Abstract
Context
MicroRNAs have potential as diagnostic biomarkers and therapeutic targets in cancer. No study has evaluated the association between microRNA expression patterns and colon cancer prognosis or therapeutic outcome.
Objective
To identify microRNA expression patterns associated with colon adenocarcinomas, prognosis, or therapeutic outcome.
Design, Setting, and Patients
MicroRNA microarray expression profiling of tumors and paired nontumorous tissues was performed on a US test cohort of 84 patients with incident colon adenocarcinoma, recruited between 1993 and 2002. We evaluated associations with tumor status, TNM staging, survival prognosis, and response to adjuvant chemotherapy. Associations were validated in a second, independent Chinese cohort of 113 patients recruited between 1991 and 2000, using quantitative reverse transcription polymerase chain reaction assays. The final date of follow-up was December 31, 2005, for the Maryland cohort and August 16, 2004, for the Hong Kong cohort.
Main Outcome Measures
MicroRNAs that were differentially expressed in tumors and microRNA expression patterns associated with survival using cancer-specific death as the end point.
Results
Thirty-seven microRNAs were differentially expressed in tumors from the test cohort. Selected for validation were miR-20a, miR-21, miR-106a, miR-181b, and miR-203, and all 5 were enriched in tumors from the validation cohort (P<.001). Higher miR-21 expression was present in adenomas (P = .006) and in tumors with more advanced TNM staging (P<.001). In situ hybridization demonstrated miR-21 to be expressed at high levels in colonic carcinoma cells. The 5-year cancer-specific survival rate was 57.5% for the Maryland cohort and was 49.5% for the Hong Kong cohort. High miR-21 expression was associated with poor survival in both the training (hazard ratio, 2.5; 95% confidence interval, 1.2-5.2) and validation cohorts (hazard ratio, 2.4; 95% confidence interval, 1.4-3.9), independent of clinical covariates, including TNM staging, and was associated with a poor therapeutic outcome.
Conclusions
Expression patterns of microRNAs are systematically altered in colon adenocarcinomas. High miR-21 expression is associated with poor survival and poor therapeutic outcome.
COLON ADENOCARCINOMA IS A major cause of cancer mortality worldwide.1 Colorectal cancer is the third most common and second leading cause of cancer death in the United States.2 Even though 5-year mortality rates have modestly declined over the last 3 decades,3 there is still a need to identify new prognostic biomarkers and therapeutic targets for this disease. Currently, chemotherapy has significant therapeutic value, but surgery is the only curative form of treatment.4
Ideal therapeutic targets should be causally associated with disease and amenable to designing therapeutic interventions, whereas ideal biomarkers should be easy to measure and have strong associations with clinical outcomes. MicroRNAs could match both criteria.5-8
MicroRNAs are 18- to 25-nucleotide, noncoding RNA molecules that regulate the translation of many genes.9 Since their discovery,10,11 microRNAs have been found to regulate a variety of cellular processes including apoptosis,12-14 differentiation,10,11,15 and cell proliferation.16 MicroRNAs may also have a causal role in carcinogenesis.5,17,18 MicroRNA expression levels are altered in most tumor types,19,20 including colon tumors.20-23 Experimental manipulation of specific microRNAs modulates tumor development in mouse-model systems.16,24-26 The prognostic potential of microRNAs has also been demonstrated for chronic lymphocytic leukemia,6 lung cancer,7 pancreatic cancer,27 and neuroblastomas.28
If aberrant microRNA expression is causal to carcinogenesis, inhibiting specific microRNAs may have therapeutic implications. Modified antisense oligonucleotides can easily be designed to specifically inhibit microRNA function.29 Antagomirs are one type of antisense oligonucleotide that has proven effective at inhibiting microRNA function in vivo in mice.30 The ease of designing specific inhibitors of microRNA function makes them candidates for therapeutic targets.
Given the therapeutic and prognostic potential for microRNAs in cancer, we evaluated microRNA profiles of colon tumors and paired nontumorous tissue to study their potential role in tumor formation, diagnosis, and response to chemotherapy in colon carcinoma.
METHODS
Tissue Collection and RNA Isolation
Pairs of primary colon tumor and adjacent nontumorous tissues came from 84 patients recruited from the University of Maryland Medical Center or Baltimore Veterans Affairs Medical Center between 1993 and 2002, and from 113 patients recruited from Queen Mary Hospital in Hong Kong between 1991 and 2000. Cases with familial adenomatous polyposis or human nonpolyposis colorectal cancer were excluded from this study.
Tissues were flash frozen after surgery. Detailed backgrounds for each tissue donor, including age, sex, clinical staging, tumor location, survival times from diagnosis, and receipt of adjuvant chemotherapy have been collected. The final date of follow-up was December 31, 2005 for the Maryland cohort and August 16, 2004, for the Hong Kong cohort. Tumor histopathology was classified according to the World Health Organization Classification of Tumor system.1 Adenoma tissue was obtained from the Cooperative Human Tissue Network. This study was approved by the Institutional Review Board of the National Institutes of Health, the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster, and the Institutional Review Board for Human Subject Research at the University of Maryland. Race was self-reported as white or black.
RNA Isolation and MicroRNA Profiling
RNA from frozen tissue samples was extracted using standard TRIZOL (Invitrogen, Carlsbad, California) methods. MicroRNA microarray profiling was performed as previously described.31 Briefly, 5 μg of total RNA was labeled and hybridized to each microRNA microarray (Ohio State microRNA microarray version 2.0, Columbus) containing quadruplicates of 389 human microRNA probes. Tumor/nontumorous pairs of tissues were profiled at the same time. Slides were scanned using a PerkinElmer ScanArray LX5K scanner (Perkin Elmer, Waltham, Massachusetts).
Microarray Analysis
The data discussed in this publication have been deposited in National Center for Biotechnology Information's (NCBI's) Gene Expression Omnibus (NCBIGEO GSE7828). The data were preprocessed by the statistical software R 2.5.0 (R Foundation for Statistical Computing, Vienna, Austria) to remove probes with higher background intensities than foreground and probes with inconsistent measurements across the quadruplicates. The data were normalized by locally weighted scatter plot smoothing LOESS and imported into Biometric Research Branch (BRB) array tools 3.5.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html) for subsequent microarray. Probes with values missing from more than 20% of the arrays were removed from the analysis leaving 230 probes. This filtering method was decided a priori to eliminate probes whose microRNAs expression levels were thought to be unreliable. Class comparison analysis using paired t tests identified microRNAs that were differentially expressed in tumors (P<.001). Class prediction algorithms in BRB array tools were used to determine whether microRNA microarray expression patterns could accurately differentiate between tumor and paired nontumor tissue. For these analyses, 3 nearest neighbors and nearest centroid algorithms were arbitrarily chosen and percent accuracy reports the percentage of tissues that were correctly identified. These algorithms were also used for quantitative reverse transcription polymerase chain reaction (RT-PCR) data in the Hong Kong validation cohort.
