Abstract
The first whole genomes to be compared for phylogenetic inference were those of mitochondria, which provided the first sets of genome-level characters for phylogenetic reconstruction. Most powerful among these characters has been the comparisons of the relative arrangements of genes, which has convincingly resolved numerous branch points, including those that had remained recalcitrant even to very large molecular sequence comparisons. Now the world faces a tsunami of complete nuclear genome sequences. In addition to the tremendous amount of DNA sequence that is becoming available for comparison, there is also a potential for many more genome-level characters to be developed, including the relative positions of introns, the domain structures of proteins, gene family membership, the presence of particular biochemical pathways, aspects of DNA replication or transcription, and many others. These characters can be especially convincing owing to their low likelihood of reverting to a primitive condition or occurring independently in separate lineages, thereby reducing the occurrence of homoplasy. The comparisons of organelle genomes pioneered the way for using such features for phylogenetic reconstructions, and it is almost certainly true, as ever more genomic sequence becomes available, that further use of genome-level characters will play a big role in outlining the relationships among major animal groups.
Keywords: genome, evolution, phylogeny, phylogenetically inferred groups, genome-level characters, gene family
1. Why do we need anything other than molecular sequence comparisons?
Over the past few decades, the comparison of nucleotide and amino acid sequences has revolutionized our understanding of the evolutionary relationships for many groups of organisms. The broader field of systematics has been reinvigorated and a generation of evolutionary biologists has come to accept that molecular sequence comparisons are an essential component for inferring phylogeny of any group. These studies have led to extensive revision of animal systematics and overturning of previous reliance on the features of the coelom and segmentation (Adoutte et al. 1999).
In the 1980s, when comparing molecular sequences for phylogenetic inference was first becoming common, some asserted with great confidence that all evolutionary relationships would soon be convincingly resolved solely with this type of data, leading to much consternation. However, some of the relationships that were equivocal in early molecular studies have remained highly recalcitrant even with much more DNA sequence data in hand. There are several potential explanations, including: (i) multiple nucleotide or amino acid substitutions may have occurred at a single site, obscuring any accumulated signal; (ii) convergent or parallel substitutions may have occurred among different lineages due to having only 4 (for nucleotides) or 20 (for amino acids) possible character states, exacerbated by convergent biases in base composition (Naylor & Brown 1998), which may even cause ever increasing confidence measures for incorrect associations with ever larger datasets (Phillips et al. 2004); (iii) the analysis may show artefactual association of the more rapidly changing lineages (Felsenstein 1978), including the attraction of long branches to the base of the in-group in association with the out-group (which is almost always a long branch; Philippe & Laurent 1998); (iv) in some cases, non-orthologous gene copies may be inadvertently compared among various lineages due to ancestral gene duplications followed by differential losses, or due to incomplete sampling; (v) differing views of scientists on alignments, exclusion sets and weighting schemes frequently cannot be arbitrated based on objective criteria and can lead to radically different phylogenetic reconstructions and (vi) the most difficult problems are when the time of shared ancestry is short relative to the subsequent time of divergence, where there has been little opportunity to accumulate signal and ample time for it to have been erased.
Molecular sequence comparison is now a mature field that has influenced the culture of systematics. Many have come to expect that the future of systematics will be dominated by creating ever more sophisticated methods for teasing a weak signal from noisy data. This causes concern that differing preferences for various methods will ensure that no consensus on many evolutionary relationships will ever be reached.
However, an alternative is possible, i.e. there may be other, less explored types of characters that could be powerful for resolving these contentious relationships. There is no doubt that comparisons of some characters have identified certain robust synapomorphies (shared and derived character states) that have supported long-standing, little contested evolutionary relationships, such as the monophyly of mammals, tetrapods and echinoderms. These synapomorphies are subjectively judged to be of the characters so unlikely to revert to an earlier condition or to occur multiple times in parallel that they could only have arisen once in the common ancestor of the group. Can new sets of characters be found that would meet these criteria to provide confident resolution of some problematic evolutionary relationships? Although there is a broad range of character types to explore, we focus here specifically on the comparison of features of genomes.
