Abstract
In adult planarians the replacement of cells lost to physiological turnover or injury is sustained by the proliferation and differentiation of stem cells known as neoblasts. Neoblast lineage relationships and the molecular changes that take place during differentiation into the appropriate cell types are poorly understood. Here we report the identification and characterization of a cohort of genes specifically expressed in neoblasts and their descendents. We find that genes with severely downregulated expression after irradiation molecularly define at least three discrete subpopulations of cells. Simultaneous BrdU labeling and in situ hybridization experiments in intact and regenerating animals indicate that these cell subpopulations are related by lineage. Our data demonstrate not only the ability to measure and study the in vivo population dynamics of adult stem cells during tissue homeostasis and regeneration, but also the utility of studies in planarians to broadly inform stem cell biology in adult organisms.
INTRODUCTION
Adult somatic stem cells (ASCs) produce descendents that differentiate to replace cells lost to physiological turnover, age, disease, and injury (Blanpain et al., 2004; Morrison et al., 1996; Morrison et al., 1997; Ohlstein and Spradling, 2006; Romanko et al., 2004; Weissman, 2000; Xie and Spradling, 2000). ASCs are normally lineage restricted in vivo such that they typically generate only the cell types found in the tissues in which they reside (Anderson et al., 2001; Morrison, 2001; Wagers et al., 2002). The specification of ASC descendents to supply a defined number and type of differentiated cells depends on the function and age of the organ, and the demands imposed by physiology, disease or unexpected trauma. A detailed mechanistic understanding of how ASCs are regulated to generate distinct lineages under normal and aberrant conditions remains a central unresolved issue of developmental biology and regenerative medicine.
Temporal and spatial coordination of changes in gene expression can produce a diverse number of differentiated cell types. For example, the sequential expression of four transcription factors in the neural stem cells of Drosophila melanogaster is critical for proper formation of the developing CNS (Grosskortenhaus et al., 2005; Isshiki et al., 2001), and co-expression of multiple transcription factors in the same ASC can affect lineage choices in the vertebrate retina (Hernandez et al., 2007; Peters and Cepko, 2002; Wang and Harris, 2005) and hematopoietic system (Iwasaki et al., 2006; Mansson et al., 2007). Additionally, specific changes in gene expression are detected as the ASCs of the mammalian intestine exit their resident microenvironment (niche), cease their proliferation and begin to differentiate along distinct lineages (Blanpain et al., 2007). In the case of tissue regeneration, ASC fate specification is likely to pose an additional level of complexity, as some ASCs may be specified toward a distinct lineage only after a major challenge to the system occurs (e.g., injury), instead of contributing to physiological turnover (Ito et al., 2005).
The ASCs of the planarian Schmidtea mediterranea provide an interesting model for studying both stem cell function and lineage commitment during homeostasis and regeneration. The adult flatworm contains a large population of mitotically active ASCs, known as neoblasts (Randolph, 1892), which generate a constant supply of progeny to sustain the high rate of physiological somatic cell turnover (Pellettieri and Sánchez Alvarado, 2007). Planarians display unique plasticity, as small fragments removed from almost anywhere in their bodies can regenerate entire animals (Morgan, 1898), indicating that ASCs can produce all the different cell types found in the adult flatworm. Historically, the term neoblast has been used to describe the planarian stem cells based solely on their morphology (Hori, 1982, 1991; Hyman, 1951), and therefore, a longstanding question in planarian biology is whether all the ASCs are totipotent, or if the population is molecularly heterogeneous.
The development of cell (Newmark and Sánchez Alvarado, 2000; Robb and Sanchez Alvarado, 2002) and molecular (Newmark et al., 2003; Sánchez Alvarado et al., 2002; Zayas et al., 2005) reagents has facilitated the study of stem cells in planarians. Planarian ASCs express conserved regulators of stem cell function in other organisms (Guo et al., 2006; Reddien et al., 2005b), and are widely distributed throughout the mesenchyme of the animal, yet absent in the pharynx and the region anterior to the photoreceptors. Pulse experiments with the thymidine analog BrdU have demonstrated that ASCs are initially the only cells which incorporate BrdU, but that over time the labeled progeny migrate to post-mitotic tissues (e.g., anterior region of the animal), and integrate into differentiated tissues (e.g., epidermis) (Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b). Exposure to γ-irradiation effectively ablates neoblasts, which prevents planarians from maintaining physiologic homeostasis and regenerating after injury (Bardeen, 1904; Dubois, 1949; Reddien et al., 2005a). Two populations of irradiation-sensitive cells with neoblast morphology can be isolated using Fluorescence Activated Cell Sorting (FACS) (Hayashi et al., 2006; Reddien et al., 2005b), yet it is still unclear how the isolated irradiation-sensitive cells relate to those observed with BrdU labeling studies performed in vivo. Despite significant advances, the lack of a defined panel of markers that label discrete cell types has prevented a detailed molecular and lineage characterization of the ASCs in planarians.
In this study, we defined the expression profiles of wild type and irradiated animals, and examined expression levels and spatial distribution of the most severely affected genes both in FACS-purified cells and by whole-mount in situ hybridizations. Lineage tracing experiments using a combination of BrdU and in situ hybridization revealed robust spatial and temporal regulation of gene expression and cell proliferation in both intact and regenerating animals. Our studies define a novel set of genes specifically expressed in either planarian ASCs or their descendants. Altogether, our findings establish S. mediterranea as a powerful experimental paradigm for the adult, in vivo study of the population dynamics of tissue stem cells during both normal physiological cell turnover and injury induced regeneration.
RESULTS
Transcriptional profiles uncover specific genes affected by irradiation
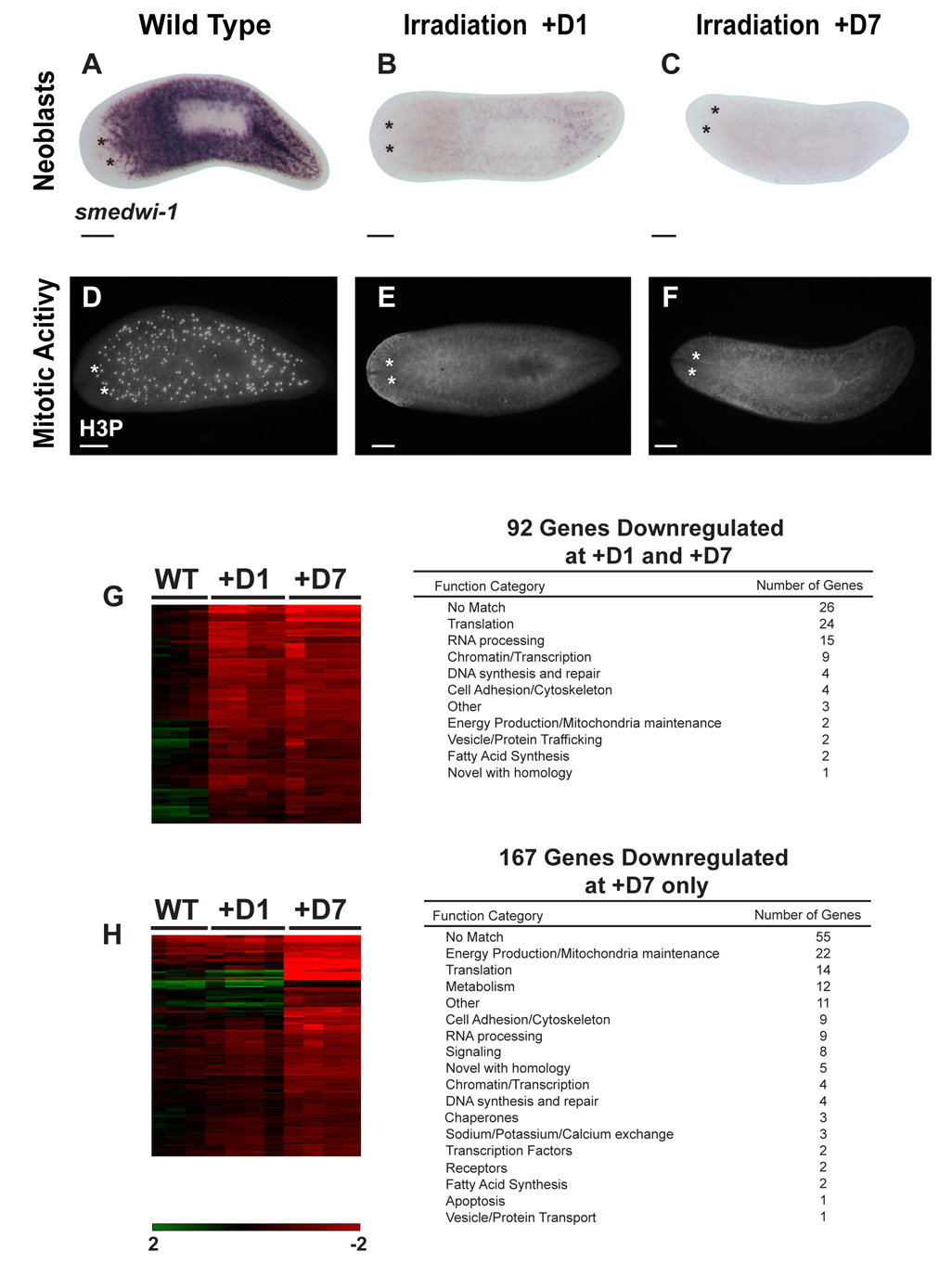
γ-irradiation of planarians results in rapid and selective elimination of neoblasts (Reddien and Sánchez Alvarado, 2004). One day after exposure to radiation, expression of smedwi-1 (Figure 1A) and mitotic activity (Figure 1D) become undetectable (Figure 1B, E). Also, no recovery of mitotic activity or neoblasts is seen by day 7 after irradiation (Figure 1C, F), yet animals appear morphologically normal and gene expression in differentiated tissues is indistinguishable from wild type (Figure S1). The specific deleterious effects of irradiation on neoblasts allowed us to hypothesize that genes downregulated by 24 hours after irradiation are likely specific to planarian ASCs or newly formed division progeny, while those with downregulated expression by day 7 but not at day 1 are likely associated with either the descendents of neoblasts or with terminally differentiated tissues undergoing turnover during this time period. To identify genes associated with planarian ASCs and their descendents, we generated a cDNA microarray containing 3,435 S. mediterranea ESTs from two different cDNA libraries (Sánchez Alvarado et al., 2002) and determined expression profiles of wild type and irradiated animals at both day 1 (+D1) and day 7 (+D7) after irradiation (see Table S1 for experimental design).
