Abstract
DTNBP1 has been recognized as a schizophrenia susceptible gene, and its protein product, dysbindin-1, is down-regulated in the brains of schizophrenic patients. However, little is known about the physiological role of dysbindin-1 in the central nervous system. We hypothesized that disruption of dysbindin-1 with unidentified proteins could contribute to pathogenesis and the symptoms of schizophrenia. GST pull-down from human neuroblastoma lysates showed an association of dysbindin-1 with the DNA-dependent protein kinase (DNA-PK) complex. The DNA-PK complex interacts only with splice isoforms A and B, but not with C. We found that isoforms A and B localized in nucleus, where the kinase complex exist, whereas the isoform C was found exclusively in cytosol. Furthermore, results of phosphorylation assay suggest that the DNA-PK complex phosphorylated dysbindin-1 isoforms A and B in cells. These observations suggest that DNA-PK regulates the dysbindin-1 isoforms A and B by phosphorylation in nucleus. Isoform C does not contain exons from 1 to 6. Since schizophrenia-related single nucleotide polymorphisms (SNPs) occur in these introns between exon 1 and exon 6, we suggest that these SNPs might affect splicing of DTNBP1, which leads to impairment of the functional interaction between dysbindin-1 and DNA-PK in schizophrenic patients.
Introduction
Dystrobrevin binding protein 1 (DTNBP1, dysbindin-1) consists of approximately 350 amino acids and was originally identified by Benson et al. [1] as a dystrobrevin-binding protein in a yeast two-hybrid screen. Dysbindin-1 attracted interest in 2002 when variations in the gene encoding it at chromosomal locus 6p22.3 were reported to be associated with schizophrenia [2], suggesting a susceptibility locus for schizophrenia. Since then, many groups have reported data that collectively support a link between schizophrenia and DTNBP1 [3]–[16]. Hence, genetic variations in the dysbindin-1 gene might be a major risk factor for schizophrenia.
Previous reports have shown that diverse high-risk single nucleotide polymorphisms (SNPs) and haplotypes could influence dysbindin-1 mRNA expression [17]–[19]. Moreover, schizophrenics demonstrate reduced dysbindin-1 mRNA expression in the frontal cortex and hippocampus [19], [20], and lower protein expression levels of dysbindin-1 have been observed post-mortem in the hippocampus of schizophrenics compared to age-matched controls [21]. Interestingly, dysbindin-1 is involved in glutamatergic [9], [21] and dopaminergic neurotransmission [22]–[24]. Collectively, this suggests that the physiological function of dysbindin-1 might be impaired in schizophrenia patients. Nevertheless, the functions of dysbindin-1 in the central nervous system (CNS) remain unclear.
To identify the proteins that interact with dysbindin-1, we examined dysbindin-1 binding proteins in lysates from human neuroblastoma cells by glutathione-S-transferase (GST) pull-down assay. We found that the DNA-dependent protein kinase (DNA-PK) complex bound to dysbindin-1 and phosphorylated dysbindin-1 in vitro. Interestingly, the functional complex interacted with dysbindin-1 in an isoform-selective manner. Dysbindin-1 isoforms A and B interacted with DNA-PK and localized in the nuclei where DNA-PK complex functions. DNA-PK phosphorylated these isoforms in cells, implying that DNA-PK regulates them by phosphorylations in nucleus. Isoform C does not interact with DNA-PK, not phosphorylated, nor localized in nucleus. These observations suggest a novel function and differences among isoforms of dysbindin-1 in mammalian cells, which could shed new light on the etiology of schizophrenia.
Results
Identification of dysbindin-1-associated proteins
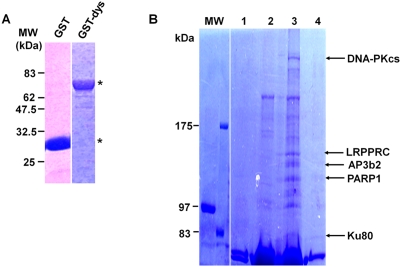
To identify proteins that interact with dysbindin-1 in neuronal cells, GST and GST-dysbindin-1 were purified and used to perform pull-down assays in lysates from human neuroblastoma cells (SH-SY5Y), mouse brain, and skeletal muscle. The purities of GST and GST-dysbindin-1 are shown in Fig. 1A. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the purified proteins, and the gel was then stained with Coomassie Brilliant Blue (CBB). As shown in Fig. 1A, the apparent molecular weights (MW) of GST and GST-dysbindin-1 were 30 and 70 kDa, respectively.
Figure 1. Identification of dysbindin-1-associated proteins.
A) Expression of the GST and GST-dysbindin-1 in E. coli (asterisk). Purified GST and GST-dysbindin-1 were separated by 10% SDS-PAGE and visualized by Coomassie Brilliant Blue (CBB) staining. B) Proteins identified as dysbindin-1-associated proteins. The purification of GST-dysbindin-1, isolation of lysates from the human neuroblastoma cell line SH-SY5Y, and the pull-down assays were performed as described in the “Materials and Methods.” Eluates of the GST- or GST-dysbindin-1 associated proteins were separated by SDS-PAGE and stained with CBB. Lanes 1 and 2 represent GST and GST-dysbindin-1 alone without cell extract, and lanes 3 and 4 show the associated proteins pulled down by incubating whole-cell extracts from SH-SY5Y cells with GST-dysbindin-1 or GST, respectively. The appropriate portions of the polyacrylamide gel containing the specific protein bands in lane 3 were analyzed by MALDI-TOF-MS as described in the “Materials and Methods.” The arrows indicate the proteins identified by MALDI-TOF-MS with a high score.
Proteins that were captured by GST or GST-dysbindin-1 were separated by SDS-PAGE and detected by CBB staining. Five proteins, which were ∼80, 115, 120, 157, and 470 kDa, respectively, were co-purified from SH-SY5Y cell lysates with GST-dysbindin-1, but not with GST alone (Fig. 1B). These protein bands were excised from the gel, subjected to in-gel trypsin digestion, and analyzed by mass assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS). They were reproducibly identified as ATP-dependent DNA helicase 2 (Ku80), poly (ADP-ribose) polymerase family, member 1 (PARP1), adaptor-related protein complex 3, beta 2 subunit (AP3b2), leucine-rich PPR-motif containing protein (LRPPRC), and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) by peptide mass fingerprinting (PMF). Interestingly, three of these proteins, Ku80, PARP1, and DNA-PKcs, are components of the DNA-PK functional complex [25], [26]. This suggests that dysbindin-1 might possibly interact with the DNA-PK complex and influence its function. In addition, we also identified several mouse dysbindin-1-associated proteins from mouse brain or skeletal muscle homogenates (Table 1). Previous studies have demonstrated an interaction of AP3b2 with dysbindin-1, which we also identified in screenings of both mouse brain and SH-SY5Y cells. Therefore, we focused on the three components of the DNA-PK complex and investigated their interactions with dysbindin-1 because they revealed the highest PMF score, they are known to physiologically function as a complex, and their interactions with dysbindin-1 have not been defined.
