprene spacer (0.32 cm path length) sandwiched between two infrasil quartz disks that are held together between two fitted copper plates and the MCD samples were injected into the cell anaerobically. The MCD sample was immediately frozen in liquid N2 and then transferred into the cryostat under a high flow of He gas. Final concentrations of all MMOH-MMOB complexes ranged from 0.4–0.6 mM MMOH. CD experiments were performed at 5 °C on a JASCO J200D spectropolarimeter equipped with a liquid-nitrogen-cooled InSb solid-state detector (near-IR, 600–2000 nm). The sample temperature was maintained by a recirculating water bath attached to the sample cell holder. Low-temperature near-IR MCD experiments (1.6–35 K) were carried out on the same spectropolarimeter, modified to accommodate an Oxford Instruments SM4000-7T superconducting magnet/cryostat. All MCD samples were checked for depolarization by comparison of the CD signal of a nickel(+)-tartrate solution placed before and after the sample. Generally, the depolarization was <7% at 1.6 K for all samples studied. Data acquisition was performed using LabView (National Instruments) software package.
All CD and MCD spectra were baseline-corrected (by measuring and subtracting zero-field baseline effects at each temperature or alternatively by changing the direction of the applied magnetic field and averaging the positive and negative field spectra). The CD and MCD spectra were simultaneously fit to Gaussian band shapes using the program PeakFit version 4 (AISN Software, Inc.). Saturation magnetization data were normalized to the maximum observed intensity and fit using in-house programs according to previously published procedures to extract ground-state parameters. VTVH MCD data were fit using a Simplex algorithm that minimizes the χ2 value (goodness of fit parameter). Two complementary approaches were employed in the analysis of VTVH MCD data: (i) a doublet model appropriate for non-Kramers systems, which incorporates the effects of a linear B-term from field-induced mixing between excited sublevels of the ground state, and (ii) a spin-projection model, where the dimer wave functions are calculated and the MCD intensity is given by the spin expectation values of the iron center being studied by VTVH MCD. Further details of both methods are described elsewhere.(40–42)
Results and Analysis
1. Excited States
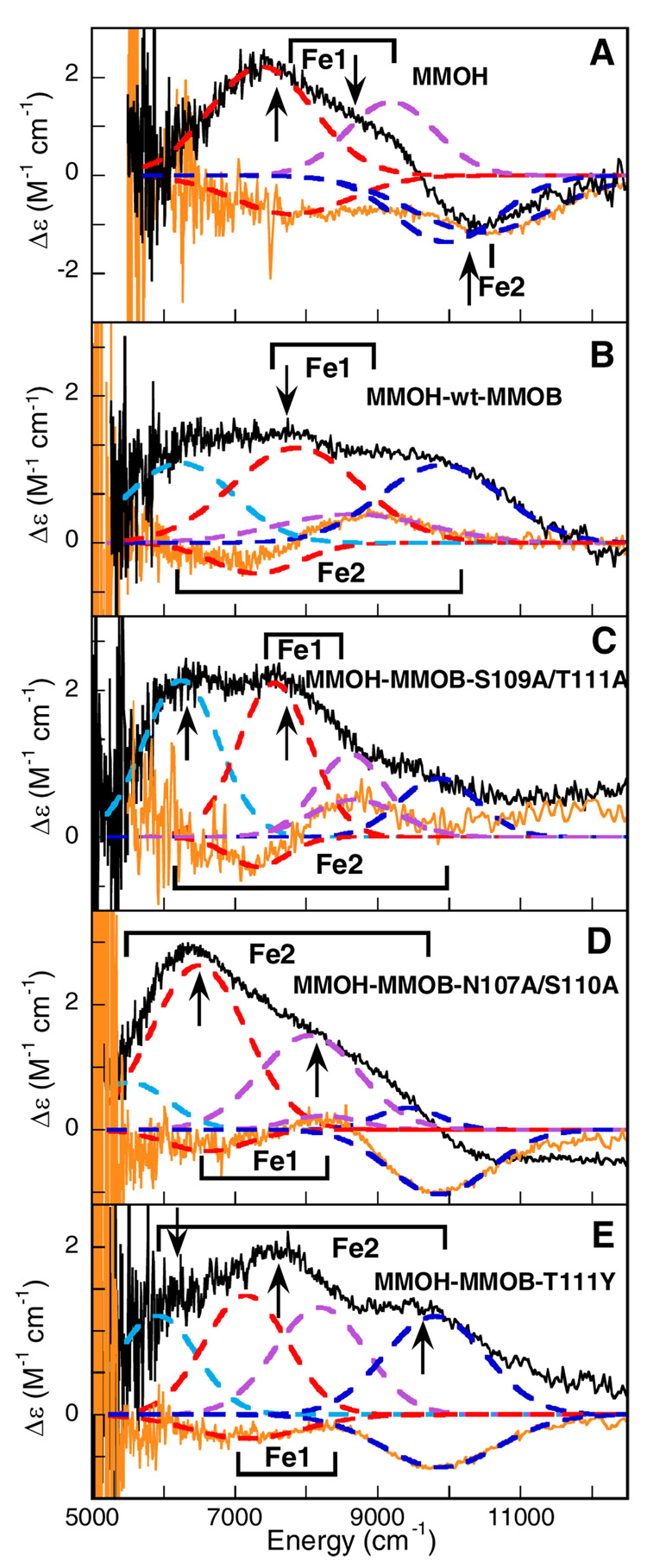
Previous studies have shown that CD/MCD spectral data in the near-IR region can be used to probe the d → d transitions of the FeII site and that this information can be correlated to the coordination number and geometry of the FeII active sites.(40) A distorted six-coordinate high-spin FeII exhibits two d → d transitions at ~10000 cm−1 split by about 2000 cm−1. A five-coordinate FeII site exhibits two transitions at ~5000 and ~10000 cm−1 for square pyramidal and at <5000 cm−1 and <10000 cm−1 for a trigonal bipyramidal FeII site. A four-coordinate tetrahedral FeII site is characterized by low-energy transitions <7000 cm−1. While CD is sensitive to the changes in the general protein environment in the vicinity of the iron center, the MCD spectral changes reflect the differences in the geometric and electronic structure of the binuclear iron center. Since each of the spectroscopic techniques has different selection rules, the observed CD/MCD transitions can have different amplitudes and signs, but their energies are similar.(40) Figure 2 presents the 5 °C CD and low-temperature MCD spectra of (A) reduced MMOH, (B) MMOH-wt-MMOB, (C) MMOH-MMOB-S109A/T111A, (D) MMOH-MMOBN107A/S110A, and (E) MMOH-MMOB-T111Y. The summary of observed CD/MCD transition energies for all MMOH-MMOB complexes is given in Table 1.
Figure 2.
CD and MCD spectra of the binuclear non-heme Fe(II)Fe(II) active site in (A) MMOH, (B) MMOH-wt-MMOB, (C) MMOH-MMOB-S109A/T111A, (D) MMOH-MMOB-N107A/S110A, and (E) MMOH-MMOB-T111Y. The experimental CD (orange solid lines) and MCD data (black solid lines) are simultaneously fit to individual Gaussian band shapes (colored --- lines). The arrows mark the energies where the VTVH MCD data were collected and the labels show the bands corresponding to Fe1 and Fe2 centers.
Table 1.
Summary of the Ligand-Field Transition Energies from the CD and MCD Gaussian Resolution Analysis for Reduced MMOH and the Reduced MMOH-wt-MMOB, MMOH-MMOB-S109A/T111A, MMOH-MMOB-N107A/S110A, and MMOH-MMOB-T111Y Complexes.
| Band energy (cm−1)a | reduced MMOH | MMOH-wt-MMOB | MMOH-MMOB-S109A/T111A | MMOH-MMOB-N107A/S110A | MMOH-MMOB-T111Y | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD | MCD | CD | MCD | CD | MCD | CD | MCD | CD | MCD | |
| 1 | (+)b 6170 |
(+) 6250 |
(+) 5520 |
(+) 5920 |
||||||
| 2 | (−) 7360 |
(+) 7360 |
(−) 7330 |
(+) 7590 |
(−) 7320 |
(+) 7560 |
(−) 6600 |
(+) 6500 |
(−) 7170 |
(+) 7160 |
| 3 | (+) 9140 |
(+) 8650 |
(+) 8680 |
(+) 8630 |
(+) 8250 |
(+) 8050 |
(+) 8200 |
|||
| 4 | (−) 9900 |
(−) 9900 |
(+) 9840 |
(+) 9850 |
(−) 9865 |
(−) 9850 |
(−) 9880 |
(+) 9800 |
||
Energies are accurate within ±200 cm−1.
Indication of the sign of the CD/MCD ligand field transitions.