Figure 1. Identification of genes affected by irradiation.
smedwi-1 in situ hybridizations in (A) wild-type, (B) 24hrs, and (C) 7 day post-irradiated animals. Antiphosphohistone H3 (H3P) immunostaining in (D) wild-type, (E) 24hrs and (F) 7 day post-irradiated animals. (G–H) Hierarchical clustering of genes downregulated by irradiation (WT n=3; +D1, +D7 n=4) (see text for details). Heat map signal values range from 2 to −2, Log Base 2. (G) ESTs permanently downregulated 24 hours post-irradiation (n=92). (H) ESTs not affected 24 hours after irradiation, but significantly downregulated 7 days post-irradiation (n=167) (150 shown). Number of unique genes and corresponding Gene Ontology Function Category are shown. Scale bars (A–F): 200µm. Day 1: +D1; Day 7: +D7.
Using Significance Analysis of Microarrays (SAM) (Tusher et al., 2001), we identified 92 genes that were statistically at least two-fold downregulated in both the +D1 and +D7 datasets over all biological replicates (n≥3) (Figure 1G). Of the differentially expressed set of genes, 66 are predicted to encode proteins with homologs in other organisms. While these genes encompass a wide variety of biological processes, they are nevertheless enriched in genes involved in translation, RNA processing, and remodeling of chromatin (Figure 1G). Importantly, among the downregulated genes are smedwi-1 and Smed-bruno-like-1, genes known to be specifically expressed in planarian ASCs (Guo et al., 2006; Reddien et al., 2005b). We also identified another class of 167 genes that showed no change in expression 1 day after irradiation, but were significantly downregulated 7 days later (Figure 1H). Of these, 112 genes code for proteins with homologs in other organisms. In contrast to the +D1 dataset, 30% of these genes are involved in energy production and metabolism. We also identified 40 genes that were significantly upregulated by day 7 after radiation exposure. These include a conserved growth factor (granulin) and a Delta/Notch signaling component (mindbomb). (Microarray datasets available as Supplemental Data).
To test the significance of the expression profile datasets, we selected 30 genes for further analysis based on the kinetics and severity of downregulation (+D1, n=21; +D7, n=9) (Table 1 and Figure S2). Our goal was to identify markers of many different radiation-sensitive cell types; therefore, we gave priority to genes with presumptive functions in regulating stem cell biology, such as cell proliferation, RNA binding, chromatin modification, protein synthesis, and also genes that may signify changes in metabolism or energy production as a cell differentiates. Our list included novel genes as well. We conclude from these experiments that the microarray analyses of wild type and irradiated animals uncovered genes with reproducible temporal downregulation profiles likely associated with distinct types of irradiation-sensitive cells, such as neoblasts and their division progeny.
Table 1. Summary of expression dynamics from radiation-sensitive genes identified by microarrays.
Letters H and NB under EST ID refer to Head- and Neoblast-enriched cDNA libraries. Alphanumerical code refers to 96-well plate coordinates. Gene Name refers to homology with proteins encoded in genomes of other organisms (BLASTP, E≤10−6). +D1 and +D7 are time points after irradiation in which downregulated expression was measured by cDNA microarrays. (UC=unchanged).
| Microarray |
WISH |
|||
|---|---|---|---|---|
| EST ID |
GENE NAME |
+D1 |
+D7 |
Expression Category |
| H.2.12c | Piwi-like protein 1 (SMEDWI-1) | down | down | 1 |
| H.39.11e | Ribonucleoside-diphosphate reductase subunit M2 (RRM2) | down | down | 1 |
| H.56.5g | Bruno-like protein 1 (BRUNOL-1) | down | down | 1 |
| NB.19.5f | Sam68-like mammalian protein 1 (SLM-1) | down | down | 1 |
| H.48.9h | Acidic leucine-rich nuclear phosphoprotein 32 family member A (pp32a) | down | down | 1 |
| H.50.4d | High mobility group protein-1 (HMG-1) | down | down | 1 |
| H.22.5f | High mobility group protein-2 (HMG- 2) | down | down | 1 |
| NB.15.8c | Chromobox protein homolog (Cbx) | down | down | 1 |
| H.9.4h | Histone deacetylase-1 (HD1) | down | down | 1 |
| NB.32.8d | Prohibitin (PHB) | down | down | 1 |
| H.8.11d | mRNA export factor THO complex subunit 4 (THOC4) | down | down | 1 |
| H.33.12d | Plasminogen activator inhibitor 1 RNA-binding protein (PAIRBP) | down | down | 1 |
| NB.10.4b | Elongation factor Tu (EF-TU) | down | down | 1 |
| NB.21.11e | Novel | down | down | 2 |
| NB.32.1g | Novel | down | down | 2 |
| NB.8.8b | L- arginine:glycine amidinotransferase (AGAT-1) | UC | down | 3 |
| H.56.4h | L- arginine:glycine amidinotransferase (AGAT-2) | UC | down | 3 |
| NB.2.5d | L- arginine:glycine amidinotransferase (AGAT-3) | UC | down | 3 |
| H.18.4a | Ras-related protein family member 10B (Ras-related) | UC | down | 3 |