To initially search for microRNAs associated with poor survival, tumor:nontumor (T:N) microRNA expression ratios were analyzed in the Maryland cohort using microarray data. Tumor:nontumor expression ratios for microRNAs were created by subtracting the log2 nontumor from the log2 tumor expression values. MicroRNAs missing more than 25% of T:N ratios were filtered out leaving 208. Expression data were dichotomized into clearly defined high and low groups to examine associations with microRNA expression and survival. Tumor:nontumor expression ratios were dichotomized with the highest tertile classified as high and the lower 2 tertiles classified as low. This cutoff was set based on associations within the test cohort prior to analyzing the validation cohort. Once set, this high-low cutoff was used universally throughout this study. To analyze associations with tumor expression and nontumor expression with survival using microarray data, the array data had to be normalized based on the day of microarray profiling to remove systematic bias introduced from the day-to-day variability observed in the microarray data acquisition. To do this, for each given day, the highest one-third expressing values were labeled high and the lowest two-thirds were labeled low, consistent with the predetermined cutoff that was used for this study.
Quantitative RT-PCR
Quantitative RT-PCR of microRNAs was performed using Taqman MicroRNA assays (Applied Biosystems, Foster City, California) according to the manufacturer's instructions with the 7500 real-time RT-PCR system (Applied Biosystems, Foster City) using expression levels of the small nuclear RNA, U6B, as the normalization control. All assays were performed in duplicate (miR-20a, miR-203) or triplicate (miR-21, miR-106a, miR-181b). Quantitative RT-PCR for miR-21, miR-106a, and miR-181b was performed by one of the investigators (A.J.S.) who was blinded to the survival outcomes and clinical data for members of the validation cohort.
In Situ Hybridization
In situ hybridization was performed with probes for human miR-21, scramble, and U6 (Exiqon, Woburn, Massachusetts) with a modified version of the manufacturer's protocol for formalin-fixed paraffin-embedded tissue written by W. Kloosterman (http://www.exiqon.com/uploads/LNA_52-_FFPE_miRNA_in_situ_protocol.pdf) on human colon tissue. Modifications included the use of polyclonal rabbit anti-DIG/HRP-conjugated antibody and DakoCytomation GenPoint Tyramide Signal Amplification System (DakoCytomation, Carpinteria, California), and VECTOR NovaRed substrate (Vector Laboratories, Burlingame, California). Images were taken on an Olympus BX40 microscope using the Olympus DP70 digital camera and DP controller software (Olympus, Champaign, Illinois).
Statistical Analysis
Expression graphs and Wilcoxon matched-pairs tests were used to analyze differences in microRNA expression between tumors and paired nontumorous tissue as well as differences between adenoma and paired nonadenoma tissue for all quantitative RTPCR data using Graphpad Prism 4.0 (Graphpad Software Inc, San Diego, California). All trend tests reported are Cuzick nonparametric test for trend across ordered groups32 and were performed using Stata 9.2 (StataCorp LP, College Station, Texas). Associations with prognosis in the validation cohort were considered statistically significant only if the P value were less than .01 to adjust for multiple comparisons testing (5 tests using a Bonferroni correction).
KaplanMeier analysis was performed with WINSTAT 2001 (R Fitch Software, Bad Krozingen, Germany). Multivariate Cox regression analysis was performed using StataCorp 9.2. For these models, we dichotomized age as 50 years or older vs younger than 50 years because the recommended screening age for colon cancer is at age 50 years; TNM staging was dichotomized based on metastasic vs nonmetastasic disease. One patient in the Maryland cohort died on the day of surgery and was included in Kaplan-Meier analysis but removed for Cox regression analysis. Analyses involving response to adjuvant therapy included only TNM stage II and III cases because treatment in stage IV is palliative care and TNM stage I cases have excellent survival prognosis regardless of therapy. Univariate Cox regression was performed on each clinical covariate to examine influence of each on patient survival. Final multivariate models were based on stepwise addition and removal of clinical covariates found to be associated with poor survival in univariate models (P<.10). A Wald statistic of P<.05 was used as the criterion for inclusion in final multivariate models. All stepwise addition models gave the same final models as stepwise removal models. All P values reported are 2-sided. All univariate and multivariate Cox regression models were tested for proportional hazards assumptions based on Schoenfeld residuals, and no model violated these assumptions.
RESULTS
MicroRNA Expression Patterns in Colon Tumors
The characteristics of the patients with incident colon adenocarcinoma in the test cohort (from Baltimore) and the validation cohort (from Hong Kong) are shown in TABLE 1. The median follow-up time was 68.0 months for the Baltimore cohort and 84.6 months for the Hong Kong cohort. The 2 cohorts were similar in TNM staging, tumor histology, and cancer-specific mortality rates. The 5-year survival rate was 57.5% for the US cohort and 49.5% for the Hong Kong cohort and were not significantly different from one another (P=.49, Kaplan-Meier test). In addition to the racial, geographic, and cultural differences between these 2 cohorts, the Baltimore cohort was considerably older (average 64.6 years vs 55.8 years) had a higher percentage of men (79% vs 50%).
Table 1.
Characteristics of Study Population and Tumorsa
| Maryland Test Cohort (n = 84) |
Hong Kong Validation Cohort (n = 113) |
|
|---|---|---|
| Recruitment area | Baltimore, Maryland | Hong Kong, China |
| Age at enrollment, y | ||
| Mean (SD) |
64.6 (10.7) |
55.8 (15) |
| Range |
32-87 |
32-84 |
| Sex, No. (%) | ||
| Men |
66 (79) |
56 (50) |
| Women |
18 (21) |
57 (50) |
| Race, No. (%) | ||
| White |
52 (62) |
0 |
| Black |
32 (38) |
0 |
| Asian |
0 |
113 (100) |
| Follow-up time, mo | ||
| Median |
68.0 |
84.6 |
| Range |
26.0-141.9 |
60.4-147.2 |
| Specific mortality rates, % | ||
| 1 year |
82.1 |
87.6 |
| 5 year |
57.5 |
49.6 |
| Tumor location, No. (%)b | ||
| Distal |
48 (59) |
90 (80) |
| Proximal |
34 (41) |
23 (20) |
| Adenocarcinoma histology, No. (%) | ||
| Adenocarcinoma |
75 (89) |
105 (93) |
| Mucinous adenocarcinoma |
8 (10) |
7 (6) |
| Adenosquamous carcinoma |
1 (1) |
0 (0) |
| Signet ring cell and mucinous |
0 |
1 (1) |
| Adjuvant chemotherapy, No. (%)c | ||
| Received |
22 (37) |
40 (35) |
| Did not receive |
37 (63) |
73 (65) |
| TNM stage, No. (%)d | ||
| I |
8 (10) |
9 (8) |
| II |
29 (34) |
37 (33) |
| III |
36 (43) |
48 (42) |
| IV | 10 (12) | 19 (17) |
Percentages may not sum to 100 due to rounding.