Table 1. Proteins identified as dysbindin-1-associated proteins by mass spectrometry.
| Bait | Prey | Identified Proteins | NCBI No. |
| Human dysbindin-1 (isoform A) | SH-SY5Y cells | DNA-PKcs | NP_008835 |
| LRPPRC | NP_573566 | ||
| AP3b2 | NP_004635 | ||
| PARP1 | NP_001609 | ||
| Ku80 | NP_066964 | ||
| Mouse dysbindin-1 | Brain | Ap3b2 | NP_067467 |
| Skeletal Muscle | Atp2a2 | NP_033852 | |
| Atp5a1 | NP_031531 | ||
| Arfgef2 | NP_001078964 | ||
| Hadha | AAH37009 | ||
| Hadhb | NP_663533 | ||
| Dapk1 | NP_083929 | ||
| Slc25a4 | AAH26925 |
For protein mass spectrometric analysis, GST fusion human dysbindin-1A and mouse dysbindin-1 were used to perform pull-down screening of SH-SY5Y cells, mouse brain, or skeletal muscle. The specific bands from the GST-dysbindin-1 lane were excised and identified by mass spectrometry and peptide mass fingerprinting. The identified proteins from respective bait and preys and their NCBI numbers are shown. These proteins were reproducibly identified by MALDI-TOF-MS with a high score.
Endogenous dysbindin-1 interacts with Ku70/80 in SH-SY5Y cells
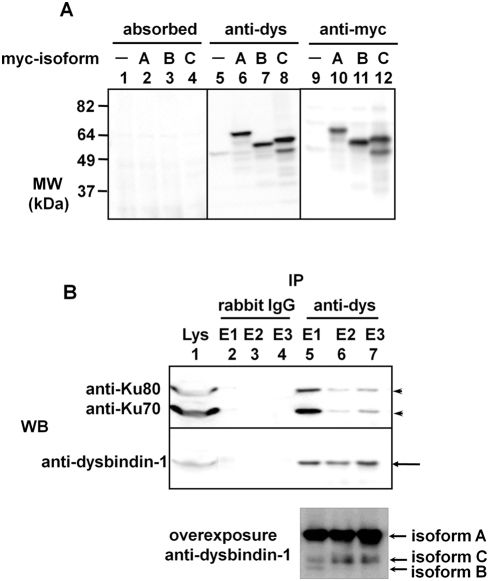
According to previous studies, it is known that Ku70 forms a heterodimer with Ku80 and is also a component of the DNA-PK complex[27]–[30]. All members of the DNA-PK complex except for Ku70 were identified as binding partners of dysbindin-1, which led us to examine the binding of Ku70/80 to dysbindin-1. To confirm the interaction between endogenous dysbindin-1 and Ku70/80, we generated a polyclonal anti-dysbindin-1 antibody by immunizing rabbits with GST-dysbindin-1 as described in the “Materials and Methods.” Three isoforms of human dysbindin-1 exist in the NCBI database. Hence, the specificity of the dysbindin-1 antibody was evaluated by immunoblotting using lysates from COS-7 cells transfected with myc-dysbindin-1s (isoforms A–C). As shown in Fig. 2A, all three isoforms of dysbindin-1 were detected by the dysbindin-1 antibody (center panel) but not by the antigen-absorbed antibody (left panel). In the absence of myc-dysbindin-1, this antibody reacted weakly with a protein band near the estimated MW of dysbindin-1. The antigen-absorbed antibody did not show immunoreactivity with proteins at this MW. Therefore, we conclude that this band was endogenous dysbindin-1 and used the dysbindin-1 antibody to detect endogenous dysbindin-1 in subsequent experiments.
Figure 2. Evaluation of the anti-dysbindin-1 antibody and co-immunoprecipitation of endogenous dysbindin-1 and Ku70/80.
A) The anti-dysbindin-1 antibody recognizes all isoforms of human dysbindin-1. The lysates of COS-7 cells overexpressing each isoform were detected by Western blotting using the anti-dysbindin-1 antibody (center panel), the antigen-absorbed antibody (left panel), or the anti-myc antibody (right panel). In the lanes 8 and 12, there were two bands, lower one of which was thought to be the degraded form of isoform C. B) Endogenous Ku70/80 co-immunoprecipitated with endogenous dysbindin-1. Immunoprecipitation was performed using SH-SY5Y cells with the anti-dysbindin-1 antibody or normal rabbit IgG (negative control). As indicated by the arrowheads, Ku70 and Ku80 immunoprecipitated with dysbindin-1 (indicated by arrows), which was pulled down with the anti-dysbindin-1 antibody (lanes 5–7). In the overexposured immunoblot, two more bands corresponding to isoforms C and B were detected, indicating that the protein levels of these isofoms were much lower than that of isoform A.
To ascertain whether endogenous dysbindin-1 interacts with Ku70/80 in neuronal cells, immunoprecipitation assays were performed in SH-SY5Y cells with the anti-dysbindin-1 antibody or with non-specific rabbit IgG (Fig. 2B). The proteins immunoprecipitated by the anti-dysbindin-1 antibody or control IgG were sequentially eluted into three fractions (E1 to E3) and resolved by SDS-PAGE. As shown in Fig. 2B, the anti-dysbindin-1 antibody immunoprecipitated dysbindin-1 and co-immunoprecipitated Ku70 and Ku80; control IgG did not immunoprecipitate dysbindin-1, Ku70, or Ku80. These data suggest that dysbindin-1 might interact with Ku70 and Ku80 in neuronal cells under physiological conditions. However, Ku70 was not identified as a dysbindin-1 binding partner in our pull-down screening. We infer that Ku70 was obscured by the robust dysbindin-1 band because the molecular weight of Ku70 is nearly equal to that of GST-dysbindin-1.
Localization of endogenous dysbindin-1 in SH-SY5Y cells
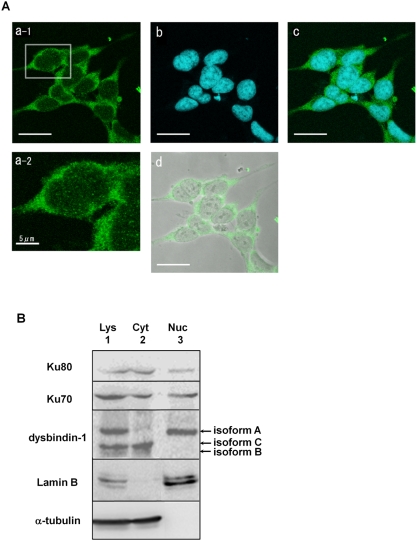
The DNA-PK complex is mainly localized to and functions in nuclei; hence, we examined whether endogenous dysbindin-1 also localized to nuclei in SH-SY5Y cells by immunocytochemistry using the anti-dysbindin-1 antibody. As shown in Fig. 3A, dysbindin-1 appeared to localize primarily to the cytoplasm, but also showed some diffuse localization in the nucleus (Fig. 3A a-2). Phase-contrast imaging showed that these cells were normal, and pre-absorption of the antibodies with antigen GST-dysbindin-1 completely abolished immunoreactivity (data not shown), confirming that endogenous dysbindin-1 exists in SH-SY5Y cells under the physiological condition.
Figure 3. Localization of endogenous dysbindin-1.
A) Localization of endogenous dysbindin-1 in SH-SY5Y cells. SH-SY5Y cells were grown on collagen-coated glass coverslips and immunostained using the anti-dysbindin-1 antibody and the secondary antibody conjugated with Alexa 488 (a-1). Nuclei were visualized by incubating with TOPRO3 (b). Nuclei and dysbindin-1 were merged in c. The boxed area is enlarged in the bottom row (a-2), which indicates existence of endogenous dysbindin-1 in nuclei. Green: dysbindin-1; Blue: TOPRO3; White: phase-contrast. Scale bar, 20 µm except for a-2, 5 µm. B) Subcellular distribution of endogenous dysbindin-1 in SH-SY5Y cells. Cytosolic and nuclear fractions were obtained from SH-SY5Y cells and identified with anti-α-tubulin and anti-lamin B antibodies. Lys: whole lysate; Cyt: cytosolic fraction; Nuc: nuclear fraction. Endogenous dysbindin-1 and Ku localized to both the nuclear and cytosolic fractions.
