Abstract
Intrinsic glomerular cells in a diabetic milieu have transcriptional activation of genes that influence the development of diabetic nephropathy. The cellular repertoire of microRNAs can regulate translation of these expressed genes into proteins. Fibronectin is a key matrix protein accumulated in excess in diabetic nephropathy. Here, we exposed cultured human and mouse mesangial cells to high glucose and transforming growth factor-β to simulate the diabetic milieu. In these conditions in vitro, as well as in mouse diabetic nephropathy models in vivo, microRNA-377 was consistently up-regulated relative to controls. Through a combination of computational and biological approaches, we identified relevant miR-377 target genes. Although fibronectin was induced by miR-377, it was not a direct target of miR-377. However, miR-377 led to reduced expressions of p21-activated kinase and superoxide dismutase, which enhanced fibronectin protein production. Thus, overexpression of miR-377 in diabetic nephropathy indirectly leads to increased fibronectin protein production; as such, miR-377 can have a critical role in the pathophysiology of this prevalent human disease.—Wang, Q., Wang, Y., Minto, A. W., Wang, J., Shi, Q., Li, X., Quigg, R. J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy.
Keywords: diabetes, microRNA, glomerulus, extracellular matrix
The incidence of diabetic nephropathy continues to rise and has now become the leading cause of end-stage kidney disease in the United States, Europe, and Japan (1, 2). Diabetic patients with end-stage kidney disease have a 5-yr survival of only 20%. As such, diabetic nephropathy is a disease with considerable worldwide impact.
The glomerulus is primarily involved in diabetic nephropathy. Key features include mesangial cell hypertrophy, apoptosis, and excessive production of the normal extracellular constituent, fibronectin (3). The pathogenesis of diabetic nephropathy is attributable to elevated ambient glucose concentrations that activate downstream pathways, among which the transforming growth factor (TGF) -β/Mothers against decapentaplegic homologue (Smad) axis is arguably the most important (3,4,5). Still, there is considerable variability among diabetic individuals, even with comparable glycemic control, in the development of diabetic nephropathy.
Intrinsic glomerular cells in a diabetic milieu have transcriptional activation of key genes that strongly influence the development and phenotype of diabetic nephropathy. Such effects can be attributable to polymorphisms in these and interacting genes. After relevant mRNA is transcribed, its stability and translation can be affected by a subset of noncoding RNAs termed microRNAs because they comprise only 21 to 23 nucleotides. MicroRNAs stably bind the 3′-untranslated regions (UTRs) of their target mRNAs when there is identity within their first 8 bases (6, 7). The degree of complementarity beyond the first 8 microRNA bases can determine whether the target mRNA is ultimately degraded or blocked from being translated, with better matches tending to undergo degradation (8).
Work from the Natarajan laboratory (9) showed that miR-192 was up-regulated in mouse mesangial cells exposed to TGF-β, which could decrease translation of the E-box repressor Zfhx1b (SIP1). Here, we explored the relevance of microRNAs in mesangial cells exposed to high glucose rather than TGF-β, as better approximating the conditions that glomerular cells are exposed to in vivo. We identified miR-377 in both human and mouse mesangial cells as highly up-regulated in these cells, and we identified downstream effects relevant to the diabetic glomerulus.
MATERIALS AND METHODS
Oligonucleotides
The sequences of all nucleic acids used to generate polymerase chain reaction (PCR) products for microarray hybridization and pcDNA-6.2-based transfectants, and for quantitative PCR (qPCR), in situ hybridization, and small-interfering RNA (siRNA) -based gene knockdowns, are listed in Table 1.
TABLE 1.
Oligonucleotides employed in these studies
| Oligonucleotide | Sequence |
|---|---|
| Primers for amplification of miR-377 targets on 3′-UTRsa | |
| P-miR-377 (left) | ACAAAAGTTGCCTTTGTGTGAT |
| P-control (left) | ACAAATGTAGCGTTAGTCTGTT |
| P-oligo-dT (right) | TTTTTTTTTTTTTTTTTTTT |
| qPCR primers | |
| P-actin (left) | AGCCATGTACGTAGCCATCC |
| P-actin (right) | CTCTCAGCTGTGGTGGTGAA |
| P-18S rRNA (left) | ATGGCCGTTCTTAGTTGGTG |
| P-18S rRNA (right) | CGCTGAGCCAGTCAGTGTAG |
| Primers for ChIP assay | |
| ChIPmir-377–1 | ACACTGTATCCTTGGCAGTGG |
| ChIPmir-377–2 | GCGGGATTTGGTACTGAAAA |
| ChIPmir-377–3 | AGGGAAAGGTCAAGGTCAGAA |
| ChIPmir-377–4 | GGGCAGATGGCTACATATTCT |
| Probes for in situ hybridizationa | |
| Probe-miR-377 | ACAAAAGTTGCCTTTGTGTGAT |
| Probe-control | ACAAATGTAGCGTTAGTCTGTT |
| siRNA | |
| SOD1 | UGAUUGGGAUUGCGCAGUAUU |
| GGACAAAUUACAGGAUUAAUU | |
| GCAGGGAACCAUCCACUUCUU | |
| AAGAGAGGCAUGUUGGAGAUU | |
| PAK1 | AAGAAGACCUCCAAUAGUAUU |
| GUAUAUACGAUCUGUGAUU | |
| GAUUGGAGCCGGCAGCAAAUU | |
| CGAAGAAAGAGCUGAUUAUUU | |
| PPM1A | ACAAUAGACUGAACCCUUAUU |
| UCACCAAUAACCAGGAUUUUU | |
| ACACGGCUGUGAUCGGUUUUU | |
| GCAAGCGGAAUGUAAUUGAUU | |
| PcDNA constructs | |
| PC-miR-377 (forward) | TGCTGAGCAGAGGTTGCCCTTGGTGAATTCGCTTTATTTATGTTGAATCACACAAAGGCAACTTTTGTT |
| PC-miR-377 (reverse) | CCTGAACAAAAGTTGCCTTTGTGTGATTCAACATAAATAAAGCGAATTCACCAAGGGCAACCTCTGCTC |
| PC-miR-337 (forward) | TCGAGAAGTTGGGGGGTGGGAACGGCGTCATGCAGGAGTTGATTGCACAGCCATTCAGCTCCTATATGATGCCTTTCTTCACCCCCTTCAA |
| PC-miR-337 (reverse) | GATCTTGAAGGGGGTGAAGAAAGGCATCATATAGGAGCTGAATGGCTGTGCAATCAACTCCTGCATGACGCCGTTCCCACCCCCCAACTTC |
Underscores indicate “locked” nucleotides.