Distal includes tumors located in or distal to the descending colon. Proximal tumors include tumors in or proximal to the splenic flexure. Tumor location was available for 82 patients in the original cohort and all those in the validation cohort.
Detailed information pertaining to receipt of chemotherapy was available for 59 patients in the test cohort and all those in the validation cohort. Chemotherapy was primarily fluorouracil-based (in forms of either intravenous fluorouracil or oral drugs including tegafur with uracil) with or without levamisole or leucovorin.
For 1 patient in the Maryland cohort, it was unclear whether that individual had stage III or IV cancer, so this patient was not included in the analysis.
We compared microRNA profiles of 84 pairs of colon tumor and adjacent nontumorous tissues in the Baltimore cohort using microRNA microarrays.31 Tumor microRNA profiles were distinctly different from nontumor profiles. Using class comparison analysis in BRB array tools, 37 independent microRNAs were found to be differentially expressed in tumors (P<.001 with a false-discovery rate<0.5%; TABLE 2). Twenty-six microRNAs were expressed at higher levels in tumors with miR-21 enriched the most at 1.8-fold. Global microRNA profiles distinguish between tumor and paired nontumorous tissue with 89% accuracy using either the 3 nearest neighbors or nearest centroid class prediction algorithms within BRB array tools (10-fold cross validation repeated 100 times), suggesting a systematic change in microRNA expression patterns during tumor formation.
Table 2.
MicroRNAs That Are Differentially Expressed in Tumors Compared With Nontumorous Tissue
| Probe From Microarray | Mature miR | P Valuea | False Discovery Rate, %b | Fold Change | Chromosomal Location |
|---|---|---|---|---|---|
| MicroRNAS With Higher Expression in Tumors | |||||
| hsa-mir-21No1 |
miR-21 |
< 1 × 10-7 |
<0.01 |
1.7 |
17q23.2 |
| hsa-mir-021-prec-17No1 |
miR-21 |
< 1 × 10-7 |
<0.01 |
1.8 |
17q23.2 |
| hsa-mir-092-prec-13092-1No2 |
miR-92 |
< 1 × 10-7 |
<0.01 |
1.4 |
13q31.3 |
| hsa-mir-222-precNo2 |
miR-222 |
1 × 10-6 |
<0.01 |
1.2 |
Xp11.3 |
| hsa-mir-181b-2No1 |
miR-181b |
2 × 10-6 |
<0.01 |
1.2 |
9q33.3 |
| hsa-mir-210-prec |
miR-210 |
1 × 10-5 |
0.03 |
1.2 |
11p15.5 |
| hsa-mir-020-prec |
miR-20a |
3 × 10-5 |
0.06 |
1.5 |
13q31.3 |
| hsa-mir-106-prec-X |
miR-106a |
3 × 10-5 |
0.06 |
1.4 |
X26.2 |
| hsa-mir-106aNo1 |
miR-106a |
4 × 10-5 |
0.06 |
1.4 |
X26.2 |
| hsa-mir-093-prec-7.1093-1 |
miR-93 |
4 × 10-5 |
0.06 |
1.2 |
7q22.1 |
| hsa-mir-335No2 |
miR-335 |
4 × 10-5 |
0.06 |
1.2 |
7q32.2 |
| hsa-mir-222-precNo1 |
miR-222 |
4 × 10-5 |
0.07 |
1.2 |
Xp11.3 |
| hsa-mir-338No1 |
miR-338 |
6 × 10-5 |
0.07 |
1.1 |
17q25.3 |
| hsa-mir-133bNo2 |
miR-133b |
7 × 10-5 |
0.08 |
1.1 |
6p12.2 |
| hsa-mir-092-prec-X092-2 |
miR-92 |
8 × 10-5 |
0.08 |
1.4 |
Xq26.2 |
| hsa-mir-346No1 |
miR-346 |
8 × 10-5 |
0.08 |
1.2 |
10q23.2 |
| hsa-mir-106bNo1 |
miR-106b |
.0002 |
0.2 |
1.2 |
7q22.1 |
| hsa-mir-135-2-prec |
miR-153a |
.0002 |
0.2 |
1.1 |
12q23.1 |
| hsa-mir-219-1No2 |
miR-219 |
.0003 |
0.2 |
1.3 |
9q34.11 |
| hsa-mir-34aNo1 |
miR-34a |
.0003 |
0.2 |
1.1 |
1p36.22 |
| hsa-mir-099b-prec-19No1 |
miR-99b |
.0004 |
0.3 |
1.1 |
19q13.41 |
| hsa-mir-185-precNo2 |
miR-185 |
.0004 |
0.3 |
1.2 |
22q11.21 |
| hsa-mir-223-prec |
miR-223 |
.0004 |
0.3 |
1.4 |
Xq12 |
| hsa-mir-211-precNo2 |
miR-211 |
.0004 |
0.3 |
1.1 |
15q13.3 |
| hsa-mir-135-1-prec |
miR-135a |
.0005 |
0.3 |
1.1 |
3p21.1 |
| hsa-mir-127-prec |
miR-127 |
.0005 |
0.3 |
1.1 |
14q32.31 |
| hsa-mir-203-precNo1 |
miR-203 |
.0005 |
0.3 |
1.4 |
14q32.33 |
| hsa-mir-212-precNo1 |
miR-212 |
.0006 |
0.4 |
1.1 |
17p13.3 |
| hsa-mir-095-prec-4 |
miR-95 |
.0007 |
0.4 |
1.2 |
4p16.1 |
| hsa-mir-017-precNo2 |
miR-17-5p |
.0007 |
0.4 |
1.3 |
13q31.3 |
| MicroRNAs With Reduced Expression in Tumors | |||||
| hsa-mir-342No2 |
miR-342 |
4 × 10-6 |
0.02 |
0.9 |
14q32.2 |
| hsa-mir-192-2/3No1 |
miR-192 |
9 × 10-6 |
0.03 |
0.7 |
11q13.1 |
| hsa-mir-1-2No2 |
miR-1 |
2 × 10-5 |
0.06 |
0.9 |
18q11.2 |
| hsa-mir-34bNo2 |
miR-34b |
5 × 10-5 |
0.07 |
0.8 |
11q23.1 |
| hsa-mir-215-precNo1 |
miR-215 |
5 × 10-5 |
0.07 |
0.7 |
1q41 |
| hsa-mir-192No1 |
miR-192 |
7 × 10-5 |
0.08 |
0.7 |
11q13.1 |
| hsa-mir-301No2 |
miR-301 |
7 × 10-5 |
0.08 |
0.7 |
17q23.2 |
| hsa-miR-324-5pNo2 |
miR-324-5p |
.0001 |
0.1 |
0.9 |
17p13.1 |
| hsa-mir-030a-precNo2 |
miR-30a-3p |
.0002 |
0.1 |
0.9 |
6q13 |
| hsa-mir-1-1No2 |
miR-1 |
.0003 |
0.2 |
0.9 |
20q13.33 |
| hsa-mir-34cNo2 |
miR-34c |
.0007 |
0.4 |
0.9 |
11q23.1 |
| hsa-mir-331No2 |
miR-331 |
.0009 |
0.5 |
0.9 |
12q22 |
| hsa-mir-148bNo2 | miR-148b | .0009 | 0.5 | 0.9 | 12q13.13 |
P values reported are the result of paired class comparison analysis of microRNA expression patterns from 84 pairs of colon adenocarcinomas and nontumorous tissue using Biometric Research Branch (BRB) array Tools 3.5.0.