Since this immunocytochemical approach could not clearly show the nuclear localization of dysbindin-1, we performed subcellular fractionation of SH-SY5Y cells followed by immunoblotting. Cytosolic and nuclear fractions were confirmed by immunoblotting using the marker protein antibodies anti-α-tubulin and anti-lamin B, respectively. As shown in Fig. 3B, endogenous dysbindin-1 was unambiguously localized to both the nuclear and cytosolic fractions, which is in accord with our immunocytochemical staining data. We also investigated the subcellular distribution of Ku70/80. Consistent with previous reports [31]–[37], Ku70/80 also localized to both the nucleus and cytosol, supporting the notion that Ku70/80 interacts with dysbindin-1 in neuronal cells. Furthermore, we identified three bands in whole-cell lysates and cytosolic fractions and a single band in the nuclear fraction. The upper, middle, and lower bands are consistent in MW with isoforms A, C, and B, respectively; hence, we hypothesize that isoform A can localize to the nucleus but not isoform C and that it might have a specific biological purpose there.
Subcellular localizations and interactions with Ku70/80 of three isoforms of dysbindin-1
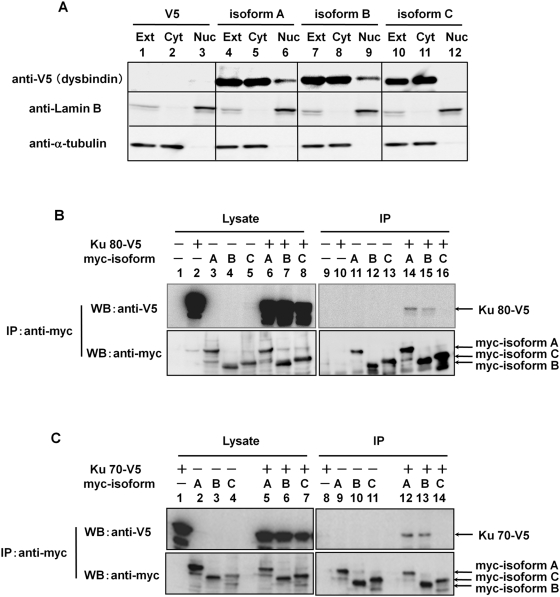
The subcellular localizations of endogenous dysbindin-1 were different among three isoforms (Fig. 3B). Therefore, to determine the localizations of the isoforms more clearly, we performed subcellular fractionation of COS-7 cells transfected with V5-dysbindin-1A, B and C. The reason why we used COS-7 cells in this overexpression experiment was that the cells were most efficiently transfected. As shown in Fig. 4A, isoforms A and B localized in both cytosol and nucleus, whereas the isoform C was exclusively found in cytosol.
Figure 4. Subcellular localization and interaction with Ku70/80 of three isoforms of dysbindin-1.
Protein extracts were prepared from COS-7 cells transfected with the plasmids as indicated. A) Subcellular distribution of three isoforms of dysbindin-1 in transfected COS-7 cells. Cytosolic and nuclear fractions were obtained from COS-7 cells and identified with anti-α-tubulin and anti-lamin B antibodies. Lys: whole lysate; Cyt: cytosolic fraction; Nuc: nuclear fraction. Isoform A and B were localized to both the nuclear and cytosolic fractions but isoform C was not. B) and C) Differences in binding specificities among dysbindin-1 isoforms. Proteins were immunoprecipitated with the anti-myc antibody and detected with the anti-V5 (upper panels) and anti-myc antibodies (lower panels). Ku70/80 were co-immunoprecipitated with isoforms A and B of dysbindin-1 but not with isoform C, indicating that Ku70/80 formed complex with dysbindin-1 in an isoform selective manner.
Next, to examine whether Ku70/80 bound to dysbindin-1 in an isoform-selective manner, we performed immunoprecipitation assays in COS-7 cells doubly transfected with respective dysbindin-1 isoforms and Ku70 or Ku80. Both of Ku70-V5 and Ku80-V5 were co-immunoprecipitated with myc-tagged isoforms A and B using the anti-myc antibody, but not with isoform C (Fig. 4B, lanes 14–16, and 4C, lanes 12–14,), suggesting that Ku70/80 bound to isoform-selectively dysbindin-1 in cells. Together with the results of subcellular fractionation (Fig. 4A), these data suggest that only isoforms of dysbindin-1 localized in the nucleus interacted with Ku70/80 there.
In vitro phosphorylation of dysbindin-1 by the DNA-PK complex
We next investigated the functional meaning of binding between dysbindin-1 and DNA-PK complex. As DNA-PK complex is known to be a serine/threonine kinase, we examined whether dysbindin-1 influences DNA-PK kinase activity by comparing the intrinsic kinase activity of the DNA-PK complex in SH-SY5Y cells transfected with myc-dysbindin-1 or empty vector. DNA-PK activities were determined by measuring incorporation of [γ-32P] into a synthetic peptide from [γ-32P]-ATP by liquid scintillation counting. The intrinsic kinase activity of the DNA-PK complex was not affected by the expression of dysbindin-1 (isoforms A, B, and C) in SH-SY5Y cells (data not shown).
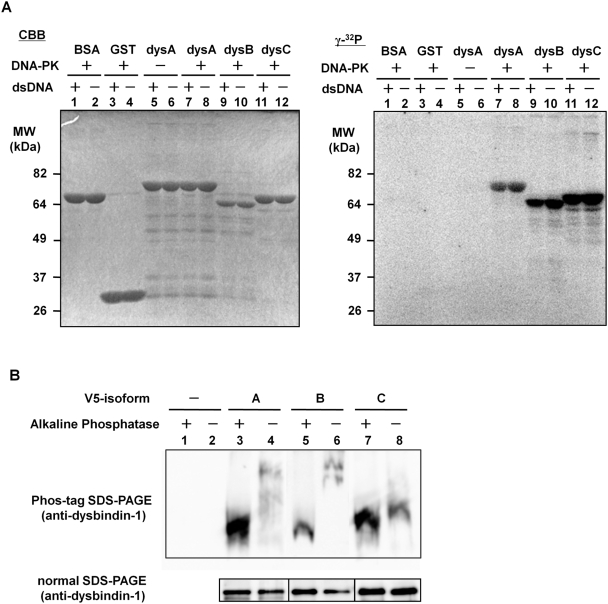
Next, we investigated whether dysbindin-1 was a substrate for phosphorylation by DNA-PK. We performed in vitro kinase assays by mixing the purified DNA-PK complex with GST or GST-dysbindin-1. After the reaction, the samples were subjected to 10% SDS-PAGE, and phosphorylated proteins were detected by incorporation of [γ-32P]. As shown in Fig. 5A, all three isoforms of dysbindin-1 were phosphorylated by DNA-PK, whereas BSA, GST, and dysbindin-1 isoform A in the absence of DNA-PK were not phosphorylated. Because DNA-PK activity is influenced by double-stranded DNA (dsDNA) [38]–[41], we next examined DNA-PK activity in the presence or absence of fragmented dsDNA (indicated by + or −). The dsDNA did not affect the phosphorylation level of dysbindin-1 (all isoforms; Fig. 5A), suggesting that phosphorylation of dysbindin-1 might be dependent on the constitutive kinase activity of DNA-PK.