1.1. Effect of wt-MMOB on MMOH
Combined CD and MCD spectra of reduced MMOH (Figure 2A) show three d → d transitions; two positive transitions (in MCD) at 7400 and 9100 cm−1, and one negative at 9900 cm−1 indicating the presence of two inequivalent five-coordinate iron centers (Note: the maximum number of LF transitions for a single FeII center above 5000 cm−1 is two).(40) The assignment of two five-coordinate irons derived from the number of d-d transitions observed experimentally, as well as their energy splitting. Although the addition of wt-MMOB to reduced MMOH (Figure 2B) significantly alters the CD/MCD spectra, resulting in four d → d transitions at 6200, 7600, 8600, and 9900 cm−1, the coordination environments of the two ferrous centers remain five-coordinate. The CD spectrum of the MMOH-wt-MMOB complex displays a positive feature at 8600 cm−1 which is not observed in the CD spectrum of MMOH, while the MCD spectrum shows a new positive band at 6200 cm−1. In addition, the MCD band at 9900 cm−1 changes sign upon wt-MMOB binding. Based on the changes in the CD/MCD spectra, the bands at 7600 and 8600 cm−1 were assigned to the Fe1 center, while the two transitions that were more substantially perturbed, at 6200 and 9900 cm−1, were attributed to Fe2 (the definitive assignment of the iron transitions follows from the VTVH MCD analysis; vide infra). These data clearly indicated that binding of wt-MMOB induces a conformational (from CD) and structural (from MCD) change in the active site of MMOH, which correlates with the observed enhancement of O2 reactivity of the binuclear iron active site without change in the coordination number. The MCD changes were attributed predominantly to LF perturbations of Fe2.
1.2. MMOH-MMOB-S109A/T111A
The CD spectrum of the MMOH-MMOB-S109A/T111A complex shown in Figure 2C exhibits two transitions: a negative feature at 7320 cm−1 and a positive one at 8680 cm−1, both of which also appear in the CD spectrum of MMOH-wt-MMOB. The corresponding MCD spectrum displays four positive bands at 6250, 7560, 8630 and 9850 cm−1. The four overlapping CD/MCD transitions are comparable to the transitions observed in the MMOH-wt-MMOB complex. This indicates that MMOB-S109A/T111A variant binding causes conformational changes, exhibited by CD, and structural changes, exhibited by MCD, in the active site of reduced MMOH similar to wt-MMOB binding, where only the Fe2 center is significantly perturbed in MCD. Consequently, the two complexes exhibit increased reactivity with O2, with both having similar rate constants for formation of intermediate P in the absence as well as presence of substrate.
1.3. MMOH-MMOB-N107A/S110A
The CD spectrum of the MMOH-MMOB-N107A/S110A complex (Figure 2D) displays three bands (a negative feature at 6600 cm−1, a positive one at 8250 cm−1, and a negative one at 9865 cm−1). The positive CD peak at ~8000 cm−1 may also be present in MMOH, but not resolved due to an overlap with the flanking negative peaks. This indicates that binding MMOB-N107A/S110 shifts the ligand field transition energies relative to MMOH, but does not induce the same conformational change in MMOH caused by wt-MMOB (or MMOB-S109A/T111A). While the overall MCD spectrum of the MMOH-MMOB-N107A/S110A complex resembles the MMOH spectrum with respect to general shape and MCD intensity distribution, there are significant changes in transition energies. The MCD spectrum of this complex can be Gaussian-resolved into four transitions at 5520, 6500, 8050, and 9865 cm−1 (Figure 2D), again consistent with two inequivalent distorted five-coordinate iron sites. However, in contrast to MMOH, the MCD transition at 9865 cm−1 in the MMOH-MMOB-N107A/S110A complex is positive (see Supporting Information, Figure S1) and the lowest energy band, not observed in MMOH (<5000 cm−1), moves to higher energy (5520 cm−1). These perturbed transitions correlate with the Fe2 center, while the remaining two d-d transitions at 6500 and 8050 cm−1 then correlate to the Fe1 center. These CD/MCD data demonstrate that binding of MMOB-N107A/S110A to reduced MMOH induces conformational and structural changes in the MMOH active site different from those observed upon wt-MMOB binding. This results in a site that remains more similar to MMOH, consistent with the reduced reactivity of this complex with O2.
1.4. MMOH-MMOB-T111Y
The CD spectrum of the MMOH-MMOB-T111Y complex (Figure 2E) shows two negative bands at 7170 cm−1 and 9880 cm−1, and is, overall, very similar to the CD spectrum of reduced MMOH (Figure 2A). The low-temperature MCD spectrum of the MMOH-MMOB-T111Y complex displays four positive transitions at 5920, 7160, 8200, and 9900 cm−1, again indicative of two inequivalent five-coordinate ferrous centers. The transitions at 7160 and 8200 cm−1 are assigned to the Fe1 center, while the remaining two bands are attributed to the Fe2 center. All four d-d transitions observed in the MCD are very similar in energy to those in the MMOH-wt-MMOB complex. Therefore, the MCD data indicate that adding wt or T111Y MMOB induces virtually identical geometric changes in the MMOH active site. However, since the CD spectra of MMOH in the presence of the T111Y variant is different from that of the MMOH complex with wt-MMOB, there must be a conformational difference in the vicinity of the active site that correlates with the relatively low reactivity of the MMOH-MMOB-T111Y complex with O2.
In summary, the CD spectral changes indicate that the active sites of the two reactive complexes (MMOH-wt-MMOB in Figure 2B and MMOH-MMOB-S109A/T111A in Figure 2C) are similar, undergoing a specific conformational change associated with increased O2 reactivity (rate constant for compound P formation for both complexes is >200 s−1). In contrast, the CD spectra of the MMOH-MMOB-N107A/S110A and the MMOH-MMOB-T111Y complexes (Figures 2D and 2E) are similar to that of MMOH (Figure 2A), reflecting the lack of this conformational change and their low O2 reactivity (rate constant for P formation is ~2 s−1 for N107A/S110A and ~3 s−1 for the T111Y variant). The MCD data further reveal that the two reactive complexes, MMOH-wt-MMOB and MMOH-MMOB-S109A/T111A, undergo significant and similar structural changes upon the addition of the B-component, which dominantly affect their Fe2 centers. For the low reactivity complexes, MMOH-MMOB-N107A/S110A has limited structural perturbation relative to MMOH, while MMOH-MMOB-T111Y has a structural change more similar to wt-MMOB but no conformational shift. Thus, both a structural change to the Fe(II) site (shown by MCD) and a conformational shift in the vicinity of the active site (shown by CD) correlate with increased O2 reactivity.
2. Variable-Temperature Variable-Field (VTVH) MCD: Ground-State Analysis
The MCD intensity of reduced MMOH and MMOH in the presence of wt-MMOB and MMOB variants increases with decreasing temperature and increasing magnetic field, indicating that the observed MCD transitions are C-terms and that the ground states of these various complexes are paramagnetic doublets. In order to obtain the ground state spin Hamiltonian parameters (Zero Field Splitting (ZFS) parameters D and E of each Fe(II) and the exchange coupling parameter J between the metal ions), VTVH MCD data were obtained for all species and fit utilizing two approaches (the doublet and spin projection models) based on the electronic structure of a coupled binuclear ferrous system.(40) These models were used to (i) obtain the ground-state parameters, (ii) identify which Fe center is associated with a particular MCD transition, and (iii) estimate the polarization of the electronic transition.
