Abstract
Telomerase activity is developmentally regulated in mammals. Here we examine telomerase activity in plants, whose development differs in fundamental ways from that of animals. Using a modified version of the telomere repeat amplification protocol (TRAP) assay, we detected an activity in extracts from carrots, cauliflower, soybean, Arabidopsis, and rice with all the characteristics expected for a telomerase synthesizing the plant telomere repeat sequence TTTAGGG. The activity was dependent on RNA and protein components, required dGTP, dATP, and dTTP, but not dCTP, and generated products with a seven nucleotide periodicity. Telomerase activity was abundant in cauliflower meristematic tissue and undifferentiated cells from Arabidopsis, soybean, and carrot suspension cultures, but was low or not detectable in a sampling of differentiated tissues from mature plants. Telomerase from cauliflower meristematic tissues exhibited relaxed DNA sequence requirements, which might reflect the capacity to form telomeres on broken chromosomes in vivo. The dramatic differences in telomerase expression and their correlation with cellular proliferation capacity mirror changes in human telomerase levels during differentiation and immortalization. Hence, telomerase activation appears to be a conserved mechanism involved in conferring long-term proliferation capacity.
Keywords: DNA primer, PCR, cell proliferation, soybean, cauliflower
Telomeres are nucleoprotein structures at the ends of chromosomes that are essential for maintaining the integrity of the genome. The primary mechanism for generating and sustaining telomere DNA in eukaryotes is telomerase (1). A ribonucleoprotein with a reverse transcriptase activity, telomerase synthesizes the G-rich strand of telomere DNA using an internal RNA subunit as a template (2). This DNA addition compensates for the loss of terminal sequences that occurs during replication by the conventional cellular machinery (3, 4).
Widespread among eukaryotes, telomerase has been detected in protozoa, yeast, amphibians, and mammals (5–11). Telomerase activity is developmentally regulated in humans (12, 13). In the differentiated human soma, telomeres shorten in a phenomenon attributed to the lack of detectable telomerase activity in these cells (14, 15). Once telomeres shorten below a critical threshold, they appear to lose the capacity to effectively cap chromosomes and activate a damaged DNA response pathway that causes cell-cycle arrest (12). Hence, telomeres have been proposed to represent a biological clock that determines life span (16). A subset of normal human tissues express telomerase, including the regenerating tissues of the blood and epidermis (17–19) and the germ line (20). In addition, telomerase activity can be found in most human primary malignancies (13, 21).
The confinement of telomerase expression to the permanently regenerating tissues of the soma is not a feature shared by all organisms. In mice, many somatic tissues have detectable telomerase activity (11, 22) and telomeres do not shorten (23). However, more recent studies indicate that telomerase levels are higher in immortalized mouse cells than in somatic tissues (24, 25). It has been speculated that the presence of telomerase in normal mouse cells reflects the ease of immortalization (22). Taken together, the data argue that telomerase expression is necessary for sustained tumor growth and for the lifelong proliferation capacity of normal regenerative tissues.
Numerous reports, beginning with the elegant genetic studies of Barbara McClintock (26), are consistent with the expression of telomerase in at least a subset of plant tissues (reviewed in ref. 27). Broken maize chromosomes can be “healed” in the embryo, but not in the endosperm (26). Presumably, telomerase, active in the embryo but not in the endosperm, stabilizes the fractured chromosomes by de novo telomere formation. Several recent studies indicate that broken chromosomes in wheat and barley acquire telomere repeats (28–31), but the de novo addition appears to be confined to gametogenesis or early zygote development (30). Chromosome ends in most plant species are comprised of TTTAGGG repeats (32, 33). As in humans, plant telomere DNA tracts are dynamic and subject to developmental regulation. Barley telomeres shorten during differentiation and aging, but increase in undifferentiated callus cultures (34).
In this paper, we report the identification of telomerase activity in a wide variety of angiosperms. The activity is abundant in undifferentiated cells, but is low or absent in non-proliferating tissues. These findings indicate that telomerase activation is a conserved mechanism involved in conferring long-term proliferation capacity.
MATERIALS AND METHODS
Preparation of Plant Extracts.