MicroRNA microarray
Primary cultures of normal human mesangial cells (NHMCs; Lonza, Allendale, NJ, USA) were grown in mesangial cell growth medium (Lonza) containing 5 mM or 22.5 mM glucose. The medium was refreshed each 2 days, and the cells were harvested after 1 wk. The total RNAs were extracted with TRIzol (Invitrogen Corporation, Carlsbad, CA, USA), and the microRNAs were further enriched by PureLink miRNA isolation Kit (Invitrogen) and hybridized to the Ncode Multi-Species miRNA Microarray Kit (Invitrogen), according to manufacturer’s instructions.
Identification of miR-377 targets
Total RNA prepared from NHMCs was reverse transcribed to cDNA. The 3′-UTRs of potential miR-377 target genes were amplified from this cDNA by PCR using P-miR-377 and P-oligo-dT as primers (Table 1). As a control performed in parallel, the P-miR-377 primer was replaced by an irrelevant oligonucleotide (P-control). PCR products were labeled (Bioprimer DNA labeling System, Invitrogen) and hybridized with human expression microarrays (GeneChip Human Gene 1.0 ST Array; Affymetrix, Santa Clara, CA, USA). Data were first extracted with Affymetrix GeneChip Expression Console followed by DNA-Chip Analyzer (http://www.dchip.org) to generate a normalized signal for each gene’s exon on the array. Specific hybridization of miR-377 relative to control probe was identified statistically with corrections for the multiple comparisons. To be considered as a potential target gene of miR-377, statistical differences (P<0.05) were required of all probe sets to the 3′ end of the sequence. In the microarray, these could also be in a 5′ direction for those in reverse orientation on the array.
Studies with diabetic mice
Diabetes occurring spontaneously in female nonobese diabetic (NOD/Lt) mice (10) and induced in normal C57BL/6 mice by multiple subdiabetogenic doses of streptozotocin (11) was studied; each strain develops renal disease with features of human diabetic nephropathy (11, 12). Controls were littermate NOD/Lt mice that failed to develop hyperglycemia (blood glucose concentrations >20 mM by repeated measurements) and C57BL/6 mice given citrate vehicle at the same time and schedule as streptozotocin. In some instances, glomeruli were isolated magnetically after intra-arterial injection of Dynabeads (13), and then either studied directly for protein expression or used to culture mesangial cells using standard techniques (14). All animal procedures were approved by the University of Chicago Institutional Animal Care and Use Committee.
qPCR
MicroRNAs were quantified by qPCR (TaqMan MicroRNA Assay, Applied Biosystems, Foster City, CA, USA), according to the instructions supplied by the manufacturer. Mouse small nucleolar 234 and human U18 genes were used as internal controls and for normalization. Messenger RNA expression levels were analyzed by qRT-PCR using a SYBR Green-based detection system (Applied Biosystems). Actin mRNA and 18S rRNA were used as internal controls and for normalization. Reactions were performed on a 7900 HT Fast Real-Time PCR System (Applied Biosystems).
In situ hybridization
Locked nucleic acid-based in situ detection of miRNA in kidney sections was performed essentially as described (15), except that paraffin-embedded sections were used in place of cryosections. Briefly, kidney samples were fixed at 4°C in methyl Carnoy’s solution, cold-processed, embedded in paraffin wax, and cut into 4-μm sections. After deparaffinization and hydration of tissue sections, slides were incubated in phosphate-buffered saline (PBS) for 10 min and then treated with 10 μg/ml Proteinase K (Roche Diagnostic Systems, Indianapolis, IN, USA) in 50 mM Tris, pH 7.5, 5 mM ethylene diamine tetraacetic acid for 6 min; washed in PBS for 10 min; and then fixed in 4% paraformaldehyde in PBS for 15 min. Slides were incubated with 0.25% acetic anhydride in 0.1 M triethanolamine for 10 min, washed in PBS for 5 min, and prehybridized in hybridization buffer [50% formamide, 5× standard saline citrate (SSC), 5× Denhardt’s solution, 200 μg/ml yeast RNA, 500 μg/ml salmon sperm DNA, 0.2% Roche blocking reagents, 0.25% 3-[(3-cholamidopropyl) diethylammonio]-1 propane sulfonate (CHAPS), and 0.1% Tween] at room temperature for 4–6 h. Hybridization with 20 nM 3′-digoxygenin-labeled oligoribonucleotide probe (DIG Oligonucleotide 3′-end-labeling kit; Roche) in fresh hybridization buffer was performed at 50°C for 18 h. Slides were washed 4 times (15 min each) in 2× SSC at 50°C, rinsed twice with 0.1% Triton X-100 in PBS for 10 min, and blocked for 1 h in 10% sheep serum and 0.1% Triton X-100 in PBS. Slides were incubated with preabsorbed alkaline phosphatase-conjugated anti-digoxigenin antibodies in PBS with 0.1% Triton X-100 overnight. Sections were washed in PBS with 0.1% Triton X-100 3 times, each for 10 min, followed by rinsing twice for 5 min each with development buffer (100 mM Tris, pH 9.5; 50 mM MgCl2; 100 mM NaCl; and 0.1% Tween 20), and then incubated with 350 μg/ml 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt and 75 μg/ml nitroblue tetrazolium chloride in alkaline phosphatase (AP) buffer in the dark. High-resolution images at ×200 were taken from 10-wk-diabetic and control renal cortices; these were viewed by an observer masked to their origins to evaluate relevant staining patterns.
Western blot analysis
Cells were lysed in RIPA buffer (150 mM NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) with protease inhibitors (Roche). Protein concentrations were determined by bicinchoninic assay (Pierce Biotechnology, Rockford, IL, USA). Proteins were separated on 10% SDS-PAGE gels under reducing conditions and transferred to polyvinylidene difluoride membranes. The membranes were probed with antibodies to fibronectin (Sigma Chemical, St. Louis, MO, USA), collagen IV, PPM1A, SOD1, SOD2 (Abcam, Cambridge, MA, USA), PAK1, Bcl-XL, TGF-β (Cell Signaling Technology, Danvers, MA, USA), Smad7 (Imgenex Corp., San Diego, CA, USA), and then with secondary antibodies as detailed previously (16). Membranes were stripped and reprobed with anti-α-actin antibody (Sigma) and secondary antibody for data normalization.
Gene knockdown with siRNA
Four separate siRNAs targeting SOD1, PAK1, and PPM1A were used in these experiments (Dharmacon, Lafayette, CO, USA) (Table 1). The siRNAs were transfected at a concentration of 20 nM via electroporation (Amaxa, Gaithersburg, MD, USA).