2.1. MMOH
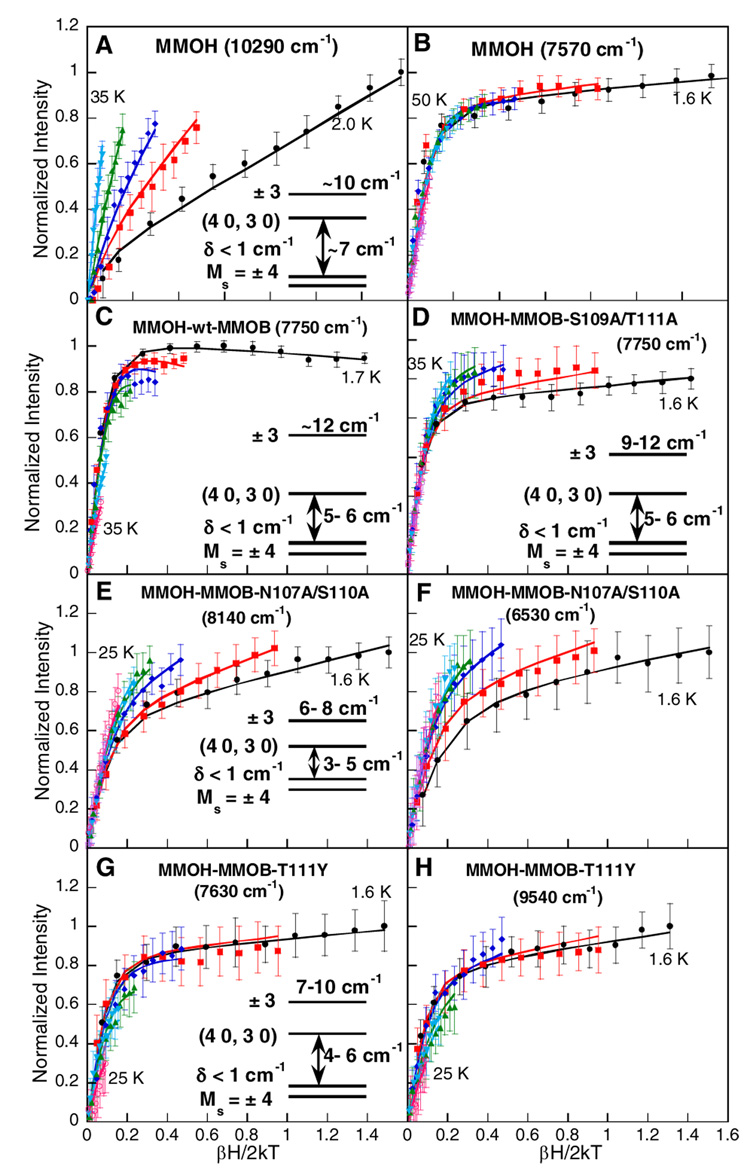
VTVH MCD saturation magnetization data for reduced MMOH measured at 7570 cm−1 (arrow in Figure 2A, peak position: 7360 cm−1; Table 1) and 10290 cm−1 (arrow in Figure 2A, peak position: 9900 cm−1; Table 1) are shown in Figure 3A and B. A comparison of these data shows a large difference in the nesting behavior of the isotherms. The band at 7570 cm−1 (Figure 3B) exhibits rapid saturation indicative of a system with a large g∥, while isotherms collected at 10290 cm−1 (Figure 3A) show a large nesting behavior suggesting either a change in the ground state properties (i.e. small g∥) or a different polarization of that transition (vide infra). Our previous analysis of the VTVH MCD data for the transition at 7570 cm−1 (using the doublet model) demonstrated that the ground state doublet |4, ±4〉 (Stot, Mstot) is lowest in energy, with g∥= 14.7 and δ < 1.0 cm−1, and that the two iron centers are weakly ferromagnetically coupled (J = 0.2 – 0.4 cm−1).(17) Furthermore, both metal ions were shown to have negative ZFS of different magnitudes (−4 ± 3 cm−1 and −9 ± 6 cm−1).
Figure 3.
Saturation magnetization behavior of the MCD signal for reduced MMOH at (A) 10290 cm−1 and (B) 7570 cm−1, (C) reduced MMOH-wt-MMOB at 7750 cm−1, (D) reduced MMOH-MMOB-S109A/T111A at 7750 cm−1, (E) reduced MMOH-MMOB-N107A/S110A complex at 8140 cm−1 and (F) 6530 cm−1, and (G) reduced MMOH-MMOB-T111Y complex at 7630 cm−1 and (H) 9540 cm−1. The intensity amplitude for a range of magnetic fields (0–7.0 T) at a series of fixed temperatures is plotted as a function of βH/2kT. The fits to the VTVH MCD data (solid lines) were obtained using equation (1) from reference 67 with the parameters given in Table 2. The resultant energy level diagrams are presented in the insets (Note: different transitions belonging to the same complex have the same energy level diagrams).
Here, the analysis of the transition at 7570 cm−1 agrees with the previous study and shows that the VTVH MCD data measured at 10290 cm−1 can be fit with the doublet model resulting in ground state parameters similar to those determined for the lower energy transition (Table 2; parameters for the lower energy transition). The two transitions have been further analyzed using the spin-projection model to understand the origin of the differences in saturation behavior.(42) A reasonable fit to the VTVH MCD data taken at 10290 cm−1 can only be obtained with the larger value of |−D| for the iron center associated with this ligand field transition (Figure S2). In contrast, the transition at 7570 cm−1 is not sensitive to the magnitude of the ZFS parameters (Figure S3). VTVH MCD data have also been collected at 8470 cm−1 (corresponding to the transition with a peak position at 9140 cm−1; Table 1). A nesting behavior similar to that measured at 7570 cm−1 is observed (Figure S4). The similar VTVH MCD behavior of the bands at 7570 and 9140 cm−1, which is very different from that of the 9900 cm−1 data, supports the association of these bands with the same Fe, Fe1 in Figure 1. The larger absolute value of |−D| assigned to the transition at 9900cm−1 indicates that an iron center with a more distorted geometry must be associated with this MCD transition. Based on the crystal structure, the more distorted iron center is Fe2 containing the bidentate carboxylate (Glu243). Thus the transition at 9900cm−1 is assigned to Fe2.
Table 2.
Summary of the VTVH MCD Fit Parameters for the Ground and Excited States of Reduced MMOH (at 7570 cm−1), Reduced MMOH-wt-MMOB (at 7750 cm−1), Reduced MMOH-MMOB-S109A/T111A (at 7750 cm−1), Reduced MMOH-MMOB-N107A/S110A (at 8140 cm−1), and Reduced MMOH-MMOB-T111Y (at 7630 cm−1).
| MMOH | MMOH-wt-MMOB | MMOH-MMOB-S109A/T111A | MMOH-MMOB-N107A/S110A | MMOH-MMOB-T111Y | |
|---|---|---|---|---|---|
| δ (cm−1) | <~1.0 | 0.6796 | 0.1172 | 0.8102 | 1.0420 |
| g∥(ground) | 14.7 | 16.2 | 16.3 | 14.4 | 16.1 |
| Asatlim | 0.3416 | 2.0860 | 1.7604 | 1.3073 | 1.6984 |
| B(%Asatlim) | 0.5503 | 0.6750 | 0.9091 | 4.2374 | 1.1925 |
| g⊥ | 0.12 | 0.09 | 0.04 | 0 | 0 |
| Mz/Mxy | 0.09 | ~0 | ~0 | 0.042 | 0.1014 |
| ES1 (cm−1) | 7–8 | 5–6 | 5–6 | 3–5 | 4–6 |
| g∥1(excited) | 0 | 0 | 0 | 0 | 0 |
| ES2 (cm−1) | ~10 | ~12 | 9–12 | 6–8 | 7–10 |
| g∥2(excited) | 12 | 12 | 12 | 12 | 12 |
2.2. MMOH-wt-MMOB
Previously collected VTVH MCD data for the reduced MMOH-wt-MMOB complex(17) are again consistent with those collected at 7750 cm−1 (arrow in Figure 2B, Figure 3C) and have indicated that the two ferrous centers are ferromagnetically coupled, similar to MMOH, with J = 0.3 – 0.5 cm−1. However, the ground state parameters of one of the iron centers is altered (g∥ = 16, and D1 = D2 = −6 ± 3 cm−1; Table 2 and Table 3). The ground state of this complex is still a doublet with Ms = ±4, but relative to MMOH the energy separation of the two excited state sublevels increases (compare energy level inserts in Figures 3A and 3C). Wt-MMOB binding perturbs one of the iron sites, Fe2, leading to the two iron sites becoming more similar in terms of the magnitudes of their ZFS parameters. The assignment of the perturbed bands to the Fe2 site is consistent with the model of the docked structure of the MMOH-MMOB complex,(32) suggesting that the Fe2 ligands are most likely to be perturbed by MMOB binding. However, since both the sign and magnitude of the ferromagnetic coupling do not change upon binding of wt-MMOB, the active site maintains a µ-1,1 bridging carboxylate.
Table 3.
Summary of Spin-Hamiltonian Parameters for reduced MMOH, MMOH-wt-MMOB, MMOH-S109A/T111A, MMOH-MMOB-N107A/S110A, and MMOH-MMOB-T111Y Using the Doublet Model with Spin-Hamiltonian Analysis and the Spin-Projection Model. D1 is the ZFS of the iron center associated with the transition energy shown in the brackets.