False discovery rate is calculated by BRB array tools. The false discovery rate of 0.5% predicts that this list is 99.5% accurate.
We next performed a preliminary analysis to identify whether any of the 37 differentially expressed (P<.001) microRNAs were associated with cancer survival. We analyzed individual microRNA T:N expression ratios for associations with poor prognosis in the Maryland test cohort. Tumor:nontumor microRNA expression ratios were classified as high based on highest tertile. We searched for any microRNA for which high T:N ratios were associated with cancer survival (P<.05) using Cox regression. Five microRNAs satisfied these criteria. High expression of miR-20a (hazard ratio [HR], 2.2;95% confidence interval [CI], 1.1-4.6; P=.03), miR-21 (HR, 2.5; 95% CI, 1.2-5.0; P=.01), miR-106a (HR, 2.3; 95% CI, 1.1-4.5; P=.02), miR-181b (HR, 2.0; 95% CI, 1.0-3.9; P=.04), and miR-203 (HR, 3.1; 95% CI, 1.5-6.4; P=.003) were each associated with poor survival and were selected for further analysis.
To validate overall differences in microRNA expression between tumor and nontumorous tissue, we measured the expression levels of these 5 microRNAs with quantitative RT-PCR in tumor and paired nontumorous tissue in the independent validation cohort of 113 patients with incident colon cancer recruited from Hong Kong, China (Table 1). MiR-20a (2.3-fold), miR-21 (2.8-fold), miR106a (2.4-fold), miR-181b (1.4-fold), and miR-203 (1.8-fold) were all expressed at higher levels in tumors (P<.001, Wilcoxon matched-pairs test; TABLE 3). Most tumors (89% for miR-20a, 87% for miR-21, 90% for miR-106a, 71% for miR-181b, and 74% for miR-203) had higher expression of these microRNAs than paired nontumorous tissue. Expression patterns for these 5 microRNAs distinguish tumor vs paired nontumor status with 96% or 98% accuracy based on 3 nearest neighbors or nearest centroid algorithms, respectively (10-fold cross validation, repeated 100 times).
Table 3.
Expression of MicroRNAs in Colon Adenocarcinoma Tumors and Colon Adenomas
| microRNA | Average Difference in Threshold Cyclea |
SD (Difference in Threshold Cycle) |
Fold Changeb |
P Valuec |
|---|---|---|---|---|
| MicroRNA Expression in Tumors vs Paired Nontumorous Tissue From the Hong Kong Validation Cohortd | ||||
|
miR-20a |
1.18 |
0.97 |
2.3 |
<.001 |
|
miR-21 |
1.47 |
1.20 |
2.8 |
<.001 |
|
miR-106a |
1.25 |
0.94 |
2.4 |
<.001 |
|
miR-181b |
0.47 |
1.03 |
1.4 |
<.001 |
|
miR-203 |
0.83 |
1.40 |
1.8 |
<.001 |
| MicroRNA Expression in Adenoma vs Paired Nonadenoma Tissuee | ||||
|
miR-20a |
-0.11 |
0.97 |
0.9 |
.82 |
|
miR-21 |
0.64 |
0.90 |
1.6 |
.006 |
|
miR-106a |
0.28 |
1.22 |
1.2 |
.19 |
|
miR-181b |
0.30 |
1.24 |
1.2 |
.27 |
| miR-203 | 0.77 | 1.98 | 1.7 | .14 |
Threshold cycle is the unit of measurement in quantitative reverse transcription polymerase chain reaction (RT-PCR) to measure relative gene expression. Average (tumor change in threshold cycle minus paired nontumor change in threshold cycle) or average (adenoma change in threshold cycle minus paired nonadenoma change in threshold cycle) from quantitative RT-PCR. Positive values indicate higher expression in tumor tissue.
Calcluated by 2average difference in threshold cycles.
Wilcoxon matched pairs test.
For the tumor/nontumor comparisons, 113 pairs of tissues were used for miR-20a and miR-203 while 111 pairs of tissue were used for miR-21, miR-106a, and miR-181b.
For all adenoma/nonadenoma comparisons, 18 pairs of tissue were used.
High miR-21 Expression and Prognosis
Colon adenocarcinomas from 89% to 93% of the patients in this study were of a typical histology. A minority of tumors were of mucinous adenocarcinoma (8 of 84 patients [10%] in the US cohort; 7 of 113 patients [6%] in the Hong Kong cohort), adenosquamous carcinoma (1 of 84 patients [1%] in the US cohort), or signet ring cell carcinoma (1 of 113 patients [1%] in the Hong Kong cohort) histologies (Table 1). Different subtypes of adenocarcinomas can be associated with different clinical outcomes, including survival prognosis.33 To remove potential confounding associated with histology, we excluded all patients (9 in the US cohort; 8 in the Hong Kong cohort) with mucinous adenocarcinomas, adenosquamous carcinomas, and signet ring cell carcinomas from the initial analysis.
We found high T:N expression ratios for miR-20a, miR-21, miR-106a, miR-181b, and miR-203 to be associated with poor survival in the Maryland test cohort. These associations could be due to microRNA expression levels in the tumor tissue, the surrounding nontumorous tissue, or a combination of both. To distinguish these possibilities, we analyzed the association of microRNA expression in tumors and paired nontumorous tissues separately. High expression levels in tumors (based on highest tertile) for miR-20a (HR, 2.7; 95% CI, 1.3-5.8; P=.01), miR-21 (HR, 2.5; 95% CI, 1.2-5.2; P=.01), miR-106a (HR, 2.4; 95% CI, 1.2-5.1; P=.02), miR-181b (HR, 3.2; 95% CI, 1.6-6.7; P=.002), and miR-203 (HR, 3.3; 95% CI, 1.5-7.1; P=.001) were each associated with a poor survival in the Maryland test cohort. No significant association with microRNA expression in nontumorous tissue was observed for any of the 5 microRNAs.
The associations with microRNA expression and survival in the test cohort were made in the context of a microarray experiment in which we were evaluating the dichotomized expression of 37 microRNAs. To validate these findings, we used quantitative RT-PCR to measure tumor- and nontumor-expression levels for these 5 microRNAs in the Hong Kong validation cohort and analyzed associations with prognosis. We dichotomized high and low expression for each microRNA based on highest tertiles, consistent with our methods in the test cohort. High miR-21 tumor expression was associated with poor prognosis in the Hong Kong validation cohort (P=.001, Kaplan-Meier log-rank test) while expression in nontumorous tissue was not (FIGURE 1), consistent with associations in the Maryland test cohort. We did not find statistically significant associations with prognosis and expression of miR-20a, miR-106a, miR-181b, or miR-203 in this cohort.