Chromatin immunoprecipitation
For chromatin immunoprecipitation (ChIP) assays, antibodies to acetyl-histone 3 and 4 were employed using a kit from Upstate Cell Signaling (Millipore, Billerica, MA, USA). The manufacturer’s instructions were followed with some modifications. Briefly, primary cultures of mouse mesangial cells were cultured to confluence in 5 or 22.5 mM glucose medium, followed by cross-linking with 1% formaldehyde and lysis with SDS. Chromatin was sonicated and precleared with protein A agarose/salmon sperm D solution followed by specific immunoprecipitation with anti-acetyl-histone 3 or 4 antibodies. PCR was performed on specifically recovered DNA using primers designed upstream of miR-377 in a relatively CpG-rich region (Table 1). PCR products were fractionated by agarose gel electrophoresis, and ethidium bromide-stained bands were visualized under UV light. In all experiments, PCR with input DNA was used as a positive control.
Generation of stable mesangial cell transfectants
To clone the miR-377 hairpin, complementary oligonucleotides were used to form dsDNA with 5′-overhangs, which was then ligated to linearized pcDNA6.2-GW and pcDNA6.2-GW/EmGFP (Invitrogen). Further oligonucleotides corresponding to the miR-337 hairpin were annealed and cloned into pcDNA6.2-GW/EmGFP or pcDNA6.2-GW/EmGFP-miR-377 through Bam H1 sites to generate pcDNA6.2-GW/EmGFP-mir-337 and pcDNA6.2-GW/EmGFP-mir-337/mir-377. The sequences of all clones were validated by sequencing prior to use. Plasmids were transfected with Lipofectamine 2000 (Invitrogen) with stably transfected cell lines selected by growth in DMEM containing 5.5 mM glucose and 2 μg/ml blastacidin. Confirmation was achieved by fluorescence microscopy for the presence of the green fluorescent protein (GFP) and qPCR for the expression of the microRNAs.
RESULTS
MicroRNA expression in mesangial cells in diabetic conditions
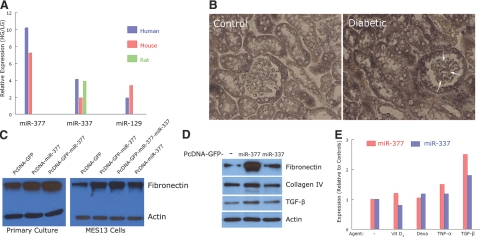
Initial work focused on microRNA species differentially expressed in human mesangial cells cultured in medium containing 22.5 mM glucose to mimic conditions in diabetic nephropathy. In the microarray expression profile, human miR-377, miR-337, and miR-129 exhibited the greatest change on exposure to high glucose (Fig. 1A). The probes for the homologous mouse and rat microRNAs were also changed to a similar extent, presumably reflecting the conserved nature of microRNA. Additional qPCR studies were performed with human and mouse mesangial cells, in which high glucose led to an increase in miR-377, miR-337, and miR-129 quantities (data not shown); as with the microarray data, miR-377 showed the largest change and was the focus of further studies. Of note is that miR-192 (9) was also up-regulated in human mesangial cells exposed to high glucose (1.6±0.1-fold vs. cells in 5 mM glucose, P<0.01).
Figure 1.
A) Microarray data showing expression levels of miR-377, miR-337, and miR-129 in cultured human mesangial cells exposed to 22.5 mM glucose (HG) relative to 5 mM glucose (LG) for 1 wk (n=3 separate cultures for each condition). Data are averages from two arrays, each with probes spotted in duplicate. B) Representative photomicrographs of in situ hybridization for miR-377 in kidneys of control mice and mice with streptozotocin-induced diabetes (3 mice each) after 10 wk of consistent hyperglycemia (>350 mg/dl). Arrows depict mesangial regions. Original view, ×200. C) Effect of miR-377 on fibronectin production in primary and immortalized (MES13) cultures of mouse mesangial cells. Blots are representative of experiments performed 2 and 4 times, respectively. D) Effects of miR-377 and miR-337 on fibronectin, collagen IV, and TGF-β production in normal mouse mesangial cells. Blots are representative of experiments performed 2 or 3 times. E) Effects of 12-h exposure to (1,25)OH2-vitamin D3 (10−10 M), dexamethasone (20 nM), tumor necrosis factor α (4 ng/ml), and TGF-β (2 ng/ml) on expression of miR-377 (red) and miR-337 (blue) in normal mouse mesangial cells. Values are averages from experiments performed twice.
To examine the potential relevance of miR-377 up-regulation in diabetic nephropathy, we utilized NOD and streptozotocin-induced type 1 diabetes models. In NOD mice with 22 wk of hyperglycemia, renal cortical expression of miR-377 was increased 2.1 ± 0.9-fold relative to NOD littermates that did not develop diabetes mellitus. Histologically, miR-377 was focally present in tubular epithelial cells of control mice, whereas after 10 wk of diabetes in streptozotocin-treated mice, there was increased expression of miR-377 in these cells, as well as expression in glomeruli within both podocytes and mesangial cells (Fig. 1B).
Because mesangial cells are intimately involved in the extracellular matrix expansion occurring in diabetic nephropathy, we examined the effects of miR-377 on proteins relevant to this process. In both primary and immortalized mesangial cell cultures cultured in 5 mM glucose, overexpression of miR-377 led to increased fibronectin protein production relative to controls with vector only (Fig. 1C). The inclusion of the GFP coding sequence in front of the miR-377 sequence consistently enhanced the effects of miR-377 relative to PcDNA-miR-377 alone. While increased expression of miR-377 in mesangial cells consistently led to increased fibronectin protein, there was no effect on collagen IV expression (Fig. 1D). Typically, there was also increased TGF-β protein expression, as apparent in Fig. 1E, yet, this was not an invariable finding. Conversely, providing exogenous TGF-β increased expression of miR-377 in normal mesangial cells, whereas 1,25(OH)2-vitamin D3, dexamethasone, and tumor necrosis factor α did not alter miR-377 quantities (Fig. 1E). In these sets of experiments, we also evaluated miR-337, given its expression was increased in mesangial cells in high glucose. While TGF-β increased miR-337 expression (Fig. 1E), exogenous miR-337 did not affect production of fibronectin or collagen IV (Fig. 1D).