| Reduced MMOH (10290 cm−1) | ||
|---|---|---|
| doublet model | spin projection model | |
| J (cm−1) | +0.2 < J < +0.4 | +0.2 < J < +0.4 |
| D1 (cm−1) | −10 < D1 < −3 | −10 < D1 < −6 |
| E1/D1 (cm−1) | > 0 | 0.01–0.33 |
| D2 (cm−1) | −5 < D2 < −1 | −4 < D2 < −2 |
| E2/D2 (cm−1) | > 0 | 0.01–0.33 |
| polarization | N/A | 59.7% z, 40.3% x |
| Reduced MMOH-wt-MMOB (7750 cm−1) | ||
| doublet model | spin-projection model | |
| J (cm−1) | +0.3 < J < +0.5 | +0.3 < J < +0.5 |
| D1 (cm−1) | −9 < D1 < −3 | −6 < D1 < −3 |
| E1/D1 (cm−1) | > 0 | 0.05–0.33 |
| D2 (cm−1) | −9 < D2 < −3 | −6 < D2 < −3 |
| E2/D2 (cm−1) | > 0 | 0.1–0.33 |
| polarization | N/A | 98.4% z, 1% x, 0.6% y |
| Reduced MMOH-MMOB-S109A/T111A (7750 cm−1) | ||
| doublet model | spin-projection model | |
| J (cm−1) | +0.3 < J < +0.5 | +0.3 < J < +0.5 |
| D1 (cm−1) | −9 < D1 < −4 | −8 < D1 < −4 |
| E1/D1 (cm−1) | > 0 | 0.05–0.33 |
| D2 (cm−1) | −7 < D2 < −3 | −6 < D2 < −3 |
| E2/D2 (cm−1) | > 0 | 0.05–0.33 |
| polarization | N/A | 86.6% z, 10.8% x, 2.6% y |
| Reduced MMOH-MMOB-N107A/S110A (8140 cm−1) | ||
| doublet model | spin-projection model | |
| J (cm−1) | +0.2 < J < +0.3 | +0.2 < J < 0.35 |
| D1 (cm−1) | −10 < D1 < -2 | −10 < D1 < -2 |
| E1/D1 (cm−1) | > 0 | 0.1–0.33 |
| D2 (cm−1) | −10 < D2 < -2 | −10 < D2 < -2 |
| E2/D2 (cm−1) | > 0 | 0.1–0.33 |
| polarization | N/A | 71.3% y, 24.7% z, 4% x |
| Reduced MMOH-MMOB-T111Y (9540 cm−1) | ||
| doublet model | spin-projection model | |
| J (cm−1) | +0.2 < J < +0.4 | +0.2 < J < +0.4 |
| D1 (cm−1) | −9 < D1 < −3 | −8 < D1 < −3 |
| E1/D1 (cm−1) | > 0 | 0.005–0.33 |
| D2 (cm−1) | −8 < D2 < −3 | −8 < D2 < −3 |
| E2/D2 (cm−1) | > 0 | 0.005–0.33 |
| polarization | N/A | 70.1% z, 19.6% x, 10.3% y |
2.3. MMOH-MMOB-S109A/T111A
VTVH MCD data for the reduced MMOH-MMOB-S109A/T111A complex were collected at 7750 cm−1 and 6370 cm−1 (arrows in Figure 2C) and are shown in Figure 3D and Figure S5, respectively. The saturation behavior is similar for both transitions. The ground-state analysis of this complex results in the lowest energy doublet |4, ±4〉 (Stot, Mstot) having a g∥ = 16 and a δ < 0.6 cm−1. This is consistent with the integer spin signal at g ~16 observed in the EPR spectrum of this complex (g ~16 resonance is also observed in the other MMOB complexes; vide infra). In addition, similar to the MMOH-wt-MMOB complex two excited state sublevels at ~5 – 6 cm−1 and ~9 cm−1 are required to obtain reasonable fits (Figure 3D inset). The ground and excited-state fitting parameters for both bands are the same and are summarized in Table 2. The two metal centers are ferromagnetically coupled with a J value similar to that determined for MMOH and MMOH-wt-MMOB (+0.3 < J < +0.5 cm−1). The spin Hamiltonian analysis places restrictions on the combinations of D values for the two ferrous centers, requiring both irons to have negative ZFS of similar magnitudes, as observed in the MMOH-wt-MMOB complex.
The VTVH MCD data collected at 7750 cm−1 and 6370 cm−1 were also fit using the spin-projection model. A reasonable correlation between observed and simulated saturation data at 7750 cm−1 could only be obtained if both |−D| values were of similar magnitude (Figure S6); due to large experimental standard deviation for the band at 6370 cm−1 a unique set of fitting solutions could not be obtained from these data with respect to the magnitudes of ZFS (see Figure S7). The results for the best fit to the 7750 cm−1 band are summarized in Table 3. Similar to the MMOH-wt-MMOB complex, the two metal centers have very similar ZFS. Thus, binding of the MMOB-S109A/T111A variant affects the active site of MMOH in a manner very similar to that caused by wt-MMOB binding.
2.4. MMOH-MMOB-N107A/S110A
Figures 3E and F show VTVH MCD data collected at 8140 cm−1 and 6530 cm−1 for the reduced MMOH-MMOB-N107A/S110A complex (Figure 2D arrows). The saturation magnetization data for both transitions exhibit a pronounced nesting behavior. An investigation of the ground state properties with EPR has shown a resonance at g ~16, indicating the presence of a ground state with Ms = ±4. Consistently, the best fit to the MCD saturation data (Table 2) requires the lowest energy doublet to have a g∥ ~14.4 and δ < 1 cm−1; the first (4,0)(3,0) and second (4, ±3) excited state doublets are at 3 – 5 cm−1 and 6 – 8 cm−1 above the ground sublevel. The spin Hamiltonian analysis of these results confirms that the two iron centers in this complex are rhombic and weakly ferromagnetically coupled; the ZFS parameters (D) for both irons are negative, but different in magnitude (Table 3). The best agreement between observed MCD saturation data and simulations is obtained for |J| ~0.2 – 0.3 cm−1, indicating weak ferromagnetic coupling mediated via a µ-(1,1) carboxylate bridge (Table 3).
Both the bands at 8140 cm−1 and 6530 cm−1 were analyzed using the spin-projection model. While the experimental data do not allow the transitions to be associated with a specific iron center, the fits indicate that similar to MMOH, the magnitudes of the |−D| values for the two Fe’s are different (Figure S8 and Figure S9). The two irons in this complex resemble the two iron centers in MMOH with respect to their ZFS parameters, with the exception that this MMOB variant complex exhibits a pronounced nesting behavior at low temperature due to a very low-lying excited state at ~3 – 5 cm−1 above the ground-state sublevel that results in a larger B-term contribution to MCD intensity relative to MMOH.
2.5. MMOH-MMOB-T111Y
Saturation magnetization data for the reduced MMOH-MMOB-T111Y complex collected at 7630 cm−1, 9540 cm−1 and 6190 cm−1 (arrows in Figure 2E) are shown in Figure 3G, 3H, and Figure S10. The three transitions exhibit rapid saturation behavior, indicative of a system with a large g∥ value. Analogous to the above analyses the ground state analysis reveals the lowest energy level as an Ms = ±4 non-Kramers doublet with g∥ = 16 and δ = 0.8 cm−1; two excited state doublets at 4 cm−1 (g∥ = 0) and at ~7 – 10 cm−1 (g∥ = 12) were required to obtain a reasonable fit (see Table 2). Thus, the complex is also weakly ferromagnetically coupled (+0.2 < J < +0.4 cm−1).
The spin Hamiltonian analysis demonstrates that for both ferrous centers the D values lie between −3 cm−1 and −8 cm−1 (Table 3). Although it is not possible to distinguish between the two metal centers using the spin-projection model, best fits are obtained when the magnitudes of D1 and D2 are different (Figure S11 and Figure S12).
In summary, reduced MMOH and all four MMOH-MMOB complexes have similar ground state properties consistent with the presence of weakly ferromagnetically coupled ferrous centers linked via a µ-1,1 bridging carboxylate. The ZFS changes observed in the MMOH-wt-MMOB and the MMOH-MMOB-S109A/T111A complexes relative to MMOH are similar, consistent with the observed CD/MCD data. This suggests that only one Fe (Fe2 associated with transitions at ~6200 and ~9900 cm−1) is perturbed upon MMOB binding and its ZFS becomes similar to that of Fe1 (associated with transitions at ~7400 and ~9100 cm−1). In contrast, in MMOH and the remaining MMOB variant complexes, the two metal sites have different ZFS. In the next section, ligand field calculations are employed to correlate the experimentally observed LF transitions and ZFS of each iron center with geometric and electronic structure and its change with MMOB binding.
3. Ligand Field Calculations of Excited-State Energy Splittings
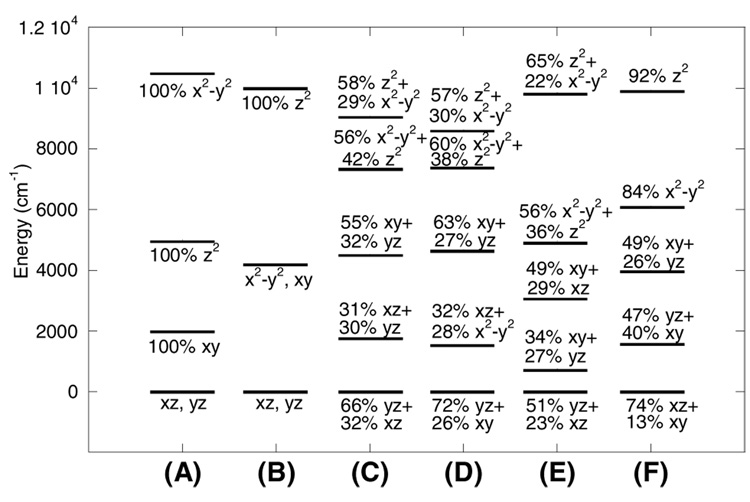
While the crystal structures for several reduced forms of MMOH have been published, no structure is yet available for the reduced MMOH-MMOB complex. To gain insight into the electronic structure of each iron center in the active site of MMOH and the effect of component B binding on this active site, the method of Companion and Komarinsky(43) was used to assess the ligand field (LF) for each ferrous center of MMOH (see Supporting Information Tables S1–S8). LF calculations facilitate the assignment of the spectral features corresponding to each of the two iron centers to the relevant metal ions in the structure of MMOH, i.e. Fe1 and Fe2 (Figure 1). Importantly, LF calculations also provide insight into structural changes that occur upon binding of MMOB to MMOH.