2. Comparisons of mitochondrial genomes have laid the foundation
The sequences from mitochondrial genes and genomes have been used extensively for phylogenetic inference, with complete mtDNA sequences being publicly available for more than 1000 animal species. (For a summary of the characteristics of animal mtDNAs, see Boore (1999).) It has been long argued (e.g. Boore & Brown 1998) that the relative arrangement (normally) of the 37 genes in animal mitochondrial genomes constitutes an especially powerful type of character for phylogenetic inference and so constitutes the first set of genome-level features to be used extensively for animal phylogeny. Briefly summarized, these genes are present in nearly all animal groups, are unambiguously homologous and can potentially be rearranged into an enormous number of states such that convergent rearrangements are very unlikely (and demonstrated to be uncommon). In the cases where it has been studied, all genes on each strand are transcribed together (Clayton 1992), so selection on gene arrangements is expected to be minimal. A summary of the evolutionary relationships convincingly demonstrated by this type of data (and in many cases left unresolved by all other studies) is found in Boore (2006), but here are a few of the more significant conclusions of deep-branch phylogenetic relationships: (i) the superphylum Eutrochozoa includes cestode platyhelminths (von von Nickisch-Rosenegk et al. 2001) and the phylum Phoronida (Helfenbein & Boore 2004); (ii) Sipuncula is closely related to Annelida rather than to Mollusca (Boore & Staton 2002); (iii) Annelida is more closely related to Mollusca than to Arthropoda (Boore & Brown 2000); (iv) Arthropoda is monophyletic and, within this phylum, Crustacea is united with Hexapoda to the exclusion of Myriapoda and Onychophora (Boore et al. 1995, 1998) and (v) Pentastomida is not a phylum, but rather a type of crustacean, and joins with Cephalocarida and Maxillopoda to the exclusion of other major crustacean groups (Lavrov et al. 2004).
3. Nuclear genomes, a treasure trove of phylogenetic characters
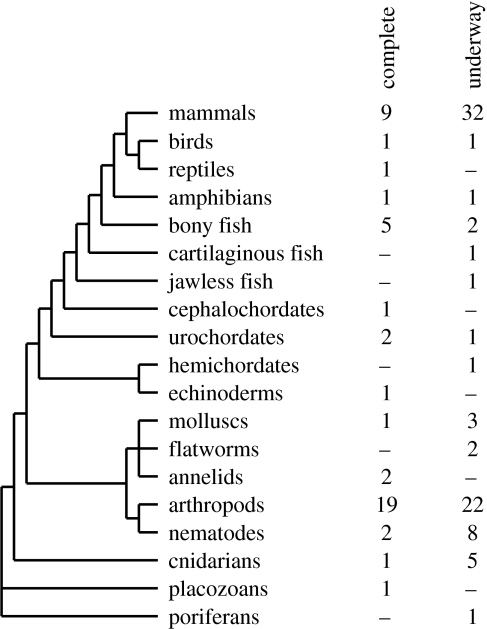
By a great margin, more DNA sequence is being generated than ever before. The facilities built and the techniques developed for sequencing the human genome are now focusing on many other organisms. The nine largest genome sequencing centres (table 1) collectively can now produce well over 170 billion nucleotides of DNA sequence per year, which would be approximately 57-fold coverage of the human genome. Imminently, there will be complete genomes of at least draft quality for many dozens of animals representing a phylogenetically diverse sample and including several equivocally placed lineages (figure 1; table 2).
Table 1.
URLs for the largest public DNA sequencing centres
| Wellcome Trust Sanger Institute | http://www.sanger.ac.uk/ |
| DOE Joint Genome Institute | http://www.jgi.doe.gov/ |
| Washington University Genome Sequencing Center | http://genome.wustl.edu/ |
| Broad Institute | http://www.broad.mit.edu/ |
| Baylor College of Medicine Genome Center | http://www.hgsc.bcm.tmc.edu/ |
| Beijing Genomics Institute | http://www.genomics.org.cn/bgi/english |
| Riken Genomic Sciences Center | http://www.riken.jp/engn/r-world/research/lab/genome/index.html |
| J. Craig Venter Institute | http://www.jcvi.org/ |
| Genoscope | http://www.cns.fr/ |
Figure 1.