Carrot (Daucus carota) and soybean (Glycine max) cell suspension cultures were grown as described (35, 36). Rice (Oryza sativa) cell suspension cultures were grown in AA2 medium as described by Buchholz et al. (37). Arabidopsis thaliana cultures were initiated by placing excised hypocotyls of 4-day-old plants on solid MS medium (38) that was supplemented with 2 mg 2,4-dichlorophenoxyacetic acid per liter and 0.05 mg kinetin per liter. The resulting callus was then transferred to liquid medium of the same composition. Developing primary roots of soybean were prepared from seeds that were sterilized in 20% bleach for 10 min, rinsed, and imbibed in tap water for 1 hr. The seeds were sandwiched between paper towels wetted in 100 μM CaCl2 and were grown in the dark for 3 to 5 days at 29°C. The terminal 15 mm of the root tips were cut off. For experiments involving developed plants, the 15 mm of root tip or stem above the cotyledons, primary and trifoliate leaves, axillary buds, and shoot apex in 3-week-old soybean plants were excised. Extracts were prepared from several tissues of cauliflower (Brassica oleracea), including floral buds, peduncles, stems, and leaves. Suspension cultures of soybean, carrot, rice, and Arabidopsis were harvested by centrifugation at 3840 × g in a Beckman JA-14 rotor for 5 min.
The excised plant tissues or cells were ground in a mortar and pestle under liquid nitrogen, suspended in 4 ml buffer W [50 mM Tris·acetate, pH 7.5/5 mM MgCl2/100 mM potassium glutamate/20 mM EGTA/1.0 mM DTT/0.1 mM phenylmethylsulfonyl fluoride/0.6 mM vanadyl ribonucleoside complex (New England Biolabs)/1.5% (wt/vol) polyvinylpyrrolidone/10% glycerol] per gram material, and centrifuged at 16,000 × g at 4°C for 15 min. For each extract, PEG 8000 (United States Biochemical) was added to a final concentration of 10%, mixed for 30 min at 4°C, and centrifuged 16,000× g for 5 min. The pellet was resuspended in a quarter of the original volume of buffer W for 30 min at 4°C, and was centrifuged for 2 min. The supernatant was stored at −80°C until use. Protein concentrations in extracts ranged from 0.25 to 2.0 mg/ml.
Telomere Repeat Amplification Protocol (TRAP) Assays.
Telomerase was detected by a modified version of the TRAP protocol (20). Primers were obtained from the Texas A&M Gene Technology Laboratory or Oligos, Etc. (Guilford, CT) and gel purified before use. Oligonucleotide sequences are shown in Table 1. The 39 μl reaction mixtures were composed of 50 mM Tris·acetate (pH 8.3), 50 mM potassium glutamate, 0.1% Triton X-100, 1 mM spermidine, 1 mM DTT, 50 μM each dNTP, 5 mM MgCl2, 10 mM EGTA, 0.4 μl [α-32P]dGTP (800 mCi/mM; 1 Ci = 37 GBq; New England Nuclear), 100 ng/μl BSA (ICN), 500 nM T4 gene 32 protein (gift of David Geidroc, Texas A&M University, College Station, TX), and 0.5 unit Taq polymerase (Fisher or Promega). After the addition of 50 ng of forward primer followed by 0.25–1.0 μg extracted plant protein, the telomerase reaction was allowed to proceed at room temperature for 45 min. Reverse primer (50 ng) was added, and the reaction mixture was covered with 50 μl paraffin oil and amplified by 30 cycles of PCR at 94°C for 30 sec, 65°C for 30 sec, and 72°C for 90 sec, with an additional 5 min 72°C extension step at the end. Samples were phenol extracted, ethanol precipitated, and resolved on 6% sequencing gels (except where noted), which were dried and subjected to autoradiography. Some samples were pretreated with 10 ng RNase A (Sigma) or H2O (mock RNase) at room temperature for 10 min before the telomerase reaction step.
Table 1.
Sequences of TRAP forward primers
| Primer sequence | Primer name |
|---|---|
| CACTATCGACTACGCGATCGG | GG(21) |
| AGGGTTTAACTACGCGATGGG | AGT-GGG(21) |
| AGGGTTTAACGCGATGGG | AGT-GGG(18) |
| GACAATCCGTCGAGCAGAGTT | TS(21) |
| AATCCGTCGAGCAGAGTT | TS(18) |
| CACTATCGACTACGCGATCAT | pBR(21) |
| CACTATCGACGCGATCAT | pBR(18) |
| CCCTAAACCCTAAACCCTAAA | (C3TA3)3 |
RESULTS
Identification of Plant Telomerase.
