Abstract
Background
KRAS and BRAF mutations appear of relevance in the genesis and progression of several solid tumor types but the co-occurrence and interaction of these mutations have not yet been fully elucidated. Using a microsatellite stable (MSS) colorectal cancer (CRC) cell line (Colo741) having mutated BRAF and KRASWT, we also aimed to investigate the KRAS-BRAF interaction. Gene expression profiles for control KRASWT, KRASG12V and KRASG12D transfected cells were obtained after cell clone selection and RT-PCR screening. Extensive qPCR was performed to confirm microarray data.
Results
We found that the KRASG12V state deregulated several genes associated to cell cycle, apoptosis and nitrogen metabolism. These findings indicated a reduced survival and proliferation with respect to the KRASWT state. The KRASG12D state was, instead, characterized by several other distinct functional changes as for example those related to chromatin organization and cell-cell adhesion without affecting apoptosis related genes.
Conclusion
These data predict that the G12D mutation may be more likely selected in a BRAF mutated context. At the same time, the presence of the KRASG12V mutation in the cells escaping apoptosis and inducing angiogenesis via IL8 may confer a more aggressive phenotype. The present results get along with the observations that CRCs with G12V are associated with a worse prognosis with respect to the WT and G12D states and may help identifying novel CRC pathways and biomarkers of clinical relevance.
Background
Normal colon epithelial cells, in their way to malignancy, may follow multiple pathways: i) the traditional adenoma-carcinoma sequence associated with chromosomal instability, in which the sequential accumulation of mutations in specific genes, including adenomatous polyposis coli (APC), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), and tumor protein p53 (TP53), drives the transition from healthy colonic epithelia through increasingly dysplastic adenoma to colorectal cancer (CRC) [1] ii) the serrated pathway leading to CRC associated with microsatellite instability (MSI), v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutations and extensive DNA methylation [2,3] and possibly, iii) a "fusion" pathway associated to methylation of the O-6-methylguanine DNA methyltransferase (MGMT), mutation of KRAS and inactivation of the gene coding for tumor protein p53 (TP53) [4].
KRAS is one of the most commonly activated oncogenes since 17% to 25% of all human tumors harbor mutations in this gene [5]. Although statistics may differ slightly from study to study, a good estimate is that in about 30–40% of CRC a mutated KRAS may be found [6-9]. Ras proteins are small guanine-nucleotide binding proteins (p21ras) involved in signal transduction with a GTPase activity, which is severely reduced when the protein is mutated in codons 12, 13 or 61. As p21ras activates downstream effectors in the GTP-bound state, reduction of this activity leads to unregulated signaling and lastly to enhanced and unregulated cell proliferation and transformation [10].
Of the multiple molecular signaling pathways initiating from KRAS, the Raf/MEK/ERK kinases and the Ras/PI3K/PTEN/Akt pathways are the best studied [11,12]. These pathways are interconnected since the mutation of genes in one pathway may influence the activity of kinases in the other pathway and both of them also interact with the TP53 pathway [12]. Because of these molecular interactions, the effects of the activation of one of these pathways may be very different in different cellular context [13,14] and may result in complex functional effects including changes in cellular proliferation, cell cycle, chromosomal instability, apoptosis, drug resistance and prognosis [8,12,15-17]. Also the role of the KRAS-BRAF interaction (being BRAF an effector of RAS in the RAF and PI3K activated pathways) is far from being understood.
Although BRAF mutations have been observed mainly in sporadic MSI CRC, they have also been detected in a small percentage of microsatellite stable (MSS) CRCs [18-22]. In particular, BRAF mutations were more often found in premalignant colon polyps and in early, rather than in advanced, CRC [18,23-25]. As concomitant KRAS and BRAF mutations are quite rare in premalignant colon polyps and early stages of CRC, they are considered as alternative or mutually exclusive mutations [26,27]. In a recent study, however, it was found that the number of concomitant KRAS and BRAF mutations increased along with the depth of the wall invasion of sporadic MSS CRC, suggesting that activation of both genes is likely to harbor a synergistic effect and that KRAS could give the tumor an invasive behavior [28]. Therefore, cells harboring a BRAF mutation would represent a model system with a genetic background well suited to study the specific contribution of activating KRAS mutations to CRC progression. To this end, we have used the colorectal adenocarcinoma cell line Colo741, which is wild-type relatively to KRAS, MSS [29] but harbors a mutation (V600E; single letter amino acid code) in the BRAF locus [30], and stably transfected these cells with constructs expressing the KRASG12V or the KRASG12D mutated coding sequences (cds) or the KRASWT. We selected these KRAS mutations since codon 12 is the most affected by point mutations in CRC (more than 90%) and because, among all mutation types in sporadic MSS CRC, G12D and G12V, they are the most frequently observed with a frequency of about 45% and 23% respectively [9].
Results
Mutated KRAS expression modifies the gene expression profile of Colo741 cells
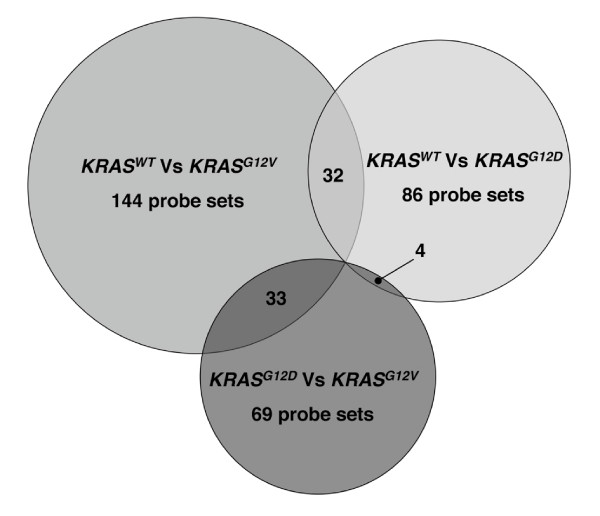
The effects of mutated KRAS on gene expression were investigated by performing GeneChip microarray studies. Data from probed and scanned arrays (two technical replicates were analyzed for the three conditions: KRASWT, KRASG12D and KRASG12V) were normalized, filtered by removing probe sets that were regarded as not expressed and then analyzed by performing a multi-class of all 6 arrays using the SAM program. A change in gene expression was considered significant if the p value was less than 0.02 and increased gene expression occurred in, at least, one out of the three conditions. We also performed a two-class unpaired comparison for KRASG12D versus KRASWT and KRASG12Vversus KRASWT to specify expression changes. We chose a 2.0 fold change cutoff. Based on these criteria, 25 probe sets were up regulated and 61 down regulated in KRASG12Dversus KRASWT. In the comparison KRASG12Vversus KRASWT, 88 probe sets resulted up regulated and 56 down regulated (see Additional File 1). Then, another two-class unpaired comparison between the mutated KRAS-expressing Colo741 found 52 and 17 probe sets respectively up- or down-regulated (see Additional File 2). In total, irrespective of the comparison, we found 229 regulated probe sets (Figure 1) corresponded to 219 unique genes.
Figure 1.
