Abstract
The eukaryotic exosome is a ten subunit 3′ exoribonuclease complex responsible for many RNA processing and degradation reactions. How the exosome accomplishes this is unknown. We show that the PIN domain of Rrp44 is an endoribonuclease. The activity of the PIN domain prefers RNA with a 5′ phosphate, suggesting coordination of 5′ and 3′ processing. We also show that the endonuclease activity is important in vivo. Furthermore, the essential exosome subunit Csl4 does not contain any essential domains, but its zinc-ribbon domain is required for exosome-mediated mRNA decay. These results suggest that specific exosome domains contribute to specific functions, and that different RNAs interact with the exosome differently. The combination of an endoribonuclease and exoribonuclease activity appears to be a widespread feature of RNA degrading machines.
Virtually all RNAs are processed from longer precursors. Although the removal of 3′ extensions appears to be a simple enzymatic reaction, eukaryotic cells contain multiple endoribonucleases and 3′ exoribonucleases{Moser, 1997 #786; Mian, 1997 #787; Deutscher, 2000 #571; Zuo, 2001 #553}. These exoribonucleases can act redundantly for some reactions, but are very specific for other functions5. In addition to these RNA processing reactions that generate mature RNAs, 3′ exoribonucleases and endonucleases also carry out mRNA degradation reactions6,7 that can be an important determinant for mRNA quality control8,9. The same RNase can process some substrates and completely degrade others. It is largely unknown what determines the specificity of different RNases for their particular substrates, or for processing versus decay.
The exosome is a major 3′ exoribonuclease that is present in all eukaryotes, but whose function has been most extensively characterized in Saccharomyces cerevisiae (yeast)10,11. The yeast exosome is responsible for the 3′-end processing of stable RNAs including rRNA, snRNA, and snoRNA, and the degradation of RNAs such as the 5′ external transcribed spacer of the rRNA precursor and aberrant mRNAs that lack a stop codon or a poly(A) tail7-14.
The core exosome contains nine subunits and shows extensive structural similarity to bacterial PNPase and archaeal exosomes (Fig. 1A)15-20. These nine subunits are all essential for viability. Six subunits have an RNase PH domain, and form a ring structure in the exosome15. Although these six subunits show clear sequence and structural similarity to RNases, all six subunit in the yeast and human exosome lack important catalytic residues and are inactive21. The six PH-ring subunits can not assemble into a PH-ring in vitro, but when the three cap proteins are added, a stable core exosome can be assembled15. The crystal structure shows that each cap protein contacts two neighboring subunits of the PH-ring. These observations suggest that the cap proteins carry out an essential structural role by bridging interactions between PH domains (Fig. 4A)15. A second possible function of the cap proteins is to bind exosome substrates, since each contains three putative RNA binding domains11,20,22. Finally, a point mutation in one of the cap proteins (Csl4) disrupts the interaction between the core exosome and one of its cofactors8. The exosome requires many cofactors, and a third possible function for the cap proteins is to provide binding sites for these cofactors.
Figure 1.
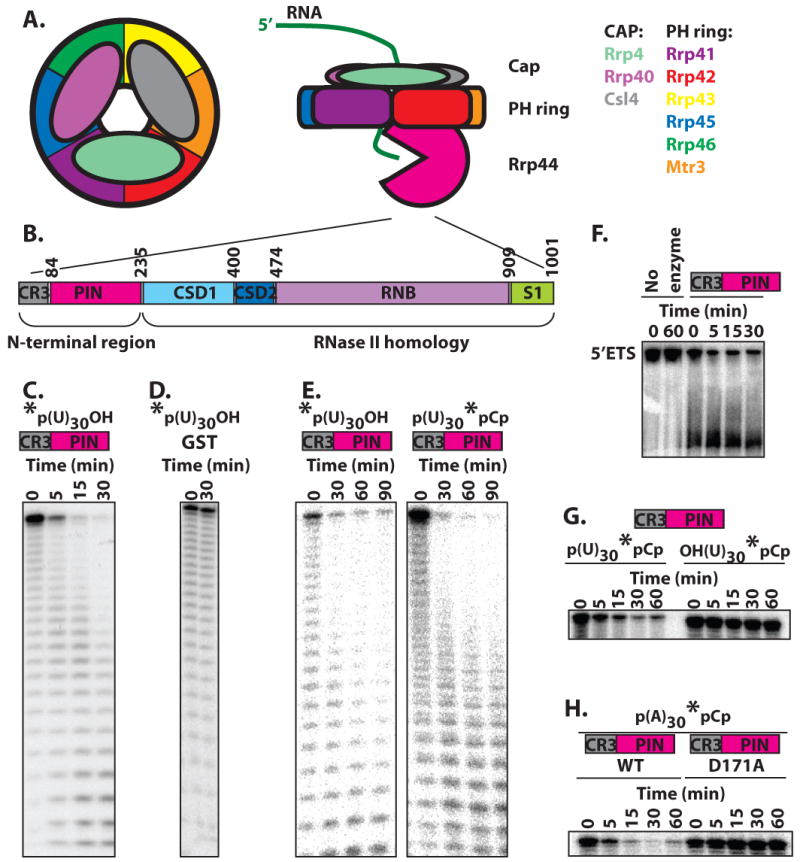
The N-terminal region of Rrp44 has ribonuclease activity. (a) The yeast exosome contains three layers. The exosome contains a PH-ring of 6 proteins with an RNase PH domain. Assembly and/or stability of the PH-ring requires three additional proteins that form a cap on one side of the ring. The exoribonuclease activity of the exosome is provided by a tenth subunit (Rrp44) that associates with the bottom of the ring. RNA substrates are thought to associate with the cap proteins, pass through the central channel in the PH-ring, and get degraded by Rrp44 after it emerges from the bottom of the ring. (b) Schematic representation of Rrp44 domains. (c) The N-terminal region of Rrp44, which contains a CR3 domain and a PIN domain, has ribonuclease activity in vitro. The N-terminal part of Rrp44 was purified as a GST-fusion recombinant protein from E. coli and incubated with 5′ end labeled U30 RNA. (d) GST by itself, purified and incubated under identical conditions does not have ribonuclease activity. (e) The N-terminal region of Rrp44 degrades both 5′ end labeled and 3′ end labeled substrates to smaller labeled oligoribonucleotides, suggesting it acts as an endonuclease. (f) The N-terminal region of Rrp44 degrades an internally labeled RNA corresponding to 5′ETS in vitro. (g) The N-terminal region of Rrp44 prefers substrates with a 5′ phosphate (left), over those with a 5′ hydroxyl (right). The apparent slight change in mobility in the right panel is an artifact of this particular gel, and does not reflect a slow degradation of the 5′ OH substrate. (h) Mutation the conserved acidic amino acid residue D171 to A in the N-terminal region of Rrp44 abolishes ribonuclease activity. The asterisk in panels C to G indicates the position of the P32 label.