Targets of miR-377 in diabetic mesangial cells
By a standard computational approach (accessed at http://microrna.sanger.ac.uk), there were 1092 potential targets of miR-377. To identify genes of particular relevance to human mesangial cells within this relatively large list, we exploited the complementarities of miR-377 core nucleotides to their 3′-UTR targets in microarray studies. Probes were generated by PCR using miR-377 cDNA and oligo-dT as primers and human mesangial cell cDNA as template; these were labeled and used for hybridization with the Affymetrix GeneChip Human Gene 1.0 ST Array (covering 28,869 genes). Our premise was that the amplified sequence in the 3′-UTR (extending 3′ from the miR-377 binding site) would hybridize with relevant 3′-UTR targets in the array. Using this biological approach, 67 of the 1092 computationally predicted targets of miR-377 were confirmed to be targets in human mesangial cells (Table 2). Hence, we were able to focus our work to a restricted and likely relevant subset of gene targets using combined computational and biological approaches.
TABLE 2.
Potential target genes of miR-377
| Actin-related protein 1 homolog A (ACTR1A) |
| A kinase anchor protein 9 (AKAP9) |
| Amyloid beta (A4) precursor-like protein 2 (APLP2) |
| Chromosome 2 open reading frame 34 (C2orf34) |
| CD2 molecule (CD2) |
| Cadherin 13 (CDH13) |
| Cadherin, EGF LAG seven-pass G-type receptor 2 (CELSR2) |
| Cytoplasmic linker associated protein 2 (CLASP2) |
| Chromosome segregation 1-like (CSE1L) |
| Colony stimulating factor 3 (CSF3) |
| Catenin (cadherin-associated protein), delta 1 (CTNND1) |
| Cullin 4A (CUL4A) |
| d-amino acid oxidase activator (DAOA) |
| Definsin, beta 108B (DEFB108B) |
| Extracellular matrix protein 2, female organ and adipocyte specific (ECM2) |
| Eukaryotic translation elongation factor 1 alpha (EEF1A1) |
| Early growth response 1 (EGR1) |
| Early growth response 2 (EGR2) |
| Eukaryotic translation initiation factor 4E (EIF4E) |
| Enhancer of zeste homolog 2 (EZH2) |
| FK506 binding protein 1B (FKBP1B) |
| Frizzled homolog 6 (FZD6) |
| GTP cyclohydrolase 1 (GCH1) |
| Guanine nucleotide binding protein-like 3 (GNL3L) |
| G protein-coupled receptor 116 (GPR116) |
| Histocompatibility complex, class I, E (HLA-E) |
| Histocompatibility complex, class I, G (HLA-G) |
| Homeobox D10 (HOXD10) |
| Isoleucyl-tRNA synthetase (IARS) |
| IBR domain containing 2 (IBRDC2) (Ring finger 144B, RNF144B) |
| Interleukin 1 family, member 6 (IL1F6) |
| Loss of heterozygosity, 11, chromosomal region 2, gene A (LOH11CR2A) |
| Motilin receptor (MLNR) |
| Mof4 family associated protein 1 (MRFAP1) |
| MutS homolog 3 (MSH3) |
| Myosin, light chain 6 (MYL6) |
| Neuron navigator 2 (NAV2) |
| NudE nuclear distribution gene E homolog 1 (NDE1) |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 (NDUFA12) |
| Non-POU domain containing, octamer-binding (NONO) |
| Nucleoproin like 2 (NUPL2) |
| Optic atrophy 1 (OPA1) |
| P21/Cdc42/Rac1-activated kinase 1 (PAK1) |
| PDZ domain containing 1 (PDZK1) |
| Protein phosphatase 1A (PPM1A) |
| Papillary renal cell carcinoma (translocation-associated) (PRCC) |
| PRP3 pre-mRNA processing factor 3 homolog (PRPF3) |
| Protein tyrosine phosphatase, receptor type, S (PTPRS) |
| Regulator of G-protein signaling 4 (RGS4) |
| Relaxin 1 (RLN1) |
| Ring finger protein 103 (RNF103) |
| Ring finger protein 135 (RNF135) |
| Ribonuclease P/MRP 30 kDa subunit (RPP30) |
| Ribosomal protein S12 (RPS12) |
| RNA terminal phosphate cyclase domain 1 (RTCD1) |
| Sterile alpha motif domain containing 7 (SAMD7) |
| Syndecan binding protein (syntenin) (SDCBP) |
| Splicing factor, arginine/serine-rich 11 (SFRS11) |
| Superoxide dismutase 2 (SOD2) |
| SON DNA binding protein (SON) |
| Actin-related protein 1 homolog A (ACTR1A) |
| Sperm specific antigen 2 (SSFA2) |
TABLE 2.
(continued)
| T-box 4 (TBX4) |
| Transmembrane protein with EGF-like and two follistatin-like domains 1 (TMEFF1) |
| Tumor necrosis factor receptor superfamily, member 21 (TNFRSF21) |
| Ubiquitin-conjugating enzyme E2 variant 2 (UBE2V2) |
| Ubiquitination factor E4B (UBE4B) |
| Zinc finger and BTB domain containing 4 (ZBTB4) |
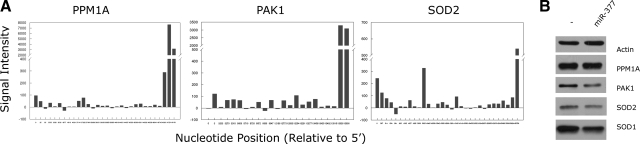
Because of limitations of scope, we concentrated on SOD2, PPM1A, and PAK1 from Table 2. Our rationale in so doing was their effects on oxidative stress (17, 18), termination of Smad signaling (19), and signaling molecules relevant to the cell cycle and cytoskeleton (20), respectively, encompassed a diversity of cellular processes each potentially relevant to matrix production, which was our primary end point in the study. The array data from these three genes are shown in Fig. 2A; although these data are specific to the genes intended for further study, they show results characteristic of all genes considered as targets; namely, specific probe binding to targets located at the 3′ end of a given gene.
Figure 2.
A) Binding of miR-377-derived probes to PPM1A, PAK1, and SOD2 sequences. Signal intensity data are relative to controls and are shown from 5′-to-3′ for each gene. B) Effects of miR-377 on NHMC expression of PPM1A, PAK1, SOD2, and SOD1 proteins relative to controls. Blots are representative of experiments performed 3–5 times.
Exogenous miR-377 added to normal mesangial cells led to reduced PAK1 and SOD2, but not PPM1A protein quantities (Fig. 2B). Of note is the related SOD1 was also predicted to be a target of miR-377 from computational but not microarray approaches. This could be attributed to a mismatch between the first 5′ nucleotide of miR-377 (i.e., the 3′ T in P-miR-377) and its 3′-UTR target sequence in SOD1 (discussed further below). Consistent with this, miR-377 also reduced SOD1 protein quantities in mesangial cells (Fig. 2B).