Figure 5. Phosphorylation of dysbindin-1 by DNA-PK complex.
A) In vitro phosphorylation of dysbindin-1 by DNA-PK complex. The purified DNA-PK complex and GST-dysbindin-1 were incubated with [γ-32P]-labeled ATP as described in the “Materials and Methods.” These samples were subjected to 10% SDS-PAGE and stained with CBB (left panel). The right panel shows the uptake of [γ-32P] ATP by phosphorylation of dysbindin-1. As shown in the left panel, the amount of BSA (lanes 1, 2), GST (lanes 3, 4), GST-dysbindin-1A (lanes 5–8), B (lanes 9, 10), and C (lanes 11, 12) were equal. Lanes 7–12 in the right panel show that the three isoforms of dysbindin-1 were phosphorylated by the DNA-PK complex. B) Phosphorylation of dysbindin-1 isoforms A and B in mammalian cells. Protein extracts were prepared as described in “Materials and Methods”. These samples were separated by Phos-tag SDS-PAGE and detected with the anti-dysbindin antibody. The phosphorylation levels of V5-dysbindin-1A and B were higher than that of C. Lanes 1 and 2: empty vector; lanes 3 and 4: V5-dysbindin-1A; lanes 5 and 6: V5-dysbindin-1B; lanes 7 and 8: V5-dysbindin-1C. The immunoblot after the normal SDS-PAGE showed that the amounts of dysbindin-1 were not altered after the dephosphorylation procedure by AP.
Phosphorylation of dysbindin-1 in mammalian cells
To examine whether dysbindin-1 is phosphorylated physiologically in cells, we analyzed three isoforms of dysbindin-1 immunopurified from Hela cells transfected with them, using Mn2+-Phos-tag SDS-PAGE. The reason why we used Hela cells in this experiment was that the kinase activity of DNA-PK was known to be very high in these cells and most of studies for the DNA-PK function were performed using the cells. As shown in Fig. 5B (lanes 4 and 6), the bands of isoforms A and B up-shifted compared to those of them treated with alkaline phosphatase (AP), indicating that isoforms A and B were predominantly phosphorylated in cells. On the other hand, isoform C were not influenced by the treatment with AP significantly, suggesting that isoform C was phosphorylated at the very low level under the physiological condition. We also observed the same phosphorylation pattern of dysbindin-1 isoforms in COS-7 cells (data not shown). This isoform selectivity of phosphorylation in cells was identical to that of binding to DNA-PK complex and nuclear localization. Combined with in vitro phosphorylation data, these observations suggested that DNA-PK complex selectively bound to and phosphorylated isoforms A and B of dysbindin-1, but not isoform C in mammalian cells.
Discussion
In this study, we identified interaction partners of dysbindin-1 in neuronal cells, mouse brain, and muscle by GST pull-down screening. Dysbindin-1 is a member of biogenesis of lysosome-related organelles complex-1 (BLOC-1) [42] and is thought to be involved in intracellular vesicular trafficking (i.e., protein sorting and vesicle docking and fusion), because genetic deletion of each component of BLOC-1 leads to disruption of intracellular vesicular trafficking in the biogenesis of lysosome-related organelles. All BLOC-1 knockout mice have phenotypes characteristic of Hermansky-pudlack syndrome (HPS), i.e., hypopigmentation of both coat and eyes due to melanosome defects and prolonged bleeding times resulting from platelet dense body defects [42]–[51]. Sandy mice in which dysbindin-1 genes are disrupted also demonstrate such phenotypes and have often been used as HPS model mice [42]. In this study, one of the novel dysbindin-1 binding partners identified was a component of the AP-3 complex, AP3b2 (Table 1). This complex appears to be functionally similar to BLOC-1 because genetic disruption of AP-3 components also results in HPS-like symptoms in mice [48], [52]–[63]. Moreover, physical and functional interactions between BLOC-1 components and AP-3 components were reported recently [64], [65]. Accordingly, we detected the binding of AP3b2 to dysbindin-1, implying that our GST pull-down screening worked well. However, dystrobrevin-1, which was reported to bind to dysbindin-1 in a yeast two-hybrid system [1], was not identified as a dysbindin-1 binding partner. Lack of detection of dystrobrevin-1 in these pull-down assays could be due to the inability to separate dystrobrevin-1 from GST-dysbindin-1 by one-dimensional SDS-PAGE.
Nonetheless, we were interested in the interaction between dysbindin-1 and the DNA-PK complex, which is involved in transcription [66], [67], DNA recombination [68], and DNA repair [38], [40], [69], [70] in many kinds of cells. The physiological role of dysbindin-1 in the nucleus remains to be elucidated, although dysbindin-1 was also reported to localize to nuclei of hippocampal neurons in vivo [21]. We demonstrated that both endogenous and exogenous dysbindin-1 bound to components of DNA-PK, Ku70, and Ku80 in neuronal cells by immunoprecipitation, immunocytochemical staining, and subcellular fractionation. Interestingly, dysbindin-1 was also phosphorylated by DNA-PK, suggesting a functional consequence of the interaction. Although phosphorylation of some substrates by DNA-PK is activated by dsDNA [38]–[41], dysbindin-1 phosphorylation by DNA-PK was not affected by the addition of dsDNA, indicating that phosphorylation of dysbindin-1 might not be involved in DNA repair. This is consistent with the idea that DNA-independent DNA-PK activity might also play an important role in transcriptional regulation besides recombination and double-stranded DNA repair as previously described[71]–[73].
Moreover, we found that three isoforms (A, B, and C) of dysbindin-1 interacted with Ku70/80 in different manners in the cells (Fig. 4). Functional differences between these isoforms have not been reported, and the isoform-dependent binding of dybindin-1 to DNA-PK may be of importance. Interestingly, the same isoform-dependency was observed in subcellular localization and phosphorylation in cells. The consistency suggested that DNA-PK complex bound to and phosphorylated dysbindin-1 in mammalian cells as well as in vitro. However, our in vitro kinase assay data showed that all three isoforms were phosphorylated by DNA-PK (Fig. 5A), implying that all purified isoforms of dysbindin-1 can bind to purified DNA-PK. We hypothesized that the distribution of dysbindin-1 in cells might be different among the isoforms and that localization differences could be the basis of their isoform-dependent interaction properties. Correspondingly, isoforms A and B were detected in considerable amounts in the nuclear fraction (Fig. 3 and 4). Hence, nuclear localization of isoforms A and B might facilitate their binding to Ku70/80. In contrast, isoform C, although abundant in total cell lysates, was not detected in the nuclear fraction. As isoform B appeared to be a minor dysbindin-1 according to its very low expression level in neuronal cells, the functional difference between isoform A and C might be of importance in terms of the physiological roles of dysbindin-1 in CNS.