Initial attempts to detect telomerase products by the conventional biochemical assay (5) in cell lysates from germinating and mature plants as well as from suspension cultures were unsuccessful. Therefore, we turned to the more sensitive PCR-based TRAP assay originally developed for detection of human telomerase activity (20). The assay works in two steps: (i) telomerase elongates a forward primer and (ii) then PCR is used to amplify the products using a reverse primer. In this case, the reverse primer, d(CCCTAAA)3, was precisely complementary to the consensus plant telomere sequence d(TTTAGGG)n. To increase the stringency of the TRAP protocol, the annealing temperature of the PCR step was increased to 65°C and the forward primer was changed to a sequence that was more efficiently used by the plant telomerases (see below). These parameters ensured that the forward primers in our study could only be extended in the PCR step if an activity in the plant extract added TTTAGGG repeats. As an initial purification step for the extracts, a 10% PEG precipitation was performed. This reduced the concentration of inhibitors to Taq polymerase or telomerase that are commonly found in crude mammalian extracts (39).
A 21-mer oligonucleotide containing two guanosine residues at the 3′ terminus, GG(21), was used as a primer to detect an enzyme in cauliflower floral buds and carrot suspension cultures that fits the criteria of telomerase. The 3′ terminus of the primer was extended by the addition of tandem repeats with a seven nucleotide periodicity (Fig. 1A). The likelihood that primer extension was a PCR artifact was discounted by the fact that no repeats were added in reactions lacking either the forward primer (Fig. 1B, lane 9) or plant extract (Fig. 1B, lane 10) during the telomerase step.
Figure 1.
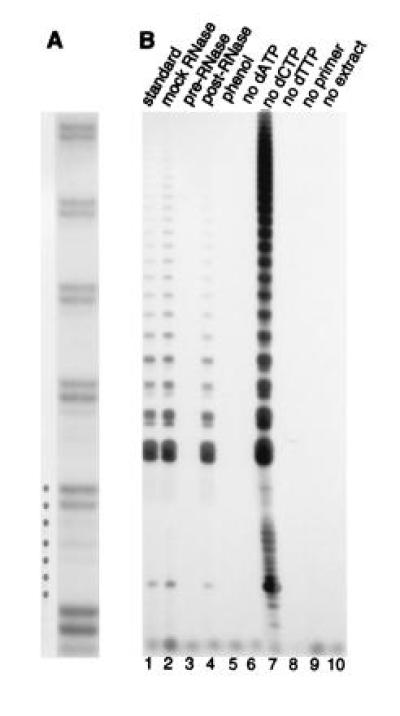
Telomerase activity in plants. (A) 250 ng of protein from cauliflower floral bud extract was assayed by TRAP using the GG(21) forward primer (see Table 1). Products were resolved on a 10% sequencing gel to reveal the periodicity of the banding profile. (B) Reactions were performed with 1.2 μg protein from carrot suspension cultures using GG(21). Lanes: 1, TRAP assay under standard conditions; 2, mock RNase treated sample; 3, sample treated with RNase A before the telomerase step; 4, sample treated with RNase after the telomerase step but prior to the PCR step; 5, sample extracted with phenol-chloroform, before the telomerase step. Lanes 6–9, reactions in which dATP, dCTP, dTTP, or the forward primer were omitted from the telomerase step, but were added just prior to the PCR step. Lane 10, carrot extract omitted.
Elongation was sensitive to low levels of RNase A (10 ng) (Fig. 1B, lane 3), a concentration of RNase that had no effect on the PCR reaction (lane 4). Phenol/chloroform treatment of the extract abolished elongation (Fig. 1B, lane 5). These data are consistent with polymerization by a ribonucleoprotein. Omission of either dATP or dTTP (Fig. 1B, lanes 6 and 8) during the telomerase step abolished elongation. Similarly, when the reaction was not supplemented with unlabeled dGTP, no elongation products were generated (data not shown). In contrast, the absence of dCTP, which would not be incorporated into d(TTTAGGG) telomeric repeats, did not result in any loss in activity (Fig. 1B, lane 7). In fact, exclusion of dCTP from the telomerase step reproducibly enhanced activity. This effect may be due to competition by dCTP for nucleotide binding sites on telomerase when all four deoxynucleotides are added. Finally, alteration of one or two nucleotides in the reverse primer repeat sequence prevented PCR amplification (data not shown). All of these properties are consistent with a plant telomerase that synthesizes TTTAGGG repeats.
DNA Sequence Requirements for the Carrot and Cauliflower Telomerases.
