Abstract
The pathogenic bacterium Shigella flexneri uses a type III secretion system to inject virulence factors from the bacterial cytosol directly into host cells. The machinery that identifies secretion substrates and controls the export of extracellular components and effector proteins consists of several inner-membrane and cytoplasmic proteins. One of the inner membrane components, Spa40, belongs to a family of proteins proposed to regulate the switching of substrate specificity of the export apparatus. We show that Spa40 is cleaved within the strictly conserved amino acid sequence NPTH and substitution of the proposed autocatalytic residue abolishes cleavage. Here we also report the crystal structure of the cytoplasmic complex Spa40C and compare it with the recent structures of the homologues from Escherichia coli and Salmonella typhimurium. These structures reveal the tight association of the cleaved fragments and show that the conserved NPTH sequence lies on a loop which, when cleaved, swings away from the catalytic N257 residue, resulting in different surface features in this region. This structural rearrangement suggests a mechanism by which non-cleaving forms of these proteins interfere with correct substrate switching of the apparatus.
Introduction
Shigella flexneri is the causative agent of bacillary dysentery in humans and is responsible for over a million deaths worldwide annually (Kotloff et al., 1999). Invasion of the colonic epithelium is dependent on a type III secretion system (T3SS) (Cossart and Sansonetti, 2004). T3SSs are found in many Gram-negative bacterial pathogens and serve as molecular injection devices to deliver bacterial virulence effector proteins into eukaryotic host target cells (Cornelis, 2006; Galan and Wolf-Watz, 2006; Blocker et al., 2008). The Shigella T3SS is encoded on a 31 kb fragment of a large virulence plasmid and consists of three major structural parts: a cytoplasmic region known as the ‘bulb’, a region spanning both bacterial membranes termed the basal body and an extracellular ‘needle’ (Blocker et al., 1999; Deane et al., 2006). The export apparatus comprises cytoplasmic and inner-membrane proteins that identify T3SS substrates and control the switching of substrate specificity during morphogenesis and upon host-cell contact (reviewed in Cornelis, 2006; Galan and Wolf-Watz, 2006). Following assembly of the basal body, early substrates are targeted to the apparatus, including the protein that polymerizes to form the hollow, extracellular needle. Upon completion of the needle, middle substrates (also known as translocators) are secreted that assemble at the tip of the needle and are involved in host-cell recognition and pore formation in the host-cell membrane. Following an activation signal, late substrates (also known as effectors) are targeted to the apparatus and translocated directly into host cells.
The S. flexneri protein Spa40 has been identified as a component of the basal body (Zenk et al., 2007) and is predicted to have a large, N-terminal transmembrane domain (Spa40TM) and a C-terminal cytoplasmic domain (Spa40C). It shares sequence homology and predicts membrane topology with YscU, an essential component of the secretion apparatus in Yersinia species (Allaoui et al., 1994). The cytoplasmic domain of YscU undergoes cleavage at a highly conserved NPTH sequence (Lavander et al., 2002) in a process that is suggested to be involved in the regulation of substrate specificity of the Yersinia T3SS (Edqvist et al., 2003). Substitution of the conserved Asn or Pro residues to Ala prevents cleavage of YscU (Lavander et al., 2002) and abolishes the export of translocators without affecting the export of effectors (Sorg et al., 2007). It is thought that cleavage of YscU results in a conformational change that allows the recognition of translocators, thus playing a role in determining the export hierarchy (Sorg et al., 2007).
T3SSs are genetically, morphologically and structurally related to the basal bodies of bacterial flagella (Blocker et al., 2003). The flagellar homologue of YscU, FlhB, plays a role in substrate specificity switching during flagella assembly (Hirano et al., 1994; Kutsukake et al., 1994; Williams et al., 1996; Fraser et al., 2003). The controlled switch between export states is believed to be mediated by conformational changes in the structure of the C-terminal cytoplasmic domain of FlhB. As is the case for YscU, this domain is consistently and specifically cleaved at the highly conserved NPTH sequence into two subdomains that remain tightly associated with each other (Minamino and Macnab, 2000a). The mechanism of FlhB cleavage has been analysed by Ferris et al. (2005) who provide evidence that the tertiary structure of FlhB plays a significant role in cleavage and propose that FlhB cleavage is an autocatalytic process.