| H.45.8g | Mitochondrial carrier protein (MCP) | UC | down | 3 |
| H.63.4c | Ornithine Decarboxylase (ODC) | UC | down | 3 |
| H.49.4f | Cytochrome p450 1A1 (CYP1A1) | UC | down | 3 |
| NB.52.12f | Novel | UC | down | 3 |
| H.18.1b | 60S ribosomal protein L21 (RPL21) | down | down | 4 |
| NB.38.10e | 60S ribosomal protein L18 (RPL18) | down | down | 4 |
| H.112.11h | Transformer-2 protein homolog (TRA-2 alpha) | down | down | 4 |
| H.101.2g | Hypothetical Protein | down | down | 4 |
| NB.21.6b | Elongation Factor 1 gamma (EF-1 gamma) | down | down | 4 |
| H.96.10c | Protein SET (SET/TAF) | down | down | 4 |
| H.21.6h | Heat Shock Protein 60 (HSP60) | UC | down | 4 |
Genes downregulated after irradiation are expressed in planarian ASCs and in discrete subsets of non-dividing cells
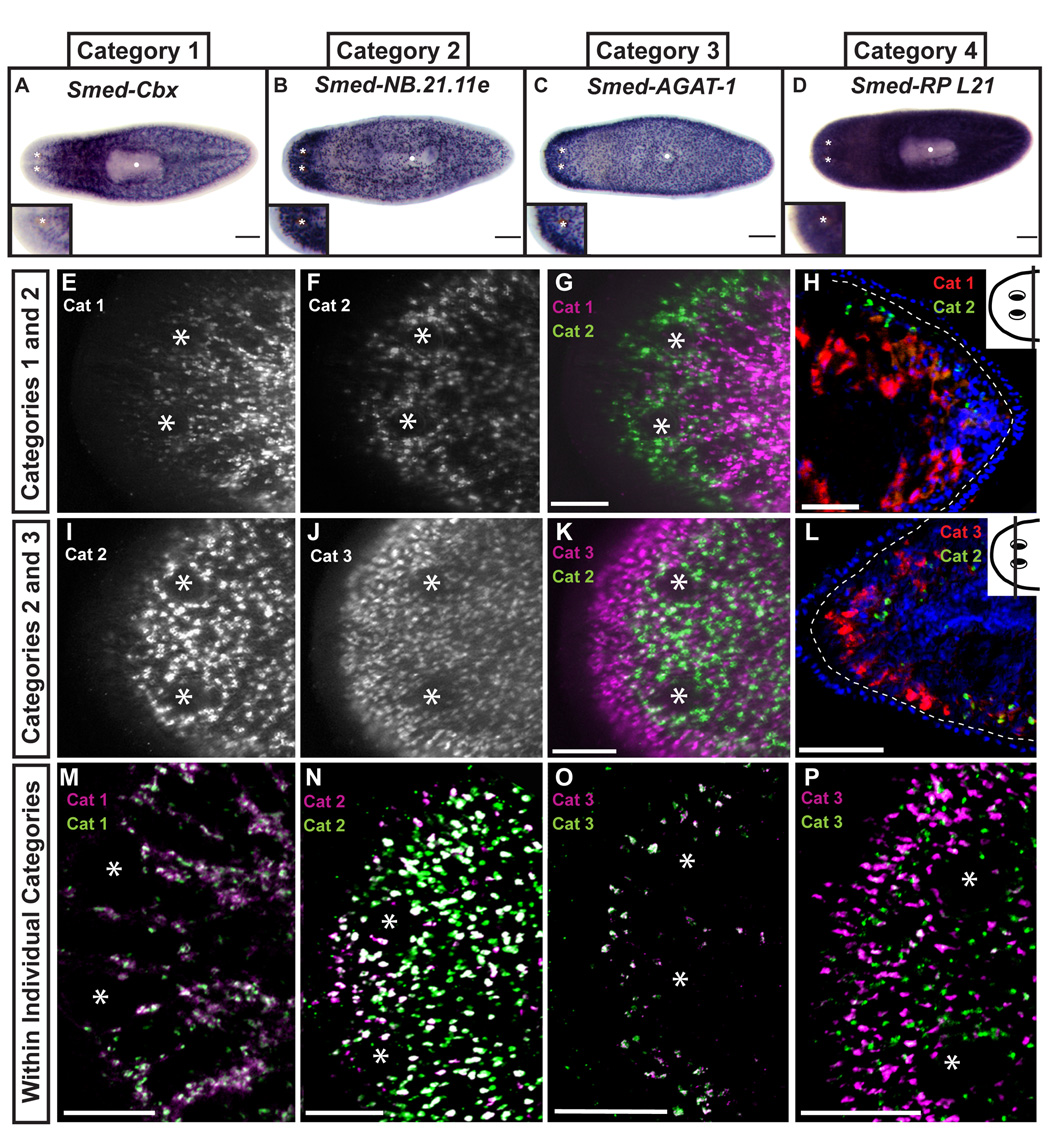
We examined the expression patterns of the 30 irradiation-sensitive genes selected by whole-mount in situ hybridization (WISH). Interestingly, the expression patterns could be catalogued into four distinct categories (Figure 2A–D and S3; Table 1). All transcripts of genes in Category 1 (n=13) were absent anterior to the photoreceptors and from the pharynx, but detected in small discrete cells throughout the animal (Figure 2A, S3) (Newmark and Sánchez Alvarado, 2000). This expression pattern is indistinguishable from that of smedwi-1 or Smed-bruno-like-1, known markers of planarian ASCs (Guo et al., 2006; Reddien et al., 2005b). Consistent with expression in dividing ASCs, the homologs of three genes in Category 1 are known to be involved in regulating proliferation in mammalian cells (Smed-RRM2-1, Smed-prohibitin-1, and Smed-pp32a-1) (Duxbury et al., 2004; Engstrom et al., 1985; Kutney et al., 2004; Nuell et al., 1991; Seo et al., 2002; Wang et al., 1999). Not all Category 1 genes are directly involved in cell cycle progression, as some are predicted to code for chromatin remodeling (Smed-Cbx-1 and Smed-HDAC-1) and nucleic acid binding (Smed-PAIRBP-1 and Smed-THOC4-1) proteins. The in situ hybridization data suggest that genes in Category 1 have an expression pattern that defines planarian ASCs.
Figure 2. Spatial distribution defines gene expression categories in the whole animal.
Thirty genes were screened by whole-mount in situ hybridization yielding four distinct gene expression patterns (see S3). Representative results shown in A–D. Insets indicate left anterior region of each animal, an area normally devoid of dividing cells. Anterior is to the left. Asterisks mark photoreceptors. White dot marks pharynx in A–D. (E–P) Single confocal sections of double fluorescent in situ hybridizations. (E–H) Category 1 (Smed-piwi-1) and 2 (Smed-NB.21.11e). (I–L) Category 2 (Smed-NB.21.11e) and 3 (Smed-AGAT-1). (H and L) Single slice confocal images of 10µm thick transverse sections counterstained with Hoechst. Dotted line demarcates basement membrane, dorsal is up. Insets mark amputation plane in whole animal. (M) Category 1 genes Smed-piwi-1 (green) and Smed-Cbx-1 (magenta); (N) Category 2 genes Smed-NB.21.11e (green) and Smed-NB.32.1g (magenta); (O) Category 3 genes Smed-AGAT-1 (green) and Smed-Ras-related (magenta); (P) Category 3 genes Smed-MCP-1 (green) and Smed-AGAT-1 (magenta). Scale Bars in A–D: 200µm; E–G, I–K: 100µm; H and L: 50 µm; M–P: 200µm.
Category 2 contained two novel genes, Smed-NB.21.11e and Smed-NB.32.1g, with downregulated expression 24hr post-irradiation (+D1 dataset), and expressed in cells that are present slightly anterior to the photoreceptors (Figure 2B, S3). Because there are no mitotically active cells anterior to the photoreceptors (Figure 1D), these genes likely mark a population of post-mitotic cells. Category 3 genes (n=8) are downregulated by day 7 after irradiation exposure and are expressed in small cells that are closer to the animal margin than the cells labeled by Category 2 genes, suggesting that Category 3 genes mark a cell type distinct from that defined by Category 2 genes (Figure 2C, Figure S3). Interestingly, the cells labeled by Categories 2 and 3 genes have increasingly peripheral expression domains, and because the division progeny of neoblasts are known to migrate through these areas (Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b), may represent cells in distinct stages of differentiation. Category 4 is composed of genes absent from the pharynx, but expressed throughout the whole animal (n=7) (Figure 2D, Figure S3). Their detection as downregulated genes in the microarray experiments is likely the result of expression in dividing cells combined with any net loss of differentiated cells to turnover in the week following irradiation. Because of their broad expression pattern, Category 4 genes are unlikely to be specific markers for neoblasts or their direct progeny and we did not study them further.