Figure 4.
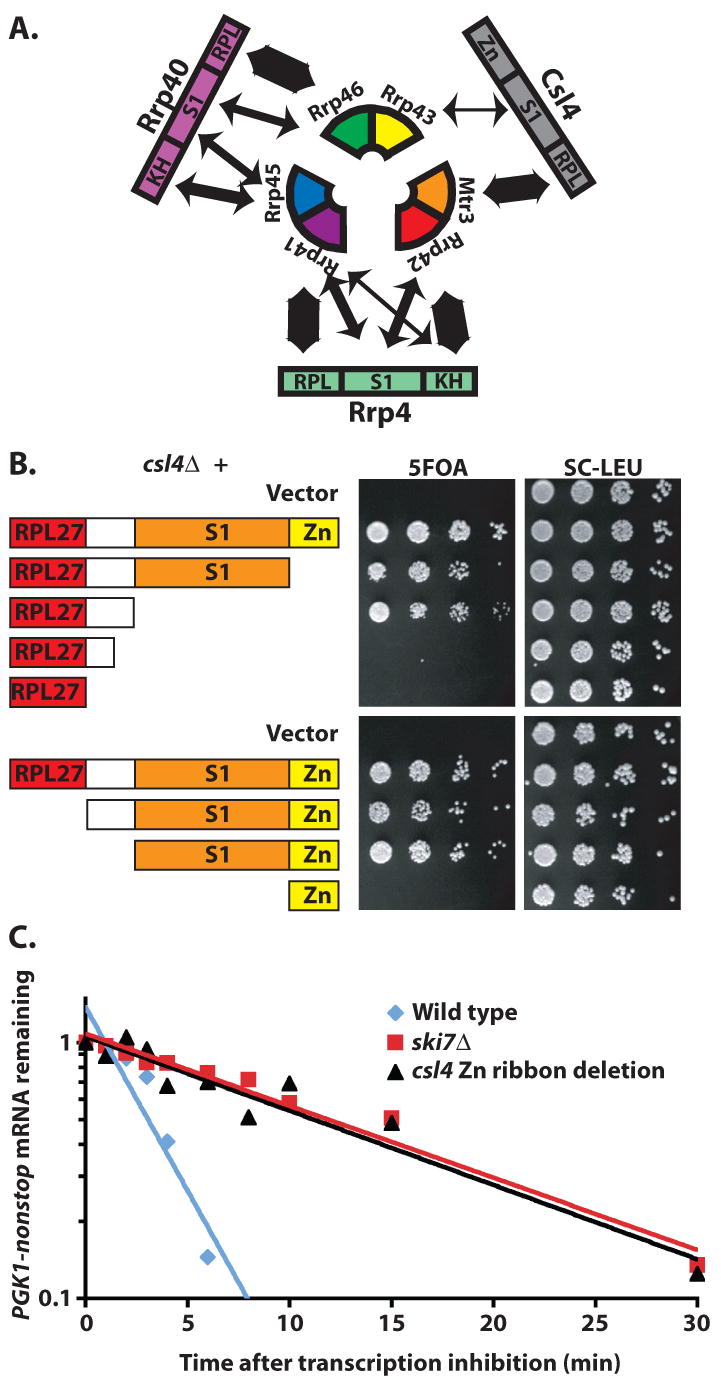
The essential Csl4 does not contain any essential domains, but its zinc-ribbon domain is required for cytoplasmic exosome-mediated mRNA decay. (a) Rrp4 and Rrp40 make extensive contacts with two neighboring subunits of the PH-ring. Csl4 makes extensive contacts with Mtr3p, but relatively small contacts with Rrp43p. The width of the double headed arrows is proportional to the extent of the buried surface calculated using Pymol. (b) The essential Csl4 does not contain any essential domains. Csl4 contains a RPL27-like domain (red), a 38 amino acid linker, not present in other eukaryotes (white), an S1 domain (Orange), and a zinc-ribbon like domain that is missing the zinc-coordinating amino acid residues (yellow). A csl4Δ strain complemented by full length CSL4 on a plasmid with a URA3 marker was transformed with LEU2 plasmids encoding the depicted truncations. Growth on 5FOA indicates that truncated Csl4 can carry out the essential function of the exosome. Similar results were obtained with a GAL∷csl4 strain (Supplemental Fig. 4 online). (c) The zinc-ribbon domain of Csl4 is required for exosome-mediated mRNA decay. The decay rate of PGK1pG-nonstop mRNA was measured in a wild-type strain (half life = 2 minutes), a strain lacking the zinc-ribbon domain of Csl4 (half life = 10 minutes), and a ski7Δ strain (half life = 10 minutes). Shown is the average value from two independent experiments. The conclusion that the zinc-ribbon domain of Csl4 is required for exosome-mediated mRNA decay was confirmed using two other approaches (Supplemental Fig. 7 online).