We tested the primer specificity of the telomerases from carrots and cauliflower. All known telomerase activities show a preference for G-rich DNA and will most efficiently extend primers whose 3′ ends base pair with the RNA template (2). TRAP assays using the C-rich telomere repeat strand sequence as forward primer failed to be extended by either the carrot or the cauliflower telomerase (Fig. 2 A, lane 8, and B, lane 8). In contrast, primers with dG residues on their 3′ ends were efficiently elongated (Fig. 2 A, lanes 1 and 2, and B, lanes 1 and 2). We saw no obvious enhancement of primer utilization when we included a G-rich copy of the plant telomere repeat on the primer 5′ end (Fig. 2 A, lane 2, and B, lane 2). However, primer length had a striking effect on extension by plant telomerases. 18-mer primers were not recognized by the carrot telomerase (Fig. 2A, lanes 3 and 4). This included the 18-mer TS primer (Fig. 2A, lane 4), which is the standard primer used in mammalian TRAP assays (20). However, when three nucleotides were added onto the TS primer 5′ terminus, it became an efficient substrate for elongation (Fig. 2A, lane 5).
Figure 2.

DNA primer specificity of two plant telomerases. (A) Primer specificity for carrot telomerase. Protein samples (1.2 μg) from carrot suspension cultures were assayed by TRAP using different forward primers as indicated (see Table 1). (B) Primer specificity for the cauliflower telomerase. Samples (250 ng) from cauliflower floral bud extract were assayed with the primers indicated. (C) Model for the predicted plant telomerase RNA templating domain and inferred alignment of primers. The plant templating sequence in based on the telomerase RNAs from humans and ciliates (40, 41). Nucleotides in boldface type in the DNA primers are capable of forming Watson–Crick base pairs with the RNA template. The underlined nucleotides in the RNA template indicate possible positioning of the 3′ terminal nucleotide in the pBR (21) primer.
A similar result was obtained with the cauliflower telomerase; 18-mer primers (Fig. 2B, lanes 3, 4, and 7) were used less efficiently than the 21 nucleotide versions (Fig. 2B, lanes 2, 5, and 6). Interestingly, the pBR primer whose 3′ terminus contains only a single nucleotide that can potentially pair with the plant telomerase RNA template (see below) was extended as well as a primer bearing three 3′ terminal dG residues (Fig. 2B, compare lanes 2 and 6). The pBR primer was not extended by the carrot telomerase (Fig. 2B, lanes 6 and 7), suggesting that the cauliflower telomerase had a more relaxed sequence specificity than the carrot enzyme.
Telomerase enzymes that synthesize six or eight nucleotide repeated sequences generate products with a corresponding banding periodicity, the strongest bands in the profile reflecting enzyme-primer rearrangement as multiple repeats are generated (2). The permutation of the banding profile is determined by where the primer 3′ terminus initially aligns on the RNA template for the first polymerization cycle. As expected, reaction products with carrot (Fig. 2A, lanes 1, 2, and 5) and cauliflower (Fig. 2B, lanes 1, 2, 5, and 6) extracts generated elongation profiles that were offset from one another when 21-mer primers having different 3′ terminal sequences were used.
A model showing the predicted alignment of four primers on the cauliflower telomerase RNA template is presented in Fig. 2C. This model accounts for the one nucleotide offset in the banding profiles from reactions with GG(21) and AGT-GGG(21) (Fig. 2B, lanes 1 and 2) and the three nucleotide offset between GG(21) and TS(21) reaction products (Fig. 2B, lanes 1 and 3). Interestingly, the extension profile of pBR(21) is offset by five nucleotides from the TS(21) primer and two nucleotides from GG(21). Hence, the 3′ terminal dT on the pBR(21) primer does not seem to align opposite an rA residue in the template. Rather, the primer appears to be positioned across from an rU residue (underlined in Fig. 2C), allowing the primer to be extended initially by three dG residues. The cauliflower telomerase also extended a primer terminating in a dC residue, which cannot form a base pair with any nucleotides in the telomerase RNA templating domain (data not shown). Delivery of a primer into the telomerase polymerization site in the absence of Watson–Crick base paired alignment on the RNA template has been documented for the Euplotes telomerase (ref. 42; J. Bednenko, M. Melek, and D.E.S., unpublished work). Such a mechanism may enable telomerase to form telomeres de novo on broken chromosome ends (see below).
Telomerase Expression Patterns in Plants.