Roles of miR-377 in diabetic mesangial cells
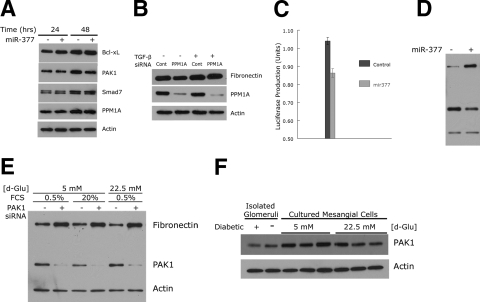
Although the quantities of PPM1A were unaffected by exogenous miR-377, we still considered this as a viable target of miR-377, particularly given its documented relevance to diabetes and the effects of TGF-β (19, 21). In normal mesangial cells to which synthetic miR-377 was added 24 h previously, there was reduced PAK1 but not PPM1A protein over a 24-h culture in medium containing 0.5% FCS (Fig. 3A). As potential downstream targets of these two proteins (19, 22), Bcl-xL and Smad7 were unaffected by miR-377 in these experiments. To further evaluate any effects of miR-377 on PPM1A and related processes, normal mesangial cells were exposed to TGF-β and/or PPM1A-specific siRNA. In these short-term experiments, TGF-β increased fibronectin expression, which was unaffected by knockdown of PPM1A (Fig. 3B). Thus, these data show that despite being predicted as a target of miR-377 in mesangial cells, PPM1A appears to be unaffected by miR-377. Furthermore, they argue against the importance of PPM1A (and potentially Smad inactivation) to limit TGF-β-stimulated fibronectin production (23, 24).
Figure 3.
A) Effects of miR-377 on mesangial cell expression of Bcl-xL, PAK1, Smad7 and PPM1A proteins. Indicated are elapsed times after miR-377 transfection. Blots are representative of experiments performed 2–3 times. B) Effects of PPM1A gene knockdown on TGF-β-induced fibronectin production in normal mesangial cells. Blots are representative of experiments performed 4 times. C) MicroRNA-377 reduces expression of luciferase in a reporter containing the 3′-UTR of PAK1. Data are expressed as means ± se of experiments performed in triplicate 24 h following cotransfection. D) MicroRNA-377 increases expression of fibronectin and decreases PAK1 in normal mesangial cells. Blots are representative of experiments performed 4 times. The protein mobilities in the gel are the same as in E. E) Effects of glucose and FCS concentrations and PAK1 gene knockdown on fibronectin expression in normal mesangial cells. Blots are representative of experiments performed twice. F) Expression of PAK1 in glomeruli isolated from 10-wk diabetic and control mice, and normal mesangial cells cultured in 5 and 22.5 mM glucose for 6-wk duration.
In contrast to PPM1A, miR-377 effectively reduced PAK1, SOD1, and SOD2 protein quantities (cf. Fig. 2B). To confirm that miR-377 targeted PAK1, a standard luciferase reporter assay was performed in which the 3′-UTR of PAK1 was inserted in a pGL3R vector. In these studies, miR-377 reduced luciferase production (as judged by its activity) relative to control microRNA (Fig. 3C). In normal mesangial cells, miR-377 decreased PAK1 expression and increased fibronectin production as before (Fig. 3D). To analyze this in the context of cellular events that might affect PAK1 protein production, mesangial cells were cultured in 5 or 22.5 mM glucose with or without limiting serum concentrations (0.5 and 20%, respectively). Low serum and glucose concentrations both increased PAK1 expression (Fig. 3E). In each instance, blocking PAK1 production with specific siRNA resulted in a considerable increase in fibronectin production, consistent with an important inhibitory role of PAK1 on extracellular matrix production in mesangial cells. Finally, the expression of PAK1 in diabetic conditions in vivo and modeled in vitro in cultured mesangial cells was evaluated. In glomeruli from diabetic mice and mesangial cells exposed to high glucose concentrations, there was reduced PAK1 protein expression compared to controls (Fig. 3F).
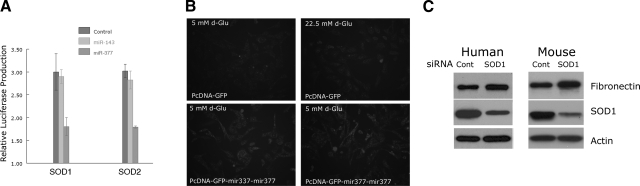
The effects of miR-377 on SOD translation, and proteins were also evaluated. In a luciferase assay in which the 3′-UTRs of SOD1 and SOD2 were included in the pGL3R vector, miR-377, but not miR-143 as control, considerably reduced luciferase activity (Fig. 4A). To evaluate effects of miR-377 on functional aspects of SODs, a fluorescence assay for oxidative stress was employed, which showed greater oxidant production in mesangial cells exposed to miR-377, comparable to those exposed to high glucose concentrations (Fig. 4B). Finally, the effect of SOD1 knockdown on fibronectin production by normal human and mouse mesangial cells was determined; in cells in which SOD1 was reduced, there was increased fibronectin production (Fig. 4C).
Figure 4.
A) MicroRNA-377 reduces expression of luciferase in a reporter containing the 3′-UTRs of SOD1 and SOD2. Data are expressed as means ± se of experiments performed in triplicate. B) Effects of miR-377 on oxidant generation as detected fluorimetrically with 2′,7′-dichlorodihydrofluorescein diacetate in normal mesangial cells. Top panels: control transfected cells cultured in 5 mM (left) and 22.5 mM glucose (right). Bottom panels: mesangial cells with miR-377 cultured in 5 mM glucose. Fluorescence micrographs are representative of experiments performed twice. C) Effects of SOD1 gene knockdown on fibronectin production in normal human and mouse mesangial cells. Blots are representative of experiments performed twice.
MiR-377 genomic structure
In attempts to identify potential transcriptional regulatory regions in the miR-377 genes, we evaluated human and mouse miR-377 genomes. Both were located in chromosome 12. Furthermore, both were clustered with miR-496 and miR-154 within 3 kb to the centromere. Consistent with their physical proximity and a shared transcriptional regulation was their expression together in states of high glucose; thus, in human mesangial cells cultured in 22.5 mM glucose, the expression of miR-496, miR-154, and miR-377 was 2.4 ± 0.7, 1.8 ± 0.4, and 2.9 ± 0.2 that of controls in 5 mM glucose, respectively. These data were largely consistent with our original microarray data, in which the expression of miR-496, miR-154, and miR-377 in human mesangial cells in 22.5 mM glucose was 1.6 ± 0.2, 1.3 ± 0.1, and 10.7 ± 0.7 that of controls in 5 mM glucose, respectively. We evaluated the relevance of a CpG-rich region to the 5′ of this cluster by PCR using ChIP with antibodies to acetylated histone 3 or 4 as a template. Yet, even in conditions of transcriptional activity (i.e., exposure to high glucose), the acetylated histone-containing chromatin did not contain these DNA regions (data not shown). Besides this CpG island, we could not identify other transcriptional regulatory regions within several kilobases of this microRNA cluster to explain the stimulatory effects of glucose and TGF-β.