The method of Companion and Komarinsky utilizes the electrostatic crystal field approximation, where ligands are treated as point charges that induce perturbations on the FeII d-orbital energy levels. The Hamiltonian matrix, used for the calculation of the d-orbital energy levels with spatial coordinates from a relevant crystal structure, and the ligand field parameters α2 and α4 for each ligand have been described previously.(40) The parameters α2 and α4 for oxygen- and nitrogen-based ligands coordinated to Fe(II) are determined experimentally using crystallographically and spectroscopically well-characterized ferrous model systems.(44, 45) The α2 and α4 parameters used for carboxylates, solvent molecules and histidines, together with the corresponding FeII-ligand bond lengths are given in Supporting Information Tables S1–2 and figure S2–figure S5.
3.1. Ligand Field of MMOH
The multiple crystal structures available for reduced MMOH exhibit some degree of variation with respect to the number of water molecules coordinated to the two ferrous centers (one or two waters coordinated to Fe1 and one or no water on the Fe2 site; Figure 1). Our earlier study used LF calculations to investigate general trends and the effects of different geometries and their specific distortions on the excited state splittings for two Fe(II) centers.(17) While this study evaluated the splitting of the excited state LF transitions for two ferrous centers in the context of their geometries, no analysis was presented on the specific changes in the active site of MMOH induced by MMOB component binding. Here the previous LF calculations are extended to correlate possible changes in the active site of MMOH induced by MMOB binding to changes in the observed LF transitions for the two irons. A ligand field model of the reduced MMOH active site was constructed using the coordinates from the crystal structure with the PDB code 1MTY as shown in Figure 1. Other crystal structures of reduced MMOH were also examined in preliminary LF calculations, but the selected structure results in the best agreement between experimental and calculated d-orbital excited state splittings (see Supporting Information, Table S8).
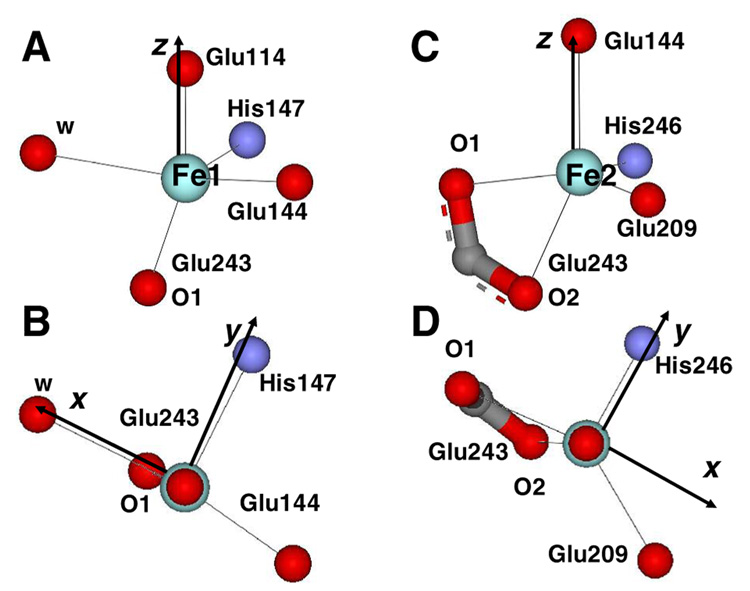
The geometry of the Fe1 site (Figure 4A–B) is distorted between the limits of square pyramidal and trigonal bipyramidal.(46) Designating the Fe1-O1Glu114 bond (1.90 Å) as the z-axis and Fe1-NHis147 as y, ligands NHis147, Owat and OGlu144 lie within the xy plane (Figure 4B), with both oxygen ligands tilted towards the y-axis (∠Owat-Fe1-OGlu144 = 166.6°; Figure 4B). The O1 ligand of residue Glu243 is shifted in −z direction away from the axial OGlu114 ligand. The net result of these distortions is a stabilization of the dz2 orbital and a destabilization of the dx2-y2 orbital, thus giving rise to a small energy splitting of the two associated d-d transitions. The energies and compositions of the relevant molecular orbitals are given in Table 4 and Figure 5C. The calculated d-d transitions are estimated to be 7360 and 9073 cm−1, with an excited state splitting of ~1500 cm−1. These values are in good agreement with the spectral data assigned to Fe1 (7500 cm−1 and 9100 cm−1 with an excited state splitting of 1600 cm−1; Table 4).
Figure 4.
(A) Side view and (B) top view for the orientation of the Fe1 site in reduced MMOH. (C) Side view and (D) top view for the orientation of the Fe2 site in reduced MMOH. The coordinate system obtained from the ligand field calculations is indicated.
Table 4.
Summary of experimental spectral features for Fe1 and Fe2 centers in reduced MMO complexes and comparison to the Calculated Ligand Field Transition Energies for Reduced MMOH, MMOH-wt-MMOB, MMOH-MMOB-N107A/S110A, MMOH-MMOB-T111Y, and MMOH-MMOB-S109A/T111A Complexes.
| Reduced Complex | Fe1 LF transitions (cm−1) | Fe2 LF transitions (cm−1) | D1 (cm−1) | D2 (cm−1) | ||
|---|---|---|---|---|---|---|
| MMOH | −4 | −10 | ||||
| (exp.) | 7360 | 9140 | - | 9900 | −3 | −9 |
| (calc.) | 7360 | 9073 | 4930 | 9860 | ||
| MMOH-wt | −6 | −6 | ||||
| MMOB | −5 | −5 | ||||
| (exp.) | 7590 | 8650 | 6170 | 9840 | ||
| (calc.) | 7420 | 8635 | 6121 | 9939 | ||
| MMOH- | −5 | −5 | ||||
| MMOB- | −8 | −6 | ||||
| S109A/T111Aa | −6 | −8 | ||||
| (exp.) | 7560 | 8630 | 6250 | 9850 | ||
| (calc.) | - | - | - | - | ||
| MMOH- | −3 | −10 | ||||
| MMOB- | −4 | −10 | ||||
| N107A/S110A | −3 | −9 | ||||
| (exp.) | 6500 | 8050 | 5520 | 9865 | ||
| (calc.) | 6560 | 8140 | 5400 | 9940 | ||
| MMOH- | −8 | −6 | ||||
| MMOB- | −6 | −8 | ||||
| T111Y | ||||||
| (exp.) | 7160 | 8200 | 5920 | 9800 | ||
| (calc.) | 6900 | 8400 | 5900 | 10000 | ||
The ligand field transitions for MMOB-S109A/T111A variant were not calculated since its transitions are identical to those of wt-MMOB.
Figure 5.
Results of ligand field calculations showing calculated d-orbital energy level diagrams of (A) an ideal 5-coordinate square pyramidal geometry, (B) an ideal 5-coordinate trigonal bipyramidal geometry, (C) Fe1 in MMOH, (D) Fe1 in MMOH-wt-MMOB, (E) Fe2 in MMOH, and (F) Fe2 in MMOH-wt-MMOB. Wave function descriptions use Fe1-OGlu114 bond as z axis and the Fe1-water bond as the x axis in Fe1, and Fe2-OGlu144 bond as z axis and the Fe1-NHis246 bond as the x axis in Fe2.
The geometry of the Fe2 site (Figure 4C) is also distorted between the limits of square pyramidal and trigonal bipyramidal. Here, the Fe2-O2Glu144 bond is designated as the z-axis, the Fe2-NHis246 as y, and ligands NHis246, O1Glu243 and OGlu209 span the xy plane (Figure 4C). In contrast to the Fe1 site, the planar oxygen ligands are less tilted towards the y-axis (NHis246-Fe2-OGlu209 = 120.5°, O1Glu243-Fe2-OGlu209 = 143.3°), adopting a more regular trigonal bipyramidal geometry (Figure 4D). The O2Glu243 (from µ-1,1 bridging Glu243) occupies a position approximately opposite the apical O2Glu144 ligand (Figure 4D). In comparison to the Fe1 site, the Fe2 site possesses a distinctly more square pyramidal character, resulting in an increased excited state splitting, as initially predicted.(17) The LF calculations give two transitions at 4930 cm−1 and 9860 cm−1, leading to an energy splitting of 4930 cm−1 (Table 4). The low energy transition is below the detection limit (~5000 cm−1) of the CD spectrophotometer, but the higher energy band observed at ~9900 cm−1, assigned to Fe2, is in agreement with the calculations (Table 4), with an excited state energy splitting for Fe2 of ~5000 cm−1.