Figure 1. High miR-21 Expression in Tumors and Poor Survival in Patients With Typical Adenocarcinoma Histology.
This analysis excludes patients with either mucinous adenocarcinoma or adenosquamous carcinoma histology. A, MicroRNA microarrays were used to measure microRNA expression levels of tumors and nontumorous tissues. Tissues with undetectable expression of miR-21 based on microarray data were excluded. High miR-21 expression was classified according to the highest tertile (2.6-fold to 7.9-fold higher than nontumor). B, The association of high miR-21 expression in tumors with poor prognosis is validated in an independent cohort. Expression levels of miR-21 were measured by quantitative reverse transcription polymerase chain reaction. High expression is based on the highest tertile (3.3-fold to 8.7-fold higher than nontumor). Log-rank P values are from Kaplan-Meier analysis.
Multivariate Cox proportional hazards analysis was used to further evaluate the association of miR-21 expression in tumors with prognosis in both the Maryland test cohort and the Hong Kong validation cohort (TABLE 4) to evaluate the potential formiR-21 expression as a prognostic biomarker. The dichotomized miR-21 expression values were not associated with age, sex, race, or tumor location (Fisher exact test). In univariate analysis for the Maryland test cohort, high expression of miR-21 in tumors (HR, 2.5; 95% CI, 1.2-5.2; P=.01) and TNM staging (HR, 3.5; 95% CI, 1.6-7.9; P=.002) was associated with prognosis while age, sex, race, and tumor location were not. In the final multivariate model, which included miR-21 expression and TNM staging, high miR-21 expression in tumors was associated with a poor survival prognosis independent of tumor staging (HR, 2.7; 95% CI, 1.3-5.5; P=.008; Table 4).
Table 4.
Univariate and Multivariate Cox Regression Analysis of miR-21 Expression Levels and Overall Cancer Survival in Subjects With Colon Adenocarcinomaa
| Univariate Analysis |
Multivariate Analysisb |
|||
|---|---|---|---|---|
| Characteristic | HR (95% CI) | P Value | HR (95% CI) | P Value |
| Maryland Test Cohort | ||||
| miR-21 expression (n=71)c | ||||
| Low |
1.0 [Reference] |
.01 |
1.0 [Reference] |
.008 |
| High |
2.5 (1.2-5.2) |
2.7 (1.3-5.5) |
||
| TNM stage | ||||
| I-II |
1.0 [Reference] |
.002 |
1.0 [Reference] |
.002 |
| III-IV |
3.5 (1.6-7.9) |
3.7 (1.6-8.3) |
||
| Age at enrollment, y | ||||
| <50 |
1.0 [Reference] |
.52 |
||
| ≥50 |
0.7 (0.2-2.3) |
|||
| Sex | ||||
| Women |
1.0 [Reference] |
.57 |
||
| Men |
1.4 (0.5-3.9) |
|||
| Race | ||||
| White |
1.0 [Reference] |
.97 |
||
| Black |
1.0 (0.5-2.1) |
|||
| Tumor location (proximal/distal) |
||||
| Distal |
1.0 [Reference] |
.26 |
||
| Proximal |
0.6 (0.3-1.4) |
|||
| Hong Kong Validation Cohort | ||||
| miR-21 expression (n=103)c | ||||
| Low |
1.0 [Reference] |
.002 |
1.0 [Reference] |
.002 |
| High |
2.4 (1.4-3.9) |
2.4 (1.4-4.1) |
||
| TNM stage | ||||
| I-II |
1.0 [Reference] |
<.001 |
1.0 [Reference] |
<.001 |
| III-IV |
4.7 (2.4-9.5) |
4.7 (2.4-9.5) |
||
| Age at enrollment, y | ||||
| <50 |
1.0 [Reference] |
.14 |
||
| ≥50 |
1.5 (0.9-2.6) |
|||
| Sex | ||||
| Women |
1.0 [Reference] |
.29 |
||
| Men |
1.4 (0.8-2.3) |
|||
| Tumor location | ||||
| Distal |
1.0 [Reference] |
.27 | ||
| Proximal | 0.7 (0.3-1.4) | |||
Abbreviations: CI, confidence interval; HR, hazard ratio.
Cases with mucinous adenocarcinoma, adenosquamous carcinoma, or signet ring cell carcinomas were excluded from this analysis.
Multivariate analysis used stepwise addition and removal of clinical covariates found to be associated with survival in univariate models (P < .10) and final models include only those covariates that were significantly associated with survival (Wald statistic, P < .05). For both final models, only miR-21 expression and TNM staging were included.
High expression in tumors for all miRNAs was defined based on the highest tertile. MicroRNA expression was measured with miRNA microarrays for the Maryland cohort and with quantitative reverse transcription polymerase chain reaction with the Hong Kong cohort.
In the Hong Kong validation cohort, the dichotomized values for miR-21 expression in tumors were not significantly associated with age, sex, tumor histology, or tumor location (Fisher exact test). High miR-21 expression in tumors (HR, 2.4; 95% CI, 1.4-3.9; P=.002) and TNM staging (HR, 4.7; 95% CI, 2.4-9.5; P<.001) were significantly associated with survival in univariate models while age, sex, and tumor location were not (Table 4). In the final multivariate Cox regression model, including miR-21 expression and TNM staging, high miR-21 expression in tumors was associated with poor survival prognosis (HR, 2.4; 95% CI, 1.4-4.1; P=.002) independent of other clinical covariates, consistent with findings in the Maryland test cohort.
miR-21 Expression in Colon Adenomas
Adenomas represent a precursor stage for colon adenocarcinomas.34 We tested miR-20a, miR-21, miR-106a, miR-181b, and miR-203 expression levels by quantitative RT-PCR in 18 pairs of adenoma and adjacent nonadenoma tissue that were acquired from the Cooperative Human Tissue Network. Using only 18 pairs of tissues reduced the power to detect differences in these analyses and may result in false-negatives. However, miR-21 was significantly enriched at 1.6-fold higher (P = .006, Wilcoxon matched-pairs test; Table 3). Adenoma tissue expressed higher levels of in 15 of 18 matched pairs.
Tumor Stages and miR-21 Expression
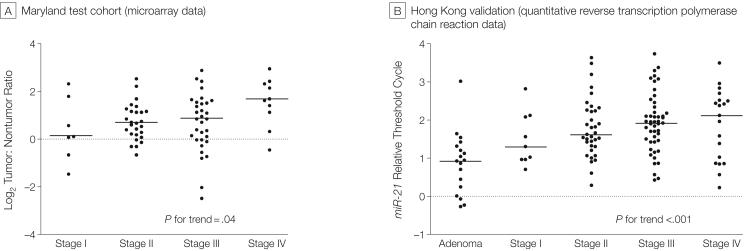
If miR-21 expression is causal to the progression of colon cancer, expression of miR-21 may be associated with more advanced stages of the disease. Patients were stratified based on the diagnosis of adenoma and TNM staging, in which adenoma was considered the least advanced and TNM stage IV was most advanced. Adenomas expressed lower levels of miR-21 than tumors from the validation cohort (P<.001, Mann-Whitney test). More advanced tumors expressed higher levels of miR-21 using either microarray data from the Maryland test cohort (P=.04, test for trend) or the quantitative RT-PCR data from the Hong Kong validation cohort (test for trend, P<.001; FIGURE 2).