Recently, the structures of the cleaved and non-cleavable forms of the cytoplasmic domains of the homologous proteins Escherichia coli EscU and Salmonella typhimurium SpaS were determined (Zarivach et al., 2008). These structures confirmed the autocatalytic mechanism proposed by Ferris et al. (2005) and provided the first structural insight into the subtle conformational change arising from the autocleavage event. Here we report the crystal structure of the S. flexneri Spa40C complex and compare it with EscUC and SpaSC. These structures highlight the tight association of the cleaved cytoplasmic subdomains and reveal that the conformational change upon cleavage involves only the movement of the PTH loop. The surface exposure of this loop and its orientation in the cleaved form suggest that it is the surface features in this region, rather than the cleavage event itself, that are important for binding partner proteins. Comparison of Spa40C with the recent structures of EscUC and SpaSC as well as mapping of important functional mutations from several species provides insight into the potential roles of Spa40 in regulation of substrate secretion.
Results and discussion
Whole-cell lysate from overproduction of the cytoplasmic domain of Spa40 (Spa40C) revealed three major bands (Fig. 1A, lane 1). Purification of the soluble fraction revealed that the two lower-molecular-weight bands co-elute upon purification, the third band being insoluble (Fig. 1A, lane 2). N-terminal sequencing of the purified complex revealed that the lower band corresponds to the N-terminal portion of Spa40C starting at residue D207. The sequence for the upper band begins at P258 within the conserved NPTH sequence. Mass spectrometry data for the purified complex (6382 ± 2 and 10872 ± 2 Da) corresponded to the expected masses (6382 and 10871 Da) following cleavage of Spa40C on the N-terminal side of Pro258 and confirmed that there was no modification to the newly formed termini, consistent with the cleavage mechanism proposed by Ferris et al. (2005) and Zarivach et al. (2008). These data confirm the identification of the lower and upper bands as Spa40CN (residues 207–257) and Spa40CC (residues 258–342) respectively, and establishes that Spa40C is cleaved at the highly conserved NPTH sequence. As the C-terminal His-tag is only present on Spa40CC, co-elution from the Ni-NTA and size-exclusion columns established that Spa40CN and Spa40CC form a stable complex following cleavage. Uncleaved Spa40C is not present in the soluble fraction but is found entirely in inclusion bodies. Purification of Spa40C under denaturing conditions revealed that Spa40CN did not co-elute with the Spa40CC domain, identifying that the folded state of these proteins is required for their association (Fig. 1A, lane 3).
Fig. 1.
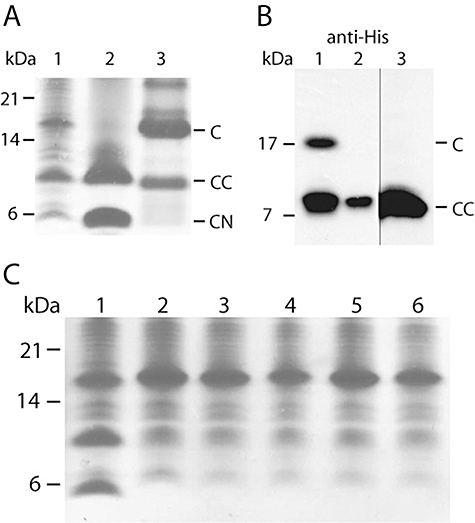
Cleavage and self-association of Spa40.
A. Coomassie-stained SDS-PAGE of Spa40C whole-cell lysate (lane 1), after native purification by affinity and size-exclusion chromatography (lane 2) and after purification under denaturing conditions (lane 3).
B. Western blot using an antibody against the His6 tag of whole-cell lysate from overproduction of Spa40C (lane 1) and Spa40FL (lane 2). Western blot of the membrane fraction from overproduction of Spa40FL (lane 3).
C. Coomassie-stained SDS-PAGE of whole-cell lysate from overproduction of wild-type sequence Spa40C (lane 1) and mutant forms of Spa40C: Asn→Ala (lane 2), Asn→Leu (lane 3), Asn→His (lane 4), Asn→Asp (lane 5), Asn→Gln (lane 6).