3.2. Ligand Field of the MMOH-MMOB complex
The spectroscopic and computational data presented here have demonstrated that both irons in the active site of MMOH are coordinatively unsaturated, providing, in principle, two vacant positions (one on each iron) for dioxygen binding. However, as mentioned above, in the absence of MMOB reduced MMOH is relatively unreactive with O2 (the rate of formation of the peroxo intermediate P is <1 s−1). It thus follows that the access of O2 to the irons is limited, and that some structural rearrangement in the MMOH active site may be necessary in order to increase reactivity. In order to examine the effects of MMOB on the MMOH active site, LF calculations were combined with spectroscopic and crystallographic data to develop a plausible structural model for the regulation of O2 reactivity. Inspection of the MMOH active site structure (Figure 1) indicates that the most direct approach for dioxygen binding is via apical positions opposite the two histidine ligands His147 (Fe1; Figure 4B) and His 246 (Fe2; Figure 4D). While access of O2 to Fe1 is unhindered, two connected obstructions, OGlu209 and an H2O hydrogen-bonded to it, mask its access to Fe2. EPR data suggest that, although Fe1 seems to be accessible, it alone does not readily bind O2 to form a superoxo intermediate as evident from the persistence of g ~16 signal in the slow reaction of MMOH with O2 in the absence of MMOB.(11, 21) Past DFT calculations examining the energetics of formation of a monodentate superoxo (end-on-bound O2 to one iron center) vs. the peroxo intermediate in ribonucleotide reductase(47) show that the end-on-bound superoxide Fe(III)-Fe(II) species is much higher in energy (thermodynamically unstable by 40 kcal/mol) relative to the bridged cis-µ-1,2 peroxo Fe(III)-Fe(III) species.(47) This suggests that Fe1 in MMOH may be unreactive towards O2 on its own due to the poor super-exchange pathway of the carboxylate bridges, which do not allow for stabilization of the superoxide through the second electron transfer to O2 from Fe2 (bridging carboxylates are present in both MMO and ribonucleotide reductase). This assessment is in accord with a recent DFT study by Han and Noodleman(48) favoring the bridged cis-µ-1,2 structure as the preferred model for P intermediate in the MMO-catalyzed reaction.
CD/MCD data show that the addition of MMOB affects predominantly the Fe2 site of MMOH, while changes in the Fe1 site are minor (Table 4), supporting a mechanism where Fe2 must be made accessible for a bridged peroxo intermediate to form. Thus, reorientation of Glu209, through movement of the ∠Fe1-Fe2-Glu209, emerges as the likely coordinate relevant to the modulation of MMOH reactivity. Upon increasing this angle to 135°, the Fe2 site adopts a less distorted square pyramidal conformation as compared to the MMOH resting state, with the NHis246 as the apical ligand (Figure 4D). This arrangement would greatly improve O2 access to Fe2. LF calculations performed for the Fe2 site with ∠Fe1-Fe2-Glu209 = 135° give transitions at 6160 cm−1 and 9900 cm−1 (excited state splitting = 3740 cm−1), in agreement with the experimental energies of 6170 cm−1 and 9860 cm−1 for the MMOH-wt-MMOB complex (Figure 5F and Table 4).(49)
An analysis of the molecular orbital coefficients for Fe1 shows that the ground state redox active d orbital has yz as its predominant character, independent of the absence or presence of component B (Figure 5 E to F), facilitating a strong σ bonding interaction with one π* orbital of O2. In contrast, the corresponding d orbital on Fe2 changes its character from predominantly yz to predominantly xz upon binding of MMOB (Figure 5). This change in character of the redox active orbital of Fe2 would enable effective π overlap with O2. Consequently, MMOB binding activates the biferrous site for the formation of a peroxo intermediate (compound P).
Discussion
While resting reduced MMOH is relatively unreactive with dioxygen, the addition of wild-type MMOB increases its reactivity ~1000-fold.(10) Here, we have used spectroscopy combined with Ligand Field Theory to probe the specific effects of binding wt-MMOB, and several of its variants, on the biferrous active site of MMOH and to correlate these effects to their observed O2 reactivity. These data have improved our understanding of the coordination geometry of each iron site in reduced MMOH, resolving the number of water molecules coordinated to the Fe1 center. The CD/MCD data are consistent with the presence of two five-coordinate, weakly ferromagnetically coupled ferrous ions in MMOH, both in the absence and presence of MMOB. Additionally, the LF calculations allow the assignment of a set of specific spectral features to each iron center in the crystal structure of MMOH, confirming that Fe1 (Figure 1) correlates with LF transitions at 7500 and 9100 cm−1 and Fe2 with LF transition at <5000 and 9900 cm−1 (Table 1). The weak ferromagnetic coupling (J = 0.3–0.4 cm−1) supports the presence of a µ-1,1 carboxylate bridge. Both iron sites can be best described as having distorted geometries between the trigonal bipyramidal and square pyramidal limits (Figure 4). On Fe1, the mixing of the dx 2–y2 and dz2 orbitals leads to a small d-d excited state splitting (~1600 cm−1). For the Fe2 site, the distortion towards square pyramidal is more pronounced as evidenced by the larger excited state splitting and larger negative ZFS.
CD/MCD measurements indicate that the electronic and geometric structures of MMOH are perturbed upon binding of MMOB (Table 1). While binding of wt MMOB induces a conformational change in the active site of MMOH, the coordination numbers of the two iron centers are not affected and the metal ions are still weakly ferromagnetically coupled (~0.4 cm−1), indicating that the µ-1,1 carboxylate bridge remains. The observed structural change is associated primarily with the Fe2 center, resulting in a decreased LF splitting of its transitions (from >4900 to ~3700 cm−1), while negligible change is observed in the Fe1 center (Table 4). The net effect of wt-MMOB binding results in the Fe2 center becoming more similar to the Fe1 center, both in its electronic and geometric structure.
Ligand field calculations show that structural changes associated with the terminal glutamate on Fe2 can reproduce the experimentally observed d-d transition energies of the MMOH-MMOB complexes, providing insight into the mechanism of the regulation of O2 reactivity. The best agreement between spectroscopic data and LF calculations is obtained by increasing the Fe-Fe-Glu209 angle (Figure 6), resulting in a more open structure that would enable dioxygen to bridge the site by coordinating to the vacant positions opposite the His ligands on each iron, thus forming the bridged peroxo Fe(III)-Fe(III) species.(47, 48) The invoked flexibility and likely perturbation of Glu209 residue (from our CD/MCD results and LF calculations) as one of the main factors contributing to regulation of MMO reactivity is in agreement with the proposed models arising from the EPR spin-labeling(38) and chemical cross-linking experiments(32) that have demonstrated that the likely interface between MMOH and MMOB allows interaction with the MMOH helices containing Glu209.(32) The LF analysis further indicates that the reorientation of the Glu209 residue changes the redox active orbital of Fe2, which may facilitate efficient overlap with the orthogonal O2 π* orbitals. Both of these findings are in agreement with recent DFT calculations performed on the reduced active site of MMOH(50) and appear necessary for efficient reaction of O2 with the diiron site.
Figure 6.
Proposed effect of component B on the active site structure of the reduced MMOH active site.
These active site perturbations nicely explain most of the CD/MCD data obtained for the three variant complexes. The ligand field change on Fe2, shown by the perturbation of the outer peaks in the MCD spectra, correlates with an increase in the reactivity of MMOH bound by wt-MMOB and the MMOB-S109A/T111A variant. Likewise for MMOB-N107A/S110A, where there is no change in O2 reactivity upon binding of this variant to MMOH, there is no perturbation at the Fe2 center observed in the MCD spectrum. In the MMOB-T111Y variant, however, a change similar to that induced by the wt-MMOB is observed in the MCD even though the O2 reactivity of MMOH is not increased upon binding, indicating the geometric and electronic structural change at Fe2 is not sufficient for O2 reactivity. Binding this variant to MMOH does not, however, induce the conformational change reflected in the CD spectrum for wt-MMOB binding. This suggests that binding wt-MMOB and the S109A/T111A variant to MMOH, which both exhibit similar perturbations in their CD spectra, increases reactivity by modulating the access of O2 to the active site as well as by altering the geometric and electronic structure of the Fe2 center. An earlier study had also suggested that the conformational change associated with MMOB binding was necessary to allow substrate molecules to interact with the active site pocket.(17, 18) Additionally, conformational changes in the vicinity of the MMOH diiron cluster have been invoked to account for the 5-fold decrease in the deuterium kinetic isotope for the reaction of intermediate Q with methane when MMOB is replaced by mutated forms similar to those used in this study.(34, 37) Therefore, these studies support a two-fold role for MMOB, both of which must occur for efficient reactivity: (i) regulation of O2 and substrate access to the site (from CD) and (ii) activation of the diferrous site for O2 reaction through LF changes on Fe2 (from MCD).