DISCUSSION
There is increasing evidence that microRNAs are involved in the pathogenesis of metabolic diseases such as diabetes mellitus (25). For example, miR-375 regulates insulin secretion in the pancreatic beta cell (26), which is further modulated by the effects of miR-124a and miR-96 on components of exocytosis pathways (27). The kidney is frequently involved in diabetes with considerable negative consequences. Because mesangial cells are primarily affected in diabetic nephropathy and excessive glucose concentration is a primary driving force in this disease, our simple goal here was to determine whether exposure of mesangial cells to high ambient glucose concentrations affected their expression of microRNAs. We found that miR-377 was reliably overexpressed in human mesangial cells exposed to high glucose levels. In addition, miR-377 was associated with increased expression of the matrix protein, fibronectin that is accumulated in excess in diabetic nephropathy. Not only did miR-377 expression rise together with fibronectin, it could replace high glucose (or TGF-β) to stimulate mesangial cells to produce fibronectin protein. Besides examining events relevant to diabetic nephropathy in human mesangial cells, we performed follow-up experiments in two separate mouse models of diabetic nephropathy in which the expression of miR-377 was consistently up-regulated. Thus, miR-377 is likely to be relevant in diabetic nephropathy.
Although the regulatory influences on microRNA transcription are still incompletely defined, it does appear there are similarities to mRNA, among which the pattern of histone protein acetylation is relevant (28, 29). In both human and mouse genomes, miR-377 neighbors miR-496 and miR-154, both of which were increased together with miR-377 in mesangial cells exposed to high glucose concentrations. Computationally, these three microRNAs shared only four potential target genes (G-protein coupled receptor 113, potassium channel V2, centrosomal protein 152 kDa, and TM2 domain-containing 1), although miR-154 also was predicted to target SOD2 as did miR-377. We could not find transcriptional sites in these chromosomal regions using computational and biological approaches. The details behind transcriptional regulation of miR-377 remain undefined. Nonetheless, that all three were up-regulated by high glucose has considerable implications for diabetes and diabetic nephropathy.
MicroRNAs are relatively promiscuous, at least when considering the many potential mRNAs they complement. Even using algorithm-based approaches with theoretical and empirical constraints, the numbers of potential targets remaining can still be daunting and relatively difficult to examine with standard biological experimentation. Biological processes have considerable specificity to their expression of mRNAs, and proteins that depend on the tissue/cell examined and can change over time. Thus, we attempted to limit target genes of miR-377 to those relevant to our studies. As a surrogate for microRNA-mRNA hybridization in the mesangial cell, we relied on hybridization of their respective cDNAs, together with oligo-dT in a PCR reaction (akin to performing 3′-rapid amplification of cDNA ends). Subsequently, we used a contemporary human microarray with a relatively even distribution of targets to identify specific 3′-UTRs (among the nearly 30,000 genes on the array) complemented by the miR-377 PCR product. Because these are not equivalent biological processes, our approach only suggests targets affected by miR-377 in a mesangial cell. This was well illustrated by the findings with SOD1 and SOD2, both predicted to be miR-377 target genes and confirmed experimentally. These have nearly identical 3′-UTR targets; yet, the 3′ Thymidine in P-miR-377 complemented SOD2 but not SOD1. While this is relatively inconsequential in microRNA targeting (as complementarity of the early bases is key), this is critical for PCR priming. As such, there were no specific PCR products generated for SOD1, and it was not predicted as a target by this technique. To overcome this in the future, we propose the use of degenerate nucleotides in the 3′ positions of microRNA PCR primers.
Using this microarray approach, we reduced the number of potential miR-377 targets from 1092 to 67. From this shortened list, we concentrated on PPM1A, PAK1, and SOD2, as was discussed earlier. The potential for the remaining 64 genes in Table 2 to serve as targets of miR-377 was not systematically studied, though it is certainly conceivable there are those within this list that do have relevance to diabetic nephropathy.
It is clear that the TGF-β/Smad axis is important in diabetic nephropathy. Work by Lin et al. (19, 30, 31) has shown that PPM1A plays an important role in limiting TGF-β signaling through dephosphorylation of receptor-regulated Smads 2/3. Thus, our initial hypothesis was that miR-377 targeted PPM1A and relieved its inhibitory effects on Smad signaling and fibronectin gene transcription. However, by a combination of approaches, we could conclusively dismiss this as relevant in our mesangial cell culture model system. This also illustrates the point that combined in silico and microarray approaches can suggest microRNA targets that are not relevant in a given biological context.
In contrast, PAK1 held up as a target of miR-377 through a combination of biological assays. Like microRNAs, PAK1 has a variety of actions, the sum of which can account for its final effects in a given cell (32). For example, PAK1 can activate mitogen-activated kinases (MAPKs) and nuclear factor (NF) -κB members (33), including via direct activation of MAPK kinase 1 (34, 35) and NF-κB-inducing kinase (36). Besides prosurvival actions related to these two major signaling pathways, PAK1 can also promote cell survival via phosphorylation of Raf1, ultimately antagonizing the proapoptotic Bad (22); these actions can be blocked by the proapoptotic p110C protein (37). Given its important effects on cellular survival, aberrant PAK1 expression is found in some cancers, with the extent of protein expression directly correlating with tumor grade (20). PAK1 also can affect cellular metabolism through its phosphorylation and activation of NADPH oxidase, phosphoglycerate mutase, and phosphoglucomutase (30). In our studies, we showed that reducing PAK1 protein quantities with miR-377 (or gene-specific siRNA) led to increased fibronectin accumulation. In follow-up studies, we found decreased quantities of PAK1 protein in diabetic glomeruli and mesangial cells cultured in high-glucose medium, circumstantial evidence that reduced PAK1 activity is relevant in diabetic nephropathy. The consequence of miR-377-mediated suppression of PAK1 could, therefore, include altered activities of MAPK, NF-κB, and/or BCL pathway members, ultimately leading to loss of tightly control fibronectin production, along with other potential effects of relevance to diabetic nephropathy (38,39,40). The detailed molecular mechanisms behind PAK1 in diabetic nephropathy are a topic of our ongoing studies.