To determine if transcripts from Categories 1 through 3 were either expressed in the same or different cells, we used double fluorescent in situ hybridization (FISH) and focused our analyses on the anterior-most region of the animal. Minimal, if any, overlap was observed between the Category 1 planarian ASC marker smedwi-1 and genes in Categories 2 and 3 (4.7±0.6% and 2.8±0.9%, respectively; n=4 animals, ≥760 cells) (Figure 2E–G, Figure S4, and supplemental movies 1, 2). Moreover, inspection of mitotic ASCs with an antibody against the phosphorylated form of histone H3 (H3P; Figure 1D) revealed that less than 1% of these cells expressed Category 2 (0.07±0.07% for Smed-NB.21.11e) or Category 3 (0.06±0.05% for Smed-AGAT-1) markers (n=5 animals, ≥1231 cells) (Figure S5). In contrast, co-expression of Category 2 and 3 genes was apparent in some cells. 44±6.8% of cells expressing the Category 2 gene Smed-NB.21.11e expressed low levels of the Category 3 gene Smed-AGAT-1 (n=8 animals, ≥1894 cells) (Figure 2I–K, supplemental movie 3). However, nearly 70% (68.3±3.5%) of cells expressing Smed-AGAT-1 were negative for Smed-NB.21.11e, suggesting that Category 2 expression represents a transition state before progression to a Category 3 state. Additionally, while ASCs were found deep in the mesenchyme (Figure 2H), cells expressing Category 2 or Category 3 genes were present in a more superficial location below the basement membrane (Figure 2L). While patterns of gene expression define the distinct categories, we found subtle differences in expression from genes within the same category (Figure 2M–P), reflecting a previously unappreciated level of heterogeneity and cellular complexity. This was most evident for genes in Category 3, which vary in the amount of overlap (Figure 2O–P), suggesting cells in the same location of the animal can differ in their gene expression. We conclude from these data that Category 1 represents genes specifically expressed in mesenchymal ASCs, while genes in Categories 2 and 3 label distinct subsets of post-mitotic cells that occupy the periphery of the animal along the anteroposterior and dorsoventral axes (see Figure 6A for diagram).
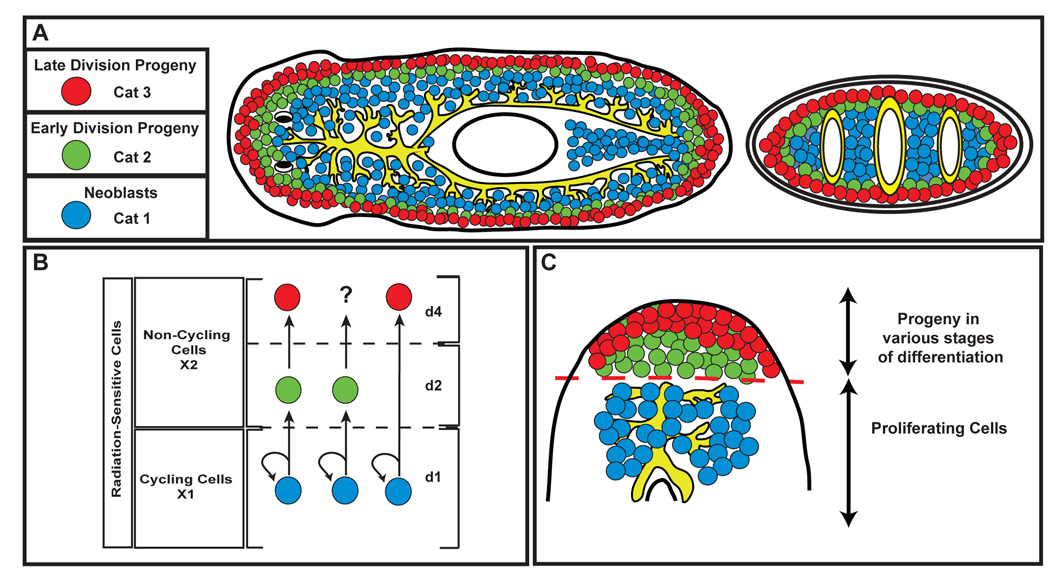
Figure 6. Planarian ASCs and their descendents.
(A) Distribution of expression patterns observed for Category 1, 2, and 3 genes. Representative genes for each category are listed. (B) Lineage determination in planarian ASCs. Three different scenarios for changes in gene expression during migration are depicted. (C) Population dynamics of ASCs and descendents during regeneration. Proliferating ASCs are restricted to the area below the amputation plane, and give rise to descendents which migrate into the blastema tissue and differentiate into the appropriate cell types. Dashed red line: amputation plane.
The defined molecular markers reveal heterogeneity in isolated irradiation-sensitive cells
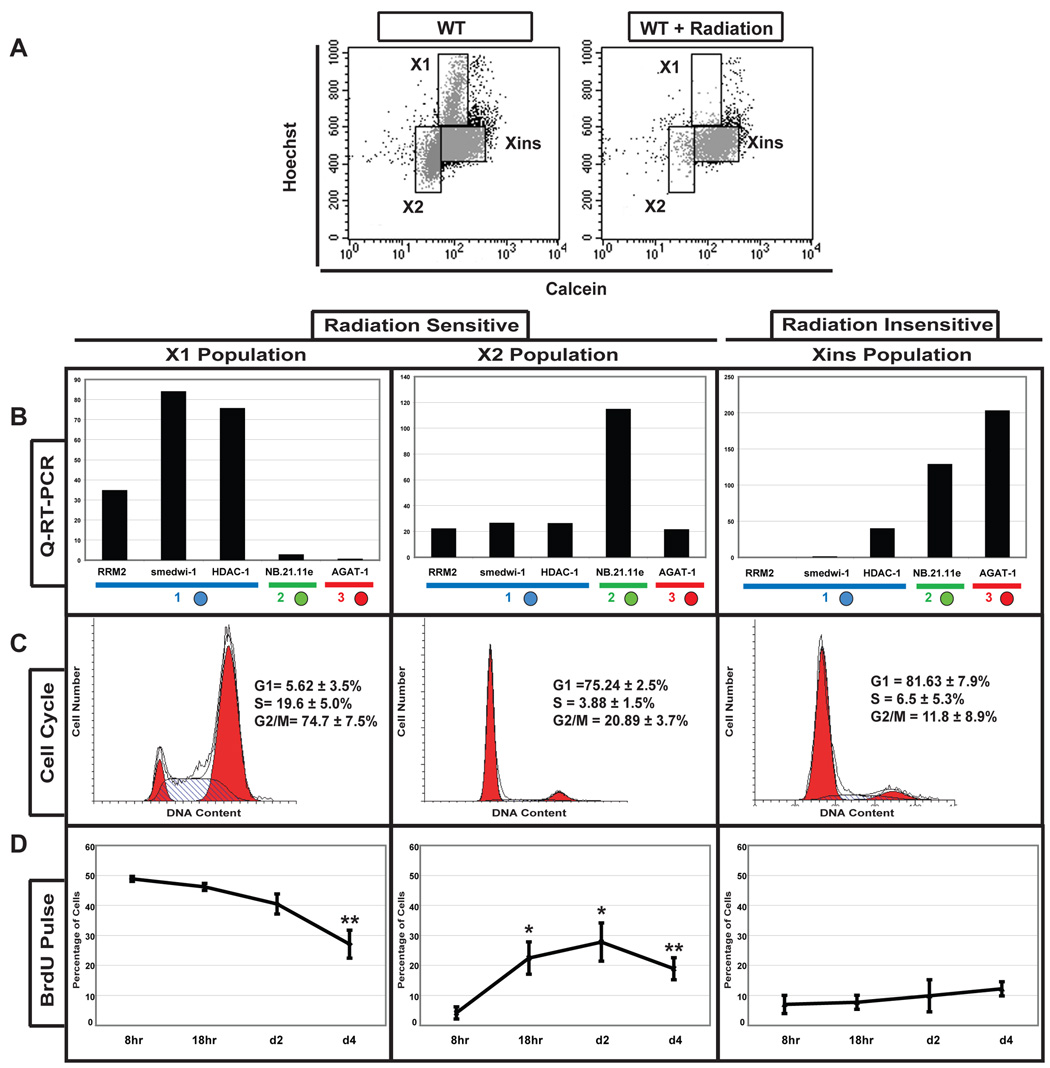
Fluorescence activated cell sorting (FACS) can be used to isolate two populations of irradiation-sensitive planarian cells (i.e., X1 and X2) that differ in their DNA content (Figure 3A) (Hayashi et al., 2006; Reddien et al., 2005b). Because little is known about the heterogeneity within these populations or how these isolated cells relate to studies performed in vivo, we used genes from the different expression categories representing markers of ASCs and post-mitotic cells to examine the composition of the irradiation-sensitive cell populations. First, we used quantitative RT-PCR (qRT-PCR) and in situ hybridization to examine the expression of the different category markers within the isolated irradiation-sensitive cells (Figure 3B and Table S2). Genes in Category 1 showed a high level of expression in the X1 fraction of cells with ≥2N DNA content, with the percentage of cells that expressed each marker varying from 30–80%. We also detected low expression levels of Category 1 genes within 23–44% cells of the X2 population, suggesting the ASCs observed in the whole animal are present in both irradiation-sensitive cell populations. Category 2 and 3 markers of post-mitotic cells were also expressed in the X2 irradiation-sensitive cells (8.5% and 12%, respectively), yet absent from the X1 population. The FISH experiments demonstrated little overlap in expression between the category markers in the whole animal, indicating that the X2 irradiation-sensitive cell population contains a mix of ASCs and at least two post-mitotic cell types.