In summary, the spectroscopic data and LF calculations presented here have provided insight into the role of MMOB in the regulation of O2 reactivity of MMOH. Reduced resting MMOH is relatively unreactive towards dioxygen which appears to result from the steric hindrance within the active site due to Glu209 on Fe2 (Figure 1) coupled with poor overlap between the redox active orbitals on the two irons and the π* orbitals of O2. Binding of MMOB to MMOH induces a geometric rearrangement in the Fe2 site (Figure 6) allowing O2 to effectively bridge the two irons and a more global conformational change that may control access of O2 and substrate to the site. Both the conformational change and the ligand field perturbation at Fe2 appear to be required for the O2 reactivity of MMO.
Supplementary Material
Temperature dependence MCD data for MMOH-MMOB-N107A/S110A complex, VTVH MCD fitted data for different transitions for MMOH, MMOH-MMOB-S109A/T111A, MMOH-MMOB-N107A/S110A, and MMOH-MMOB-T111Y complexes, ligand field parameters used in the calculations of d-orbital energy levels for Fe1 and Fe2 center of reduced MMOH and the corresponding MMOH-MMOB complexes, and ligand field parameters used in LF calculations of the reduced active site of MMOH with different coordination numbers for each iron center, comparison of experimental and calculated LF transition energies for Fe1 and Fe2 sites of reduced MMOH active site. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations
- CD
circular dichroism
- MCD
magnetic circular dichroism
- VTVH MCD
variable temperature variable field MCD
- sMMO
soluble methane monooxygenase
- MMOH
hydroxylase component of soluble methane monooxygenase
- MMOB
component B of methane monooxygenase, binding is necessary for efficient O2 reactivity
- WT-MMOB
wild-type MMOB
- MMOR
reductase component of methane monooxygenase
- O, P*, P, Q, T
intermediates O, P*, P, Q, and T from the MMOH catalytic cycle
- MMOH-MMOB
complex between hydroxylase and component B of methane monooxygenase
- NADH
nicotinamide adenine dinucleotide, reduced form
- DFT
density functional theory
- LF
ligand field
- ZFS
zero-field splitting
- J
exchange coupling, coupling between the two iron sites mediated via bridging ligands
- D and E
axial and rhombic zero-field splitting parameters, probing each Fe(II) site geometry
- g∥i and g⊥i
dimer g values of a particular ith doublet
- δ
rhombic zero-field splitting of the ground-state doublet
- Mxy, Mxz, and Myz
polarizations for the directions indicated
Footnotes
Financial support for this research by NSF-Biophysics Program Grant MCB-0342807 (E.I.S.) and NIH Grant GM40466 (J.D.L.).
References
- 1.Dalton H. Oxidation of hydrocarbons by methane monooxygenase from a variety of microbes. Adv. Appl. Microbiol. 1980;26:71–87. [Google Scholar]
- 2.Wallar BJ, Lipscomb JD. Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 3.Colby J, Stirling DI, Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenase n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green J, Dalton H. Substrate specificity of soluble methane monooxygenase. Mechanistic implications. J. Biol. Chem. 1989;264:17698–17703. [PubMed] [Google Scholar]
- 5.Higgins IJ, Best DJ, Hammond RC. New findings in methane-utilizing bacteria highlight their importance in the biosphere and their commercial potential. Nature. 1980;286:561–564. doi: 10.1038/286561a0. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Lipscomb JD. Desaturation reactions catalyzed by soluble methane monooxygenase. J. Biol. Inorg. Chem. 2001;6:717–725. doi: 10.1007/s007750100250. [DOI] [PubMed] [Google Scholar]
- 7.Fox BG, Froland WA, Dege JE, Lipscomb JD. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a 3-component system with high specific activity from a type II methanotroph. J. Biol. Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 8.Lund J, Dalton H. Further characterization of the FAD and Fe2S2 redox centers of component C, the NADH: acceptor reductase of the soluble methane monooxygenase of Methylococcus capsulatus (Bath) Eur. J. Biochem. 1985;147:291–296. doi: 10.1111/j.1432-1033.1985.tb08749.x. [DOI] [PubMed] [Google Scholar]
- 9.Green J, Dalton H. Protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath). A novel regulator protein of enzyme activity. J. Biol. Chem. 1985;260:15795–15801. [PubMed] [Google Scholar]
- 10.Liu Y, Nesheim JC, Lee S-K, Lipscomb JD. Gating effects of component B on oxygen activation by the methane monooxygenase hydroxylase component. J. Biol. Chem. 1995;270:24662–24665. doi: 10.1074/jbc.270.42.24662. [DOI] [PubMed] [Google Scholar]
- 11.Fox BG, Surerus KK, Münck E, Lipscomb JD. Evidence for a µ-oxo bridged binuclear iron cluster in the hydroxylase component of methane monooxygenase. Mössbauer and EPR studies. J. Biol. Chem. 1988;263:10553–10556. [PubMed] [Google Scholar]
- 12.Elango N, Radhakrishnan R, Froland WA, Wallar BJ, Earhart CA, Lipscomb JD, Ohlendorf DH. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 1997;6:556–568. doi: 10.1002/pro.5560060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-heme iron hydroxylase that catalyzes the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 14.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Geometry of the soluble methane monooxygenase catalytic diiron center in two oxidation states. Chem. Biol. 1995;2:409–418. [PubMed] [Google Scholar]
- 15.Rosenzweig AC, Brandstetter H, Whittington DA, Nordlund P, Lippard SJ, Frederick CA. Crystal structure of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins. 1997;29:141–152. [PubMed] [Google Scholar]
- 16.Whittington DA, Lippard SJ. Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometric variability at the dinuclear iron active site. J. Am. Chem. Soc. 2001;123:827–838. doi: 10.1021/ja003240n. [DOI] [PubMed] [Google Scholar]
- 17.Pulver S, Froland WA, Fox BG, Lipscomb JD, Solomon EI. Spectroscopic studies of the coupled binuclear non-heme iron active site in the fully reduced hydroxylase component of methane monooxygenase: Comparison to deoxy and deoxy-azide hemerythrin. J. Am. Chem. Soc. 1993;115:12409–12422. [Google Scholar]
- 18.Pulver SC, Froland WA, Lipscomb JD, Solomon EI. Ligand field circular dichroism and magnetic circular dichroism studies of component B and substrate binding to the hydroxylase component of methane monooxygenase. J. Am. Chem. Soc. 1997;119:387–395. [Google Scholar]
- 19.Lee S-K, Nesheim JC, Lipscomb JD. Transient intermediates of the methane monooxygenase catalytic cycle. J. Biol. Chem. 1993;268:21569–21577. [PubMed] [Google Scholar]
- 20.Lee S-K, Fox BG, Froland WA, Lipscomb JD, Münck E. A transient intermediate of the methane monooxygenase catalytic cycle containing an Fe(IV)-Fe(IV) cluster. J. Am. Chem. Soc. 1993;115:6450–6451. [Google Scholar]
- 21.Lee SY, Lipscomb JD. Oxygen activation catalyzed by methane monooxygenase hydroxylase component: Proton delivery during the O-O bond cleavage steps. Biochemistry. 1999;38:4423–4432. doi: 10.1021/bi982712w. [DOI] [PubMed] [Google Scholar]
- 22.Liu KE, Valentine AM, Wang DL, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. Kinetic and spectroscopic characterization of intermediates and component interactions in reactions of methane monooxygenase from Methylococcus capsulatus (Bath) J. Am. Chem. Soc. 1995;117:10174–10185. [Google Scholar]
- 23.Shu LJ, Nesheim JC, Kauffmann K, Münck E, Lipscomb JD, Que L., Jr An Fe(IV)-O2 diamond core structure for the key intermediate Q of methane monooxygenase. Science. 1997;275:515–518. doi: 10.1126/science.275.5299.515. [DOI] [PubMed] [Google Scholar]
- 24.Brazeau BJ, Lipscomb JD. Kinetics and activation thermodynamics of methane monooxygenase hydroxylase compound Q formation and reaction with substrates. Biochemistry. 2000;39:13503–13515. doi: 10.1021/bi001473l. [DOI] [PubMed] [Google Scholar]
- 25.Lipscomb JD, Que L., Jr MMO: P450 in wolf's clothing? J. Biol. Inorg. Chem. 1998;3:331–336. [Google Scholar]