Western blot analysis of whole-cell E. coli lysate following overproduction of full-length Spa40 (Spa40FL) revealed a band corresponding to the same mass as the cleaved Spa40CC domain identified in Spa40C preparations (Fig. 1B, lanes 1 and 2). Following cell fractionation, this band was found to be enriched in the membrane fraction (Fig. 1B, lane 3), suggesting that the observed cleavage is maintained in the context of the full-length, membrane-localized protein. This result, using heterologous overproduction of Spa40FL in E. coli, is in agreement with the observation that Spa40 isolated from preparations of the S. flexneri intact basal body has a molecular weight of approximately 33 kDa, representing the Spa40TM plus Spa40CN domains following cleavage of Spa40CC (Zenk et al., 2007).
Cleavage at the conserved NPTH site is abolished by replacement of the Asn with Ala in the Yersinia and flagellar homologues of Spa40 (Lavander et al., 2002; Ferris et al., 2005). This substitution was generated in Shigella Spa40C by site-directed mutagenesis and analysis of whole-cell E. coli lysate revealed that the lower bands representing the cleaved products are not present (Fig. 1C). Affinity purification, under native conditions, of Spa40C(N257A) followed by N-terminal sequencing identified only the uncleaved Spa40C (data not shown). Additional non-conservative substitutions with Leu or His and conservative substitutions to Asp or Gln also resulted in the production of only uncleaved Spa40C (Fig. 1C). The absence of cleaved product for these mutants confirms the essential role of the Asn side-chain in the cleavage reaction.
Crystals were grown of the purified Spa40C complex. Data sets were collected for two different P1 crystal forms, both containing one protein molecule in the asymmetric unit (Table 1). The structure of Spa40C was solved by molecular replacement and refined against the higher-resolution data. The model comprises residues 237–257 of Spa40CN (residues 207–236 were present in the construct but not visible in electron density) and 258–338 of Spa40CC (the last four residues, 339–342, were not visible in electron density). The structure was refined using data to 2 Å with residuals Rwork and Rfree of 17.8% and 22.7% respectively (Table 1).
Table 1.
Data collection and refinement statistics (values in parentheses are for the highest-resolution shell).
| Crystal form 1 | Crystal form 2 | |
|---|---|---|
| Data collection | ||
| X-ray source | ESRF ID23-2 | ESRF ID23-1 |
| Detector | MarMosaic 225 CCD | ADSC 315 CCD |
| Wavelength (Å) | 0.873 | 1.040 |
| Space group | P1 | P1 |
| Unit-cell dimensions | ||
| a (Å) | 24.6 | 25.0 |
| b (Å) | 30.5 | 30.8 |
| c (Å) | 32.0 | 32.1 |
| α (°) | 103.6 | 102.5 |
| β (°) | 110.1 | 111.0 |
| γ (°) | 104.8 | 94.3 |
| Resolution limits (Å) | 27–2.25 (2.37–2.25) | 30–2.0 (2.11–2.00) |
| Measured reflections | 6347 | 14093 |
| Unique reflections | 3481 | 5433 |
| Completeness (%) | 91.1 (85.1) | 93.2 (93.3) |
| Multiplicity | 1.8 (1.7) | 2.6 (2.7) |
| Rmerge | 0.098 (0.31) | 0.109 (0.399) |
| Rpim | 0.098 (0.310) | 0.084 (0.307) |
| Average I/σ(I) | 5.4 (2.2) | 3.8 (1.7) |
| Wilson B-value (Å2) | 25.7 | 21.6 |
| Refinement | ||
| Resolution range (Å) | 30–2.0 (2.05–2.00) | |
| No. working set refl.s | 5181 (414) | |
| No. free set refl.s | 252 (20) | |
| R | 0.178 (0.267) | |
| Rfree | 0.227 (0.292) | |
| Number of atoms (protein/water) | 806/29 | |
| Number of atoms with alternate conformations (protein/water) | 3/0 | |
| r.m.s.d. bond length (Å) | 0.010 | |
| r.m.s.d. bond angle (°) | 1.105 | |
| Mean B factor (protein/water; Å2) | 23.3/30.6 | |
| Ramachandran plot, residues in | ||
| Favoured regions (%) | 98.0 | |
| Allowed regions (%) | 2.0 | |
The structure of Spa40C consists of a central β-sheet surrounded by four α-helices (Fig. 2A). The structured region of Spa40CN consists of a single α-helix (α1) followed by a single β-strand (β1) that lies at the centre of the structure and forms one strand of the five-stranded β-sheet. The structure of Spa40CC starts at P258 and consists of four β-strands and three α-helices that wrap around β1 of Spa40CN. The NPTH sequence lies on a loop between β1 and β2, the PTH region having flipped away from N257, revealing that the cleavage of the N257-P258 peptide bond results in a conformational change to this loop region (Fig. 2B). P258 moves ∼10 Å from the catalytic asparagine residue and both P258 and T259 are less well-ordered than the surrounding residues, as indicated by higher isotropic temperature factors. The overall structure of Spa40C and the movement of the PTH loop are very similar to that seen in the cleaved structures of the homologues from E. coli, EscU and S. typhimurium, SpaS (possessing ∼30% sequence identity, 1.26 ± 0.10 and 1.00 ± 0.09 Å r.m.s.d. over 92 Cα residues respectively) (Zarivach et al., 2008). The autocleavage mechanism requires a specific conformation of the NPTH loop, this conformation being stabilized by the correctly folded Spa40C domain.