Figure 2. miR-21 Expressed at Higher Levels in Colon Adenocarcinomas With Increasing Expression in More Advanced Tumors.
A, MicroRNA microarrays were used to measure miR-21 expression levels in the Maryland test cohort. Dot plots represent miR-21 log2 (tumor:nontumor ratios) for paired tissues as calculated from microRNA microarrays from the original cohort. Values greater than 0 indicate tumors with expression values higher than nontumorous tissue. Tissue types have been ordered from TNM stage I to stage IV tumors. Bars indicate median value. The Cuzick nonparametric test for trend was used to evaluate trends. B, miR-21 is expressed at higher levels in more advanced tumors. Dot plots represent miR-21 relative threshold cycle values from quantitative reverse transcription polymerase chain reaction for adenoma and tumor expression levels, each has been normalized to paired nonadenoma or nontumorous tissue, respectively. Relative threshold cycle values greater than 0 indicate expression at levels higher than nontumorous (or nonadenoma) tissue. Tissue types have been ordered from adenoma to stage I through IV tumors. Horizontal bars indicate median expression value. The Cuzick nonparametric test for trend was used to evaluate trends.
miR-21 Expression in Colonic Epithelial Cells
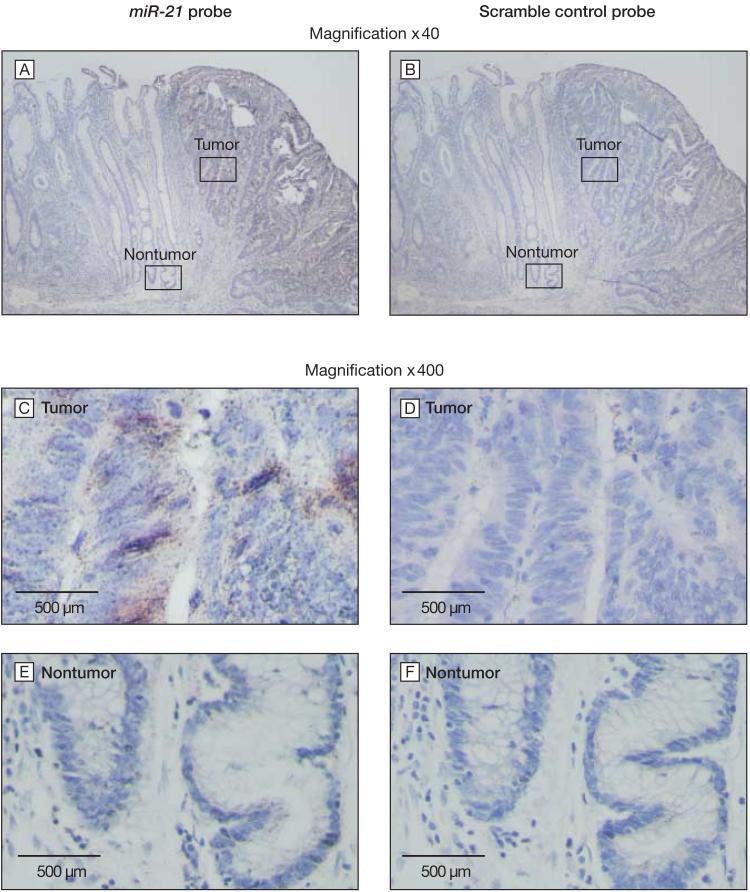
Although we found that high expression of miR-21 in tumors was associated with a worse survival outcome, these experiments did not identify the cells within a tumor that expressed miR-21. To identify these cells, we used in situ hybridization to visualize miR-21 expression in tumor and adjacent nontumor tissue (FIGURE 3). The miR-21 is expressed at higher levels in colonic epithelial cells in human tumor tissue, consistent for a role for miR-21 overexpression within tumor cells during colon carcinogenesis.
Figure 3. In Situ Hybridization of miR-21 in Colon Tumors.
In situ hybridization for miR-21 was optimized to distinguish high (brown) and low expression of miR-21. The 3' DIG-labeled probe was hybridized and detected with a polyclonal anti-DIG antibody (DakoCytomation) using amplification with the GenPoint Tyramide Signal Amplification System (DakoCytomation) using Vector NovaRed (Vector Laboratories) as the substrate. The slide was counterstained with Mayer's hematoxylin. A, Colonic epithelial cells in human tumor express higher levels of miR-21 compared with adjacent nontumorous tissue. C, Colonic epithelial cells in tumor tissue express significant amounts of miR-21, at high magnification. E, Nontumor tissue shows no significant expression of miR-21 at the same magnification. B, D, F, The scramble control probe shows no significant staining at low or high magnification in serial sections of tumor and nontumor tissue, as expected.
miR-21 Expression Levels and Therapeutic Outcome
We analyzed associations with miR-21 expression and therapeutic outcomes in stage II and III cancer patients treated with adjuvant chemotherapy. Information on the administration of adjuvant chemotherapy was available for 47 of 65 stage II or III patients in the Maryland test cohort and for all patients in the Hong Kong validation cohort. In both cohorts chemotherapy regimens were primarily fluorouracil based (in forms of either intravenous 5-fluorouracil or oral drugs including tegafur with uracil [UFT]) with or without generics. Only patients with typical adenocarcinoma histology were used for this analysis, leaving 20 of 42 stage II or III patients who had received chemotherapy in the Maryland cohort. For those who received chemotherapy, high miR-21 expression in tumors predicted worse overall survival (HR, 4.3; 95% CI, 1.1-16.4; P= .03) giving preliminary support that high miR-21 is associated with poor therapeutic outcome.
For the Hong Kong validation cohort, 77 individuals with stage II or III cancer with typical adenocarcinoma histology were used for this analysis. Among the 36 patients who received adjuvant chemotherapy, high miR-21 expression in tumors was associated with a poor response to therapy (HR, 3.5; 95% CI, 1.1-11.6; P= .04), consistent with observations in the Maryland cohort. Additionally, among the 25 patients with stage III cancer who received adjuvant chemotherapy, high miR-21 expression was associated with poor survival (HR, 3.9; 95% CI, 1.2-12.9; P=.03). Analyses using cancer relapse as an end point resulted in similar associations with high miR-21 expression in tumors predicting a more rapid disease recurrence in patients with TNM stage III cancer who received adjuvant chemotherapy (HR, 3.5; 95% CI, 1.0-11.5; P=.04).