Venn diagrams obtained by SAM analysis. Global comparison among the genes regulated by the KRASG12D, KRASG12V and KRASWT isoforms expressed by transfection in Colo741 cells. The number of probe sets associated to the co-regulated genes is reported in the overlapping areas.
Analysis of gene expression profiles
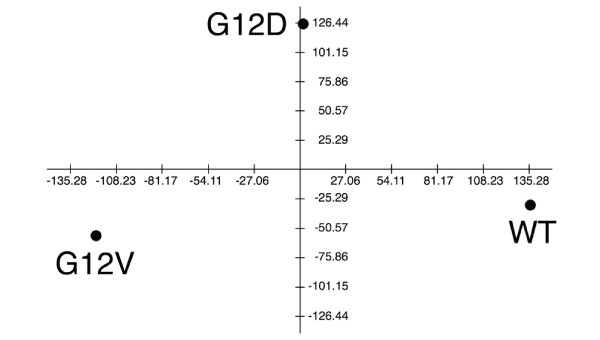
All the differentially 229 expressed probe sets, after creating a single sample by averaging the individual expression values from technical replicas, were then analyzed with the software tool TIGR MeV. Analysis of mutated KRAS-expressing Colo741 clones by PCA revealed a distinct partition among those expressing the recombinant KRASWT and those expressing the KRASG12D and KRASG12V (Figure 2). Points in the two-dimensional plot represented the samples. The distance between any pair of points was related to the similarity between the two observations in high-dimensional space. Samples that were near each other in the plot were similar in a large number of variables, i.e., expression level of individual genes. Conversely, samples that were far apart in the plot were different in a large number of variables.
Figure 2.
Microarray analysis performed with TIGR MeV program: principal component analysis. Microarray analysis of Colo741 cell clones transfected with constructs expressing the KRASG12D (G12D), KRASG12V (G12V) and KRASWT (WT) isoforms. Probe sets associated to dysregulation of gene expression levels among the three groups were identified using SAM (see Materials and Methods). The corresponding values from two independent microarray analysis were averaged. Principal component analysis (PCA) is shown to provide the 2D projections onto the plane spanned by the two principal components for the three different KRAS profiling data sets.
Differential expression among the three KRAS conditions was visualized by hierarchical clustering (HCL) which generates a tree (dendogram) to group similar objects together (Figure 3).
Figure 3.
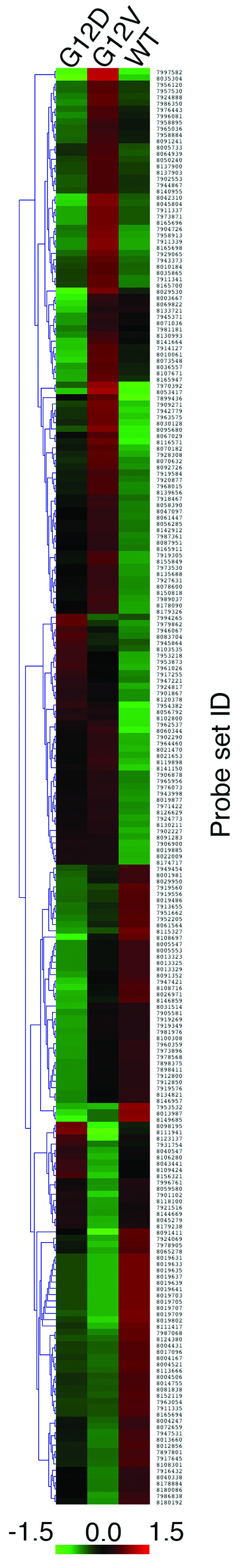
Microarray analysis performed with TIGR MeV program: hierarchical clustering. Heat map visualization obtained by hierarchical clustering (HCL). Ratios for each probe relative to the mean value (calculated from the two independent microarray analysis for each condition) were used to rearrange the gene list on the basis of their expression pattern. Probes corresponding to genes with similar regulation trend were placed close to each other. The color-ratio bar indicates intensity of gene up-regulation (red), down-regulation (green) and no change (black).
Expression patterns and biological pathways specifically identified by EASE in KRAS-expressing clones
To gain a more mechanistic understanding of the main processes affected by the KRAS, the EASE score [31] was used to identify Gene Ontology (GO) functional categories, which were significantly over-represented (see Additional Files 1 and 2). After filtering the results to avoid redundant and/or generic categories, statistically significant GO terms associated with KRAS-regulated genes were found (Table 1). This analysis identified cell cycle arrest and apoptosis genes as being the most affected ones by KRASG12V. When genes regulated by KRASG12D (see Additional File 1) were subjected to EASE analysis, genes involved in cellular component organization and biogenesis were identified. Finally, we chose to examine the 69 probe sets differently regulated between mutated KRAS-expressing Colo741 (see Additional File 2) in order to compare the specific effects of the two oncogenes KRASG12V and KRASG12D. Again, we used the EASE score to perform ontological categorization and KEGG pathway analysis. These analyses identified genes associated to immune system processes and to the biosynthesis of steroids as the most affected ones (Table 1).
Table 1.
Gene Ontology analysis and KEGG pathway analysis of KRAS isoform-expressing Colo741 cell clones.
| System Gene Category – Term | Count | % | P-Value | Genes |
| KRASG12V Vs KRASWT | ||||
| GO biological process | ||||
| Cell cycle arrest | 6 | 6.7 | 5.50E-05 | DDIT3, DHCR24, GADD45A, IL8, PPP1R15A, SESN2 |
| Apoptosis | 13 | 14.6 | 3.90E-04 | ANXA1, APOE, GADD45A, IFIH1, IL24, DDIT3, DDIT4, DHCR24, PMAIP1, PPP1R15A, SEMA6A, TNFRSF19, TRIB3 |
| KEGG pathway | ||||
| Nitrogen metabolism | 3 | 3.4 | 1.50E-02 | ASNS, CTH, GLS |
| KRASG12D Vs KRASWT | ||||
| GO biological process | ||||
| Cellular component organization and biogenesis | 12 | 27.3 | 5.10E-02 | CRYAB, EHD2, FHOD1, HIST1H1A, HSPB1, LIN7C, MAP3K11, PCDHB5, PCDHB16, SEMA6A, SLC7A11, SMCHD1 |
| KRASG12D Vs KRASG12V | ||||
| GO biological process | ||||
| Immune system process | 14 | 32.6 | 3.50E-07 | BST2, CDK6, IFITM3, IFIT1, IFI27, IFI44, IL8, IL24, MICA, OAS1, OAS2, OAS3, SYK, S100B |
| Sterol metabolic process | 6 | 14 | 1.10E-06 | APOE, FDFT1, HMGCR, HMGCS1, IDI1, SC4MOL |
| KEGG pathway | ||||
| Biosynthesis of steroids | 4 | 9.2 | 5.70E-05 | FDFT1, IDI1, HMGCR, SC4MOL |
Gene name symbols used are those approved by the Human Genome Organisation Gene Nomenclature Committee http://www.genenames.org/.