Fig. 2.
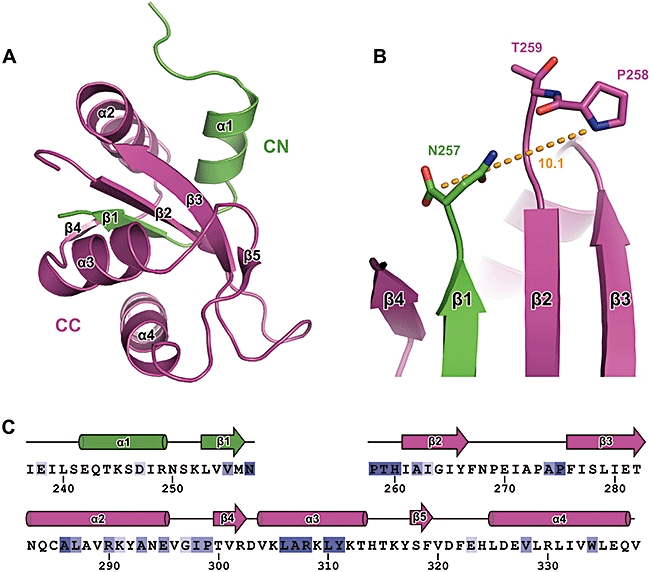
The structure of Spa40C.
A. Ribbon diagram of Spa40C showing Spa40CN (green) and Spa40CC (purple) with α-helices and β-strands labelled.
B. Illustration of the flip of the cleaved PTH loop away from the catalytic N257. The distance between the carbonyl carbon atom of N257 and the amine nitrogen atom of P258 is shown in Å (orange).
C. Sequence of Spa40C coloured by conservation across species (dark blue being most conserved) using the multiple sequence alignment from Zarivach et al. (2008). The positions and labelling of α-helices (cylinders) and β-strands (arrows) are illustrated above the sequence.
The striking feature of these structures is that Spa40CN forms part of the core of the fold with the conserved hydrophobic residues L253 to M256 (LVVM) completely buried by the surrounding Spa40CC structure. For this reason, the cleavage at the NPTH site does not result in a large rearrangement of the domains but is restricted to a conformational change of the PTH loop only. Although cleavage does not result in a major rearrangement of the Spa40CN and Spa40CC domains with respect to each other, the structural changes in the NPTH loop region significantly alter this face of the molecule. Upon cleavage, the creation of charged terminal residues alone alters the properties of Spa40. However, the surface exposure of the NPTH loop and the movement of the cleaved PTH loop away from N257 result in several changes to the surface of Spa40C (Fig. 3A). In particular, the side-chains of P258, T259 and H260 are exposed on the surface in an orientation that would not be possible in the uncleaved protein. The movement of the loop also exposes underlying residues that now form part of the new surface. Mutations that abolish cleavage will hold the PTH loop in a conformation that will bury these residues and thus alter the surface properties of Spa40C. Indeed, Zarivach et al. (2008) showed that the structure of the non-cleaving mutants of EscU can fold correctly and the only structural difference in these mutants is the orientation of the PTH loop. For these reasons, it is likely that it is not the act of cleaving the N–P bond but the result of that cleavage, which is the change in surface exposure of specific residues, that is critical. Therefore, the altered function of the non-cleaving forms of these proteins is probably the result of the loss of binding or recognition of other T3SS components.