Figure 3. Molecular Analyses of FACS-purified irradiation-sensitive cells.
(A) Wild type (WT) and irradiated planarian (+D7) flow cytometry (Hoechst/Calcein). Populations X1 and X2 disappear after irradiation. Xins designates the irradiation insensitive population. (B) Quantitative RT-PCR of X1, X2 and Xins cells. Blue, green, and red circles represent Category 1, 2, and 3 genes, respectively. Gene expression levels are relative to the ubiquitously expressed GAPDH. (C) Flow cytometric cell cycle profile of sorted X1, X2 and Xins populations stained with propidium iodide. Percentages of cells within each phase of the cell cycle are shown (n=6). (D) BrdU incorporation into X1, X2 and Xins populations at defined times after BrdU administration by feeding. Error bars are SEM. Student’s t-test was used for statistical comparisons to 8hr time point (*p<0.03, **p<0.01).
To determine if the observed heterogeneity within the isolated irradiation-sensitive cells could be attributed to cell cycle status, we examined the cell cycle profile and kinetics of BrdU incorporation for both the irradiation-sensitive (X1 and X2) and irradiation insensitive (Xins) FACS populations (Figure 3C, D). Consistent with increased Hoechst fluorescence suggesting greater than 2N DNA content, the X1 cell population encompassed a large portion of cycling cells, with 19.6±5% in S and 74.7±7.5% in G2/M (Figure 3C). In contrast, the X2 and irradiation-insensitive Xins populations are mainly comprised of non-cycling (G0/G1) cells (75.2±2.5% and 81.63±7.9%, respectively). Consistently, the X1 cells were also the first to incorporate BrdU (Figure 3D). Eight hours after a single pulse of BrdU was delivered to the whole animal, 49±0.8% of cells in the X1 fraction were labeled (Figure 3D). In contrast, only 4±2% of the X2 and 7±3% of the Xins cells had detectable levels of BrdU labeling (Figure 3D). By 18 hours the percentage of labeled X2 cells increased to 22.5±5.4% (p<0.03), while no increase was detected in the Xins fraction (Figure 3D). Four days after BrdU administration the percentage of labeled X1 cells significantly decreased to 27±4.6% (p<0.01) and the amount in the X2 fraction remained constant (18.9±3.7%). Our results conclusively demonstrate that the cycling ASCs in the X1 fraction can give rise to a portion of the cells in the X2 population. Our data also indicates that the isolated cell populations are not of sufficient purity to address whether the BrdU positive cells in the X2 fraction represent ASCs re-entering the cell cycle or their post-mitotic progeny on a path to differentiation. Therefore, we performed BrdU lineage tracing and gene expression analyses in vivo to resolve the lineage relationships that must exist between ASCs and their committed descendents.
Neoblast progeny express Category 2 and 3 genes at distinct spatial and temporal points during in vivo differentiation
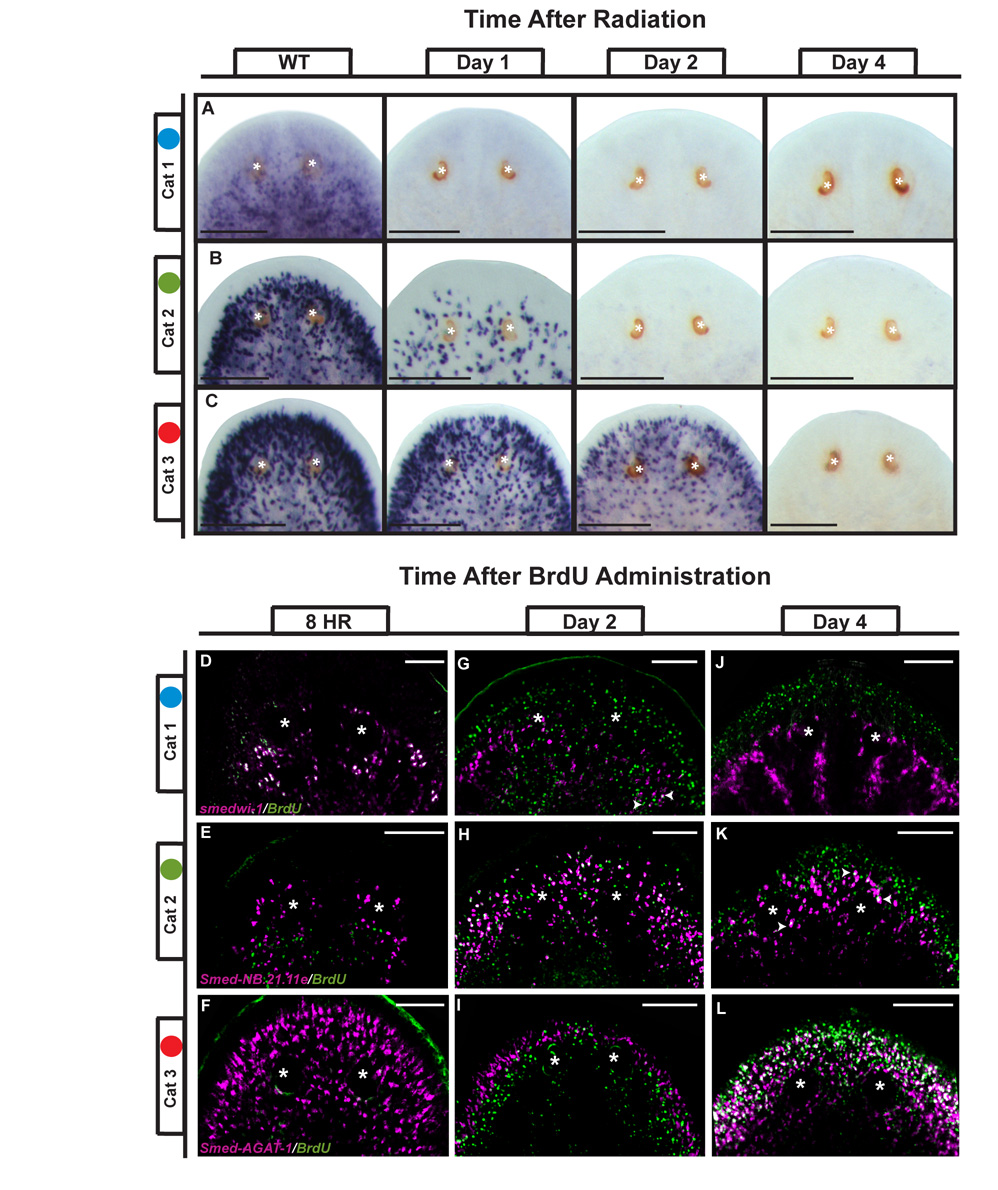
Our data show that genes in Categories 1, 2 and 3 are downregulated at different times after irradiation (Table 1 and Figure S6) and are expressed in discrete subsets of irradiation-sensitive cells with increasing superficial and peripheral expression domains (Figure 2 and Figure 3B, Table S2). To determine if the persistence of gene expression after irradiation corresponded with the location of the cell in the animal, we performed WISH time courses using a representative gene from each of the three categories. Consistent with a rapid loss of dividing ASCs after exposure to irradiation, the expression of the Category 1 gene Smed-Cbx-1 disappeared within 24hrs after irradiation (Figure 4A). In contrast, the expression of the Category 2 gene Smed-NB.21.11e transcript did not disappear until day 2 (Figure 4B), while transcripts of the Category 3 gene Smed-AGAT-1 persisted until days 3–4 (Figure 4C). Interestingly, the temporal order of downregulation correlated with the spatial distribution of these transcripts in the animal: the more peripheral the location of the cell, the longer the gene expression persisted after irradiation.
Figure 4. Changes in gene expression at defined spatial and temporal boundaries during tissue homeostasis.
Anterior portion of the animal is shown. (A–C) Genes from Categories 1 (Smed-Cbx-1), 2 (Smed-NB.21.11e), and 3 (Smed-AGAT-1) disappear at different rates after exposure to irradiation as visualized by whole-mount in situ hybridization. (D–L) Single pulse of BrdU delivered by feeding combined with in situ hybridization of Categories 1, 2 and 3 genes as indicated. Single slice confocal images with gene expression in red and BrdU staining in green at 8 hours (D–F), 2 days (G–I), and 4 days (J–L). Arrowheads indicate double positive cells. Asterisks mark the photoreceptors. Scale Bars in A–C: 200µm; D–L: 100µm.