This reconstruction of the major branches of animal evolution is used to plot the numbers of taxa with complete genome sequences done and underway. The taxonomic ranks shown are arbitrary, split for illustration, but not meant to be consistent among the major groups, and the taxa listed do not comprehensively cover all of life. Branch lengths hold no meaning. While opinions may differ on particular genomes as to whether they are complete versus needing more work, and whether they are well enough along to consider them ‘underway’, it is clear that soon there will be a large and phylogenetically broad sampling of genome sequences.
Table 2.
Complete nuclear genome sequencing projects done and underway as summarized in figure 1. (Asterisk indicate genomes currently funded to only low coverage.)
| taxonomy | organism | common name |
|---|---|---|
| complete | ||
| Chordata, Mammalia | Bos taurus | cow |
| Canis familiaris | dog | |
| Homo sapiens | human | |
| Macaca mulatta | rhesus macaque | |
| Monodelphis domestica | opossum | |
| Mus musculus | mouse | |
| Ornithorhynchus anatinus | duck-billed platypus | |
| Pan troglodytes | common chimpanzee | |
| Rattus norvegicus | Norway rat | |
| Chordata, Aves | Gallus gallus | red jungle fowl (chicken) |
| Chordata, Sauria | Anolis carolinensis | green anole |
| Chordata, Amphibia | Xenopus tropicalis | western clawed frog |
| Chordata, Teleostei | Danio rerio | zebrafish |
| Gasterosteus aculeatus | stickleback | |
| Oryzias latipes | Japanese killifish | |
| Takifugu rubripes | Japanese pufferfish | |
| Tetraodon nigroviridis | green spotted pufferfish | |
| Chordata, Cephalochordata | Branchiostoma floridae | amphioxis |
| Chordata, Urochordata | Ciona intestinalis | sea squirt |
| Ciona savignyi | sea squirt | |
| Echinodermata, Echinozoa | Strongylocentrotus purpuratus | purple sea urchin |
| Mollusca, Bivalvia | Lottia gigantea | owl limpet |
| Annelida, Oligochaeta | Helobdella robusta | leech |
| Annelida, Polychaeta | Capitella capitata | bristle worm |
| Arthropoda, Diptera | Aedes aegypti | mosquito (carrying yellow fever) |
| Anopheles gambiae | mosquito (carrying malaria) | |
| Drosophila ananassae, D. erecta, D. grimshawi, D. melanogaster, D. mojavensis, D. persimilis, D. pseudoobscura, D. sechellia, D. simulans (8), D. virilis, D. willistoni, D. yakuba | fruitfly | |
| Arthropoda, Coleoptera | Tribolium castaneum | red flour beetle |
| Arthropoda, Hymenoptera | Apis mellifera | honeybee |
| Arthropoda, Lepidoptera | Bicyclus anynana | butterfly |
| Heliothis virescens | cotton bollworm | |
| Arthropoda, Crustacea | Daphnia pulex | water flea |
| Nematoda, Chromadorea | Caenorhabditis briggsae | roundworm |
| Caenorhabditis elegans | roundworm | |
| Brugia malayi | filarial roundworm | |
| Cnidaria, Anthozoa | Nematostella vectensis | starlet sea anemone |
| Placozoa | Trichoplax adhaerens | none |
| in progress | ||
| Chordata, Mammalia | Callithrix jacchus | marmoset |
| Cavia porcellus | guinea pig | |
| Choloepus hoffmanni* | Hoffmann's two-toed sloth | |
| Cynocephalus volans* | flying lemur | |
| Dasypus novemcinctus* | nine-banded armadillo | |
| Dipodomys panamintinus* | kangaroo rat | |
| Echinops telfairi* | lesser Madagascar hedgehog | |
| Equus caballus | horse | |
| Erinaceus europaeus* | brown-breasted hedgehog | |
| Felis catus | cat | |
| Lama glama* | llama | |
| Loxodonta africana* | elephant | |
| Macropus eugenii* | tammar wallaby | |
| Manis pentadactyla* | pangolin | |
| Microcebus murinus* | grey mouse lemur | |
| Myotis lucifugus | little brown bat | |
| Ochotona princeps* | pika | |
| Oryctolagus cuniculus* | rabbit | |
| Otolemur garnettii* | bushbaby | |
| Papio anubis | baboon | |
| Pongo pygmaeus abelii | orang-utan (Sumatran) | |
| Pongo pygmaeus pygmaeus | orang-utan (Bornean) | |