Fig. 3.
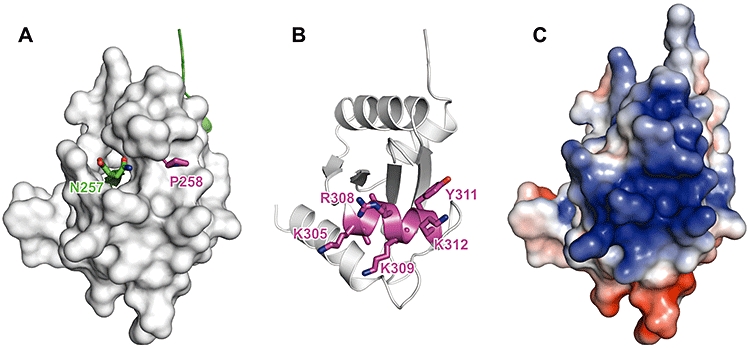
Surface features of Spa40C.
A. The surface of Spa40CC (white) and a ribbon representation of Spa40CN (green) are shown. The side-chain atoms of N257 and P258 are shown as sticks.
B. Ribbon diagram of Spa40C in the same orientation as for A showing helix α3 (purple) and the side-chain atoms of residues K305–K312.
C. Electrostatic surface of Spa40 showing the highly conserved positive patch (blue) surrounding the NPTH region. For calculation of electrostatics of those residues where the side-chain orientation could not be assigned because of a lack of electron density (R302, K305, F322, E323, Q337 and V338), the side-chains were modelled in the highest probability rotamer conformation that avoided clashes. Electrostatic surfaces were calculated using apbs (Baker et al., 2001) and are displayed on the molecular surface (plotted at ± 3 kT e−1).
The region from residue L306 to Y311 is highly conserved across all species and with the homologue from flagella, FlhB, possessing the consensus sequence LARXLY. This can be mapped to helix α3 which lies directly below the NPTH loop region (Fig. 3B). The hydrophobic residues in this region are buried and likely to be important for the correct positioning of the helix. In Spa40, the face of this helix that is exposed on the surface is composed of residues K305, R308, K309 and K312, resulting in a highly positively charged patch (Fig. 3C). The high conservation of residues involved in the positioning of this helix and the proximity of this charged patch to the NPTH loop suggest that it may play a role in the recruitment or binding of T3SS components. Although several of these charged residues are conserved in closely related species, such as Salmonella, only R308 is conserved in all Spa40 homologues and in FlhB. This may indicate that as species diverged, the core fold was conserved, but the surface features, and therefore the interaction faces, evolved to satisfy other requirements, such as specificity between T3SS and flagellar substrates in those species that possess both.
In order to ensure correct assembly of the T3SS, the substrate specificity of the secretion apparatus must be switched upon completion of the needle, or hook in flagella. It is well established for both flagella and the Yersinia T3SS that this switch is regulated by two proteins. The first protein belongs to the Spa40 family (YscU in Yersinia and FlhB in flagella) and the second is responsible for determining the length of the needle/hook (Spa32 in Shigella, YscP in Yersinia and FliK in flagella). In the wild-type situation, interactions between the C-terminal domains of these proteins are thought to be responsible for the switching mechanism (reviewed in Cornelis, 2006; Ferris and Minamino, 2006). Functional knockouts of Spa32, YscP or FliK result in extended needle/polyhook structures (Hirano et al., 1994; Tamano et al., 2002; Journet et al., 2003). In both Yersinia and flagellar systems, mutations have been identified in the genes for YscU/FlhB that suppress the phenotype of the YscP/FliK knockout (Kutsukake et al., 1994; Williams et al., 1996; Edqvist et al., 2003). Several of these mutations affect residues that are well conserved across species and with flagellar FlhB. We have mapped the position of these mutations onto our structure of Spa40 (Fig. 4A). The YscU residues A268 and V292, equivalent to Spa40 A262 and A286, when mutated to Phe and Thr respectively, suppress the YscP knockout. It is clear from the structure that insertion of these larger side-chains would disrupt the packing of β2 and α2 in the region of the NPTH loop. It is likely that this perturbation would interfere with the folding of Spa40C and so may affect the ability of the NPTH loop to adopt the conformation required for efficient cleavage. This is in agreement with the observation that the equivalent mutations in FlhB are resistant to the cleavage process (Minamino and Macnab, 2000a).