To test whether genes in Categories 1 through 3 reflected the order of gene expression during the transition from neoblast to post-mitotic progeny, we labeled dividing cells in vivo with a single pulse of BrdU followed by detection of the expression of representative genes from Categories 1, 2, and 3 at different time points. Eight hours after administration of BrdU, 99.3±0.5% of cells that had incorporated the thymidine analog expressed the Category 1 ASC marker smedwi-1 (n=6 animals, 736/741 cells) (Figure 4D). In contrast, less than 1% of cells expressing Smed-NB.21.11e or Smed-AGAT-1 were double labeled by BrdU at this time (n=4 animals, 3/1582 cells; n=5 animals, 6/1682 cells, respectively) (Figure 4E, F, S7). By the second day after administration of the BrdU pulse, only 30.2±4.2% of BrdU positive cells expressed smedwi-1 (n=5 animals, 419/1385 cells). In contrast, the progeny of the dividing cells had migrated slightly anterior to the photoreceptors, where they no longer expressed smedwi-1, but instead expressed the Category 2 marker Smed-NB.21.11e (Figure 4G, H). The anterior migration corresponded to an increase in the number of BrdU positive cells that expressed Category 2 and 3 markers (Figure S7). Four days after BrdU administration, BrdU labeled post-mitotic progeny have migrated further toward the anterior margin, where an increased number expressed the Category 3 marker Smed-AGAT-1 (Figure 4J–L and S7). These data indicate that the progeny of neoblasts express distinct genes at defined temporal and spatial points during differentiation.
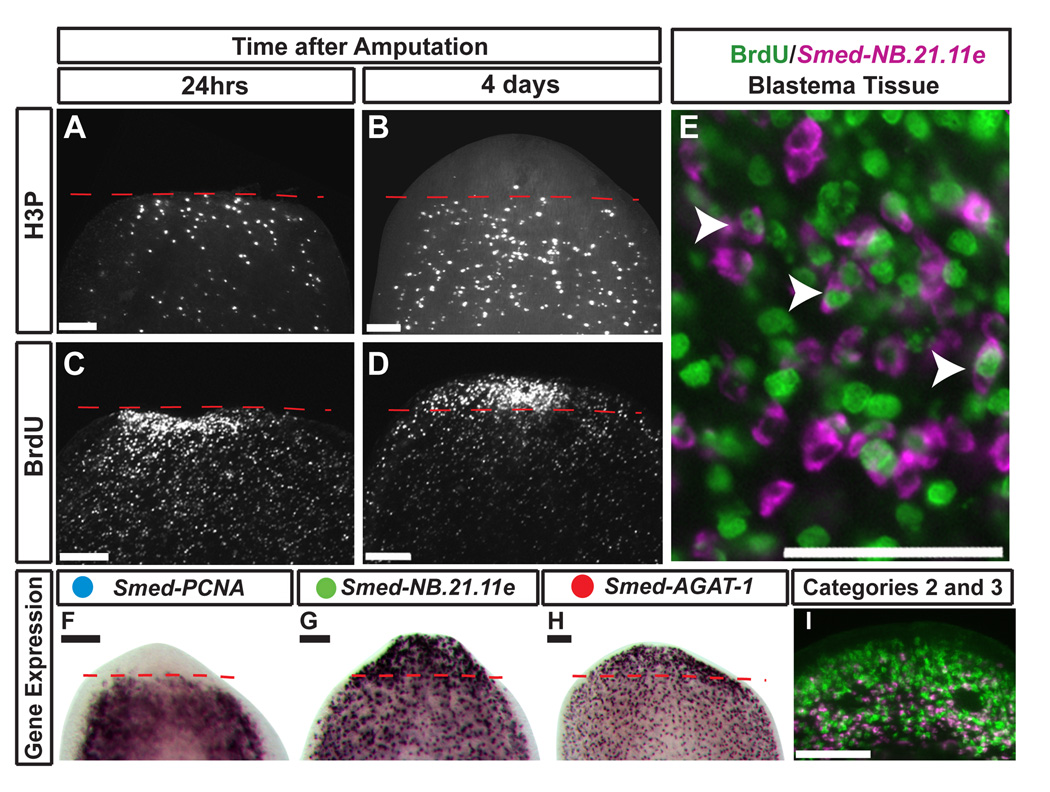
Injury to planarians such as amputation triggers a burst of neoblast proliferation (Baguña, 1976; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005a) that results in the formation of the regeneration blastema (Hori, 1991, 1997). To test whether the lineage observed during homeostasis is maintained during regeneration, we labeled dividing ASCs with a pulse of BrdU and investigated differentiation events following amputation. Neoblasts accumulated at the wound site 24 hours after amputation (Figure 5A, C). Four days later, BrdU-positive cells were concentrated in the cephalic blastema (Figure 5D), yet this area is devoid of proliferation, indicated by the absence of H3P staining and Smed-PCNA expression (Figure 5B, F). Consistent with their distribution under normal homeostatic conditions, the cells expressing the Smed-NB.21.11e and Smed-AGAT-1 were strongly enriched in the new tissue lacking mitotic activity (Figure 5G–I). Post-mitotic BrdU-positive cells in the new blastema tissue expressed Smed-NB.21.11e (Figure 5E), confirming that the descendents of the dividing ASCs contribute to the formation of new tissue after amputation. Additionally, we observe that Category 2 and 3 expression domains have been re-established by day 4 (Figure 5I). In sum, we conclude that a defined lineage from stem cell to committed post-mitotic neoblast progeny exists both during normal tissue homeostasis and during regeneration.
Figure 5. Lineage determination and population dynamics during anterior regeneration.
(A–G) Anterior blastemas of trunk fragments after amputation. Red dashed line marks amputation plane. Anti-phosphohistone H3 (H3P) immunostaining at 1 (A) and 4 days (B) after amputation. Injection of a single pulse of BrdU followed by amputation 18 hours later, and fixed at 1 (C) and 4 days (D) after amputation. (E) Single confocal slice image of simultaneously labeled BrdU nuclei (green) and Smed-NB.21.11e positive cells (magenta) in the blastema. Arrowheads show coincident staining. (F–I) Gene expression for Smed-PCNA (F), Smed-NB.21.11e (G), Smed-AGAT-1 (H), and Smed-NB.21.11e (magenta) and Smed-AGAT-1 (green) (I) in regeneration blastemas 4 days post-amputation. Image in (I) is a single confocal slice of only the regeneration blastema. Scale Bars in A–D, F–I: 100µm; E: 50 µm.
DISCUSSION
Adult planarians provide a unique way to study the mechanisms used by ASCs to generate the appropriate number and types of differentiated cells lost during cell turnover and amputation. Because little was known about the molecular changes that occur during commitment of stem cells in this organism, we identified and characterized a set of genes associated with ASCs and their earliest descendents. The new panel of molecular markers uncovered in this study provides a way to monitor distinct subsets of cells in vivo, allowing us to begin to address the complex population dynamics that occur during homeostasis and injury repair. Moreover, we define a lineage of committed ASC descendents by changes in gene expression during migration and differentiation. Our results significantly expand on earlier expression analysis of irradiated animals from a related planaria species (Rossi et al., 2007), and provide the most comprehensive gene expression survey of planarian ASCs and their committed progeny to date.
Identification of genes expressed in planarian stem cells and their post-mitotic descendents
Comparing expression profiles of planarians at different times after irradiation yielded a valuable list of genes associated with stem cells and their division progeny. The irradiation-sensitive genes examined in this study produced three general categories of expression patterns that can be used to distinguish between ASCs and subsets of their post-mitotic descendents (Figure 6A). These data validate the microarray results and provide new insight into the cellular complexity of the planarian stem cell compartment. Our results suggest that the 92 genes with downregulated expression 24hrs after irradiation (13 of which are also affected by radiation in related planarian species; Rossi et al., 2007) are largely associated with the ASCs and warrant further study. Given that many of the proteins coded by the identified genes have homologs in other organisms, these studies are likely to provide valuable insights into the mechanisms regulating metazoan stem cells.