One important difference between the yeast exosome and PNPase and the archaeal exosome is that the yeast exosome contains a tenth essential subunit (Rrp44), which is homologous to bacterial RNase II (Fig. 1A+B), and is responsible for the 3′ exoribonuclease activity of the exosome21,23-26. Importantly, while Rrp44 is an essential gene, a point mutation that inactivates its exoribonuclease activity in vitro is viable21, suggesting that either the exoribonuclease activity may not be a central feature of the exosome, or the point mutant protein retains partial exoribonucleolytic activity in vivo.
The N-terminus of Rrp44 contains a CR3 domain and a PIN domain (Fig. 1B). The CR3 domain is a small domain of unknown function that contains three conserved cysteine residues. PIN domains were initially thought to have some regulatory or signaling function, but recent crystal structures of PIN domains revealed a putative active site with similarity to RNase H27,28. Indeed, some PIN domains have endoribonuclease activity, including the SMG6 protein, which is involved in nonsense-mediated decay27-32. PIN domains contain four acidic amino acid residues (Asp91, Glu120, Asp171 and Asp198 in Rrp44) and mutations in the third residue abolish activity of PIN domains in vitro and in vivo29,33,34.
In this paper we show that the Rrp44 PIN domain is an active endoribonuclease, revealing that the exosome does not only have exoribonuclease activity, but also endoribonuclease activity. Furthermore, we further characterize the functions of the cap proteins of the yeast exosome, and show that while Rrp4 and Rrp40 might have a largely structural role, Csl4 does not appear to have an important role in exosome structure, but instead appears to have specific domains that are required for specific functions.
Results
The exosome contains an endoribonuclease domain
To test whether the Rrp44 PIN domain contains endonuclease activity, we purified a truncated Rrp44 as a GST fusion from E. coli. This truncated Rrp44 contains the PIN domain, but lacks the RNB domain, and has robust nuclease activity in vitro on several RNA substrates (Fig. 1), while GST by itself does not (Fig. 1D). The activity level of the truncated Rrp44 was similar to the activity level of the full length protein (although under different conditions; Supplemental Fig. 1 online). Figure 1H shows that mutation of the third conserved acidic residue (Asp171) in the Rrp44 PIN domain abolished the endoribonuclease activity, as has been shown for some other PIN domains either in vitro or in vivo29,33,34. We conclude that the PIN domain of Rrp44 is a nuclease.
Other PIN domains and the related RNases H act as endonucleases. To test whether the Rrp44 PIN acts as an endonuclease we used 5′ or 3′ labeled substrates. An exonuclease will yield only labeled mononucleotides with one of these substrates. In contrast, the PIN domain yielded a very similar pattern of labeled oligonucleotides with each substrate showing that it is an endonuclease (Fig. 1E). Interestingly, the PIN domain was dramatically more active on substrates with a 5′ monophosphate than on substrates with a 5′ hydroxyl (Fig. 1G), which may have important implications (see discussion). As expected for an endonuclease, the PIN domain also had some activity on circular U30 (Supplemental Fig. 1B online). However, degradation of circular RNA was inefficient, probably because it lacks a free 5′ phosphate. All of these results indicate that the Rrp44 PIN domain is an active endoribonuclease, as has been shown for SMG629,32.
Either the endoribonuclease or exoribonuclease activity is sufficient for exosome function
To address whether this endonuclease activity is important for exosome function, we generated truncations and point mutants of Rrp44. C-terminal truncations that removed the RNase II homology, but left the CR3 and PIN domains, allowed for slow growth (Fig. 2A; Supplemental Fig. 2 online). Previously it was shown that a point mutation inactivating the catalytic activity of the RNB domain is viable21. The residual growth we observed upon deletion of the RNB domain rules out the possibility that the mutated RNB domain was viable due to some low level activity in vivo. Instead, our results show that exoribonuclease activity is not a critical feature of the exosome core. C-terminal truncations that also removed part of the PIN domain, or truncation from the N-terminus, did not support any growth. The smallest viable Rrp44 allele we generated includes the CR3 and PIN nuclease domains (amino acids 1-235). We conclude that the PIN domain is essential for exosome function, but we can not exclude the alternative that the PIN domain (also) functions separate from the exosome.
Figure 2.