Superoxide dismutases are a group of metal-containing enzymes that catalyze the reduction of reactive oxygen species (ROS) to less toxic moieties (17). The widely distributed copper/zinc SOD (SOD1) comprises 90% of the total SOD, while manganese SOD (SOD2) is responsible for ROS reduction in mitochondria (18). There is considerable evidence for roles of ROS and SODs in diabetes mellitus and diabetic nephropathy (41). Excessive glucose concentrations result in generation of ROS through the mitochondrial electron-transport chain (41), which is compounded in diabetes by coexistent SOD deficiencies (42, 43). Acceleration of diabetic renal injury has been observed in SOD1 knockout mice (44), while overexpression of human SOD1 can limit features of diabetic nephropathy in mice with streptozotocin-induced type 1 diabetes, as used here (45), as well in spontaneously occurring type 2 diabetes in the leptin receptor-mutant db/db mouse (46). Overexpression of SOD2 can correct a variety of hyperglycemia-induced diabetic complications in target cells (41); this extends to cultured glomerular mesangial cells, in which overexpression of SOD2 can suppress glucose-induced extracellular matrix production (47). In this study, SOD1 and SOD2 were predicted and validated to be the targets of miR-377. We showed that miR-377 negatively regulates SOD1 and SOD2 protein expression, while overexpression of miR-377 alone enhances oxidative stress in mesangial cells and leads to increased fibronectin protein accumulation.
MicroRNAs regulate gene expression through their targeting 3′-UTR sequences. Each microRNA has hundreds of potential targets, yet, it recognizes a portion of these in time and space to play an important role in regulating development, differentiation, and metabolism through regulation of translation, which is typically but not always suppressive (48). Here, we have shown that miR-377 is a relevant microRNA induced in mesangial cells exposed to high glucose concentrations in vivo and in vitro. MiR-377 targets and suppresses translation of important mesangial cell proteins, including SOD1, SOD2, and PAK1. This leads to enhanced susceptibility to oxidant stress and accumulation of the extracellular matrix protein, fibronectin. Hence, miR-377 is positioned to have a critical role in the mesangial cell response to the diabetic milieu (Fig. 5).
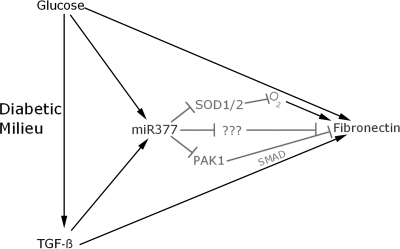
Figure 5.
Potential roles of miR-377 in diabetic nephropathy. Glucose and TGF-β are critical components of the glomerular diabetic milieu, which stimulate production of miR-377 and fibronectin, the latter partially via oxidative stress (O2) and Smad activation. Centrally placed in the diabetic mesangial cell is miR-377, which reduces production of SOD1/2 and PAK1 proteins, as well as other potential candidates from among those listed in Table 2. Up-regulated miR-377 ultimately results in increased fibronectin protein production in diabetic mesangial cells.
Acknowledgments
This work was supported in its entirety by an innovation grant from the Juvenile Diabetes Research Foundation. Funding from the National Institutes of Health (R21DK057684 and U24DK058820) was essential for our earlier supporting studies in diabetic nephropathy.
References
- Rychlik I, Miltenberger-Miltenyi G, Ritz E. The drama of the continuous increase in end-stage renal failure in patients with type II diabetes mellitus. Nephrol Dial Transplant. 1998;13:6–10. doi: 10.1093/ndt/13.suppl_8.6. [DOI] [PubMed] [Google Scholar]
- Ismail N, Becker B, Strzelczyk P, Ritz E. Renal disease and hypertension in non-insulin-dependent diabetes mellitus. Kidney Int. 1999;55:1–28. doi: 10.1046/j.1523-1755.1999.00232.x. [DOI] [PubMed] [Google Scholar]
- Mason R M, Wahab N A. Extracellular matrix metabolism in diabetic nephropathy. J Am Soc Nephrol. 2003;14:1358–1373. doi: 10.1097/01.asn.0000065640.77499.d7. [DOI] [PubMed] [Google Scholar]
- Haneda M, Koya D, Isono M, Kikkawa R. Overview of glucose signaling in mesangial cells in diabetic nephropathy. J Am Soc Nephrol. 2003;14:1374–1382. doi: 10.1097/01.asn.0000064500.89551.76. [DOI] [PubMed] [Google Scholar]
- Sheetz M J, King G L. Molecular understanding of hyperglycemia’s adverse effects for diabetic complications. JAMA. 2002;288:2579–2588. doi: 10.1001/jama.288.20.2579. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W P, Wienholds E, Ketting R F, Plasterk R H. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi J J, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter E H, Prochazka M, Coleman D L. The non-obese diabetic (NOD) mouse. Am J Pathol. 1987;128:380–383. [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sun L, Wang Y, Ning G, Minto A W, Kong J, Quigg R J, Li Y C. Renoprotective role of the vitamin D receptor in diabetic nephropathy. Kidney Int. 2008;73:163–171. doi: 10.1038/sj.ki.5002572. [DOI] [PubMed] [Google Scholar]
- Doi T, Hattori M, Agodoa L Y, Sato T, Yoshida H, Striker L J, Striker G E. Glomerular lesions in nonobese diabetic mouse: before and after the onset of hyperglycemia. Lab Invest. 1990;63:204–212. [PubMed] [Google Scholar]
- Takemoto M, Asker N, Gerhardt H, Lundkvist A, Johansson B R, Saito Y, Betsholtz C. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161:799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigg R J, Morgan B P, Holers V M, Adler S, Sneed A E, Lo C F. Complement regulation in the rat glomerulus: Crry and CD59 regulate complement in glomerular mesangial and endothelial cells. Kidney Int. 1995;48:412–421. doi: 10.1038/ki.1995.309. [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li Y C, Wang J, Kong J, Qi Y, Quigg R J, Li X. miR-17–92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor R, Mittal S, Iqbal J. Superoxide dismutase applications and relevance to human diseases. Med Sci Monit. 2002;8:RA210–RA215. [PubMed] [Google Scholar]