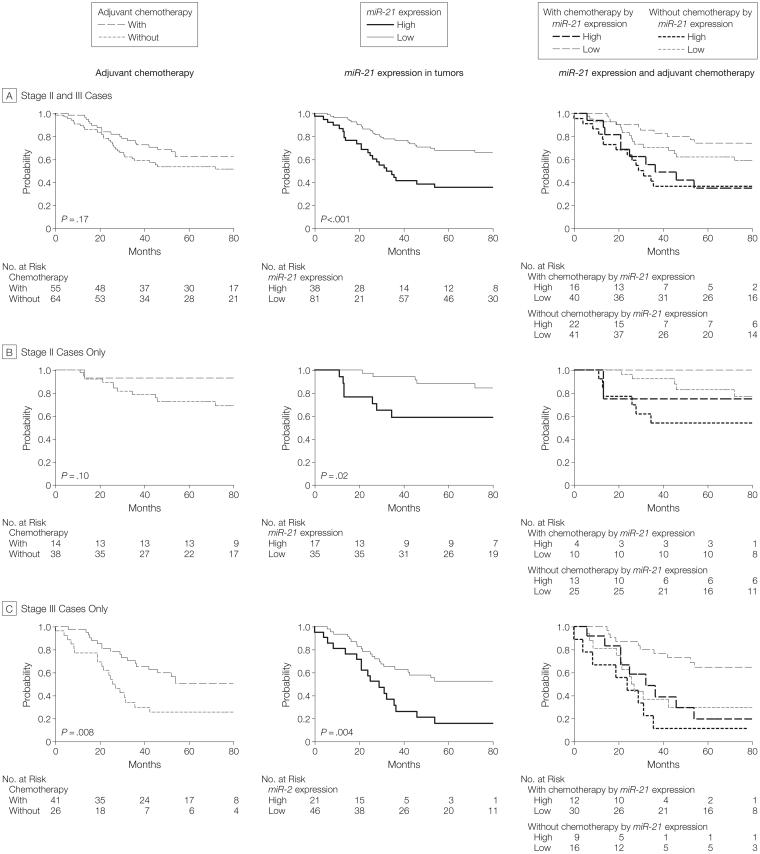
Both cohorts showed similar associations of high miR-21 expression in tumors with therapeutic outcomes, although neither cohort was large enough to perform a fully stratified analysis. Although these are 2 different populations, the similarity of the associations with miR-21 expression, prognosis, and therapeutic outcome allowed for pooled analysis using both cohorts. Kaplan-Meier analysis of the pooled cohorts demonstrated that high miR-21 expression was associated with a poor prognosis in either stage II (P=.02) or stage III (P=.004) patients (FIGURE 4) further indicating its potential as a prognostic biomarker. Receipt of adjuvant chemotherapy was beneficial for patients with either stage II or stage III cancer, although this was only significant for stage III patients. For individuals who received adjuvant therapy, high miR-21 expression was associated with a poor therapeutic outcome in patients with stage II or III cancer (P=.003, Kaplan-Meier log rank) or in patients with stage III cancer alone (P=.007, Kaplan-Meier log rank; Figure 4). Multivariate Cox regression demonstrated that high miR-21 expression predicted poor prognosis (HR, 3.0; 95% CI, 1.7-5.4; P<.001) and treatment with adjuvant chemotherapy was associated with better survival (HR, 0.4; 95% CI, 0.2-0.8; P=.004) independent of other clinical covariates (TABLE 5).
Figure 4. Combined Analysis of Maryland Test Cohort and Hong Kong Validation Cohort Examining Associations Between miR-21 Expression in Tumors and Receipt of Adjuvant Chemotherapy With Prognosis.
This analysis includes all patients with TNM stage II or III cancer except those with mucinous adenocarcinoma or adenosquamous carcinoma histologies. A, For the 119 patients with stage II or III cancer, high miR-21 expression is associated with poor survival for those who received chemotherapy (P=.003). B, Among the 52 patients with stage II cancer, associations between high miR-21 expression and prognosis were not statistically significant in 14 (26.9%) of the individuals who received chemotherapy (P=.11). C, For all 67 patients with TNM stage III cancer, high miR-21 expression was significantly associated with poor survival among those who received chemotherapy (P=.007).
Table 5.
Univariate and Multivariate Cox Regression Analysis of miR-21 Expression, Receipt of Adjuvant Chemotherapy, and Cancer Survival in Patients With Stage II or III With Adenocarcinoma in both Maryland and Hong Kong Cohortsa
| Univariate Analysis |
Multivariate Analysisb |
|||
|---|---|---|---|---|
| Characteristics | HR (95 CI) | P Value | HR (95% CI) | P Value |
| miR-21 expression (n=119)c | ||||
| Low |
1.0 [Reference] |
.001 |
1.0 [Reference] |
<.001 |
| High |
2.6 (1.5-4.5) |
3.0 (1.7-5.4) |
||
| Adjuvant chemotherapy | ||||
| Did not receive |
1.0 [Reference] |
.21 |
1.0 [Reference] |
.004 |
| Received |
0.7 (0.4-1.2) |
0.4 (0.2-0.8) |
||
| TNM stage | ||||
| II |
1.0 [Reference] |
.001 |
1.0 [Reference] |
<.001 |
| III |
3.2 (1.7-6.1) |
5.2 (2.6-11) |
||
| Tumor location | ||||
| Distal |
1.0 [Reference] |
.02 |
1.0 [Reference] |
.007 |
| Proximal |
0.4 (0.2-0.8) |
0.3 (0.1-0.7) |
||
| Age at enrollment, y | ||||
| <50 |
1.0 [Reference] |
.32 |
||
| ≥50 |
1.4 (0.7-2.5) |
|||
| Sex | ||||
| Women |
1.0 [Reference] |
.44 | ||
| Men | 1.3 (0.7-2.2) | |||
Abbreviations: CI, confidence interval; HR, hazard ratio.
Patients with TNM stage II or III cancer with typical adenocarcinoma histology were included in this analysis.
Multivariate analysis used stepwise addition and removal of clinical covariates found to be associated with survival in univariate models (P < .10) and final models include only those covariates that were significantly associated with survival (Wald statistic, P < .05). miR-21 expression, receipt of adjuvant therapy, TNM staging, and tumor location were included in final multivariate model.
High expression in tumors for all miRNAs was defined based on the highest tertile. Race was not associated with poor prognosis. MicroRNA expression was measured with miRNA microarrays for the Maryland cohort and with quantitative reverse transcription polymerase chain reaction with the Hong Kong cohort.