Non-redundant functional categories, number of genes contained within each category, percentages ranked by the degree of over-representation in the category as determined by EASE (P-value) and gene members found to be modulated by the KRAS isoforms are shown. Redundant categories with similar gene members were removed to yield a single representative category.
Quantitative RT-PCR validated the Microarray data
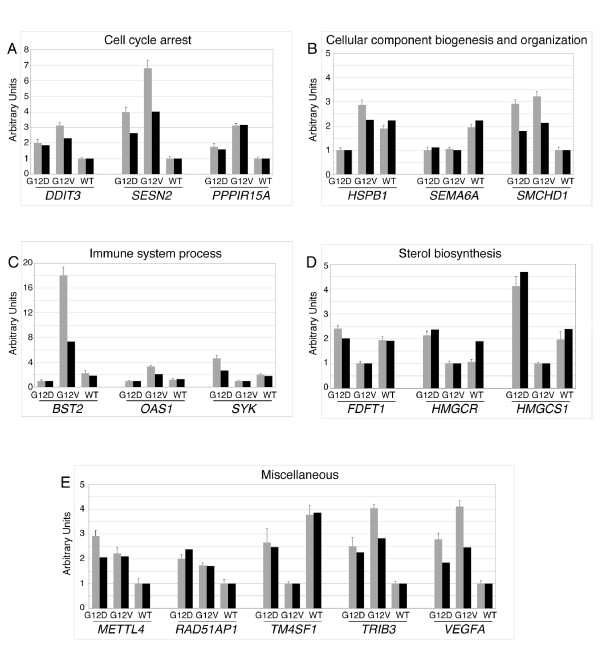
To verify and validate the GeneChip microarray data, we performed real-time RT-PCR (Figure 4) on a subset of seventeen KRAS-modulated genes. RNA samples subjected to RT-PCR were identical to those used for the microarray analysis. In particular, we confirmed the regulated expression patterns of genes chosen on the basis of their association with cell cycle arrest processes [DNA-damage-inducible transcript 3 (DDIT3), sestrin 2 (SESN2), and protein phosphatase 1, regulatory (inhibitor) subunit 15A (PPP1R15A)], cellular component organization and biogenesis [heat shock 27 kDa protein 1 (HSPB1), semaphorin 6A (SEMA6A) and structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1)], immune system processes [bone marrow stromal cell antigen 2 (BST2), 2',5'-oligoadenylate synthetase 1 (OAS1) and spleen tyrosine kinase (SYK)], as well as genes known to be regulated during sterol biosynthesis [farnesyl-diphosphate farnesyltransferase (FDFT1), 3-hydroxy-3-methylglutaryl-Coenzyme A reductase (HMGCR) and 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (HMGCS1)]. All validation results, on changes in mRNA expression of these genes, were proved to be regulated coherently with the GeneChip microarray data (Figure 4). In summary, the two methodologies (real time PCR and microarray) produced highly consistent results, which provided a good level of assurance regarding the validity of the microarray data.
Figure 4.
Real-Time RT-PCR validation of microarray data. Real-Time RT-PCR analysis performed on Colo741 cell clones transfected with constructs expressing the KRASG12D (G12D), KRASG12V (G12V) and KRASWT (WT) isoforms to validate the microarray data. This was accomplished on randomly selected genes from Table 1 and showed, in arbitrary units, KRAS isoform-dependent regulation of cell cycle arrest genes (A), of cellular component organization and biogenesis genes (B), of immune system process genes (C) or sterol metabolic process genes (D). Other KRAS isoform-regulated genes associated to miscellaneous functions and randomly selected from Tables S1 and S2, are shown in (E). Real-Time RT-PCR and microarray data are respectively indicated by gray and black bars. Expression levels are relative to the expression of the housekeeping Ribosomal protein L19 gene (RPL19). Standard deviations of Real-Time RT-PCR data are indicated as vertical bars. Gene name symbols used are those approved by the Human Genome Organisation Gene Nomenclature Committee http://www.genenames.org/.
Discussion
The purpose of this study was to investigate by a genome wide transcription profiling approach the specific gene expression modulation due to the two most frequently occurring KRAS mutations in CRC (respectively, G12V and G12D) in BRAF mutated CRC-derived cells. The gene expression profiles associated to these mutations transfected in the host CRC Colo741 cells, characterized by mutated BRAF and wild type KRAS and evaluated versus the transfected WT isoform, were not overlapping and could be clear-cut discriminated. Furthermore, the Principal Component Analysis performed in this study made it evident that the highest degree of change in gene expression was to be associated to the G12V state.
Both G12D and G12V states appeared to co-regulate genes associated to biological processes which are highly correlated to cancer. The expression of KRASG12V, in particular, modulated a series of genes involved in cell cycle control, apoptosis and nitrogen metabolism so that these cells are likely to undergo cell death and/or lower proliferation with respect to the cells expressing the KRASWT isoform. Thus, it appears at first sight that the expression of KRASG12V might explain why, at least at the early stages of CRC genesis, concomitant KRAS and BRAF mutations are rarely occurring. However, BRAFV600E was shown to induce genomic instability promoting the acquisition of additional genetic defects [32] and, in the specific context of MSS CRC, concomitant BRAF and KRAS mutation occurrence was shown to increase with colon wall invasion and metastases [28]. Interestingly, Costa and coworkers reported very recently a strong link between tumor recurrence, distant metastases, survival and BRAFV600E plus RAS mutation in thyroid carcinoma [33]. It is tempting to suggest that specific KRAS and BRAF mutation interaction may have a role to modulate gene expression profiling in specific tumor types (either MSS, or MSI, or CIN) toward a more aggressive phenotype. In our view, it is not a single gene or a given genetic system that may control tumor progression. In fact, the gene expression patterns appear modulated by the genome context coupled with the effects of different gene mutations [34].
Our data also showed that IL8 was upregulated by KRASG12V. With this in mind, we considered of high interest previous reports showing that HRASG12V-induced IL8 expression plays a critical role in tumor growth and angiogenesis [35], that the degree of its expression was associated with the CRC induction and progression including the development of liver metastases [36,37], and that IL8 was a central element in CRC-specific gene network [38]. We are therefore tempted to speculate that the presence of KRASG12V in those cells which might escape apoptosis may confer an aggressive phenotype by inducing angiogenesis via IL8 and possibly facilitate metastasis.
Our results also showed that the genes regulated by the KRASG12Disoform were related to the cellular component organization and biogenesis but not to apoptosis nor cell stress but instead it downregulated indeed at least two genes coding for chaperone proteins (HSPB1 and CRYAB). Since we showed that the G12V mutation generated more stressful conditions favoring cell cycle arrest and apoptosis than the G12D, it appears more likely that the BRAF mutation is associated with G12D. Interestingly, this observation appears in agreement with the data reported by Costa and collaborators in thyroid carcinoma [33] and by Oliveira and coworkers investigating the CRC [28].