What allows planarian ASCs to maintain an undifferentiated state in the adult organism? Studies of stem cells in other organisms suggest that one such mechanism may be the ability to modify the chromatin state within the cell (Molofsky et al., 2005; Xi and Xie, 2005). Post-translational modifications occur on the tails of histones to control the accessibility of transcription and other regulatory factors to certain genes (Jenuwein and Allis, 2001). Therefore, it is thought that molecules which can modify chromatin state may be enriched in stem cell populations to maintain their potential. In this study, we identified several different chromatin modifying factors expressed in neoblasts, including Smed-Cbx-1 (Chromobox), Smed-HDAC-1, and Smed-HMGB-1, -2. Interestingly, the mammalian homolog of HDAC has been reported to form specific repressive transcriptional complexes with known regulators of potential in embryonic stem cells (Liang et al., 2008). RNAi of Smed-HDAC-1 and Smed-Cbx-1 produced a phenotype that mimics the effects of irradiation, indicating these molecules regulate neoblast function (Reddien et al., 2005a). The identification of this class of molecules in planarian ASCs, combined with the ability to perturb their expression in the adult, sets the stage for the in vivo study of how specific chromatin modifications direct stem cell potential during cell turnover and regeneration.
Irradiation sensitive cell populations in planarians are molecularly heterogeneous
After exposure to irradiation, two populations of cells which differ in DNA content (X1 and X2) are ablated resulting in an inability of planarians to maintain homeostasis and/or generate new tissues after amputation (Hayashi et al., 2006; Higuchi et al., 2007; Higuchi and Levin, 2007; Reddien et al., 2005b). Previous work demonstrated that 96% of X1 cells express the conserved cell division protein Cyclin B (Reddien et al., 2005b), supporting our flow cytometry cell cycle findings (Figure 3C) showing this population to be highly enriched in actively dividing cells (94% S/G2M). Our data also show that 49±0.8% of the X1 cells incorporate BrdU by 8hr and suggest that planarian ASCs enter the cell cycle at a rapid rate (Figure 3D). These results are consistent with previous measurements of cell cycle entry and fraction of labeled mitosis (FLM) experiments which demonstrated that at any given time ~2% of neoblasts enter the cell cycle and ~50% of the mitoses are labeled after 7hr (Newmark and Sánchez Alvarado, 2000). Quantitative-RTPCR analysis of X1 cells showed high levels of expression of Category 1 ASC markers, and combined with BrdU analyses, demonstrates that isolated X1 cells are the dividing ASCs observed in the whole animal.
Little is known about the FACS-defined X2 population of irradiation-sensitive cells. We dissected the composition of the X2 population using markers for distinct subsets of irradiation-sensitive cells defined in vivo, providing the first molecular characterization of cells that previously could only be described based solely on morphological attributes (Higuchi et al., 2007; Hori, 1982; Hyman, 1951). Our data demonstrate that the X2 population is heterogeneous, containing a mix of stem cells and at least two distinct subpopulations of post-mitotic progeny. Interestingly, the percentage of irradiation-sensitive cells expressing each of the stem cell markers varied (Table S2). Further characterization will be required to determine if the molecular variation detected in the X2 population reflects changes in expression during exit and re-entry to the cell cycle, or whether subsets of ASCs expressing different combinations of genes and possibly varied differentiation potentials may exist.
Given the large degree of cell and molecular heterogeneity uncovered in the irradiation-sensitive cell populations, the lineage relationships of the FACS-isolated cell populations remain difficult to address. Our data clearly show that a portion of the cells in the X2 population is derived from the dividing X1 ASCs. These results are consistent with the observed reduction in X2 cells after experimental elimination of the X1 population (Oviedo and Levin, 2007). Because we observed both stem cells and post-mitotic cells in the X2 population, it was not possible to determine whether the BrdU-labeled cells represent ASCs about to re-enter the cell cycle or their differentiating progeny. These results demonstrate that FACS criteria and gene expression of isolated cells alone are not adequate to establish lineage relationships between planarian ASCs and their division progeny.
The in vivo lineage relationships of three molecularly defined cell populations
The lack of markers for discrete cell types and a defined lineage in planaria have limited attempts to address fundamental questions about the molecular changes that occur in neoblasts and their division progeny during the process of cell turnover and regeneration. By identifying irradiation-sensitive genes and observing their expression pattern in the whole animal, we resolved at least three distinct types of cells with increasingly peripheral expression domains. We established their lineage relationships by combining BrdU pulse experiments with the detection of gene expression during cell turnover and regeneration. Previous BrdU pulse experiments demonstrated that over time it is possible to observe both the labeled post-mitotic progeny of neoblasts and their migration to the anterior region of the animal (Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b). Our data support a model whereby the in vivo transition from cycling ASC to a non-dividing progeny during cell turnover is accompanied by changes in gene expression that occur at specific times and locations during migration (Figure 6B). We show that ASCs labeled at an early time point after a pulse of BrdU are present posterior to the photoreceptors and express Category 1 markers. Around day two after the single BrdU pulse, the early fate of one lineage of ASC descendents is defined by expression of the Category 2 markers. The fourth day after the pulse the labeled ASC descendents have migrated further toward the periphery and express the Category 3 marker Smed-AGAT-1. Interestingly, expression of Smed-NB.21.11e appears to be transitory, as the number of BrdU positive cells that express the Category 3 marker continued to increase by day 4, suggesting the progeny sequentially express Category 2 and 3 markers during differentiation and migration. Future experiments will determine if all progeny must transition through a Category 2 state to become Category 3 or if these cell types represent distinct lineages (Figure 6B).
The panel of markers defined in this study allowed us to determine the contribution of either neoblasts or their descendents to the formation of new tissue during regeneration (Figure 6C). The ability to monitor gene expression within labeled ASC descendents after injury provided us with a new level of resolution to describe the molecular events used to generate the “regenerative cells” observed by ultrastructural morphology in the blastema tissue (Hori, 1982, 1997). Our data suggest that changes in gene expression specify different cell types during the migration of ASC descendents from the pre-existing tissue to the newly made post-mitotic tissue. While the expression domains of the post-mitotic progeny are modeled as mutually exclusive, it is possible that progeny transition through differentiation phases more rapidly after a stimulus such as amputation.
Many of the lineage markers identified are also required for neoblast function
The in situ and lineage analyses allowed us to associate known RNAi phenotypes with expression in subsets of undifferentiated, irradiation-sensitive cells. Consistent with prior findings, several genes in Category 1 are known to play key roles in neoblast function (e.g., Smed-HDAC-1, Smed-Cbx-1 and Smed-Bruno-like-1) (Reddien et al., 2005a; Guo et al., 2006). RNAi of other genes in this category such as Smed-RRM2, which is expressed in 30% of the isolated dividing ASCs, prevented continued homeostasis or the formation of new tissue after amputation (Table S3). In mammals, RRM2 is specifically expressed in S phase (Engstrom et al., 1985), and is required for cell cycle progression (Duxbury et al., 2004; Kittler et al., 2004), suggesting Smed-RRM2 is a functional regulator of planarian ASCs entering the cell cycle. In contrast, the Category 3 gene Smed-AGAT-1 is expressed in differentiating ASC descendents and is necessary for tissue maintenance and formation of new tissue during regeneration (Reddien et al., 2005a). L-arginine:glycine amidinotransferase (AGAT) is required for in the synthesis of creatine (Braissant et al., 2005), which plays important roles in the interconversion of ATP and ADP to maintain adequate energy levels (Lee et al., 1998). Interestingly, upregulation of enzymes necessary for creatine synthesis has been associated with the ability to regenerate damaged skeletal muscles in the dystrophin-mutant mdx mouse (McClure et al., 2007; Nakayama et al., 2004). The committed ASC descendents expressing Smed-AGAT-1 are present near the musculature and nervous system (Figure S8), placing them in a privileged position to provide creatine to cells and tissues that normally use large amounts of this molecule, but do not synthesize it, i.e., muscle and CNS (Lee et al., 1998). Using distinct types of committed descendents to synthesize and secrete creatine may provide the planarian with an advantage during periods with large swings in energy consumption, such as during regeneration and prolonged starvation.
Conclusion
The molecular characterization of planarian stem cells and their descendents reported here demonstrates the feasibility of carrying out complex lineage analyses of a collectively totipotent ASC population. These studies are important for uncovering conserved biological properties underpinning the regulation of stem cell populations in adult animals, and pave the way for a systematic elucidation of how neoblasts are able to generate derivatives of all three germ layers, including the germ line. Future studies will expand the collection of irradiation-sensitive genes through both the use of genome-wide microarrays and the systematic silencing of their functions using RNAi to further increase the number and resolution of lineage markers, as well as to identify key regulatory molecules dictating cell fate decisions of the planarian ASCs. Altogether, our results establish S. mediterranea as a model system for in vivo studies of lineage and cell population dynamics of adult stem cells during tissue homeostasis and regeneration.