In contrast to animals cells, cell division in plants is localized to a few discrete regions called meristems. Cell division at the apical meristems at the ends of the root and shoot increase the length of the plant. The vascular cambium, a cylindrical layer of tissue running along the vertical axis of the plant, also carries out cell division to increase the girth (43). Since plant telomere length declines in differentiated cells (34), we predicted that the expression of telomerase would be localized to meristematic tissues in mature plants. Nonproliferative or quiescent tissues such as the leaves and the axillary buds were not expected to demonstrate telomerase activity.
To test this hypothesis, a 10-fold dilution series of several different soybean extracts was assayed to compare the in vitro telomerase activities of different samples over a linear range of activity (17). In the mature plant, telomerase activity was detected in the root tips (Fig. 3A, lanes 7–9) and, at a lower level, in the stem (lanes 10–12), consistent with the presence of the root apical meristem and the vascular cambium in these two regions. TRAP failed to detect any activity in the primary leaves (Fig. 3A, lanes 13–15), the trifoliate leaves (lanes 16–18), or the axillary buds (lanes 19–21). In contrast to the root tip (Fig. 3A, lanes 7–9), telomerase levels were reproducibly low or not detectable in the shoot apex (Fig. 3A, lanes 22–24, and data not shown). Telomerase activity in 3-day-old primary roots (Fig. 3A, lanes 4–6) was substantially higher than in any tissues in the mature plant. This observation is consistent with increased cell division in the germinating plant. By far, the highest levels of telomerase were found in the soybean suspension culture (Fig. 3A, lanes 1–3).
Figure 3.
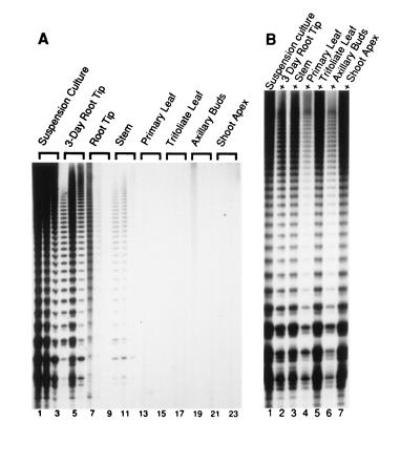
Regulation of telomerase expression in soybean. (A) A dilution series of each extract in 1.0, 0.1, and 0.01 μg protein samples from a variety of soybean tissues was assayed to compare the amount of activity between samples. In the 3-day-old root tip samples, the higher level of telomerase activity in a reaction with 0.1 μg protein versus 1 μg of protein is consistent with the presence of an inhibitor. The autoradiograph was developed after a 10-day exposure. (B) Mixing experiments with soybean extracts. TRAP assays were performed with 1 μg suspension culture alone (lane 1) or mixed with an equal amount the extracts indicated.
Mixing experiments verified that the failure to detect telomerase in particular tissues was not caused by inhibitors to Taq polymerase. Extracts from suspension culture were assayed in the presence of equal amounts of protein from soybean plant extracts. Most of the extracts had no effect on extension by the suspension culture (Fig. 3B, lanes 2, 3, and 5), including the shoot apex (lane 7). A slight inhibition of the soybean suspension culture was observed in the presence of primary leaf and axillary bud extracts (Fig. 3B, lanes 4 and 6). However, this observation cannot account the complete lack of telomerase activity in extracts from these differentiated tissues.
The lack of detectable activity in the soybean shoot apex was surprising. However, we suspected that the inability to detect activity was due to a low level of meristematic cells relative to other cell types in the extract. Therefore, we examined a third type of meristematic tissue, floral buds from cauliflower. The cauliflower head is a massive inflorescence containing many small floral meristems supported by a branched peduncle. A 10-fold dilution series of extracts prepared from cauliflower floral buds demonstrated a high amount of telomerase activity (Fig. 4, lanes 1–3). The amount of telomerase was approximately 10-fold lower in the peduncles (Fig. 4, lanes 4–6) and 100- to 1000-fold lower in the stem (lanes 10–12) and leaf (lanes 7–9). We are unable to ascertain whether the telomerase activity we detected in the stem and leaves is due to a low level of expression or to a residual amount of telomerase in cells that were recently derived from the vascular cambium. Mixing experiments (Fig. 4, lanes 13–15) revealed that the lower levels of telomerase in other tissues were not due to telomerase or Taq polymerase inhibitors.
Figure 4.