It is known that both KRAS mutations under study greatly reduce the KRAS GTPase activity, locking the protein in a constitutively active state [39] To our knowledge, however, the crystal structures of these two KRAS isoforms have not yet been determined and compared. Therefore, possible different protein-protein interactions and affinities of interactions with downstream KRAS effectors cannot be ruled out. Interestingly, these mutations were shown to affect differently the structural conformation of the highly related HRAS protein, suggesting that differences between the switch I region of G12D and G12V Ras could modify interactions with downstream effectors [39].
The present sets of genes modulated by the two KRAS mutations investigated were quite different (as analyzed in details in the following) and may be partly explaining the differences observed in other studies addressing their correlation with in vitro invasion properties [40], survival of CRC patients [8] and chromosomal instability and aneuploidy [41].
Among the present downregulated genes in the two KRAS mutations with respect to the WT isoform, we observed SPARC, TRPM1, SEMA6A and ENO2. Reduced levels of the gene coding for SPARC was associated with therapy-refractory CRC [42] and inactivation of SPARC was related to rapid progression of CRC [43]; TRPM1 expression was found to decline with an increased degree of aggressiveness of the melanoma [44]; downregulation of SEMA6A was observed in ovarian carcinoma cell lines resistant to several chemotherapeutic drugs [45] and its extracellular domain was shown to be able to inhibit angiogenesis [46]. Concerning ENO2, literature reports indicated upregulation in cancer [47,48] rather than downregulation, as it occurred in our mutant transfected cells, but additional studies are needed to find any specific correlation of this subunit of the enolase in CRC and KRAS/BRAF mutations.
For the other genes that presently resulted co-regulated by the two mutated KRAS isoforms with respect to the WT isoform, we could not find in the literature a correlation with cancer. Consequently, we suggest that these genes deserve a future careful investigation as they might represent possible novel CRC markers.
Among the genes upregulated by the KRASG12V, we observed DDIT3, PPP1R15A, SESN2, APOE, DDIT3, DDIT4 ASNS, and CTH. All these genes are known to be induced by a variety of stressors (including unfolded protein, endoplasmic reticulum stress, DNA damage, oxidative stress, amino acid deprivation, acidosis) [49-58] and since IL24 which was also upregulated in our study, is able to activate the unfolded protein response [59] and cell cycle arrest [60], it is very likely that the expression of KRASG12V in cells already expressing a mutated BRAF unleashes a cascade of events leading to cell stress and hence possibly to apoptosis and inhibition of cell cycle progression. It is interesting to note that it was recently demonstrated that HRASG12V but not BRAF V600E engages a rapid cell-cycle arrest mediated by the endoplasmic stress response in melanocytes [61]. The upregulation of transcription of genes related to stress response resulting from our experiment is in agreement with previous reports, albeit obtained with fibroblasts expressing HRASG12V, suggesting that Ras is part of the stress sensing machinery [62]. Furthermore, the downregulation in cells transfected with KRASG12V of genes involved in sterol metabolic processes appeared to us worth to note. Interestingly, there is a large number of published studies showing that products of the mevalonate pathway are essential to the post-translational processing and function of nuclear lamins, small G proteins (including Ras), and growth factor receptors constituting a survival pathway that when inhibited induces apoptosis and inhibits angiogenesis [63-65]. The downregulation of the DHCR24 in the cells transfected with the KRASG12V isoform with respect to the WT isoform goes in the same line since this gene was reported to be associated to resistance to oxidative stress-induced apoptosis [66]. On the other hand, the upregulation of TNFRSF19 may suggest that a caspase independent cell death may take place in these cells, as already shown for 293T cells [67] and that this same gene might promote cell growth as reported for melanoma cells [68]. The significance of an upregulation of genes implicated in the immune processes in our Colo741 expressing the KRASG12V with respect to KRASG12D was less clear.
Among the genes associated with the KRASG12D we observed two genes, HIST1H1A [69] and SMCHD1[70], related to chromatin organization; PCDHB5 and PCDHB16 [71] and LIN7C [72] implicated in cell-cell adhesion; CRYAB [73] and FHOD1 [74] in cytoskeleton organization; EHD2 [75] in receptor internalization; SLC7A11 [76] in amino acid transport; HSPB1 [77,78] in cellular stress response; MAP3K11 [79] in invasive activity; SEMA6A [80] in axon guidance and retrograde signaling. Given the variety of processes that may be affected by the KRASG12D isoform gene expression modulation, the interpretation of our data appears quite complex. Nevertheless, since reduction of MAP3K11 and increase of LIN7C have been shown to facilitate respectively the in vitro invasive activity [79] and the oncogenic activation of PI3K [72], we suggest that the concordant modulation of these genes by the KRASG12D isoform in Colo741 cells may be functional to an higher aggressive phenotype with respect to the sole presence of the BRAF mutation. The downregulation of SEMA6A points to the same direction (see also above). Moreover, the downregulation of EHD2 may lead to increasing the growth signaling at the cell membrane by reduction of the internalization of growth receptors. Similarly, the downregulation of the proteins involved in cell-cell adhesion (PCDHB5 and PCDHB16) could possibly promote cell delamination and cell migration.
A further comment on our present expression data is that Colo741 cells expressing the KRASG12D isoform were not apoptosis-prone or stressed cells since they did not upregulate genes induced by cellular stressors with respect to cells expressing the KRAS WT isoform. Conversely, they downregulated indeed at least two genes coding for chaperone proteins (HSPB1 and CRYAB).
In relationship with all these observations and list of genes that we have specifically considered, it is clear that specific experiments are definitely required to better clarify the role of these genes in our present CRC model.
Conclusion
Our data support the hypothesis that the presently investigated KRAS mutations elicit, in the host BRAF mutated cells under study, biological consequences which may help explaining previous observations in CRC and contribute to identify novel pathways and biomarkers of potential clinical relevance.
Methods
Generation of stable Colo741 cell clones expressing WT, G12V and G12D KRAS mutants
The procedures for the generation of Colo741 cell clones stably transfected with construct coding for the KRAS WT and mutant isoforms, as well as the experiments needed to assess their expression and the activation of the KRAS pathway were performed according to standard protocols (see Additional Files 3, 4, 5 and 6).
RNA extraction and quality analysis
Total RNA was isolated using RNeasy® MinElute columns (Qiagen). RNA concentration and purity were determined from measuring absorbance at 260 and 280 nm; 2 μg total RNA was run on a 1% denaturing gel and 100 ng were loaded on the 2100 Bioanalyzer (Agilent, Palo Alto, CA) to verify RNA integrity.
Amplification of RNA and array hybridization
According to the recommendations of the manufacturer, 100 ng of total RNA was used in the first-round synthesis of double-stranded cDNA. The RNA was reverse transcribed using a WT cDNA synthesis and amplification kit (Affymetrix UK Ltd., High Wycombe, UK). The resulting biotin-labeled cRNA was purified using an IVT clean-up kit (Affymetrix) and quantified using a UV spectrophotometer (A260/280; Beckman, Palo Alto, CA). An aliquot (15 μg) of cRNA was fragmented by heat and ion-mediated hydrolysis at 94°C for 35 minutes. Fragmented cRNA, run on the Bioanalyzer (Agilent Technologies, Santa Clara, CA) to verify the correct electropherogram, was hybridized in a hybridization oven (16 hours, 45°C) to a Human Gene 1.0 ST array (Affymetrix) representing whole-transcript coverage. Each one of the 28,869 genes is represented on the array by approximately 26 probes spread across the full length of the gene, providing a more complete and more accurate picture of gene expression than the 3' based expression array design. The washing and staining procedures of the arrays with phycoerythrin-conjugated streptavidin (Invitrogen) was completed in the Fluidics Station 450 (Affymetrix). The arrays were subsequently scanned using a confocal laser GeneChip Scanner 3000 7G and the GeneChip Command Console (Affymetrix).