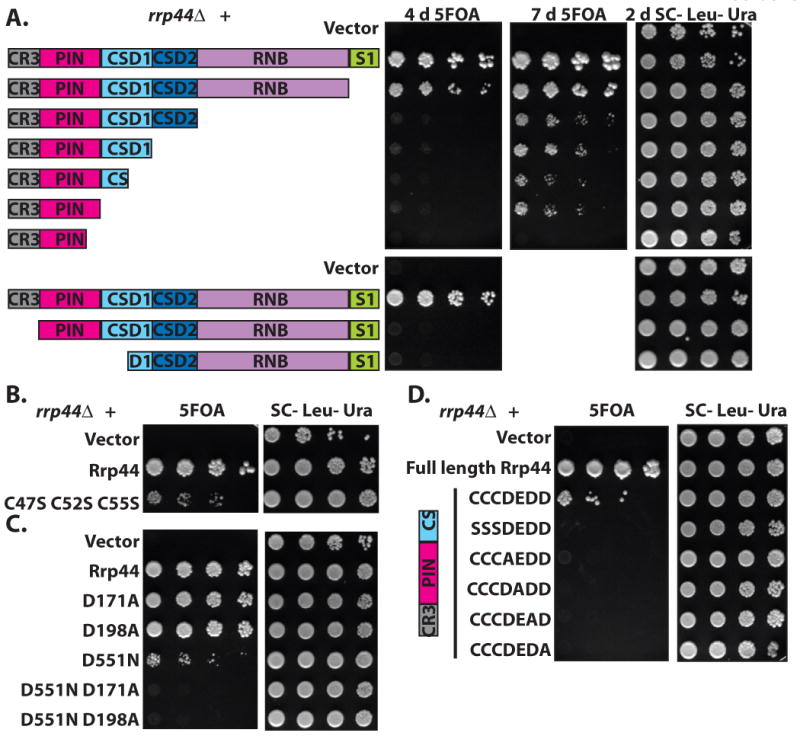
The PIN active site is important for exosome function. (a) The N-terminal region of Rrp44 is required and sufficient for viability. An rrp44Δ strain complemented by full length RRP44 on a plasmid with a URA3 marker was transformed with LEU2 plasmids encoding each of the depicted truncations. Growth on 5FOA indicates that truncated Rrp44 can carry out the essential function of the exosome. The smallest complementing Rrp44 allele includes amino acids 1-235. Similar results were obtained with a GAL∷rrp44 strain (Supplemental Fig. 2 online) (b) Mutation of the three conserved cysteine residues in the CR3 domain to serine reduces growth. (c) Mutations in the PIN domain active site residues of Rrp44 do not have a large affect on growth, but are lethal in combination with a mutation of the active site residue of the RNB domain. Similar results were obtained for the D91A and E120A mutations (Supplemental Fig. 3 online) (d) Mutations of the PIN domain active site residues are lethal in combination with a truncation that removes the RNB domain. CCDEDD indicates Cys47, Cys52, Cys55, Asp91, Glu120 Asp171, and Asp198, which where mutated in the last 5 rows as indicated.
To further address the importance of the CR3 and PIN domains, we made point mutants in conserved residues. Mutating the three conserved cysteines of the CR3 domain caused slow growth (Fig. 2B), thus both N-terminal truncations and point mutants indicate an important role for the CR3 domain. In contrast, the D171A point mutation that inactivates the PIN domain (or mutations in the other three conserved acidic residues) does not have an obvious growth phenotype (Fig. 2C; Supplemental Fig. 3 online). However, combining the viable PIN mutations with either the viable D551N or a viable C-terminal truncation resulted in no growth (Fig. 2C and D; Supplemental Fig. 3 online). These results suggest that either active site is sufficient for viability, but simultaneous inactivation of both catalytic sites results in a non-functional exosome.
The exosome carries out both RNA processing and RNA degradation reactions. Endonuclease activity might be well-suited for processing reactions, but exonuclease activity would be more appropriate for degradation reactions. We therefore tested the hypothesis that the PIN domain of Rrp44 is sufficient for exosome-mediated RNA processing reactions, while the RNB domain is required for exosome-mediated RNA decay. Figure 3A shows that truncations lacking the RNase II homologous domain accumulate the 5′ ETS and 7S precursors to 5.8S rRNA. Similarly, 7S processing and 5′ ETS degradation intermediates accumulated due to the D551N mutation in the RNB domain. We did not observe any similar degradation or processing intermediates due to the D171A mutation in the PIN domain, and the effect of the combination of D551N with D171A appeared similar to the D551N single mutant (Fig. 3B). Thus, both degradation and processing functions of the exosome require the RNB domain. An alternative explanation for the presence of two catalytic sites is that it increases the efficiency of the overall reaction. Under this hypothesis the endo- and exonuclease domains do not have separate substrates, but both may act together on most or all exosome substrates.
Figure 3.