| Procavia capensis* | hyrax | |
| Pteropus vampyrus* | large flying fox (megabat) | |
| Sorex araneus* | shrew | |
| Spermophilus tridecemlineatus | thirteen-lined ground squirrel | |
| Sus scrofa | pig | |
| Tenrec ecaudatus | lesser hedgehog | |
| Tupaia belangeri | tree shrew | |
| Tursiops truncatus* | bottle-nosed dolphin | |
| Tarsius syrichta | tarsier | |
| Vicugna pacos | alpaca | |
| Chordata, Aves | Taeniopygia guttata | zebra finch |
| Chordata, Amphibia | Xenopus laevis | African clawed frog |
| Chordata, Teleostei | Oreochromis niloticus | tilapia |
| Salmo salar | Atlantic salmon | |
| Chordata, Chondrichthyes | Callorhinchus milii* | elephant shark |
| Chordata, Hyperoartia | Petromyzon marinus* | sea lamprey |
| Chordata, Urochordata | Oikopleura dioica | larvacean |
| Hemichordata, Enteropneusta | Saccoglossus kowalevskii | acorn worm |
| Mollusca, Bivalvia | Mytilus californianus | California sea mussel |
| Mollusca, Gastropoda | Aplysia californica | California sea hare |
| Biomphalaria glabrata | snail | |
| Platyhelminthes, Trematoda | Schistosoma mansoni | blood fluke |
| Platyhelminthes, Turbellaria | Schmidtea mediterranea | planarian |
| Arthropoda, Diptera | Culex pipiens | common house mosquito |
| Glossina morsitans | tsetse fly | |
| Drosophila americana, D. auraria, D. equinoxialis, D. hydei, D. littoralis, D. mercatorum, D. mimica, D. miranda, D. novamexicana, D. repleta, D. silvestris | fruitfly | |
| Arthropoda, Hemiptera | Acyrthosiphon pisum | pea aphid |
| Rhodnius prolixus | kissing bug | |
| Arthropoda, Hymenoptera | Nasonia giraulti | wasp (parasitic) |
| Nasonia longicornis | wasp (parasitic) | |
| Nasonia vitripennis | wasp (parasitic) | |
| Arthropoda, Lepidoptera | Bombyx mori | silkworm |
| Arthropoda, Phthiraptera | Pediculus humanus corporis | body louse |
| Arthropoda, Chelicerata | Ixodes scapularis | deer tick |
| Tetranychus urticae | spider mite | |
| Nematoda, Chromadorea | Caenorhabditis brenneri | roundworm |
| Caenorhabditis japonica | roundworm | |
| Caenorhabditis remanei | roundworm | |
| Haemonchus contortus | barber pole worm | |
| Heterorhabditis bacteriophora | roundworm | |
| Pristionchus pacificus | roundworm | |
| Nematoda, Enoplea | Trichinella spiralis | trichinosis roundworm |
| Cnidaria, Anthozoa | Acropora millepora* | milli coral |
| Acropora palmata* | elkhorn coral | |
| Porites lobata | lobe coral | |
| Cnidaria, Hydrozoa | Hydra magnipapillata* | hydra |
| Hydractinia symbiolongicarpus* | none | |
| Porifera, Demosponge | Reniera sp. | sponge |
In these genomic data are many higher-order features, beyond the linear sequences, that constitute genome-level characters that are potentially useful for phylogenetic reconstruction, including: (i) gene content, including components of multiunit complexes such as the ribosome, splicosome, DNA replication machinery, or oxidative phosphorylation enzymes and the presence versus the absence of particular biochemical pathways (e.g. de Rosa et al. 1999; Fitz-Gibbon & House 1999; Snel et al. 1999, 2005; House & Fitz-Gibbon 2002; Huson & Steel 2004); (ii) the relative arrangements of genes (Boore & Brown 1998); (iii) movements of genes among intracellular compartments (i.e. plastid, mitochondrion, nucleus; e.g. Nugent & Palmer 1991); (iv) insertions of segments of DNA, including transposons and numts (Fukuda et al. 1985; Richly & Leister 2004); (v) variation in intron positions (e.g. Qiu et al. 1998); (vi) secondary structures of rRNAs or tRNAs (e.g. Murrell et al. 2003); (vii) details of genome-level processes, such as the rearrangements that generate antibody diversity (Frieder et al. 2006) and (viii) deviations from the ‘universal’ genetic code (Telford et al. 2000; Santos et al. 2004). Many others are likely to be found.