- Johnson F, Giulivi C. Superoxide dismutases and their impact upon human health. Mol Aspects Med. 2005;26:340–352. doi: 10.1016/j.mam.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang Y Y, Su Y, Wrighton K H, Long J, Hu M, Davis C M, Wang J, Brunicardi F C, Shi Y, Chen Y G, Meng A, Feng X H. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Gururaj A E, Barnes C J. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- Schiffer M, Schiffer L E, Gupta A, Shaw A S, Roberts I S, Mundel P, Bottinger E P. Inhibitory smads and TGF-β signaling in glomerular cells. J Am Soc Nephrol. 2002;13:2657–2666. doi: 10.1097/01.asn.0000033276.06451.50. [DOI] [PubMed] [Google Scholar]
- Jakobi R, Moertl E, Koeppel M A. p21-activated protein kinase gamma-PAK suppresses programmed cell death of BALB3T3 fibroblasts. J Biol Chem. 2001;276:16624–16634. doi: 10.1074/jbc.M007753200. [DOI] [PubMed] [Google Scholar]
- Schnaper H W, Hayashida T, Hubchak S C, Poncelet A C. TGF-beta signal transduction and mesangial cell fibrogenesis. Am J Physiol Renal Fluid Electrolyte Physiol. 2003;284:F243–F252. doi: 10.1152/ajprenal.00300.2002. [DOI] [PubMed] [Google Scholar]
- Wu D T, Bitzer M, Ju W, Mundel P, Bottinger E P. TGF-beta concentration specifies differential signaling profiles of growth arrest/differentiation and apoptosis in podocytes. J Am Soc Nephrol. 2005;16:3211–3221. doi: 10.1681/ASN.2004121055. [DOI] [PubMed] [Google Scholar]
- Poy M N, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes Metab. 2007;9:67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- Poy M N, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald P E, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman J M, Chuang J C, Coetzee G A, Jones P A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer N. Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Lin X, Chen Y, Meng A, Feng X. Termination of TGF-β superfamily signaling through SMAD dephosphorylation—a functional genomic view. J Genet Genomics. 2007;34:1–9. doi: 10.1016/S1673-8527(07)60001-0. [DOI] [PubMed] [Google Scholar]
- Duan X, Liang Y Y, Feng X H, Lin X. Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1 in the bone morphogenetic protein signaling pathway. J Biol Chem. 2006;281:36526–36532. doi: 10.1074/jbc.M605169200. [DOI] [PubMed] [Google Scholar]
- Zhao Z S, Manser E. PAK and other Rho-associated kinases—effectors with surprisingly diverse mechanisms of regulation. Biochem J. 2005;386:201–214. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost J A, Swantek J L, Stippec S, Yin M J, Gaynor R, Cobb M H. Stimulation of NF-κB activity by multiple signaling pathways requires PAK1. J Biol Chem. 2000;275:19693–19699. doi: 10.1074/jbc.M909860199. [DOI] [PubMed] [Google Scholar]
- Slack-Davis J K, Eblen S T, Zecevic M, Boerner S A, Tarcsafalvi A, Diaz H B, Marshall M S, Weber M J, Parsons J T, Catling A D. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol. 2003;162:281–291. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E R, Eblen S T, Catling A D. MEK1 activation by PAK: a novel mechanism. Cell Signal. 2007;19:1488–1496. doi: 10.1016/j.cellsig.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Foryst-Ludwig A, Klar S, Schweitzer K, Naumann M. The PAK1 autoregulatory domain is required for interaction with NIK in Helicobacter pylori-induced NF-κB activation. Biol Chem. 2006;387:79–86. doi: 10.1515/BC.2006.011. [DOI] [PubMed] [Google Scholar]
- Chen C, Yan J, Sun Q, Yao L, Jian Y, Lu J, Gu J. Induction of apoptosis by p110C requires mitochondrial translocation of the proapoptotic BCL-2 family member BAD. FEBS Lett. 2006;580:813–821. doi: 10.1016/j.febslet.2005.12.097. [DOI] [PubMed] [Google Scholar]
- Kang B P, Frencher S, Reddy V, Kessler A, Malhotra A, Meggs L G. High glucose promotes mesangial cell apoptosis by oxidant-dependent mechanism. Am J Physiol Renal Fluid Electrolyte Physiol. 2003;284:F455–F466. doi: 10.1152/ajprenal.00137.2002. [DOI] [PubMed] [Google Scholar]
- Lin C L, Cheng H, Tung C W, Huang W J, Chang P J, Yang J T, Wang J Y. Simvastatin reverses high glucose-induced apoptosis of mesangial cells via modulation of Wnt signaling pathway. Am J Nephrol. 2008;28:290–297. doi: 10.1159/000111142. [DOI] [PubMed] [Google Scholar]
- Baumgartner-Parzer S M, Wagner L, Pettermann M, Grillari J, Gessl A, Waldhausl W. High-glucose–triggered apoptosis in cultured endothelial cells. Diabetes. 1995;44:1323–1327. doi: 10.2337/diab.44.11.1323. [DOI] [PubMed] [Google Scholar]
- Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- Ramakrishna V, Jailkhani R. Evaluation of oxidative stress in Insulin Dependent Diabetes Mellitus (IDDM) patients. Diagn Pathol. 2007;2:22. doi: 10.1186/1746-1596-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura K, Nagasaka A, Hayashi R, Makino M, Nagata M, Kakizawa H, Kobayashi T, Fujiwara K, Kato T, Iwase K, Shinohara R, Kato K, Itoh M. Changes in superoxide dismutase activities and concentrations and myeloperoxidase activities in leukocytes from patients with diabetes mellitus. J Diabetes Complications. 1999;13:264–270. doi: 10.1016/s1056-8727(99)00053-7. [DOI] [PubMed] [Google Scholar]
- DeRubertis F R, Craven P A, Melhem M F. Acceleration of diabetic renal injury in the superoxide dismutase knockout mouse: effects of tempol. Metabolism. 2007;56:1256–1264. doi: 10.1016/j.metabol.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Craven P A, Melhem M F, Phillips S L, DeRubertis F R. Overexpression of Cu2+/Zn2+ superoxide dismutase protects against early diabetic glomerular injury in transgenic mice. Diabetes. 2001;50:2114–2125. doi: 10.2337/diabetes.50.9.2114. [DOI] [PubMed] [Google Scholar]
- DeRubertis F R, Craven P A, Melhem M F, Salah E M. Attenuation of renal injury in db/db mice overexpressing superoxide dismutase: evidence for reduced superoxide-nitric oxide interaction. Diabetes. 2004;53:762–768. doi: 10.2337/diabetes.53.3.762. [DOI] [PubMed] [Google Scholar]
- Craven P A, Phillips S L, Melhem M F, Liachenko J, DeRubertis F R. Overexpression of manganese superoxide dismutase suppresses increases in collagen accumulation induced by culture of mesangial cells in high-media glucose. Metabolism. 2001;50:1043–1048. doi: 10.1053/meta.2001.25802. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz J A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]