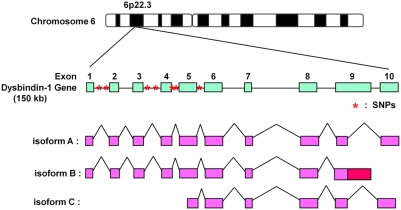
What is the difference between dysbindin-1 isoforms A and C? Figure 6 shows the amino acid sequences of the spliced isoforms of dysbindin-1. It appears that isoform C is an N-terminally truncated form of isoform A. Hence, the N-terminal region of isoform A could possibly localize to nuclei and bind Ku70/80. To our surprise, almost all of the schizophrenia-related SNPs are found in the introns between exon 1 and exon 6, which are in isoforms A and B but not in C (Fig. 5B) [2]–[8], [10], [12]–[14], [17], [18]. This led us to hypothesize that the schizophrenia-related SNPs in the coding region of the dysbindin-1 gene might affect its splicing variations and lead to a reduction in isoform A and an increase in the content of isoform C, which could be defective with regard to interaction with the DNA-PK complex.
Figure 6. Schematic representation of the human dysbindin-1 gene and its three isoform structures.
The gene encoding dysbindin-1 located at chromosomal locus 6p22.3, and its several genetic variations are associated with schizophrenia. These variations (SNPs and haplotypes) are located in intron or promoter regions, and almost all are located in the N-terminus of the gene. Red asterisks indicate major SNPs. Three isoforms (A, B, and C) of human dysbindin-1 were reported by NCBI. Isoform A encodes the longest isoform, and isoform B contains an additional segment in the coding region compared to isoform A. Isoform C contains an alternate splice site in the 5′ coding region and uses a downstream start codon, compared to isoform A; this isoform has a shorter N-terminus compared to isoform A.
In conclusion, in this study we describe for the first time a functional interaction between dysbindin-1 and the DNA-PK complex and show a functional difference in dysbindin-1 isoforms. This newly acquired information provides a basis for the novel hypothesis that alternative splicing of dysbindin-1 due to schizophrenia-related SNPs may underlie the etiology of schizophrenia. Further studies that focus on the relationship between schizophrenia-related SNPs and dysbindin-1 splice variants and the expression levels of the three isoforms in the brains of post-mortem schizophrenic patients would be helpful in understanding the role of dysbindin-1 in schizophrenia.
Materials and Methods
Plasmid Construction
Three isoforms of human dysbindin-1 cDNA and mouse dysbindin-1 cDNA were cloned from a human fetal brain cDNA library (BD Biosciences) and a mouse brain cDNA library by polymerase chain reaction (PCR), respectively. These cDNAs were subcloned into pcDNA3.1 (Invitrogen) or pGEX-4T-3 (GE Healthcare), which contained His-tag×6 at the C-terminus. The myc-dysbindin-1 contained myc-tag×6 at the N-terminus of dysbindin-1, and V5-dysbindin-1 contained a V5-tag at the C-terminus of −1. GST-dysbindin-1 was able to express GST protein at the N-terminus and His-tag×3 at the C-terminus of dysbindin-1. The full-length human Ku80 and Ku70 were cloned from a human fetal brain cDNA library by PCR. The cDNAs of Ku80 and Ku70 were subcloned into a pcDNA3.1 expression vector that contained a V5-tag in the C-terminus of the cDNA. All constructs were confirmed by sequencing using a fully automated DNA sequencer (Beckman Coulter).
Cell Culture and Transient Transfection
The cell lines SH-SY5Y and COS-7 were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Invitrogen) in a 5% CO2 atmosphere at 37°C. SH-SY5Y cells were plated in collagen-coated dishes and were harvested at 100% confluency for GST pull-down assay and immunoprecipitation analysis. COS-7 cells were transiently transfected with plasmid constructs using FuGENE 6 transfection reagents (Roche Applied Science). After 48 h of transfection, the cells were harvested for immunoprecipitation.
Expression and Purification of GST and GST-dysbindin-1
A pGEX expression vector containing human (isoform A) or mouse dysbindin-1 was transformed into BL21 (DE3) strain. An overnight culture of the transformant in Luria Broth (LB) medium was diluted and shaken at 37°C until the OD600 reached 0.3–0.5; 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was then added. During induction by IPTG, the culture was shaken at 27°C for 3 h. The bacterial cells were collected by centrifugation (1,600×g for 20 min), washed with phosphate-buffered saline (PBS), and suspended in sonication buffer [50 mM Tris-HCl (pH 7.0), 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.2 mM PMSF, and 1/1000 volume of protease inhibitor cocktail (Sigma-Aldrich)]. The suspended Escherichia coli were fractured four times by a FRENCH® pressure cell press (Ohtake Works, Co.), and Triton-X 100 was added to a final concentration of 1%. The suspension was then incubated for 30 min on ice. The lysates were centrifuged (16,000×g for 30 min), and the supernatant was subjected to affinity purification using glutathione Sepharose 4B beads (GE Healthcare) and Talon Metal Affinity Resin (BD Biosciences), according to the manufacturer's protocol. All subsequent steps were performed at 4°C. The quantity and purity of the proteins was assessed by SDS-PAGE; Coomassie Brilliant Blue (CBB) was used to stain the separated proteins.
GST Pull-Down Assay
The SH-SY5Y cells were homogenized in lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% (w/v) Triton-X 100, 0.2 mM PMSF, and 1/1000 volume of protease inhibitor cocktail] and incubated for 30 min on ice. Mouse brain or skeletal muscle (2 g each) was homogenized in lysis buffer using a Hitachi homogenizer and incubated for 30 min on ice. The lysate was centrifuged (16,000×g for 30 min), and the supernatant was precleared with glutathione Sepharose 4B beads for 1 h. The precleared lysate was incubated with equivalent amounts of GST and GST-dysbindin-1, respectively, for 3 h at 4°C. After incubation, the beads were washed four times with lysis buffer, and the bead-bound proteins were eluted by boiling for 5 min in 2×SDS sample buffer. The precipitated proteins were separated by SDS–PAGE, and the specific bands in the GST-dysbindin-1 lane were analyzed by MALDI-TOF-MS.
Protein Identification by Mass Spectrometry
Following electrophoresis, proteins were stained with colloidal CBB. The protein bands of interest were excised from the gel, cut into small pieces, dehydrated with acetonitrile (ACN) for 10 min, and dried completely in a vacuum centrifuge. DTT-containing buffer (10 mM DTT in 100 mM NH4HCO3) was added to the gel pieces, and the gel pieces were incubated for 1 h at 56°C. After the samples had cooled to room temperature, the DTT-containing buffer was replaced with iodoacetamide-containing buffer (55 mM iodoacetamide 100 mM NH4HCO3), and the gel pieces were vortexed for 45 min at room temperature. The gel pieces were then washed with 100 mM NH4HCO3 buffer and dehydrated by ACN several times repeatedly and dried in a vacuum centrifuge. The gel pieces were then incubated in trypsin (Promega)-containing buffer (12.5 ng/µL trypsin in 50 mM NH4HCO3 and 5 mM CaCl2) in an ice-cold bath. After 45 min, the protein digestion was performed overnight at 37°C. Digestion was stopped by the addition of 0.1% TFA in 50% (v/v) ACN/water. Peptides were extracted by the addition of 50 mM NH4HCO3 in 50% ACN, with three changes (20 min per extraction) at room temperature, and concentrated. The peptides in the extract were purified from the supernatant by absorption onto ZipTipC18 (Millipore) according to the manufacturer's instructions. After five washes with 0.1% TFA in water (v/v), bound peptides were eluted with 10 µL of saturated matrix-solution (R-cyano-4-hydroxy-cinnamic acid, Sigma-Aldrich) in 0.1% TFA (v/v) in 75% (v/v) ACN/water. Then, 0.3 ml of each eluted sample were spotted on the target plate repeatedly and dried at room temperature. MALDI-TOF-MS was performed on an AXIMA-CFR mass spectrometer (Shimadzu). MALDI peptide spectra were calibrated using several peaks of self-digested trypsin and matrix ion as internal standards. The data were analyzed using the MASCOT search program (Matrix Science, London, UK). The peptide masses were compared to the NCBI database for identification of the intact proteins.