Of course, the reliability of these features can only be assessed by the study of their consistency with other characters, and several are already suspect. For example, convergent gene losses may be common as organisms independently evolve smaller genomes or no longer experience selection for maintaining a particular biochemical pathway; in contrast, convergent gain of genes seems much less likely. Independent evolution of smaller genomes may also lead to parallel losses of the most expendable structures in the RNA or protein genes. There is a certain time horizon that limits the usefulness of any particular type of character; for example, once retroelements degrade in the sequence beyond the point where the insertion can be reliably inferred to be of single origin, the insertion is no longer useful as a phylogenetic character. Certain changes in the genetic code and in the tRNA secondary structures of mitochondria are known to have occurred convergently (although occasional homoplasy has not disqualified the use of either morphological characters or molecular sequence comparisons). There is also a problem in the case of closely spaced sequential internodes where random partitioning of polymorphisms, including those of genome-level characters, can lead to incorrect inference of phylogeny (e.g. Salem et al. 2003; see Boore (2006) for additional caveats and precautions).
Already there have been important insights gained from comparing such features, including: (i) tarsiers have been shown to be the sister group to the clade of monkeys and apes rather than the prosimians based on the patterns of SINE element integration (Schmitz et al. 2001); (ii) patterns of SINE and LINE insertions have also supported the monophyly of toothed plus baleen whales, that hippopotamuses are the sister group to cetaceans, that camels are the most basal cetartiodactyls (Nikaido et al. 1999), and that river dolphins are paraphyletic (Nikaido et al. 2001); (iii)animal interphylum relationships have been clarified by the comparisons of the gene membership within Hox clusters (de Rosa et al. 1999) and (iv) a study of the presence of spliceosomal introns supports the monophyly of Actinopterygia and clarifies several relationships within the group, including the basal position of bichirs (Venkatesh et al. 1999). For further discussion, see Murphy et al. (2004), Okada et al. (2004) and Boore (2006).
4. What are the advantages of using these genome-level characters?
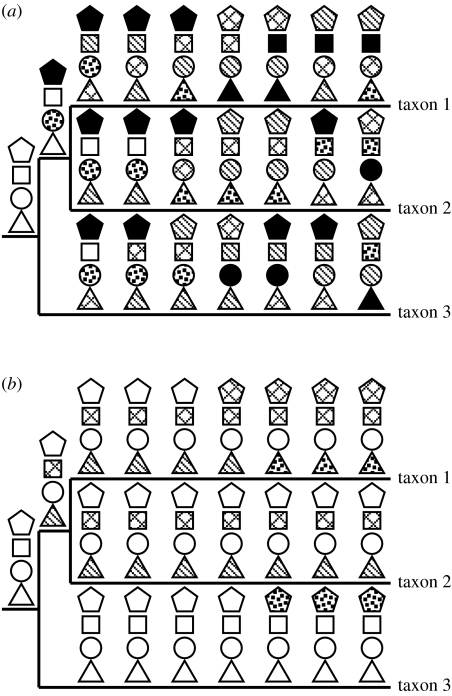
In general, these types of features would be expected to change in a saltatory, non-clocklike manner. This may seem, at first, to be wrong-headed, since great effort has been expended for many studies to identify clocklike characters, to enable accurate molecular clock estimates of time of divergence. But it is this aspect that makes these genome-level characters especially useful for addressing the most difficult branch points, those with a short time of shared history followed by a long period of divergence, as mentioned above. It is for resolving these relationships that clocklike behaviour guarantees failure, since the ratio of signal to noise will closely match the ratio of the two time periods. Rather it is the least clocklike characters that are expected to prevail, where an occasional and abrupt change may have occurred and then remain (figure 2). Admittedly, the concomitant disadvantage is that, typically, many such characters must be examined in order to find those that happened to have changed during the period of shared ancestry and so marking the relationship (see Boore (2006) for further analysis and discussion).