EXPERIMENTAL PROCEDURES
Exposure to gamma-irradiation
Asexual clonal line CIW4 of S. mediterranea were maintained and used as described (Gurley et al., 2008; Reddien et al., 2005a). Planarians were exposed to 100 Gray (Gy) of gamma-irradiation using a J.L. Shepherd and Associates model 30, 6,000 Ci Cs137 instrument at approximately 5.90 Gy/min (17min).
cDNA microarray experiments
Microarray data sets have been deposited into the Gene Omnibus Database (accession number GSE11503) and uploaded into the Schmidtea mediterranea Genome Database (SmedGD), a searchable database (http://smedgd.neuro.utah.edu) containing all available data associated with the planarian genome, including predicted and annotated genes, ESTs, protein homologies, gene expression patterns, and RNAi phenotypes (Robb et al., 2007).
Whole-mount in situ hybridization
Animals were fixed and processed as previously reported (Gurley et al., 2008) and automated whole-mount in situ hybridization experiments were performed as described (Sánchez Alvarado et al., 2002). PCNA sequence accession number: EU856391
Immunostaining
Immunostaining with anti-phosphohistone H3 (H3P) was performed as previously reported (Reddien et al., 2005a).
BrdU experiments
BrdU (Molecular Probes) was injected (3–5 injections of 300nl 10mg/ml) or fed (5mg/mL) in liver paste to animals (Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b).
Flow Cytometry
Dissociation of planarians into single cells, cellular labeling, and isolation of FACS-sorted cells were performed as described previously (Reddien et al., 2005b).
Quantitative Real Time RT-PCR
Reverse transcription reactions were carried out with total RNA isolated from FACS-sorted cells using the reverse transcription portion of the qRT-PCR kit (Superscript III Platinum Two-Step qRT-PCR; Invitrogen, Carlsbad, CA) to produce cDNA.
Supplementary Material
The supplemental data include 8 figures, 4 tables, and detailed experimental procedures.
ACKNOWLEDGMENTS
We thank Dr. Bret Pearson and members of the Sánchez Laboratory for scientific discussions, suggestions, and comments, and Brian Daley for assistance with microarrays. This work was supported by NIH-NIGMS RO-1 GM57260 and the Howard Hughes Medical Institute to ASA. GTE was supported by NIH Developmental Biology Training Grant 5T32 HD07491.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson DJ, Gage FH, Weissman IL. Can stem cells cross lineage boundaries? Nature medicine. 2001;7:393–395. doi: 10.1038/86439. [DOI] [PubMed] [Google Scholar]
- Baguña J. Mitosis in the intact and regenerating planarian Dugesia mediterranea I. Mitotic studies during growth, feeding, and starvation. J Exp Zool. 1976;195:53–64. [Google Scholar]
- Bardeen CR, Baetjer FH. The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool. 1904;1:191–195. [Google Scholar]
- Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Braissant O, Henry H, Villard AM, Speer O, Wallimann T, Bachmann C. Creatine synthesis and transport during rat embryogenesis: spatiotemporal expression of AGAT, GAMT and CT1. BMC Dev Biol. 2005;5:9. doi: 10.1186/1471-213X-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F. Contribution á l ‘ètude de la migration des cellules de regeneration chez les Planaires dulcicoles. C R Hebd Seances Acad Sci. 1949;229:747–749. [PubMed] [Google Scholar]
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. The Journal of biological chemistry. 1985;260:9114–9116. [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hernandez J, Matter-Sadzinski L, Skowronska-Krawczyk D, Chiodini F, Alliod C, Ballivet M, Matter JM. Highly conserved sequences mediate the dynamic interplay of basic helix-loop-helix proteins regulating retinogenesis. The Journal of biological chemistry. 2007;282:37894–37905. doi: 10.1074/jbc.M703616200. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Hayashi T, Hori I, Shibata N, Sakamoto H, Agata K. Characterization and categorization of fluorescence activated cell sorted planarian stem cells by ultrastructural analysis. Dev Growth Differ. 2007 doi: 10.1111/j.1440-169X.2007.00947.x. [DOI] [PubMed] [Google Scholar]
- Hori I. An Ultrastructural Study of the Chromatid Body in Planarian Regenerative Cells. Journal of electron microscopy. 1982;31:63. [Google Scholar]
- Hori I. Role of fixed parenchyma cells in blastema formation of the planarian Dugesia japonica. The International journal of developmental biology. 1991;35:101–108. [PubMed] [Google Scholar]
- Hori I. Cytological approach to morphogenesis in the planarian blastema. II. The effect of neuropeptides. Journal of submicroscopic cytology and pathology. 1997;29:91–97. [PubMed] [Google Scholar]
- Hyman LH. The Invertebrates: Platyhelminthes and Rhynchocoela. Vol. 2. McGraw-Hill Book Company, Inc.; 1951. [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature medicine. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Kutney SN, Hong R, Macfarlan T, Chakravarti D. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. The Journal of biological chemistry. 2004;279:30850–30855. doi: 10.1074/jbc.M404969200. [DOI] [PubMed] [Google Scholar]
- Lee H, Kim JH, Chae YJ, Ogawa H, Lee MH, Gerton GL. Creatine synthesis and transport systems in the male rat reproductive tract. Biol Reprod. 1998;58:1437–1444. doi: 10.1095/biolreprod58.6.1437. [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature cell biology. 2008 doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- McClure WC, Rabon RE, Ogawa H, Tseng BS. Upregulation of the creatine synthetic pathway in skeletal muscles of mature mdx mice. Neuromuscul Disord. 2007;17:639–650. doi: 10.1016/j.nmd.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH. Experimental studies of the regeneration of Planaria maculata. Arch Entw Mech Org. 1898;7:364–397. [Google Scholar]
- Morrison SJ. Stem cell potential: can anything make anything? Curr Biol. 2001;11:R7–R9. doi: 10.1016/s0960-9822(00)00033-6. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nature medicine. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. 1997;124:1929–1939. doi: 10.1242/dev.124.10.1929. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Nara N, Kawakita Y, Takeshima Y, Arakawa M, Katoh M, Morita S, Iwatsuki K, Tanaka K, Okamoto S, et al. Cloning of cDNA encoding a regeneration-associated muscle protease whose expression is attenuated in cell lines derived from Duchenne muscular dystrophy patients. The American journal of pathology. 2004;164:1773–1782. doi: 10.1016/S0002-9440(10)63735-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Reddien PW, Cebria F, Sánchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci U S A. 2003;100 Suppl 1:11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Nuell MJ, Stewart DA, Walker L, Friedman V, Wood CM, Owens GA, Smith JR, Schneider EL, Dell' Orco R, Lumpkin CK, et al. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Molecular and cellular biology. 1991;11:1372–1381. doi: 10.1128/mcb.11.3.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134:3121–3131. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annual review of genetics. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Peters MA, Cepko CL. The dorsal-ventral axis of the neural retina is divided into multiple domains of restricted gene expression which exhibit features of lineage compartments. Dev Biol. 2002;251:59–73. doi: 10.1006/dbio.2002.0791. [DOI] [PubMed] [Google Scholar]
- Randolph H. The regeneration of the tail in lumbriculus. J Morphol. 1892;7:317–344. [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Robb SM, Sánchez Alvarado A. Identification of immunological reagents for use in the study of freshwater planarians by means of whole-mount immunofluorescence and confocal microscopy. Genesis. 2002;32:293–298. doi: 10.1002/gene.10087. [DOI] [PubMed] [Google Scholar]
- Romanko MJ, Rola R, Fike JR, Szele FG, Dizon ML, Felling RJ, Brazel CY, Levison SW. Roles of the mammalian subventricular zone in cell replacement after brain injury. Prog Neurobiol. 2004;74:77–99. doi: 10.1016/j.pneurobio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, Mannini L, Batistoni R, Wang E, Gremigni V. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol. 2007;8:R62. doi: 10.1186/gb-2007-8-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. The Journal of biological chemistry. 2002;277:14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002;297:2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501–3510. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–1489. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290:328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci U S A. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplemental data include 8 figures, 4 tables, and detailed experimental procedures.