Antibodies
The rabbit polyclonal antibody to dysbindin-1 was generated by injecting rabbits subcutaneously with 1.8 mg of purified GST-dysbindin-1 (mouse) protein from E. coli using the standard immunization protocol. The antiserum was immunoaffinity-purified using a column in which MBP (maltose binding protein)-dysbindin-1 (mouse) proteins were coupled using Affi-Gel® 15 (Bio-Rad) according to the manufacturer's protocol. Cyclic incubation of the IgG fraction of the antiserum overnight was followed by the elution of affinity-purified antibodies with 100 mM glycine-HCl (pH 2) and neutralized with 1 M Tris-HCl (pH 9). Affinity-purified antibodies were supplemented with 10 mM NaN3, stored at 4°C, and diluted 1∶1000 for immunoblotting. The anti-Ku70 and Ku80 antibodies were gifts of Dr. Y. Hosoi, Niigata University. The following mouse monoclonal antibodies were purchased from the vendors indicated in parentheses: anti-myc, anti-V5 and anti-lamin B (Invitrogen) and anti-α-tubulin (Sigma-Aldrich). The followning rabbit polyclonal antibodies were purchased from the vendors indicated in parentheses: anti-V5 (Millipore) and anti-myc (cell signaling).
Immunoblotting
Samples were separated by SDS–PAGE and transferred to PVDF membranes (Immobilon-P; Millipore). The membranes were blocked with 5% skim milk in PBS with 0.05% Tween®20 (TPBS) for 1 h at room temperature and then incubated for 2 h at room temperature or overnight at 4°C with primary antibodies in 5% skim milk. After washing, the membranes were incubated for 45 min with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling) at room temperature. The immunoreactive bands were visualized by enhanced chemiluminescence (ECL) and scanned by LAS 3000 (Fuji film co. LTD).
Immunoprecipitation
COS-7 cells were transfected with myc-dysbindin-1 and V5-tagged constructs of Ku70 or Ku80 using FuGENE 6. Cells from 10-cm plates were homogenized in 1 ml of lysis buffer [50 mM Tris-HCl (pH 7.5) containing 150 mM NaCl, 1 mM EDTA, 1% (w/v) Triton X-100, 0.2 mM PMSF, and protease inhibitor cocktail]. The lysates were then centrifuged at 16,000×g for 30 min at 4°C. The supernatant was precleared with protein G Sepharose 4 fast flow beads (GE Healthcare) for 1 h, and then incubated with 1 µl of anti-myc or V5 antibody for 2 h. The beads (15 µl) were added to the lysate and incubated for 1 h. After the beads were washed four times with lysis buffer, the precipitates were analyzed by SDS–PAGE and immunoblotted with either the anti-myc or anti-V5 antibody.
The immunoprecipitations of endogenous dysbindin-1 and Ku70/80 were performed using the anti-dysbindin-1 antibody linked to gel beads (Seize® Primary Immunoprecipitation Kit, PIERCE). The anti-dysbindin-1 antibody and normal rabbit IgG (control) were coupled to the gel according to the manufacturer's protocol. Lysates from SH-SY5Y cells were mixed with the antibody-coupled gel or with control IgG-coupled gel overnight at 4°C. The gels were washed four times with lysis buffer, and immunoprecipitated proteins were eluted by 100 mM glycine-HCl (pH 2.8). The eluted proteins were analyzed by SDS-PAGE and immunoblotted with anti-Ku70, -Ku80, and -dysbindin-1 antibodies, respectively.
Immunofluorescence
Cells grown on collagen-coated glass coverslips were washed with PBS and fixed with ice-cold methanol (−20°C) for 20 min. The cells were permeabilized in PBS containing 0.5% (w/v) Triton X-100 for 10 min and blocked in PBS containing 5% normal goat serum (NGS), 0.02% Triton X-100, and 20% glycerol for 1 h at room temperature. Incubations with the primary antibody were performed overnight at 4°C in PBS containing 2% NGS, 0.02% Triton X-100, and 20% glycerol. The cells were washed and then incubated with the appropriate secondary antibodies [Alexa 488 donkey anti-rabbit IgG (1∶1000); Alexa 568 donkey anti-rabbit IgG (1∶1000)] in PBS containing 2% NGS, 0.02% Triton X-100, and 20% glycerol for 1 h. Nuclei were visualized by incubating with TOPRO3 (Invitrogen). After washing with PBS, the cover slips were mounted using Mowiol (Calbiochem, La Jolla, CA). Z-stacks of four to ten images were acquired on a Zeiss LSM510 meta laser scanning confocal microscope (Carl Zeiss, Jena, Germany). Brightest point projections of the Z-stacks were used for image analysis.
Cell Fractionation
Briefly, cells were supplemented with hypotonic buffer [10 mM Tris-HCl (pH 7.2), 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 1 mM NaF, 1 mM DTT, 0.2 mM PMSF, and 1/1000 volume of protease inhibitor cocktail] and were scraped, passed through a 27-gauge needle ten times, and centrifuged at 100×g for 10 min at 4°C. The supernatant was the cytosolic fraction. The resulting crude nuclear pellets were suspended in cell lysis buffer [50 mM HEPES (pH 7.5), 10% glycerol, 0.5% Triton X-100, 150 mM NaCl, 1 mM DTT, 0.2 mM PMSF, and 1/1000 volume of protease inhibitor cocktail] and centrifuged at 16,000×g for 60 min at 4°C. The final nuclear pellets were dissolved in RIPA buffer [50 mM HEPES (pH 7.5), 1% Triton X- 100, 0.1% SDS, 150 mM NaCl, 1% deoxycholatic sodium, 1 mM NaF, 1 mM DTT, 0.2 mM PMSF, and 1/1000 volume of protease inhibitor cocktail] and sonicated on ice.
In Vitro Kinase Assay
DNA-PK activity of SH-SY5Y cells transfected with myc-dysbindin-1 was assayed using a synthetic peptide (EPPLSQEAFADLWKK) and [γ-32P]-ATP according to the methods of Hosoi [74]. Cell lysate (5 µl) and peptide substrate (5 µg) were mixed in the kinase reaction buffer [20 mM HEPES-NaOH (pH 7.2), 100 mM NaCl, 5 mM MgCl2, 50 µM [γ-32P]-ATP, 1 mM DTT, and 0.5 mM each of NaF and β-sodium glycerophosphate]. The reaction mixture was incubated at 37°C for 20 min, and the reaction was stopped by addition of 300 mM phosphoric acid. The reaction mixture was spotted onto a P81 paper disk (Whatman), washed in 15% phosphoric acid, and counted in a liquid scintillation counter (Beckman Coulter). Radioactivity was defined as the counts per minute of 32P incorporated in the presence of DNA. The counts per minute of 32P incorporated in the absence of DNA were used as a control.
DNA-PK activity was assayed using purified GST-dysbindin-1, DNA-PK (Promega), and [γ-32P]-ATP. Purified GST-dysbindin-1 (10 µg) and DNA-PK (16 U) were mixed in kinase reaction buffer and incubated at 37°C for 20 min. The reaction was stopped by addition of SDS-sample buffer. To detect phosphorylated proteins, the reaction products were separated on 10% polyacrylamide gels, and the gels were stained with CBB. The gels were then dried, and phosphoproteins were detected by autoradiography using BAS 2500 (Fuji film co.LTD).