Figure 2.
Illustration of why clocklike characters (a) may be less informative than non-clocklike characters (b) when the internode between the subsequent lineage splits is short. Each of the four shapes is meant to be a character with states indicated by patterning. In (a), the circles and triangles are not informative and the squares and pentagons are homoplasious. The two changes accumulated in the common ancestor of taxa 1 and 2 (for the pentagons and circles), which were at one point synapomorphies, have been erased by the subsequent changes. In (b), the changes are rarer and saltatory. The pentagons and triangles are not informative and the circles are constant, but the squares are informative for uniting taxa 1 and 2.
5. What about clades without representative genome sequences?
This enormous dataset provides a new class of characters that could lead to definitive resolution of some branches of the tree of life, not only for these taxa but also for others where targeted study for identified characters could be fruitful. As shown in figure 1, whole-genome sampling will include many major lineages, but not all. It seems unlikely that there will soon be available a whole-genome sequence of a gastrotrich or a loriciferan, for example. Fortunately, we can use the genomes in hand to identify sets of genome-level characters that can be diagnostic for the relationships of related groups without genome projects. One could, for example, then determine the gene order using Southern hybridization or probe a large DNA insert library (i.e. in BAC or fosmid vectors) to find a clone to sequence for the region of interest of the genome. Gene rearrangements, losses and duplications can also be identified using comparative genomic hybridization (CGH) chips with tiled large-insert clones, as has been done for a sampling of diverse human populations (Sharp et al. 2005) and more broadly across the great apes (Locke et al. 2003) or using the arrays of oligonucleotides (representational oligonucleotide microarray analysis, ROMA; Sebat et al. 2004).
6. What are the main challenges that are before us?
First, we must increase the representation of the understudied groups of animals for large-scale genomic sequencing. There is no reason to believe that taxa that have been traditionally studied intensively, i.e. those with higher species richness, greater breadth of niche occupation, more important roles in pathogenesis or amenability to laboratory experimentation, will be more informative towards the goals of understanding broad patterns of the evolution of animals and their genomes. Second, we need to have a codification of nomenclature for the genes, which is based on the assessment of orthology (Dehal & Boore 2006). The renaming of genes to indicate orthology is not feasible because it would render large bodies of literature difficult to interpret and because scientists who study the model organisms, and who have largely done the naming, are invested in their parochial nomenclature. Thus, the solution must be a lexicon superimposed on these names already in place. Third, a system must be devised for codifying the genome-level characters themselves for entry into the databases and matrices for broad comparisons. Finally, we need for the community to devise the standards of interpretation and analysis, such as the use of cladistic reasoning rather than associating taxa by similarity alone (Boore 2006). Then, it seems probable that the genome-level characters will provide the best dataset for convincingly reconstructing relationships for some of the most hotly contended nodes in the tree of life and establishing a framework for all organismal relationships.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Evolution of the animals: a Linnean tercentenary celebration’.