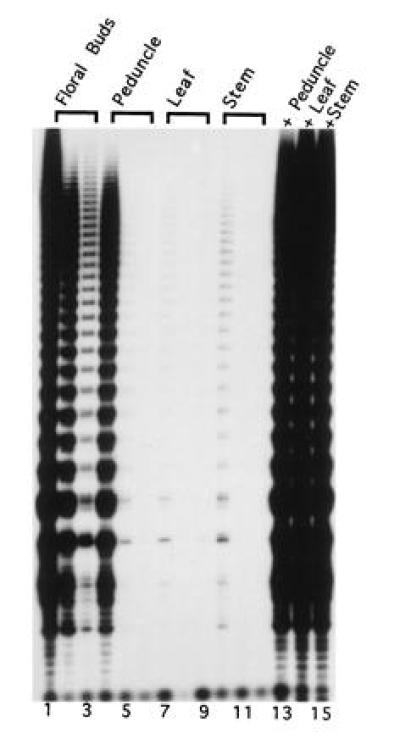
Regulation of telomerase expression in cauliflower. A dilution series of 0.5, 0.05, and 0.005 μg protein samples from the indicated cauliflower extract was assayed. Mixing experiments were also performed combining 0.5 μg cauliflower floral bud extract protein an equal amount of extract from the other samples (lanes 13–15). The autoradiograph was developed after a 10-day exposure.
Significantly, telomerase was much more abundant in the meristematic tips of cauliflower than in any of the other plants we assayed, including soybean, carrot, and pea (data not shown). Moreover, telomerase activity in whole cell cauliflower extracts or in isolated nuclei was comparable to the activity found in suspension cultures (data not shown). Without exception, suspension cultures displayed very high levels of telomerase. We tested several, including extracts from carrot (Figs. 1 and 2A), soybean (Fig. 3), Arabidopsis, and rice (data not shown). The presence of telomerase in suspension cultures correlates with previous results describing an increase in telomere length in barley callus cultures (34).
DISCUSSION
Given the striking correlations between telomerase activity and proliferation capacity in mammals, we expected that analysis of plant telomerase might yield further insight into this relationship. Plants have a more plastic developmental pattern than animals. In contrast to animal cells, many plant cells are totipotent, and individual cells, in the presence of phytohormones such as auxins and cytokinins, can be induced to undergo differentiation into mature plants. Other hormone regimes can lead to dedifferentiation of mature root, stem, and leaf tissue into immortal suspension cultures. Most of the cells in plant explants achieve immortality, unlike animal explants where most cells die in a crisis, leaving only a few immortal cells to continue the line. Plants also differ from animals in that their germ line is not specified until very late in development. If the shoot apex, which normally will give rise to reproductive organs, is removed, new shoots will proliferate and assume reproductive functions. Such developmental plasticity is distinct from the more deterministic pattern in animals, and is essential for survival of plants in environments where growing points can be destroyed by herbivores.
Using a modified version of the TRAP assay, we detected an activity with all the characteristics expected for telomerase in extracts from a variety of plants. The activity was dependent on both RNA and protein components in the extracts and generated products with a seven nucleotide periodicity that were consistent with TTTAGGG repeats. The plant telomerase displayed a strong dependence on primer length, extending 21-mer primers more efficiently than 18-mer primers. Further studies are needed to determine whether the requirement for longer primers is a consequence of our PCR assay conditions or reflects the need for the enzyme to form stable contacts with DNA sequences upstream of the primer 3′ terminus, as reported for other telomerases (42, 44–47).
One surprising finding in this study was the ability of the telomerase from cauliflower floral buds to initiate DNA synthesis in the absence of Watson–Crick base paired primer alignment on its RNA template. Although the human telomerase requires that its primers contain a minimum of two to four base pairs of 3′ terminal complementarity to the RNA template for in vitro elongation (45), Tetrahymena and Euplotes telomerases can initiate synthesis in the absence of such base pairing (42, 44). In the ciliate Euplotes, this phenomenon is developmentally regulated: telomerase from cells undergoing programmed chromosome fragmentation and de novo telomere formation performs this reaction, while the enzyme from vegetatively growing cells does not (J. Bednenko, M. Melek, and D.E.S., unpublished work). The ability of a plant telomerase to catalyze de novo telomere formation in vitro may provide a biochemical basis for new telomere formation frequently observed on broken chromosome ends early in wheat and maize development (26, 30).