GeneChip microarray analysis and data normalization
Affymetrix raw data files [cell intensity (CEL) files] were used as input files in expression console environment (Affymetrix). Briefly, CEL files were processed using the Robust Multi-Array Analysis (RMA) procedure [81], an algorithm that is publicly available at http://www.bioconductor.org. The RMA method was used to convert the intensities from the multiple probes of a probe set into a single expression value with greater precision and reduced background noise (relying on the perfect match probes only and thus ignoring the mismatch probes) and then to normalize by sketch quantile normalization. Quality assessments were also performed in the expression console environment. This procedure, based on various metrics, allowed us to identify a chip as an outlier (see for details Quality assessment of exon and gene arrays http://www.affymetrix.com/support/technical/whitepapers/exon_gene_arrays_qa_whitepaper.pdf. Significance Analysis of Microarrays (SAM), Principal Component Analysis (PCA) of variance and Hierarchical Clustering (HCL), after mean scaling and log2 transformation, were performed with the software tool of The Institute for Genomic Research (TIGR) MeV http://www.tigr.org/software/tm4/mev.html[82].
Individual genes with different expression levels, among the three groups, were identified using SAM [83]. The false discovery rate expressed as q-value was used to evaluate statistical significance, and its threshold was set at 0.02 (2%). For comparison purposes, an arbitrary filter was applied excluding all genes that did not exhibit a difference in expression of at least 2-fold. Genes differentially expressed were investigated using 1) a multiclass analysis to test differences among the three groups of cells and 2) a two-class analysis within each pair groups to specify expression changes.
We used PCA to reduce the complexity of high-dimensional data and to simplify the task of identifying patterns and sources of variability in these large data sets.
The results from SAM were visualized using HCL [84].
All the microarray information has been submitted to the National Center for Biotechnology Information Gene Expression Omnibus web site http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12398.
Pathways identification by Expression Analysis Systemic Explore (EASE)
Gene lists from Affymetrix results were examined using the EASE program, accessible via http://david.abcc.ncifcrf.gov/. EASE is a customized stand-alone software application with statistical functions for discovering biological themes within gene lists. This software assigns genes of interest into functional categories based on the Gene Ontology database (GO, http://www.geneontology.org/index.shtml) and uses the Fisher's exact test statistics to determine the probability of observing the number of genes within a list of interest versus the number of genes in each category on the array. A more detailed analysis of the genes' association with physiological pathways was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/pathway.html). Each identified process was confirmed through PubMed/Medline.
RT-PCR analysis
Starting from about 1 μg of total RNA, cDNA was synthesized by using an Oligo(dT)20, random hexamers mix and a Superscript III first-strand synthesis system supermix for RT-PCR (Invitrogen). cDNAs were diluted 5 – 20 times, then subjected to PCR analysis.
Relative quantification was performed by real-time quantitative RT-PCR (qPCR) sybrgreen using the ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA) following manufacturer's instructions. The housekeeping gene ribosomal protein L19 (RPL19) was used as the endogenous control for normalization because, in the microarray data, it showed in all conditions the steadiest expression when normally used housekeeping genes were compared.
To avoid possible signal production from potential contaminating genomic DNA, specific primers for each gene were designed across a common exon-exon splice junction by the Primer Express software (Applied Biosystems) (see Additional File 7). Dissociation curve analysis defined the specificity of the products by the presence of a single dissociation peak on the thermal melting curve.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MMo designed research, performed research, analyzed data, wrote paper; EB, MMa, AD FR and CTS performed research; WG designed research, analyzed data, wrote paper; PC designed research, performed research, analyzed data, wrote paper.
Supplementary Material
Table S1. Probe sets significantly regulated by KRASG12V and KRASG12D Vs KRASG12WT in Colo741 cell clones. Probe sets ID, with relative gene symbols or Ensembl Transcript ID, significantly regulated by KRASG12V or KRASG12D in Colo741 cell clones, as determined by using SAM software with multi-class analysis. Q-values were calculated using the two-class, unpaired, option with the additional requirement of at least a 2-fold change in gene expression, relatively to KRASWT-expressing clones.
Table S2. Probe sets ID significantly regulated in KRASG12V Vs KRASG12D transfected Colo741 cell clones. Probe sets ID, with relative gene symbols or Ensembl Transcript ID, significantly regulated in the comparison between the clones bearing the KRAS mutations (G12V and G12D). Q-values were calculated as in table S1.
Supplemental Methods. Methods for; generation of KRAS expression constructs, cells and cell culture, immunolocalization, establishment and selection of stable cell clones and Western blot analysis.
Figure S1. Expression and localization of recombinant KRAS proteins. In vitro transcription/translation of the empty vector (lane 1), KRASWT (lane 2), KRASG12V(lane 3) and KRASG12D (lane 4), recombinant chimera RED2:KRASWT (lane 5) and luciferase (control) expressing plasmids (lane 6) (A). Arrow heads point to the position of molecular mass standards whose sizes are expressed in kDa (A). Cells after 48 hours from transfection with CFP-KRASG12V construct (B-G). Immunolocalization by an anti-pan-Ras antibody (F, G) and an Alexa514-conjugated secondary antibody giving a green signal (C, D, F, G,), direct visualization of the cyan signal pseudocolored in red (B, D, E, G) and direct visualization of the signal displayed by the nuclear dye 7-Amino Actinomycin D pseudocolored in blue (D, G). The merged fluorescence signals are shown in (D, G). Magnification of the area in the white rectangles are shown in the lower left insets of (E-G). Arrow heads point to some of the cell membrane regions displaying colocalization of the pan-RAS and CFP-KRASG12V signals. Gamma adjustment was applied to each panel to adapt color rendering in the CMYK process. Scale bar = 50 mm.
Figure S2. Screening by semiquantitative RT-PCR analysis of Colo741 cell clones transfected with constructs expressing KRASWT (WT), KRASG12D (G12D) and KRASG12V (G12V) mRNA isoforms. For each construct both cell clones expressing the transgene (1D8-3C7-4D3) and cell clones not expressing the transgene (2G7-1H2-1B3) are shown. The latter were discarded from further experiments. For all the assayed samples, a reverse transcription PCR assays, performed omitting the reverse transcriptase, was also carried out to exclude from further analysis clones yelding a KRAS amplicon resulting from a plasmid integrated in the genomic DNA (false positive samples). In this example no amplicons were detected in minus reverse transcriptase assays. Primers for GAPDH were used to normalize the results.