Mn2+-Phos-tag SDS-PAGE and Immunoblotting
The samples were prepared from HeLa cells transfected with V5-dysbindin-1 (isoform A, B and C) by immunoprecipitating with the anti-V5 antibody. These samples were divided in half. One half was treated with alkaline phosphatase (AP) (indicated by plus) and the other wasn't (indicated by minus). These samples were separated by Mn2+-Phos-tag SDS-PAGE [75] and detected by immunoblotting with the anti-dysbindin-1 antibody according to manufactures' instruction. In the Mn2+-Phos-tag SDS-PAGE, Phos-tag acrylamide binds to phosphates in the gel and makes phosphorylated proteins migrate more slowly than unphosphorylated forms of ones.
Acknowledgments
We thank Dr. Yoshihisa Matsumoto for Ku70/80 antisera. Special thanks to the member of the laboratory.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The Ministry of Health, Labor, and Welfare, Japan (to S.I.), and the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to E.F.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem. 2001;276:24232–24241. doi: 10.1074/jbc.M010418200. [DOI] [PubMed] [Google Scholar]
- 2.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwab SG, Knapp M, Mondabon S, Hallmayer J, Borrmann-Hassenbach M, et al. Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am J Hum Genet. 2003;72:185–190. doi: 10.1086/345463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang JX, Zhou J, Fan JB, Li XW, Shi YY, et al. Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Mol Psychiatry. 2003;8:717–718. doi: 10.1038/sj.mp.4001287. [DOI] [PubMed] [Google Scholar]
- 5.Van Den Bogaert A, Schumacher J, Schulze TG, Otte AC, Ohlraun S, et al. The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. Am J Hum Genet. 2003;73:1438–1443. doi: 10.1086/379928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Oord EJ, Sullivan PF, Jiang Y, Walsh D, O'Neill FA, et al. Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol Psychiatry. 2003;8:499–510. doi: 10.1038/sj.mp.4001263. [DOI] [PubMed] [Google Scholar]
- 7.Funke B, Finn CT, Plocik AM, Lake S, DeRosse P, et al. Association of the DTNBP1 locus with schizophrenia in a U.S. population. Am J Hum Genet. 2004;75:891–898. doi: 10.1086/425279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, et al. Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry. 2004;55:971–975. doi: 10.1016/j.biopsych.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, et al. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet. 2004;13:2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- 10.Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, et al. Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1). Arch Gen Psychiatry. 2004;61:336–344. doi: 10.1001/archpsyc.61.4.336. [DOI] [PubMed] [Google Scholar]
- 11.Gornick MC, Addington AM, Sporn A, Gogtay N, Greenstein D, et al. Dysbindin (DTNBP1, 6p22.3) is associated with childhood-onset psychosis and endophenotypes measured by the Premorbid Adjustment Scale (PAS). J Autism Dev Disord. 2005;35:831–838. doi: 10.1007/s10803-005-0028-3. [DOI] [PubMed] [Google Scholar]
- 12.Tochigi M, Zhang X, Ohashi J, Hibino H, Otowa T, et al. Association study of the dysbindin (DTNBP1) gene in schizophrenia from the Japanese population. Neurosci Res. 2006;56:154–158. doi: 10.1016/j.neures.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Duan J, Martinez M, Sanders AR, Hou C, Burrell GJ, et al. DTNBP1 (Dystrobrevin binding protein 1) and schizophrenia: association evidence in the 3′ end of the gene. Hum Hered. 2007;64:97–106. doi: 10.1159/000101961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosato S, Ruggeri M, Bonetto C, Bertani M, Marrella G, et al. Association study of dysbindin gene with clinical and outcome measures in a representative cohort of Italian schizophrenic patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144:647–659. doi: 10.1002/ajmg.b.30484. [DOI] [PubMed] [Google Scholar]
- 15.Turunen JA, Peltonen JO, Pietilainen OP, Hennah W, Loukola A, et al. The role of DTNBP1, NRG1, and AKT1 in the genetics of schizophrenia in Finland. Schizophr Res. 2007;91:27–36. doi: 10.1016/j.schres.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Vilella E, Costas J, Sanjuan J, Guitart M, De Diego Y, et al. Association of schizophrenia with DTNBP1 but not with DAO, DAOA, NRG1 and RGS4 nor their genetic interaction. J Psychiatr Res. 2008;42:278–288. doi: 10.1016/j.jpsychires.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Bray NJ, Buckland PR, Owen MJ, O'Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- 18.Bray NJ, Preece A, Williams NM, Moskvina V, Buckland PR, et al. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- 19.Weickert CS, Straub RE, McClintock BW, Matsumoto M, Hashimoto R, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61:544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 20.Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumamoto N, Matsuzaki S, Inoue K, Hattori T, Shimizu S, et al. Hyperactivation of midbrain dopaminergic system in schizophrenia could be attributed to the down-regulation of dysbindin. Biochem Biophys Res Commun. 2006;345:904–909. doi: 10.1016/j.bbrc.2006.04.163. [DOI] [PubMed] [Google Scholar]
- 23.Murotani T, Ishizuka T, Hattori S, Hashimoto R, Matsuzaki S, et al. High dopamine turnover in the brains of Sandy mice. Neurosci Lett. 2007;421:47–51. doi: 10.1016/j.neulet.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 24.Iizuka Y, Sei Y, Weinberger DR, Straub RE. Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J Neurosci. 2007;27:12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 26.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 27.Weaver DT. Regulation and repair of double-strand DNA breaks. Crit Rev Eukaryot Gene Expr. 1996;6:345–375. doi: 10.1615/critreveukargeneexpr.v6.i4.20. [DOI] [PubMed] [Google Scholar]
- 28.Jin S, Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. Embo J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu G. Role of the Ku autoantigen in V(D)J recombination and double-strand break repair. Curr Top Microbiol Immunol. 1996;217:113–132. doi: 10.1007/978-3-642-50140-1_8. [DOI] [PubMed] [Google Scholar]
- 30.Chu G. Double strand break repair. J Biol Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 31.Fewell JW, Kuff EL. Intracellular redistribution of Ku immunoreactivity in response to cell-cell contact and growth modulating components in the medium. J Cell Sci. 1996;109 (Pt 7):1937–1946. doi: 10.1242/jcs.109.7.1937. [DOI] [PubMed] [Google Scholar]
- 32.Grawunder U, Finnie N, Jackson SP, Riwar B, Jessberger R. Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. Eur J Biochem. 1996;241:931–940. doi: 10.1111/j.1432-1033.1996.00931.x. [DOI] [PubMed] [Google Scholar]
- 33.Koike M, Matsuda Y, Mimori T, Harada YN, Shiomi N, et al. Chromosomal localization of the mouse and rat DNA double-strand break repair genes Ku p70 and Ku p80/XRCC5 and their mRNA expression in various mouse tissues. Genomics. 1996;38:38–44. doi: 10.1006/geno.1996.0589. [DOI] [PubMed] [Google Scholar]
- 34.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 35.Koike M, Miyasaka T, Mimori T, Shiomi T. Subcellular localization and protein-protein interaction regions of Ku proteins. Biochem Biophys Res Commun. 1998;252:679–685. doi: 10.1006/bbrc.1998.9368. [DOI] [PubMed] [Google Scholar]