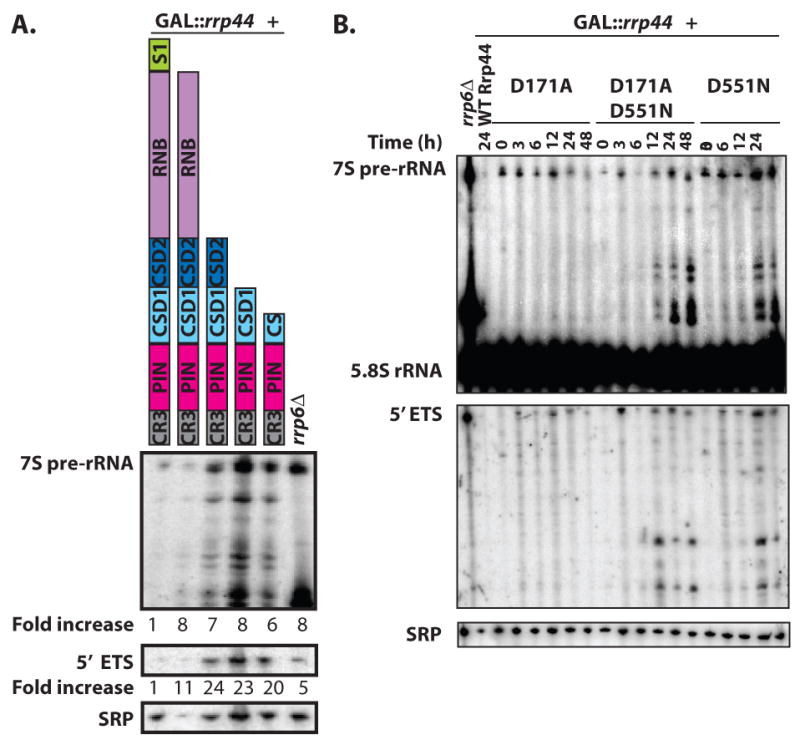
The RNB domain of Rrp44 is required for both RNA processing and RNA degradation activities of the exosome. (a) The indicated Rrp44 truncations were expressed in a GAL∷rrp44 yeast strain. RNA was isolated from cultures grown in dextrose (to inhibit expression of the GAL∷rrp44 gene), and analyzed by northern blotting with probes that hybridize to the 7S precursor of 5.8S rRNA (top panel), the 5′ ETS of the rRNA (middle panel), and the RNA subunit of the signal recognition particle (bottom panel). The relative levels of the 7S pre-rRNA and the 5′ETS were normalized for loading using the SRP signal and are indicated under the top and middle panels. Shown is a representative experiment. (b) RRP44 alleles containing the D171A, the D551N, or the double mutant D171A D551N where introduced into a GAL∷rrp44 yeast strain. Cultures where first grown in media containing galactose, and then incubated in media containing glucose for the indicated time (in hours), and analyzed by northern blotting with probes that hybridize to the 5.8S rRNA (top panel), the 5′ ETS of the rRNA (middle panel), and the RNA subunit of the signal recognition particle (bottom panel). Shown is a representative experiment.
The essential Csl4 does not contain an essential domain
To gain further insight into the structure-function relationship of the exosome, we next focused on the function of the three “cap” proteins. The Csl4 subunit contains an N-terminal domain which resembles RPL27, a central S1 domain, and a C-terminal domain that shows structural similarity to zinc ribbons, although the zinc-coordinating residues have been mutated. In addition, the yeast Csl4 contains a 38 amino acid linker that is absent in Csl4 from humans and most other eukaryotes. The first 22 amino acid residues of this inserted sequence include ten glutamate and two aspartate residues. To test which domains of Csl4 are essential, we made several truncations. Strikingly, each of the domains could be removed, and either the RPL27-like or S1+Zn-ribbon domains are sufficient for the essential function of the exosome (Fig. 4B; Supplemental Fig. 4 online). Thus, the essential Csl4 does not appear to have an essential domain.
The other two cap proteins, Rrp4 and Rrp40 each contain an N-terminal RPL27-like domain, a central S1 domain, and a C-terminal KH domain. In contrast to Csl4, eight truncations of Rrp4 or Rrp40 each failed to complement rrp4Δ and rrp40Δ (Supplemental Fig. 5 online). Thus, the observation that the essential Csl4 does not contain any essential domains is specific for Csl4. Furthermore, the function of Rrp4 or Rrp40 could not be provided by chimeric proteins where one domain of Rrp4 was replaced by the paralogous Rrp40 domain (or vice versa), or by co-expressing two parts of Rrp4 (Supplemental Fig. 6 online). Overall, these results are consistent with Rrp4 and Rrp40 providing important bridging contacts for the PH-ring15. However, Csl4 does not appear to share this function.
The Zinc-ribbon like domain of Csl4 is required for exosome-mediated mRNA decay
Although the zinc-ribbon domain of Csl4 is not required for the essential exosome function(s), three assays each indicate that it is required for cytoplasmic exosome-mediated mRNA decay. First, PGK1-nonstop mRNA is normally rapidly degraded, but is stabilized upon inactivation of the cytoplasmic exosome (e.g. ski7Δ). The Csl4 truncation that deletes the zinc ribbon domain results in a defect similar to ski7Δ (Fig. 4C). Second, the zinc ribbon is also required for the rapid degradation of the his3-nonstop mRNA (Supplemental Fig. 7 online). Third, a role of the exosome in general mRNA decay can be monitored through synthetic lethality with dcp1-2 mutation, and this assay also indicates that the zinc ribbon of Csl4 is required for exosome-mediated mRNA decay (Supplemental Fig. 7 online). Therefore, the zinc-ribbon domain of Csl4 is not required for exosome structure or the essential exosome function(s), but is required for cytoplasmic exosome-mediated mRNA decay.
Discussion
The exosome contains an endoribonuclease activity that is stimulated by a 5′ phosphate
We show for the first time that Rrp44 contains a previously overlooked endoribonuclease domain – the PIN domain. Two observations explain why it has previously been reported that Rrp44 with a mutation in the RNB domain does not have any nuclease activity in vitro21,24. First, those assays were carried out under conditions favorable for the activity of RNB domains (i.e. Tris buffer and Mg++ ions). Under similar conditions we also failed to detect significant activity (Supplemental Fig. 1 online). Second, we observed robust endonuclease activity on truncated proteins, but much less activity on full length protein or any truncations that did not remove CSD1 (data not shown). This latter observation suggests that CSD1 and perhaps other domains of the exosome regulate the activity of the PIN domain. Nevertheless, the activity level of the truncated Rrp44 under conditions where PIN domains are generally active (i.e. HEPES buffer and Mn++ ions), was similar to the activity level of the full length protein under conditions for the RNB domain (Supplemental Fig. 1 online). This suggests that although the activity was not previously detected, it is a robust activity that is likely important in vivo under some circumstances or on some substrates that remain to be determined.