References
- Adoutte A, Balavoine G, Lartillot N, de Rosa R. Animal evolution—the end of the intermediate taxon? Trends Genet. 1999;15:104–108. doi: 10.1016/s0168-9525(98)01671-0. doi:10.1016/S0168-9525(98)01671-0 [DOI] [PubMed] [Google Scholar]
- Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. doi:10.1093/nar/27.8.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J.L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006;21:439–446. doi: 10.1016/j.tree.2006.05.009. doi:10.1016/j.tree.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Boore J.L, Brown W.M. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. doi:10.1016/S0959-437X(98)80035-X [DOI] [PubMed] [Google Scholar]
- Boore J.L, Brown W.M. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 2000;17:87–106. doi: 10.1093/oxfordjournals.molbev.a026241. [DOI] [PubMed] [Google Scholar]
- Boore J.L, Staton J. The mitochondrial genome of the sipunculid Phascolopsis gouldii supports its association with Annelida rather than Mollusca. Mol. Biol. Evol. 2002;19:127–137. doi: 10.1093/oxfordjournals.molbev.a004065. [DOI] [PubMed] [Google Scholar]
- Boore J.L, Collins T.M, Stanton D, Daehler L.L, Brown W.M. Deducing arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. doi:10.1038/376163a0 [DOI] [PubMed] [Google Scholar]
- Boore J.L, Lavrov D, Brown W.M. Gene translocation links insects and crustaceans. Nature. 1998;392:667–668. doi: 10.1038/33577. doi:10.1038/33577 [DOI] [PubMed] [Google Scholar]
- Clayton D.A. Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore J.L. A phylogenomic gene cluster resource: the phylogenetically inferred groups (PhIGs) database. BMC Bioinform. 2006;7:201. doi: 10.1186/1471-2105-7-201. doi:10.1186/1471-2105-7-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa R, Grenier J.K, Andreeva T, Cook C.E, Adoutte A, Akam M, Carroll S.B, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. doi:10.1038/21631 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. doi:10.2307/2412923 [Google Scholar]
- Fitz-Gibbon S.T, House C.H. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. doi:10.1093/nar/27.21.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieder D, Larijani M, Tang E, Parsa J.Y, Basit W, Martin A. Antibody diversification: mutational mechanisms and oncogenesis. Immunol. Res. 2006;35:75–88. doi: 10.1385/IR:35:1:75. doi:10.1385/IR:35:1:75 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Fukuda M, Wakasugi S, Tsuzuki T, Nomiyama H, Shimada K, Miyata T. Mitochondrial DNA-like sequences in the human nuclear genome: characterization and implications in the evolution of mitochondrial DNA. J. Mol. Biol. 1985;186:257–266. doi: 10.1016/0022-2836(85)90102-0. doi:10.1016/0022-2836(85)90102-0 [DOI] [PubMed] [Google Scholar]
- Helfenbein K.G, Boore J.L. The mitochondrial genome of Phoronis architecta—comparisons demonstrate that phoronids are lophotrochozoan protostomes. Mol. Biol. Evol. 2004;21:153–157. doi: 10.1093/molbev/msh011. doi:10.1093/molbev/msh011 [DOI] [PubMed] [Google Scholar]
- House C.H, Fitz-Gibbon S.T. Using homolog groups to create a whole-genomic tree of free-living organisms: an update. J. Mol. Evol. 2002;54:539–547. doi: 10.1007/s00239-001-0054-5. doi:10.1007/s00239-001-0054-5 [DOI] [PubMed] [Google Scholar]
- Huson D.H, Steel M. Phylogenetic trees based on gene content. Bioinformatics. 2004;20:2044–2049. doi: 10.1093/bioinformatics/bth198. doi:10.1093/bioinformatics/bth198 [DOI] [PubMed] [Google Scholar]
- Lavrov D, Brown W.M, Boore J.L. Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proc. R. Soc. B. 2004;271:537–544. doi: 10.1098/rspb.2003.2631. doi:10.1098/rspb.2003.2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D.P, Segraves R, Carbone L, Archidiacono N, Albertson D.G, Pinkel D, Eichler E.E. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. doi:10.1101/gr.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J, Pevzner P.A, O'Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. doi:10.1016/j.tig.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Murrell A, Campbell N.J, Barker S.C. The value of idiosyncratic markers and changes to conserved tRNA sequences from the mitochondrial genome of hard ticks (Acari: Ixodida: Ixodidae) for phylogenetic inference. Syst. Biol. 2003;52:296–310. doi: 10.1080/10635150390196957. [DOI] [PubMed] [Google Scholar]