- 26.Brazeau BJ, Austin RN, Tarr C, Groves JT, Lipscomb JD. Intermediate Q from soluble methane monooxygenase hydroxylates the mechanistic substrate probe norcarane: Evidence for a stepwise reaction. J. Am. Chem. Soc. 2001;123:11831–11837. doi: 10.1021/ja016376+. [DOI] [PubMed] [Google Scholar]
- 27.Wallar BJ, Lipscomb JD. Methane monooxygenase component B mutants alter the kinetic steps throughout the catalytic cycle. Biochemistry. 2001;40:2220–2233. doi: 10.1021/bi002298b. [DOI] [PubMed] [Google Scholar]
- 28.Fox BG, Liu Y, Dege JE, Lipscomb JD. Complex formation between the protein components of methane monooxygenase from Methylosinus trichosporium OB3b: Identification of sites of component interaction. J. Biol. Chem. 1991;266:540–550. [PubMed] [Google Scholar]
- 29.Paulsen KE, Liu Y, Fox BG, Lipscomb JD, Münck E, Stankovich MT. Oxidation reduction potentials of the methane monooxygenase hydroxylase component from Methylosinus trichosporium OB3b. Biochemistry. 1994;33:713–722. doi: 10.1021/bi00169a013. [DOI] [PubMed] [Google Scholar]
- 30.Liu KE, Lippard SJ. Redox properties of the hydroxylase component methane monooxygenase from Methylococcus capsulatus (Bath): Effects of protein B, reductase, and substrate. J. Biol. Chem. 1991;266:12836–12839. [PubMed] [Google Scholar]
- 31.Froland WA, Andersson KK, Lee SK, Liu Y, Lipscomb JD. Methane monooxygenase component B and reductase alter the regioselectivity of the hydroxylase component-catalyzed reactions: A novel role for protein-protein interactions in an oxygenase mechanism. J. Biol. Chem. 1992;267:17588–17597. [PubMed] [Google Scholar]
- 32.Brazeau BJ, Wallar BJ, Lipscomb JD. Effector proteins from P450(cam) and methane monooxygenase: lessons in tuning nature's powerful reagents. Biochem. Biophys. Res. Commun. 2003;312:143–148. doi: 10.1016/j.bbrc.2003.09.242. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JY, Wallar BJ, Popescu CV, Renner DB, Thomas DD, Lipscomb JD. Methane monooxygenase hydroxylase and B component interactions. Biochemistry. 2006;45:2913–2926. doi: 10.1021/bi052256t. [DOI] [PubMed] [Google Scholar]
- 34.Zheng H, Lipscomb JD. Regulation of methane monooxygenase catalysis based on size exclusion and quantum tunneling. Biochemistry. 2006;45:1685–1692. doi: 10.1021/bi051605g. [DOI] [PubMed] [Google Scholar]
- 35.Chang SL, Wallar BJ, Lipscomb JD, Mayo KH. Solution structure of component B from methane monooxygenase derived through heteronuclear NMR and molecular modeling. Biochemistry. 1999;38:5799–5812. doi: 10.1021/bi982992f. [DOI] [PubMed] [Google Scholar]
- 36.Walters KJ, Gassner GT, Lippard SJ, Wagner G. Structure of the soluble methane monooxygenase regulatory protein B. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7877–7882. doi: 10.1073/pnas.96.14.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brazeau BJ, Lipscomb JD. Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B. Biochemistry. 2003;42:5618–5631. doi: 10.1021/bi027429i. [DOI] [PubMed] [Google Scholar]
- 38.MacArthur R, Sazinsky MH, Kühne H, Whittington DA, Lippard SJ, Brudvig GW. Component B binding to the soluble methane monooxygenase hydroxylase by saturation-recovery EPR spectroscopy of spin-labeled MMOB. J. Am. Chem. Soc. 2002;124:13392–13393. doi: 10.1021/ja0279904. [DOI] [PubMed] [Google Scholar]
- 39.Sazinsky MH, Dunten PW, McCormick MS, DiDonato A, Lippard SJ. X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase. Biochemistry. 2006;45:15392–15404. doi: 10.1021/bi0618969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solomon EI, Pavel EG, Loeb KE, Campochiaro C. Magnetic circular dichroism spectroscopy as a probe of the geometric and electronic structure of non-heme iron ferrous enzymes. Coord. Chem. Rev. 1995;144:369–460. [Google Scholar]
- 41.Wei P-P, Skulan AJ, Mitić N, Yang Y-S, Saleh L, Bollinger JM, Jr, Solomon EI. Electronic and spectroscopic studies of the non-heme reduced binuclear iron sites of two ribonucleotide reductase variants: Comparison to reduced methane monooxygenase and contribution to O2 reactivity. J. Am. Chem. Soc. 2004;126:3777–3788. doi: 10.1021/ja0374731. [DOI] [PubMed] [Google Scholar]
- 42.Neese F, Solomon EI. MCD C-term signs, saturation behavior, and determination of band polarizations in randomly oriented systems with spin S ≥ 1/2. Applications to S = 1/2 and S = 5/2. Inorg. Chem. 1999;38:1847–1865. doi: 10.1021/ic981264d. [DOI] [PubMed] [Google Scholar]
- 43.Companion AL, Komarynsky MA. Crystal-filed splitting diagrams. J. Chem. Ed. 1964;41:257–262. [Google Scholar]
- 44.Solomon EI, Kirk ML, Gamelin DG, Pulver S. Bioinorganic spectroscopy. Methods Enzymol. 1995;246:71–110. doi: 10.1016/0076-6879(95)46007-1. [DOI] [PubMed] [Google Scholar]
- 45.Lever APB. Electronic spectra of dn ions, Iron(II) d6. In: Lever APB, editor. Inorganic Electronic Spectroscopy. 2nd ed. New York: Elsewier; 1984. pp. 458–467. [Google Scholar]
- 46.Two H2O molecules are potentially within bonding distance of Fe1 with Fe1-O bond lengths of 1.92 Å and 2.63 Å, respectively. In addition, the water molecule at 1.92 Å is also relatively close to the Fe2 (2.72 Å). Therefore, LF calculations were performed for three possibilities, where Fe1 and Fe2 are assumed to be either (i) both five-coordinate, (ii) six- and five-coordinate, or (iii) five- and six-coordinate, respectively. The first possibility, with two five-coordinate sites, reproduces the spectroscopic data measured for MMOH best (Table 4 and S5–S7). DFT calculations were also employed in a parallel study (Schwartz JK, Wei P-P, Mitchell KH, Fox BG, Solomon EI. J. Am. Chem. Soc. 2008;130:7098–7109. doi: 10.1021/ja800654d.) to optimize the ground state structure of the binuclear cluster in MMOH. The outcome supports the presence of two five-coordinate sites, with W1(Figure 1) bound to Fe1, while W2 optimizes to a non-bonded position 2.9 Å from Fe1, approximately opposite the histidine ligand of the Fe2 site, forming a hydrogen bond with Glu209.
- 47.Wei P-P, Skulan AJ, Wade H, DeGrado WF, Solomon EI. Spectroscopic and computational studies of the de Novo designed protein DF2t: Correlation to the biferrous active site of ribonucleotide reductase and factors that affect O2 reactivity. J. Am. Chem. Soc. 2005;127:16098–16106. doi: 10.1021/ja053661a. [DOI] [PubMed] [Google Scholar]
- 48.Han W-G, Noddleman L. Stuctural model studies for the peroxo intermediate P and the reaction pathway from P → Q of methane monooxygenase using broken-symmetry density functional calculations. Inorg. Chem. 2008;47:2975–2986. doi: 10.1021/ic701194b. [DOI] [PubMed] [Google Scholar]
- 49.Note: DFT geometry optimizations systematically varying the Fe1-Fe2-Glu209 found that an angle of 135° was necessary to mimic the spectral changes observed upon formation of the MMOH-MMOB complex. This structure was ~8 kcal/mol higher in energy than the initially optimized resting structure, a difference that MMOB binding could offset.
- 50.Schwartz JK, Wei P-P, Mitchell KH, Fox BG, Solomon EI. Geometric and electronic structure studies of the binuclear non-heme ferrous active site of toluene-4-monooxygenase: Parallels with methane monooxygenase and insight into the role of the effector proteins in O2 activation. J. Am. Chem. Soc. 2008;130:7098–7109. doi: 10.1021/ja800654d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Temperature dependence MCD data for MMOH-MMOB-N107A/S110A complex, VTVH MCD fitted data for different transitions for MMOH, MMOH-MMOB-S109A/T111A, MMOH-MMOB-N107A/S110A, and MMOH-MMOB-T111Y complexes, ligand field parameters used in the calculations of d-orbital energy levels for Fe1 and Fe2 center of reduced MMOH and the corresponding MMOH-MMOB complexes, and ligand field parameters used in LF calculations of the reduced active site of MMOH with different coordination numbers for each iron center, comparison of experimental and calculated LF transition energies for Fe1 and Fe2 sites of reduced MMOH active site. This material is available free of charge via the Internet at http://pubs.acs.org.