Our observation that Rrp44 needs either its endonuclease or exonuclease activity suggests that the endonuclease is active and important in vivo. Furthermore, this functional redundancy appears to be a general theme for a wide variety of RNases and is similar to that found for Rrp44 and Rrp6 in yeast21, Xrn1 and the exosome in yeast7,35, several combinations of Rex1, Rex2, Rex3, and Rrp6 in yeast5, and several exoribonuclease combinations in E. coli{Andrade, 2008 #827; Deutscher, 2000 #571}.
In the process of testing 5′ or 3′ labeled substrates in in vitro degradation reactions, we noticed that substrates with a 5′ phosphate are more rapidly degraded than substrates with a 5′ hydroxyl or circular substrates. This is analogous to what has been described for the bacterial RNase E, which prefers a 5′ monophosphate containing substrate, over a circular RNA or a linear RNA with a 5′ hydroxyl or 5′ triphosphate37. Despite this functional similarity, we have not detected any sequence similarity between Rrp44 and RNase E. The 5′ end stimulated activity of Rrp44 might have several important consequences for the substrate specificity of the endonuclease activity. First, PIN domains are part of a large group of enzymes and ribozymes that use two metal ion catalysis27-29,31. All of these enzymes generate a 5′ cleavage product with a 3′ hydroxyl, and a 3′ cleavage product with a 5′ monophosphate (reviewed in38). This cleavage mechanism suggests that the endonuclease activity of Rrp44 might cleave most primary transcripts relatively inefficient, but the 3′ cleavage product of an initial cleavage will have a 5′ phosphate and be a preferred substrate. Such preferred subsequent cleavage might ensure that RNA degradation is rapidly completed once it has been initiated. Second, many RNA species are processed both at the 5′ and 3′ end, and in most cases it is not clear how these events are coordinated. One possibility offered by our results is that 5′ end processing generates a 5′ monophosphate, which stimulates 3′ processing by the Rrp44 endonuclease. Testing these hypotheses in detail will require the identification of all, or most, of the endogenous substrates on the endonuclease activity of Rrp44.
A specific domain of the exosome core is required for specific exosome functions
The exosome contains ten different essential subunits containing 21 domains. How this structural complexity correlates with the multiple functions of the exosome is not clear. To begin to address these questions, we generated many deletions of domains, both in Rrp44 and in the three cap proteins. The cap proteins have been suggested to be important for exosome structure, by simultaneously interacting with two different PH-ring subunits. The finding that the RPL27-like domain and S1 domain of Csl4 are not both essential is inconsistent with Csl4 providing important bridging interactions between subunits of the PH-ring, since deletion of the S1 and Zn-ribbon domains results in Csl4 only interacting with Mtr3p (Fig. 4A). Conversely, deletion of the RPL27-like domain results in Csl4 only interacting with Rrp43p.
A high throughput screen previously identified transposon insertions in the essential CSL4 gene39. Surprisingly, these insertions are viable, which is explained by our observation that a severely truncated Csl4 protein is still functional. Similarly, T-DNA insertions into the Arabidopsis CSL4 gene are viable40. This observation was interpreted to mean that Csl4 is not an essential subunit of the Arabidopsis exosome, but our results suggest the alternative explanation that the T-DNA mutants produce a truncated Csl4 that retains some function.
A corollary of the conclusion that Csl4 is not important for exosome structure is that it must be functionally important. One possible model is that Csl4 provides important contacts with either an essential substrate RNA or an essential cofactor. If such an RNA or protein molecule makes contacts with both the RPL27-like domain and the S1 or Zn-ribbon domains, it might still be able to interact if either binding site is deleted, but not if both are deleted. Interestingly, some surface exposed conserved amino acid residues of the Csl4 Zn-ribbon domain that are not essential, become essential if the RPL27-like domain is deleted (Supplemental Fig. 4 online). This is consistent with these residues forming one binding site and the RPL27-like domain containing a second binding site for an exosome cofactor. Csl4 is also thought to provide the binding sites for Ski78, thus Csl4, and perhaps the other cap proteins, may provide important contacts for exosome cofactors, and thus be important for specific exosome functions.