COMMENT
We performed the largest study to date analyzing microRNA profiles in colon cancer tissues and the first, to our knowledge, using 2 independent cohorts. Thirty-seven microRNAs were differentially expressed in tumor tissues by microRNA microarray analysis in the Maryland test cohort. Expression patterns of all 5 tested microRNAs were validated in the Hong Kong cohort. The discriminatory power of 5 microRNAs to differentiate between tumor and nontumorous tissue suggests that predictable and systematic changes of microRNA expression patterns may occur during tumorigenesis and may be representative of sporadic colon adenocarcinomas.
miR-20a, miR-21, miR-106a, miR-181b, and miR-203 were all found to be expressed at higher levels in colon tumors, although it is uncertain whether these changes in microRNA expression patterns are merely associated with colon cancer or causal to the histological progression to cancer. Our data are consistent with published studies that provide evidence for changes in microRNA expression promoting tumor formation, especially for miR-20a and miR-21. miR-20a is part of the miR-17-92 polycistronic microRNA cluster.35 Overexpression of this cluster enhances cell proliferation in vitro36 and accelerates tumor formation in animal models.16 Enforced expression of the miR-17-92 cluster causes increased tumor size and tumor vascularization in mice by negatively regulating the anti-angiogenic thrombospondin 1 (Tsp1) protein.24 Experimental evidence also suggests that increased miR-21 expression promotes tumor development.
miR-21 is expressed at high levels in most solid tumors.20,37 Overexpression of miR-21 acts as an antiapoptotic factor in human glioblastoma cells.13 Inhibition of miR-21 inhibits cell growth in vitro and inhibits tumor growth in xenograft mouse models through an indirect down-regulation of the antiapoptotic factor, B-cell lymphoma 2 (Bcl-2).38 Studies in human cell lines have shown miR-21 can also target the tumor suppressor genes, phosphatase and tensin homolog (PTEN)39 and tropomyosin 1 (TPM1).40 These data, taken together, support a causal role for altered microRNA expression during tumorigenesis.
Adenomas represent a precursor stage of adenocarcinoma. Adenomas express high levels of miR-21. If increased miR-21 expression promotes colon tumor progression, increased expression in adenomas may be an early cellular event in the progression to cancer. Inhibiting miR-21 activity may help prevent tumor promotion in populations at high risk of colon cancer, such as individuals with familial adenomatous polyposis.41
In this study, we also demonstrated an association with microRNA expression patterns with colon cancer prognosis and therapeutic outcome. A robust association of high miR-21 expression in tumors with poor survival was observed in the Maryland test cohort and the Hong Kong validation cohort, separately. In each cohort, these associations were independent of other clinical covariates indicating that miR-21 expression may be a useful prognostic indicator, in addition to TNM staging and other clinical parameters, to help identify patients at a higher risk of terminal cancer. These observations were made in 2 independent cohorts with different racial and geographical compositions. Therefore, our observations may be broadly applicable to other populations.
High miR-21 expression in tumors was associated with a poor therapeutic outcome in both cohorts. This association may help predict the benefits of therapy in individuals whose miR-21 expression status is known and identify patients who are candidates for more aggressive initial therapies. But, if high miR-21 expression is causal to the poor therapeutic outcome, antagomirs30,42 or other antisense therapeutics that target miR-21 may prove to have therapeutic benefits in patients with tumors with high expression of miR-21.
Additional studies are required to demonstrate a causal link with miR-21 and the progression of colon cancer to determine the potential of miR-21 as either a biomarker or therapeutic target. Although a causal role is still uncertain, many of Hill's criteria for causation43 have already been met. There is a temporal relationship such that a high expression of miR-21 in tumors precedes the progression, therapeutic response, and subsequent death due to cancer. The strength and consistency of these associations has been found in 2 independent cohorts. There is a dose-response relationship such that more advanced tumors express higher levels of miR-21 in both cohorts. Published research has demonstrated biological plausibility of miR-21 causing the progression of tumors using in vitro and in vivo models. All of these are consistent with a role for miR-21 in colon carcinogenesis.
In conclusion, we found systematic differences in microRNA expression patterns between colon tumors and paired nontumorous tissue. Tumors with high expression of miR-21 was associated with poor survival outcome and poor response to adjuvant chemotherapy in 2 independent cohorts, independent of staging and other clinical covariates suggesting that miR-21 may be a useful diagnostic biomarker for colon adenocarcinomas and survival prognosis including response to therapy.
Acknowledgments
Funding/Support: This research was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health. Drs Leung, Chan, and Yuen were supported by the Research Grants Council of the Hong Kong Special Administrative Region (HKU 7498/ 06M). Drs Schetter and Zanetti were supported by the Cancer Prevention Fellowship Program, Office of Preventive Oncology, National Cancer Institute.
Role of the Sponsors: The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Footnotes
Financial Disclosures: None reported.
REFERENCES
- 1.Aaltonen LA, Hamilton SR. Pathology and Genetics of Tumours of the Digestive System. International Agency for Research on Cancer Press; Lyon, France: 2000. World Health Organization Classification of Tumors. [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Goldman E, Fisher JL. Discrepancies in cancer mortality estimates. Arch Med Res. 2006;37(4):548–551. doi: 10.1016/j.arcmed.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Bigas MA, Hoff P, Crane CH. Carcinoma of the colon and rectum. In: Kufe DW, Bast RC, Hait WN, et al., editors. Holland-Frei Cancer Medicine 7. 7th ed BC Decker Inc; Hamilton, Ontario: 2006. pp. 1369–1391. [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 7.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Waldman SA, Terzic A. Translating MicroRNA discovery into clinical biomarkers in cancer. JAMA. 2007;297(17):1923–1925. doi: 10.1001/jama.297.17.1923. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 12.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 13.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 14.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 15.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 16.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandrés E, Cubedo E, Agirre X, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and nontumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 24.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang CL, Wang BB, Bartha G, et al. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci U S A. 2006;103(49):18680–18684. doi: 10.1073/pnas.0609030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgantas RW, III, Hildreth R, Morisot S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104(8):2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67(3):976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 29.Wurdinger T, Costa FF. Molecular therapy in the microRNA era. Pharmacogenomics J. 2006;7(5):297–304. doi: 10.1038/sj.tpj.6500429. [DOI] [PubMed] [Google Scholar]
- 30.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 31.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101(26):9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuzick J. Wilcoxon-type test for trend. Stat Med. 1985;4(1):87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- 33.Kang H, O'Connell JB, Maggard MA, Sack J, Ko CY. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. 2005;48(6):1161–1168. doi: 10.1007/s10350-004-0932-1. [DOI] [PubMed] [Google Scholar]
- 34.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 35.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339(2):327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 36.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 37.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 38.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 39.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130(7):2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 40.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 41.Brosens LA, van Hattem WA, Jansen M, de Leng WW, Giardiello FM, Offerhaus GJ. Gastrointestinal polyposis syndromes. Curr Mol Med. 2007;7(1):29–46. doi: 10.2174/156652407779940404. [DOI] [PubMed] [Google Scholar]
- 42.Mattes J, Yang M, Foster PS. Regulation of microRNA by antagomirs: a new class of pharmacological antagonists for the specific regulation of gene function? Am J Respir Cell Mol Biol. 2007;36(1):8–12. doi: 10.1165/rcmb.2006-0227TR. [DOI] [PubMed] [Google Scholar]
- 43.Hill AB. Statistical Evidence and Inference. 9th ed Oxford University Press; New York, NY: 1971. [Google Scholar]