Our data indicate that soybean telomerase is highly regulated, with the greatest levels of activity in undifferentiated cells of suspension cultures. Consistent with the increased cell division associated with germinating plants, telomerase is more abundant in the primary root from 3-day-old soybean plants than in any tissues of the mature plant. Differentiated tissues such as leaves or quiescent tissues such as axillary buds exhibit extremely low or nondetectable telomerase activity. We could not detect telomerase activity in the shoot apex, although this is probably due to a low level of meristematic cells relative to other cell types in the extract. One untested possibility is that secondary products in the soybean shoot apex interfere with the extraction of telomerase.
One of the richest sources of telomerase activity we found was in the massive meristematic structure of the cauliflower head. As with soybean, other tissues in the mature cauliflower plant expressed lower levels of telomerase. The abundance of telomerase activity in cauliflower inflorescences indicates that this will be a good source of telomerase for future biochemical studies.
The plant telomerase expression profile correlates well with changes in telomere length reported during differentiation and dedifferentiation in barley (34). Early in vitro and in vivo studies of telomere length and telomerase expression in humans suggested that telomere shortening was responsible for the onset of cellular senescence (14, 15, 48). Telomerase activation was proposed to lead to immortalization by counteracting telomere decline. However, recent reports of the lack of telomere shortening in mice (23), telomerase-negative human cells lines and tumors (49, 50), and the detection of telomerase activity in some tissues of the normal soma (17, 18, 20), are inconsistent with this simple model.
As the telomere–telomerase hypothesis of aging and cancer is being refined (51, 52), the striking link between telomerase and cellular proliferation capacity remains clear (17, 18, 25). Recent studies reveal that mouse telomerase and telomerase RNA expression in tumors correlate strongly with increases in histone H4 mRNA, a marker of cellular proliferation (25). In addition, quiescent adult lymphocytes treated with mitogen stimulators transiently activate telomerase (17, 53), while telomerase is rapidly down-regulated in immortalized cells subjected to differentiation-inducing agents (54–56). The selective expression of telomerase in dividing plant cells strengthens the argument that telomerase activation is a conserved mechanism required for long-term proliferation capacity.
Note Added in Proof.
Two other groups recently reported the identification of telomerase activity in plant extracts (57, 58). Heller et al. (58) provide evidence that the activity is developmentally regulated.
Acknowledgments
We thank Titia de Lange for advice in establishing the TRAP assay, John Mullet and his group for providing materials and valuable discussions, Emily Grace for technical assistance, and Jeff Kapler and Meni Melek for critically reading the manuscript. This work was supported by National Institutes of Health Grant GM49157 to D.E.S. and a grant from the Texas A&M Interdisciplinary Research Initiative to D.E.S. and T.D.M.
Footnotes
Abbreviation: TRAP, telomere repeat amplification protocol.
References
- 1.Beissmann H, Mason J M. Adv Genet. 1992;30:185–249. doi: 10.1016/s0065-2660(08)60321-1. [DOI] [PubMed] [Google Scholar]
- 2.Greider C W. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 35–68. [Google Scholar]
- 3.Watson J D. Nature (London) New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 4.Olovnikov A M. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 5.Greider C W, Blackburn E H. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 6.Zahler A M, Prescott D M. Nucleic Acids Res. 1988;16:6953–6972. doi: 10.1093/nar/16.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shippen-Lentz D, Blackburn E H. Mol Cell Biol. 1989;9:2762–2764. doi: 10.1128/mcb.9.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn M, Blackburn E H. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 9.Mantell L L, Greider C W. EMBO J. 1994;13:3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morin G B. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 11.Prowse K R, Avilion A A, Greider C W. Proc Natl Acad Sci USA. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange T. Proc Natl Acad Sci USA. 1994;91:2882–2885. doi: 10.1073/pnas.91.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacchetti S, Counter C M. Int J Oncol. 1995;7:423–432. [PubMed] [Google Scholar]