The exosome core may have multiple ways to interact with RNA substrates
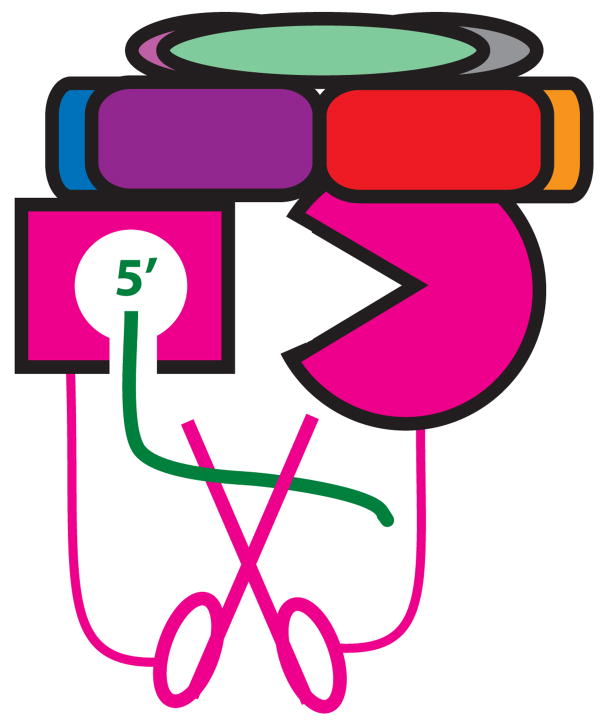
It is not clear how the exosome interacts with RNA substrates. A crystal structure of the archaeal exosome in complex with RNA19 suggests that RNA might go through the center of the PH ring as depicted in figure 1A. Our conclusion that Csl4 provides important contacts for interaction with cytoplasmic cofactors appears most consistent with the suggestion that cytoplasmic mRNA follows this route. On the other hand, it has been proposed that RNA may gain access to Rrp44 without going through the PH-ring25. The observation that the endonuclease activity is stimulated by a 5′ monophosphate indicates that Rrp44 can interact with the 5′ end of an exosome substrate. Since the 3′ exoribonuclease active site must interact with the 3′ end, Rrp44 appears to be able to interact with both ends of substrate RNAs. This is unlikely to be compatible with threading the RNA through the PH-ring. Thus our observations appear more consistent with the idea that the exosome can interact with RNA substrates in two different ways (compare fig. 1A to fig. 5). Moreover, we suggest that substrates that access Rrp44 directly might be identifiable by looking for substrates that are processed in vivo in a 5′ end dependent manner.
Figure 5.
A model of how RNA might access the 5′ end stimulated endonuclease activity of Rrp44
An endoribonuclease and exoribonuclease are combined in mRNA degrading machines throughout nature
We show for the first time that the exosome contains both an endonuclease and exonuclease activity. While this is unexpected for the exosome, it is not unique in nature. It has been suggested that the archaeal exosome also contains a PIN domain41, which could provide the archaeal exosome with an endonuclease activity. Similarly, the E. coli degradosome combines the RNase E endoribonuclease with the PNPase 3′ exoribonuclease42,43, and metazoan P-bodies combine the argonaute endoribonuclease with the Xrn1 5′ exoribonuclease, and the Ccr4, Caf1, and Pan2 3′ exoribonucleases44-49. Thus, combining endoribonucleases and exoribonucleases into one RNA degrading machine is widespread in nature, which suggests that this combination offers a fundamental advantage to the cell.
Materials and methods
Strains
The strains, plasmid and oligonucleotides we used are described in detail in supplementary material (online). The heterozygous diploids RRP44/rrp44Δ, CSL4/csl4Δ, RRP4/rrp4Δ and RRP40/rrp40Δ have been described and were obtained from Open Biosystems50. We transformed each of these strains with a URA3 plasmid that encoded the corresponding wild-type exosome subunit, We sporulated the transformants and obtained haploid progeny spores by the hydrophobic spore isolation method essentially as described51. Isolated spores were germinated to generate the deletion strains complemented with the URA3 plasmid. The GAL∷rrp44, GAL∷csl4, GAL∷rrp4 and GAL∷rrp40 strains have been previously described and were obtained from Phil Mitchell and David Tollervey10,11.
Plasmids
All yeast plasmids are low copy plasmids that contain a yeast centromere and the endogenous promoter and 3′ flanking region to control expression of the exosome subunits and were verified by sequencing.
To generate C-terminal truncations, we initially PCR amplified several hundred base pairs of the 3′ flanking region of the gene of interest and cloned it into pRS41552, which contains a LEU2 marker. We then PCR amplified the promoter and desired part of the coding region, and cloned it into the plasmid described above. N-terminal truncations were generated similarly, except that the promoter was PCR amplified and cloned first, and the truncated coding region and 3′ flanking region were added in the second step.
We generated GST fusion plasmids by PCR amplifying the RRP44 coding region from the plasmids described above, digesting the PCR product with EcoRI and XhoI, and ligating into pGEX-4T-1 (GE Healthcare).
We created site-directed mutants using the QuikChange II kit (Stratagene). Each oligonucleotide also changes a restriction site. We screened putative mutants by restriction digestion and verified them by sequencing. To combine point mutations in the PIN domain of Rrp44 with the D551N mutant in the RNB domain, we digested the plasmids with each single mutant with SacI and PshAI and ligated them together. PshAI cuts at a unique site within the CSD2 domain.
Overexpression and Purification of recombinant Rrp44
Full length and truncated Rrp44 fused to GST were expressed in the Rosetta(DE3) E. coli strain. We grew cultures at 30°C in 2 liter TB medium supplemented with 200 μg/ml ampicillin and 34 μg/ml chloramphenicol to an OD600 of 1.2 and then induced expression by addition of 0.2 mM IPTG and incubation at 18°C overnight. We harvested cells by centrifugation and froze them. Cells were thawed on ice, resuspended in 30 ml PBS buffer pH 8 with 10 mM DTT and 1 mM PMSF, and lysed using the French press. We treated the crude extracts with 5 units Benzonase (Sigma) for 30 min at 0°C, clarified the extract by 30 min centrifugation at 10,000 × g, and loaded it onto a GSTrap™ FF 1 ml column (GE healthcare) equilibrated in binding buffer. We eluted proteins with a 0-10 mM glutathione gradient in 50 mM Tris-HCl pH8.5, 10 mM DTT. We pooled fractions containing the purified protein and loaded them onto a gel filtration column (Superdex 75 10/300 GL or a Superose 12 10/300 GL - GE Healthcare, depending on the protein size) in 20 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2 and 10 mM DTT buffer at pH8. We concentrated the eluted protein by centrifugation at 15 °C with Vivaspin 500 Centrifugal Concentrators (Vivaspin). We determined protein concentrations by spectrophotometry using a Nanodrop and added 50 % (v/v) glycerol to the final fractions prior storage at -20 °C.