Figure S3. Western blotting analysis of selected Colo741 KRAS-expressing clones. Vector-transfected clones (lane 1), KRASWT (lane 2), KRASG12V (lane 3), KRASG12D (lane 4) and senescent human bone marrow stromal cells (lane 5) were subjected to immunoblotting to establish the global Ras expression (pan-Ras) (A) and the phosphorylation status of AKT (B) and ERK1/2 (C). Arrow points to the pAKT protein.
Table S3. Sequences accession numbers and primers.
Acknowledgments
Acknowledgements
The pcDNA3-KRASG12V was a kind gift from Dr. Natalia Ignatenko, Department of Cell Biology and Anatomy, University of Arizona. This work was supported by CIPE 2007-Regione Liguria (Stem Cells), "Ministero della Salute 2004" and "Compagnia di San Paolo" (project 2007.0266).
Contributor Information
Massimiliano Monticone, Email: massimiliano.monticone@cba-biotecnologie.it.
Emanuela Biollo, Email: S2661715@studenti.unige.it.
Massimo Maffei, Email: massimo.maffei@istge.it.
Alessandra Donadini, Email: alessandra.donadini@istge.it.
Francesco Romeo, Email: francesco.romeo@istge.it.
Clelia Tiziana Storlazzi, Email: c.storlazzi@biologia.uniba.it.
Walter Giaretti, Email: walter.giaretti@istge.it.
Patrizio Castagnola, Email: patrizio.castagnola@istge.it.
References
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, Barker MA, Arnold S, Simms LA, Leggett BA, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- Jass JR, Baker K, Zlobec I, Higuchi T, Barker M, Buchanan D, Young J. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a 'fusion' pathway to colorectal cancer. Histopathology. 2006;49:121–131. doi: 10.1111/j.1365-2559.2006.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005;1756:81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Kern SE, Fearon ER, Tersmette KW, Enterline JP, Leppert M, Nakamura Y, White R, Vogelstein B, Hamilton SR. Clinical and pathological associations with allelic loss in colorectal carcinoma [corrected] JAMA. 1989;261:3099–3103. doi: 10.1001/jama.261.21.3099. [DOI] [PubMed] [Google Scholar]
- Benhattar J, Losi L, Chaubert P, Givel JC, Costa J. Prognostic significance of K-ras mutations in colorectal carcinoma. Gastroenterology. 1993;104:1044–1048. doi: 10.1016/0016-5085(93)90272-e. [DOI] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, et al. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C, Westra JL, Arango D, Ollikainen M, Domingo E, Ferreira A, Velho S, Niessen R, Lagerstedt K, Alhopuro P, et al. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13:2303–2311. doi: 10.1093/hmg/ddh238. [DOI] [PubMed] [Google Scholar]
- Gibbs JB, Sigal IS, Poe M, Scolnick EM. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci USA. 1984;81:5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL, Franklin RA, McCubrey JA. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/S1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Keller JW, Franklin JL, Graves-Deal R, Friedman DB, Whitwell CW, Coffey RJ. Oncogenic KRAS provides a uniquely powerful and variable oncogenic contribution among RAS family members in the colonic epithelium. J Cell Physiol. 2007;210:740–749. doi: 10.1002/jcp.20898. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/S1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Castagnola P, Giaretti W. Mutant KRAS, chromosomal instability and prognosis in colorectal cancer. Biochim Biophys Acta. 2005;1756:115–125. doi: 10.1016/j.bbcan.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, Stephens P, Edkins S, Tsui WW, Chan AS, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–6455. [PubMed] [Google Scholar]
- Oliveira C, Pinto M, Duval A, Brennetot C, Domingo E, Espin E, Armengol M, Yamamoto H, Hamelin R, Seruca R, Schwartz S., Jr BRAF mutations characterize colon but not gastric cancer with mismatch repair deficiency. Oncogene. 2003;22:9192–9196. doi: 10.1038/sj.onc.1207061. [DOI] [PubMed] [Google Scholar]
- Wang L, Cunningham JM, Winters JL, Guenther JC, French AJ, Boardman LA, Burgart LJ, McDonnell SK, Schaid DJ, Thibodeau SN. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003;63:5209–5212. [PubMed] [Google Scholar]
- Fransen K, Klintenas M, Osterstrom A, Dimberg J, Monstein HJ, Soderkvist P. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–533. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S., Jr Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878–4881. [PubMed] [Google Scholar]
- Ikehara N, Semba S, Sakashita M, Aoyama N, Kasuga M, Yokozaki H. BRAF mutation associated with dysregulation of apoptosis in human colorectal neoplasms. Int J Cancer. 2005;115:943–950. doi: 10.1002/ijc.20957. [DOI] [PubMed] [Google Scholar]
- Calistri D, Rengucci C, Seymour I, Lattuneddu A, Polifemo AM, Monti F, Saragoni L, Amadori D. Mutation analysis of p53, K-ras, and BRAF genes in colorectal cancer progression. J Cell Physiol. 2005;204:484–488. doi: 10.1002/jcp.20310. [DOI] [PubMed] [Google Scholar]
- Ahlquist T, Bottillo I, Danielsen SA, Meling GI, Rognum TO, Lind GE, Dallapiccola B, Lothe RA. RAS signaling in colorectal carcinomas through alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia. 2008;10:680–686. doi: 10.1593/neo.08312. 682 p following 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, Duval A, Hamelin R, Machado JC, et al. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26:158–163. doi: 10.1038/sj.onc.1209758. [DOI] [PubMed] [Google Scholar]
- Suter CM, Norrie M, Ku SL, Cheong KF, Tomlinson I, Ward RL. CpG island methylation is a common finding in colorectal cancer cell lines. Br J Cancer. 2003;88:413–419. doi: 10.1038/sj.bjc.6600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsutake N, Knauf JA, Mitsutake S, Mesa C, Jr, Zhang L, Fagin JA. Conditional BRAFV600E expression induces DNA synthesis, apoptosis, dedifferentiation, and chromosomal instability in thyroid PCCL3 cells. Cancer Res. 2005;65:2465–2473. doi: 10.1158/0008-5472.CAN-04-3314. [DOI] [PubMed] [Google Scholar]
- Costa AM, Herrero A, Fresno MF, Heymann J, Alvarez JA, Cameselle-Teijeiro J, Garcia-Rostan G. BRAF mutation associated with other genetic events identifies a subset of aggressive papillary thyroid carcinoma. Clin Endocrinol (Oxf) 2008;68:618–634. doi: 10.1111/j.1365-2265.2007.03077.x. [DOI] [PubMed] [Google Scholar]