- 14.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 15.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 16.Harley C B, Homayoun V, Counter C M, Allsopp R C. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- 17.Broccoli D, Young J W, deLange T. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harle-Bachor C, Boukamp P. Proc Natl Acad Sci USA. 1996;93:6476–6481. doi: 10.1073/pnas.93.13.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor R S, Ramirez R D, Ogoshi M, Chaffins M, Piatyszek M A, Shay J W. J Invest Dermatol. 1996;106:759–765. doi: 10.1111/1523-1747.ep12345811. [DOI] [PubMed] [Google Scholar]
- 20.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 21.Harley C B, Villeponteau B. Curr Opin Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 22.Prowse K R, Greider C W. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kipling D, Cooke H J. Nature (London) 1990;347:347–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 24.Blasco M A, Rizen M, Greider C W, Hanahan D. Nat Genet. 1996;12:200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- 25.Broccoli D, Godley L A, Donehower L A, Varmus H E, de Lange T. Mol Cell Biol. 1996;16:3765–3772. doi: 10.1128/mcb.16.7.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClintock B. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards E J. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 371–387. [Google Scholar]
- 28.Wang S, Lapitan N L V. Genome. 1992;35:974–980. [Google Scholar]
- 29.Murata M, Nakata N, Yasumuro Y. Chromosoma. 1992;102:27–31. [Google Scholar]
- 30.Werner J E, Kota R S, Gill B S. Genome. 1992;35:844–848. [Google Scholar]
- 31.Tsujimoto H. J Plant Res. 1993;106:239–244. [Google Scholar]
- 32.Richards E R, Ausubel F M. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 33.Fuchs J, Brandes A, Schubert I. Plant Syst Evol. 1995;196:227–241. [Google Scholar]
- 34.Kilian A, Stiff C, Kleinhofs A. Proc Natl Acad Sci USA. 1995;92:9555–9559. doi: 10.1073/pnas.92.21.9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilde H D, Nelson W S, Booij H, de Vries S C, Thomas T L. Planta. 1988;176:205–211. doi: 10.1007/BF00392446. [DOI] [PubMed] [Google Scholar]
- 36.Horn M E, Sherrard J H, Widhold J M. Plant Physiol. 1983;72:426–429. doi: 10.1104/pp.72.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchholz, W., Teng, W., Wallace, D., Ambler, J. & Hall, T. C. (1996) Methods Mol. Biol., in press. [DOI] [PubMed]
- 38.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 39.Piatyszek M A, Kim N W, Weinrich S L, Hiyama K, Hiyama E, Wright W E, Shay J W. Methods Cell Sci. 1995;17:1–15. [Google Scholar]
- 40.Lingner J, Hendrick L L, Cech T R. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 41.Feng J, Funk W D, Wang S-S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J, Le S, West M D, Harley C B, Andrews W H, Greider C W, Villeponteau B. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 42.Melek M, Greene E C, Shippen D E. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs T. Dev Biol. 1992;153:1–15. doi: 10.1016/0012-1606(92)90087-w. [DOI] [PubMed] [Google Scholar]
- 44.Harrington L, Greider C. Nature (London) 1991;353:451–453. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 45.Morin G. Nature (London) 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 46.Lee M, Blackburn E H. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins K, Greider C W. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 48.Allsopp R C, Homayoun V, Patterson C, Goldstein S, Younglai E V, Futcher B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryan T M, Englezou A, Gupta J, Bacchetti S, Reddel R R. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murnane J P, Sabatier L, Marder B A, Morgan W F. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holt S E, Shay J W, Wright W E. Nat Biotech. 1996;14:836–839. doi: 10.1038/nbt0796-836. [DOI] [PubMed] [Google Scholar]
- 52.Autexier C, Greider C W. Trends Biochem Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 53.Counter C M, Gupta J, Harley C B, Leber B, Bacchetti S. Blood. 1995;85:2315–2320. [PubMed] [Google Scholar]
- 54.Sharma H W, Solkoloski J A, Perez J R, Maltese J Y, Sartorelli A C, Stein C A, Nichols G, Khaled Z, Telang T T, Narayanan R. Proc Natl Acad Sci USA. 1995;92:12343–12346. doi: 10.1073/pnas.92.26.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holt S E, Wright W E, Shay J W. Mol Cell Biol. 1996;16:2932–2939. doi: 10.1128/mcb.16.6.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W, Piatyszek M A, Kobayashi T, Estey E, Andreeff M, Deisseroth A B, Wright W E, Shay J E. Clin Can Res. 1996;2:799–803. [PubMed] [Google Scholar]
- 57.Fajkus J, Kovarik A, Kralovics R. FEBS Lett. 1996;391:307–309. doi: 10.1016/0014-5793(96)00757-0. [DOI] [PubMed] [Google Scholar]
- 58.Heller K, Kilian A, Piatyszek M A, Kleinhofs A. Mol Gen Genet. 1996;252:342–345. doi: 10.1007/BF02173780. [DOI] [PubMed] [Google Scholar]