- Naylor G.J.P, Brown W.M. Amphioxus mitochondrial DNA, chordate phylogeny, and the limits of inference based on comparisons of sequences. Syst. Biol. 1998;47:61–76. doi: 10.1080/106351598261030. doi:10.1080/106351598261030 [DOI] [PubMed] [Google Scholar]
- Nikaido M, Rooney A.P, Okada N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: hippopotamuses are the closest extant relatives of whales. Proc. Natl Acad. Sci. USA. 1999;96:10 261–10 266. doi: 10.1073/pnas.96.18.10261. doi:10.1073/pnas.96.18.10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido M, et al. Retroposon analysis of major cetacean lineages: the monophyly of toothed whales and the paraphyly of river dolphins. Proc. Natl Acad. Sci. USA. 2001;98:7384–7389. doi: 10.1073/pnas.121139198. doi:10.1073/pnas.121139198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent J.M, Palmer J.D. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell. 1991;66:473–481. doi: 10.1016/0092-8674(81)90011-8. doi:10.1016/0092-8674(81)90011-8 [DOI] [PubMed] [Google Scholar]
- Okada N, Shedlock A.M, Nikaido M. Retroposon mapping in molecular systematics. Methods Mol. Biol. 2004;260:189–226. doi: 10.1385/1-59259-755-6:189. [DOI] [PubMed] [Google Scholar]
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr. Biol. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. doi:10.1016/S0960-9822(98)70390-2 [DOI] [PubMed] [Google Scholar]
- Phillips M.J, Delsuc F, Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. doi:10.1093/molbev/msh137 [DOI] [PubMed] [Google Scholar]
- Qiu Y.-L, Cho Y, Cox J.C, Palmer J.D. The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature. 1998;394:671–674. doi: 10.1038/29286. doi:10.1038/29286 [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 2004;21:1081–1084. doi: 10.1093/molbev/msh110. doi:10.1093/molbev/msh110 [DOI] [PubMed] [Google Scholar]
- Salem A.-H, et al. Alu elements and hominid phylogenetics. Proc. Natl Acad. Sci. USA. 2003;100:12 787–12 791. doi: 10.1073/pnas.2133766100. doi:10.1073/pnas.2133766100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M.A.S, Moura G, Massey S.E, Tuite M.F. Driving change: the evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. doi:10.1016/j.tig.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Ohme M, Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. doi:10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- Sharp A.J, et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. doi:10.1086/431652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen M.A. Genome phylogeny based on gene content. Nat. Genet. 1999;21:108–110. doi: 10.1038/5052. doi:10.1038/5052 [DOI] [PubMed] [Google Scholar]
- Snel B, Huynen M.A, Dutilh B.E. Genome trees and the nature of genome evolution. Annu. Rev. Microbiol. 2005;59:191–209. doi: 10.1146/annurev.micro.59.030804.121233. doi:10.1146/annurev.micro.59.030804.121233 [DOI] [PubMed] [Google Scholar]
- Telford M.J, Herniou E.A, Russell R.B, Littlewood D.T.J. Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. Proc. Natl Acad. Sci. USA. 2000;97:11 359–11 364. doi: 10.1073/pnas.97.21.11359. doi:10.1073/pnas.97.21.11359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Ning Y, Brenner S. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc. Natl Acad. Sci. USA. 1999;96:10 267–10 271. doi: 10.1073/pnas.96.18.10267. doi:10.1073/pnas.96.18.10267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Nickisch-Rosenegk M, Brown W.M, Boore J.L. Sequence and structure of the mitochondrial genome of the tapeworm Hymenolepis diminuta: gene arrangement indicates that platyhelminths are derived eutrochozoans. Mol. Biol. Evol. 2001;18:721–730. doi: 10.1093/oxfordjournals.molbev.a003854. [DOI] [PubMed] [Google Scholar]