- Heng HH, Stevens JB, Lawrenson L, Liu G, Ye KJ, Bremer SW, Ye CJ. Patterns of genome dynamics and cancer evolution. Cell Oncol. 2008;30:513–514. doi: 10.3233/CLO-2008-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Rubie C, Frick VO, Pfeil S, Wagner M, Kollmar O, Kopp B, Graber S, Rau BM, Schilling MK. Correlation of IL-8 with induction, progression and metastatic potential of colorectal cancer. World J Gastroenterol. 2007;13:4996–5002. doi: 10.3748/wjg.v13.i37.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacev T, Radosevic S, Krizanac S, Kapitanovic S. Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis. 2008;29:1572–1580. doi: 10.1093/carcin/bgn164. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li X, Rao S, Wang L, Du L, Li C, Wu C, Wang H, Wang Y, Yang B. Constructing disease-specific gene networks using pair-wise relevance metric: application to colon cancer identifies interleukin 8, desmin and enolase 1 as the central elements. BMC Syst Biol. 2008;2:72. doi: 10.1186/1752-0509-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla F, Milner-White EJ, Going JJ, Birnie GD. Structural differences between valine-12 and aspartate-12 Ras proteins may modify carcinoma aggression. J Pathol. 1999;187:433–438. doi: 10.1002/(SICI)1096-9896(199903)187:4<433::AID-PATH273>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Al-Mulla F, MacKenzie EM. Differences in in vitro invasive capacity induced by differences in Ki-Ras protein mutations. J Pathol. 2001;195:549–556. doi: 10.1002/path.995. [DOI] [PubMed] [Google Scholar]
- Giaretti W, Rapallo A, Geido E, Sciutto A, Merlo F, Risio M, Rossini FP. Specific K-ras2 mutations in human sporadic colorectal adenomas are associated with DNA near-diploid aneuploidy and inhibition of proliferation. Am J Pathol. 1998;153:1201–1209. doi: 10.1016/S0002-9440(10)65664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai IT, Dai M, Owen DA, Chen LB. Genome-wide expression analysis of therapy-resistant tumors reveals SPARC as a novel target for cancer therapy. J Clin Invest. 2005;115:1492–1502. doi: 10.1172/JCI23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Kang HJ, Koh KH, Rhee H, Kim NK, Kim H. Frequent inactivation of SPARC by promoter hypermethylation in colon cancers. Int J Cancer. 2007;121:567–575. doi: 10.1002/ijc.22706. [DOI] [PubMed] [Google Scholar]
- Duncan LM, Deeds J, Cronin FE, Donovan M, Sober AJ, Kauffman M, McCarthy JJ. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 2001;19:568–576. doi: 10.1200/JCO.2001.19.2.568. [DOI] [PubMed] [Google Scholar]
- Prislei S, Mozzetti S, Filippetti F, De Donato M, Raspaglio G, Cicchillitti L, Scambia G, Ferlini C. From plasma membrane to cytoskeleton: a novel function for semaphorin 6A. Mol Cancer Ther. 2008;7:233–241. doi: 10.1158/1535-7163.MCT-07-0390. [DOI] [PubMed] [Google Scholar]
- Dhanabal M, Wu F, Alvarez E, McQueeney KD, Jeffers M, MacDougall J, Boldog FL, Hackett C, Shenoy S, Khramtsov N, et al. Recombinant semaphorin 6A-1 ectodomain inhibits in vivo growth factor and tumor cell line-induced angiogenesis. Cancer Biol Ther. 2005;4:659–668. doi: 10.1158/1535-7163.MCT-04-0290. [DOI] [PubMed] [Google Scholar]
- Paus E, Myklebust AT. Expression and interconversion of neuron-specific enolase in patient sera and extracts from small-cell lung cancer cells. Tumour Biol. 1996;17:271–280. doi: 10.1159/000217989. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen LQ, Yang GL, Li Y. Value of tumor markers in diagnosing and monitoring colorectal cancer and strategies for further improvement: analysis of 130 cases. Ai Zheng. 2007;26:1221–1226. [PubMed] [Google Scholar]
- He S, Yaung J, Kim YH, Barron E, Ryan SJ, Hinton DR. Endoplasmic reticulum stress induced by oxidative stress in retinal pigment epithelial cells. Graefes Arch Clin Exp Ophthalmol. 2008;246:677–683. doi: 10.1007/s00417-008-0770-2. [DOI] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powolny A, Takahashi K, Hopkins RG, Loo G. Induction of GADD gene expression by phenethylisothiocyanate in human colon adenocarcinoma cells. J Cell Biochem. 2003;90:1128–1139. doi: 10.1002/jcb.10733. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Shoshani T, Faerman A, Zelin E, Kamer I, Kalinski H, Gorodin S, Fishman A, Chajut A, Einat P, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- Joy Espiritu D, Mazzone T. Oxidative stress regulates adipocyte apolipoprotein E and suppresses its expression in obesity. Diabetes. 2008;57:2992–2998. doi: 10.2337/db08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Holbrook NJ. Elevated gadd153/chop expression and enhanced c-Jun N-terminal protein kinase activation sensitizes aged cells to ER stress. Exp Gerontol. 2004;39:735–744. doi: 10.1016/j.exger.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- Pan Y, Chen H, Siu F, Kilberg MS. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem. 2003;278:38402–38412. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- Lee JI, Dominy JE, Jr, Sikalidis AK, Hirschberger LL, Wang W, Stipanuk MH. HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics. 2008;33:218–229. doi: 10.1152/physiolgenomics.00263.2007. [DOI] [PubMed] [Google Scholar]
- Curthoys NP, Gstraunthaler G. Mechanism of increased renal gene expression during metabolic acidosis. Am J Physiol Renal Physiol. 2001;281:F381–390. doi: 10.1152/ajprenal.2001.281.3.F381. [DOI] [PubMed] [Google Scholar]
- Sieger KA, Mhashilkar AM, Stewart A, Sutton RB, Strube RW, Chen SY, Pataer A, Swisher SG, Grimm EA, Ramesh R, Chada S. The tumor suppressor activity of MDA-7/IL-24 is mediated by intracellular protein expression in NSCLC cells. Mol Ther. 2004;9:355–367. doi: 10.1016/j.ymthe.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Huang EY, Madireddi MT, Gopalkrishnan RV, Leszczyniecka M, Su Z, Lebedeva IV, Kang D, Jiang H, Lin JJ, Alexandre D, et al. Genomic structure, chromosomal localization and expression profile of a novel melanoma differentiation associated (mda-7) gene with cancer specific growth suppressing and apoptosis inducing properties. Oncogene. 2001;20:7051–7063. doi: 10.1038/sj.onc.1204897. [DOI] [PubMed] [Google Scholar]
- Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, Pointer JN, Gruber SB, Su LD, Nikiforov MA, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–1063. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- Stanhill A, Levin V, Hendel A, Shachar I, Kazanov D, Arber N, Kaminski N, Engelberg D. Ha-ras(val12) induces HSP70b transcription via the HSE/HSF1 system, but HSP70b expression is suppressed in Ha-ras(val12)-transformed cells. Oncogene. 2006;25:1485–1495. doi: 10.1038/sj.onc.1209193. [DOI] [PubMed] [Google Scholar]
- Fuchs D, Berges C, Opelz G, Daniel V, Naujokat C. HMG-CoA reductase inhibitor simvastatin overcomes bortezomib-induced apoptosis resistance by disrupting a geranylgeranyl pyrophosphate-dependent survival pathway. Biochem Biophys Res Commun. 2008;374:309–314. doi: 10.1016/j.bbrc.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Mo H, Elson CE. Studies of the isoprenoid-mediated inhibition of mevalonate synthesis applied to cancer chemotherapy and chemoprevention. Exp Biol Med (Maywood) 2004;229:567–585. doi: 10.1177/153537020422900701. [DOI] [PubMed] [Google Scholar]