- 36.Koike M, Awaji T, Kataoka M, Tsujimoto G, Kartasova T, et al. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J Cell Sci. 1999;112 (Pt 22):4031–4039. doi: 10.1242/jcs.112.22.4031. [DOI] [PubMed] [Google Scholar]
- 37.Endoh D, Okui T, Kon Y, Hayashi M. Hypertonic treatment inhibits radiation-induced nuclear translocation of the Ku proteins G22p1 (Ku70) and Xrcc5 (Ku80) in rat fibroblasts. Radiat Res. 2001;155:320–327. doi: 10.1667/0033-7587(2001)155[0320:htirin]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 38.Anderson CW. DNA damage and the DNA-activated protein kinase. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- 39.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 40.Rathmell WK, Chu G. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc Natl Acad Sci U S A. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suwa A, Hirakata M, Takeda Y, Jesch SA, Mimori T, et al. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci U S A. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Zhang Q, Oiso N, Novak EK, Gautam R, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermansky F, Pudlak P. Albinism associated with hemorrhagic diathesis and unusual pigmented reticular cells in the bone marrow: report of two cases with histochemical studies. Blood. 1959;14:162–169. [PubMed] [Google Scholar]
- 44.Gahl WA, Brantly M, Kaiser-Kupfer MI, Iwata F, Hazelwood S, et al. Genetic defects and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky-Pudlak syndrome). N Engl J Med. 1998;338:1258–1264. doi: 10.1056/NEJM199804303381803. [DOI] [PubMed] [Google Scholar]
- 45.Shotelersuk V, Gahl WA. Hermansky-Pudlak syndrome: models for intracellular vesicle formation. Mol Genet Metab. 1998;65:85–96. doi: 10.1006/mgme.1998.2729. [DOI] [PubMed] [Google Scholar]
- 46.Huizing M, Anikster Y, Gahl WA. Hermansky-Pudlak syndrome and related disorders of organelle formation. Traffic. 2000;1:823–835. doi: 10.1034/j.1600-0854.2000.011103.x. [DOI] [PubMed] [Google Scholar]
- 47.Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem. 2002;277:28191–28199. doi: 10.1074/jbc.M204011200. [DOI] [PubMed] [Google Scholar]
- 48.Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, et al. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1). Blood. 2003;101:4402–4407. doi: 10.1182/blood-2003-01-0020. [DOI] [PubMed] [Google Scholar]
- 49.Gwynn B, Martina JA, Bonifacino JS, Sviderskaya EV, Lamoreux ML, et al. Reduced pigmentation (rp), a mouse model of Hermansky-Pudlak syndrome, encodes a novel component of the BLOC-1 complex. Blood. 2004;104:3181–3189. doi: 10.1182/blood-2004-04-1538. [DOI] [PubMed] [Google Scholar]
- 50.Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 51.Morgan NV, Pasha S, Johnson CA, Ainsworth JR, Eady RA, et al. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). Am J Hum Genet. 2006;78:160–166. doi: 10.1086/499338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- 53.Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- 54.Feng L, Seymour AB, Jiang S, To A, Peden AA, et al. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum Mol Genet. 1999;8:323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- 55.Zhen L, Jiang S, Feng L, Bright NA, Peden AA, et al. Abnormal expression and subcellular distribution of subunit proteins of the AP-3 adaptor complex lead to platelet storage pool deficiency in the pearl mouse. Blood. 1999;94:146–155. [PubMed] [Google Scholar]
- 56.Dell'Angelica EC, Aguilar RC, Wolins N, Hazelwood S, Gahl WA, et al. Molecular characterization of the protein encoded by the Hermansky-Pudlak syndrome type 1 gene. J Biol Chem. 2000;275:1300–1306. doi: 10.1074/jbc.275.2.1300. [DOI] [PubMed] [Google Scholar]
- 57.Feng L, Rigatti BW, Novak EK, Gorin MB, Swank RT. Genomic structure of the mouse Ap3b1 gene in normal and pearl mice. Genomics. 2000;69:370–379. doi: 10.1006/geno.2000.6350. [DOI] [PubMed] [Google Scholar]
- 58.Gwynn B, Ciciotte SL, Hunter SJ, Washburn LL, Smith RS, et al. Defects in the cappuccino (cno) gene on mouse chromosome 5 and human 4p cause Hermansky-Pudlak syndrome by an AP-3-independent mechanism. Blood. 2000;96:4227–4235. [PubMed] [Google Scholar]
- 59.Kretzschmar D, Poeck B, Roth H, Ernst R, Keller A, et al. Defective pigment granule biogenesis and aberrant behavior caused by mutations in the Drosophila AP-3beta adaptin gene ruby. Genetics. 2000;155:213–223. doi: 10.1093/genetics/155.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Setaluri V. Sorting and targeting of melanosomal membrane proteins: signals, pathways, and mechanisms. Pigment Cell Res. 2000;13:128–134. doi: 10.1034/j.1600-0749.2000.130302.x. [DOI] [PubMed] [Google Scholar]
- 61.Swank RT, Novak EK, McGarry MP, Zhang Y, Li W, et al. Abnormal vesicular trafficking in mouse models of Hermansky-Pudlak syndrome. Pigment Cell Res. 2000;13(Suppl 8):59–67. doi: 10.1034/j.1600-0749.13.s8.12.x. [DOI] [PubMed] [Google Scholar]
- 62.Feng L, Novak EK, Hartnell LM, Bonifacino JS, Collinson LM, et al. The Hermansky-Pudlak syndrome 1 (HPS1) and HPS2 genes independently contribute to the production and of platelet dense granules, melanosomes, and lysosomes. Blood. 2002;99:1651–1658. [PubMed] [Google Scholar]
- 63.Huizing M, Scher CD, Strovel E, Fitzpatrick DL, Hartnell LM, et al. Nonsense mutations in ADTB3A cause complete deficiency of the beta3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr Res. 2002;51:150–158. doi: 10.1203/00006450-200202000-00006. [DOI] [PubMed] [Google Scholar]
- 64.Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, et al. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salazar G, Craige B, Styers ML, Newell-Litwa KA, Doucette MM, et al. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- 67.Cao QP, Pitt S, Leszyk J, Baril EF. DNA-dependent ATPase from HeLa cells is related to human Ku autoantigen. Biochemistry. 1994;33:8548–8557. doi: 10.1021/bi00194a021. [DOI] [PubMed] [Google Scholar]
- 68.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 69.Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. Embo J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yavuzer U, Smith GC, Bliss T, Werner D, Jackson SP. DNA end-independent activation of DNA-PK mediated via association with the DNA-binding protein C1D. Genes Dev. 1998;12:2188–2199. doi: 10.1101/gad.12.14.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ko L, Cardona GR, Chin WW. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci U S A. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ko L, Chin WW. Nuclear receptor coactivator thyroid hormone receptor-binding protein (TRBP) interacts with and stimulates its associated DNA-dependent protein kinase. J Biol Chem. 2003;278:11471–11479. doi: 10.1074/jbc.M209723200. [DOI] [PubMed] [Google Scholar]
- 74.Matsumoto Y, Suzuki N, Sakai K, Morimatsu A, Hirano K, et al. A possible mechanism for hyperthermic radiosensitization mediated through hyperthermic lability of Ku subunits in DNA-dependent protein kinase. Biochem Biophys Res Commun. 1997;234:568–572. doi: 10.1006/bbrc.1997.6689. [DOI] [PubMed] [Google Scholar]
- 75.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]