RNase assays
RNase assays contained 100-500 nM protein, 200 nM 5′ or 3′ end labeled U30 or A30 RNA or an internally labeled 5′ETS RNA transcribed by T7 RNA polymerase, and 20 mM HEPES pH 7.5, 150 mM NaCl, 3 mM MnCl2, 1 mM DTT.
Growth assays
We performed growth assays essentially as previously described9,53. Briefly, we grew strains in appropriate liquid selection media overnight at 30°C (or 23°C for temperature-sensitive mutants). We diluted these cultures in appropriate selection media to a starting OD600 of 0.2. We grew the cultures until they reached an OD of 0.8, serially diluted them in 96-well plates by a factor of five, and spotted them onto appropriate media. We generally incubated these plates for 2-3 days at 23°C, 30°C, or 37°C. To monitor the slow growth of Rrp44 truncations, we incubated the plates for up to 20 days.
Northern blotting
Stability of PGK1-nonstop mRNA8, and 5.8S rRNA processing and 5′ETS degradation54 were assayed as previously described.
Supplementary Material
Acknowledgments
We are grateful to Todd Link and Richard Brennan for help with calculating the buried surfaces between the different domains of the cap proteins and the PH-ring and Ale Klauer for technical assistance. GAL∷rrp44, GAL∷csl4, GAL∷rrp4, and GAL∷rrp40 strains were kindly provided by Phil Mitchell (University of Sheffield) and David Tollervey (University of Edinburgh). Roy Parker, Miles Wilkinson, Michelle Steiger and members of the van Hoof and Arraiano labs gave insightful comments on the manuscript. This research was supported by the Pew Scholarship Program in the Biomedical Sciences and by the National Institutes of Health (GM069900) to A.v.H.. The University of Texas at Houston Medical School-Summer Research Program supported E.G.D and M.S.-R. The work at the ITQB was supported by Fundação para a Ciência e a Tecnologia (FCT), Portugal. A.B. was a recipient of a post-doctoral fellowship from FCT, Portugal.
References
- 1.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–8. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–95. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deutscher MP, Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- 4.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–26. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–65. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–56. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs Anderson JS, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 9.Meaux S, Van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–37. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 11.Allmang C, et al. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–58. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allmang C, et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–52. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–40. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 16.Lorentzen E, et al. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–81. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 17.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–26. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 18.Navarro MV, Oliveira CC, Zanchin NI, Guimaraes BG. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J Biol Chem. 2008;283:14120–31. doi: 10.1074/jbc.M801005200. [DOI] [PubMed] [Google Scholar]
- 19.Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell. 2005;20:473–81. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–71. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 22.Chekanova JA, Dutko JA, Mian IS, Belostotsky DA. Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3′-->5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res. 2002;30:695–700. doi: 10.1093/nar/30.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–28. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–31. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang HW, et al. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci U S A. 2007;104:16844–9. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazao C, et al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–4. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 27.Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem. 2004;279:16471–8. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 28.Levin I, et al. Crystal structure of a PIN (PilT N-terminus) domain (AF0591) from Archaeoglobus fulgidus at 1.90 A resolution. Proteins. 2004;56:404–8. doi: 10.1002/prot.20090. [DOI] [PubMed] [Google Scholar]
- 29.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–25. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daines DA, Wu MH, Yuan SY. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol. 2007;189:5041–8. doi: 10.1128/JB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunker RD, McKenzie JL, Baker EN, Arcus VL. Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon. Proteins. 2008;72:510–8. doi: 10.1002/prot.22048. [DOI] [PubMed] [Google Scholar]
- 32.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promoted endonucleoytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1530. in press. [DOI] [PubMed] [Google Scholar]
- 33.Fatica A, Tollervey D, Dlakic M. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004;10:1698–701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci U S A. 2006;103:9464–9. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AW, Kolodner RD. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–27. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3′ to 5′ exonucleases in RNA degradation. Prog Nucleic Acid Res Mol Biol. 2008 doi: 10.1016/S0079-6603(08)00805-2. in press. [DOI] [PubMed] [Google Scholar]
- 37.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–3. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 38.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat Struct Mol Biol. 2008;15:1228–31. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross-Macdonald P, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–8. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 40.Chekanova JA, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–53. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–52. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 43.Py B, Causton H, Mudd EA, Higgins CF. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994;14:717–29. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–501. [PMC free article] [PubMed] [Google Scholar]
- 46.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–73. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng D, et al. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrei MA, et al. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–27. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 51.Rockmill B, Lambie EJ, Roeder GS. Spore enrichment. Methods Enzymol. 1991;194:146–9. doi: 10.1016/0076-6879(91)94012-2. [DOI] [PubMed] [Google Scholar]
- 52.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177:773–84. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol. 2000;20:8230–43. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.