- Park HJ, Zhang Y, Georgescu SP, Johnson KL, Kong D, Galper JB. Human umbilical vein endothelial cells and human dermal microvascular endothelial cells offer new insights into the relationship between lipid metabolism and angiogenesis. Stem Cell Rev. 2006;2:93–102. doi: 10.1007/s12015-006-0015-x. [DOI] [PubMed] [Google Scholar]
- Di Stasi D, Vallacchi V, Campi V, Ranzani T, Daniotti M, Chiodini E, Fiorentini S, Greeve I, Prinetti A, Rivoltini L, et al. DHCR24 gene expression is upregulated in melanoma metastases and associated to resistance to oxidative stress-induced apoptosis. Int J Cancer. 2005;115:224–230. doi: 10.1002/ijc.20885. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, Ma D. An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J Cell Sci. 2004;117:1525–1532. doi: 10.1242/jcs.00994. [DOI] [PubMed] [Google Scholar]
- Spanjaard RA, Whren KM, Graves C, Bhawan J. Tumor necrosis factor receptor superfamily member TROY is a novel melanoma biomarker and potential therapeutic target. Int J Cancer. 2007;120:1304–1310. doi: 10.1002/ijc.22367. [DOI] [PubMed] [Google Scholar]
- Lever MA, Th'ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, Maniatis T. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Latorre IJ, Chung SH, Caruana G, Bernstein A, Jones SN, Donehower LA, Justice MJ, Garner CC, Javier RT. Oncogenic function for the Dlg1 mammalian homolog of the Drosophila discs-large tumor suppressor. EMBO J. 2006;25:1406–1417. doi: 10.1038/sj.emboj.7601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard B, Ferguson C, Minajeva A, Leake MC, Gautel M, Labeit D, Ding L, Labeit S, Horwitz J, Leonard KR, Linke WA. Association of the chaperone alphaB-crystallin with titin in heart muscle. J Biol Chem. 2004;279:7917–7924. doi: 10.1074/jbc.M307473200. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ. The formin/diaphanous-related protein, FHOS, interacts with Rac1 and activates transcription from the serum response element. J Biol Chem. 2001;276:46453–46459. doi: 10.1074/jbc.M105162200. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118:4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- Lo M, Ling V, Wang YZ, Gout PW. The xc-cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br J Cancer. 2008;99:464–472. doi: 10.1038/sj.bjc.6604485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo AP, Paul C, Ducasse C, Sauvageot O, Kretz-Remy C. Small stress proteins: modulation of intracellular redox state and protection against oxidative stress. Prog Mol Subcell Biol. 2002;28:171–184. doi: 10.1007/978-3-642-56348-5_9. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Sauvageot O, Carmichael J, Diaz-Latoud C, Arrigo AP, Rubinsztein DC. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet. 2002;11:1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- Yu K, Ganesan K, Tan LK, Laban M, Wu J, Zhao XD, Li H, Leung CH, Zhu Y, Wei CL, et al. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet. 2008;4:e1000129. doi: 10.1371/journal.pgen.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostermann A, Lutz B, Gertler F, Behl C. The orthologous human and murine semaphorin 6A-1 proteins (SEMA6A-1/Sema6A-1) bind to the enabled/vasodilator-stimulated phosphoprotein-like protein (EVL) via a novel carboxyl-terminal zyxin-like domain. J Biol Chem. 2000;275:39647–39653. doi: 10.1074/jbc.M006316200. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Probe sets significantly regulated by KRASG12V and KRASG12D Vs KRASG12WT in Colo741 cell clones. Probe sets ID, with relative gene symbols or Ensembl Transcript ID, significantly regulated by KRASG12V or KRASG12D in Colo741 cell clones, as determined by using SAM software with multi-class analysis. Q-values were calculated using the two-class, unpaired, option with the additional requirement of at least a 2-fold change in gene expression, relatively to KRASWT-expressing clones.
Table S2. Probe sets ID significantly regulated in KRASG12V Vs KRASG12D transfected Colo741 cell clones. Probe sets ID, with relative gene symbols or Ensembl Transcript ID, significantly regulated in the comparison between the clones bearing the KRAS mutations (G12V and G12D). Q-values were calculated as in table S1.
Supplemental Methods. Methods for; generation of KRAS expression constructs, cells and cell culture, immunolocalization, establishment and selection of stable cell clones and Western blot analysis.
Figure S1. Expression and localization of recombinant KRAS proteins. In vitro transcription/translation of the empty vector (lane 1), KRASWT (lane 2), KRASG12V(lane 3) and KRASG12D (lane 4), recombinant chimera RED2:KRASWT (lane 5) and luciferase (control) expressing plasmids (lane 6) (A). Arrow heads point to the position of molecular mass standards whose sizes are expressed in kDa (A). Cells after 48 hours from transfection with CFP-KRASG12V construct (B-G). Immunolocalization by an anti-pan-Ras antibody (F, G) and an Alexa514-conjugated secondary antibody giving a green signal (C, D, F, G,), direct visualization of the cyan signal pseudocolored in red (B, D, E, G) and direct visualization of the signal displayed by the nuclear dye 7-Amino Actinomycin D pseudocolored in blue (D, G). The merged fluorescence signals are shown in (D, G). Magnification of the area in the white rectangles are shown in the lower left insets of (E-G). Arrow heads point to some of the cell membrane regions displaying colocalization of the pan-RAS and CFP-KRASG12V signals. Gamma adjustment was applied to each panel to adapt color rendering in the CMYK process. Scale bar = 50 mm.
Figure S2. Screening by semiquantitative RT-PCR analysis of Colo741 cell clones transfected with constructs expressing KRASWT (WT), KRASG12D (G12D) and KRASG12V (G12V) mRNA isoforms. For each construct both cell clones expressing the transgene (1D8-3C7-4D3) and cell clones not expressing the transgene (2G7-1H2-1B3) are shown. The latter were discarded from further experiments. For all the assayed samples, a reverse transcription PCR assays, performed omitting the reverse transcriptase, was also carried out to exclude from further analysis clones yelding a KRAS amplicon resulting from a plasmid integrated in the genomic DNA (false positive samples). In this example no amplicons were detected in minus reverse transcriptase assays. Primers for GAPDH were used to normalize the results.
Figure S3. Western blotting analysis of selected Colo741 KRAS-expressing clones. Vector-transfected clones (lane 1), KRASWT (lane 2), KRASG12V (lane 3), KRASG12D (lane 4) and senescent human bone marrow stromal cells (lane 5) were subjected to immunoblotting to establish the global Ras expression (pan-Ras) (A) and the phosphorylation status of AKT (B) and ERK1/2 (C). Arrow points to the pAKT protein.
Table S3. Sequences accession numbers and primers.