Fig. 4.
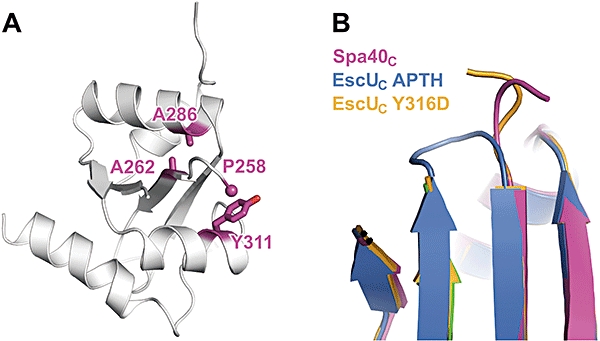
Mapping of mutations from Yersinia and flagellar homologues.
A. Position of conserved residues that are altered by mutations that suppress a YscP/FliK knockout identified in Yersinia YscU and flagellar FlhB.
B. Orientation of PTH loop in Spa40C (purple) and the EscU mutant structures EscUC(APTH) (blue) and EscUC(Y316D) (orange).
Another suppressor mutant identified in flagella and Yersinia is the mutation to aspartate of the highly conserved tyrosine residue within the LARXLY motif (Y317D in YscU, Y323D in FlhB, equivalent to Y311 in Spa40) (Williams et al., 1996; Edqvist et al., 2003). This residue lies on helix α3 and is partially exposed on the surface near the flipped P258 side-chain. This mutation does not inhibit cleavage and is likely to change the surface features of this region either directly or by interfering with the stability and/or orientation of the PTH loop. Indeed, Zarivach et al. (2008) showed that the equivalent change in EscU (Y316D) does alter the electrostatic surface features and also results in a change in the orientation of the PTH loop such that it points back towards the catalytic Asn (Fig. 4B). As this substitution does not inhibit cleavage at the NPTH site, this supports the hypothesis that it is the surface features in this region, rather than the cleavage event, that are important for function.
The mutations described above all interfere with the creation or characteristics of the NPTH surface region, but their effect must be independent of YscP/FliK as they can (partially) restore the function of a YscP/FliK knockout. There are several possible explanations for these observations. One is that the NPTH site may be the binding face for YscP/FliK and the alterations to this face mimic the bound form of YscU/FlhB, for example, by blocking the surface exposure of the NPTH region. Alternatively, the NPTH site may be involved, either directly or via an additional partner, in regulating the secretion of extracellular components of the apparatus. The cytoplasmic domains of the Spa40 homologues, YscU and FlhB, have been implicated in the recruitment of several early and middle substrates of the T3SS [YscI (Wood et al., 2008) and the translocators (Sorg et al., 2007) from Yersinia; FlgE and FlgD from flagella (Minamino and MacNab, 2000b)]. As needle assembly is proposed to involve the addition of needle monomers to the end of the growing needle and translocators are thought to assemble at the tip of this needle using a similar mechanism of binding (Deane et al., 2006; Johnson et al., 2007; Blocker et al., 2008), premature secretion of translocators would block the assembly of additional needle monomers. For this reason, an important step in correct needle assembly is the blockage of premature secretion of translocators. In this case, mutations to Spa40 that result in some recovery of needle length control, despite the absence of the length control protein, may act by interfering with this negative regulatory role for Spa40.
C-terminal truncations of FlhB have also been identified that partially suppress the phenotype of a FliK knockout. Zarivach et al. (2008) observed that the length of the C-terminal helix of EscU was similar to that of the FlhB truncation mutants while the SpaS C-terminal helix was longer. These observations, combined with the lack of an identified FliK homologue in E. coli, led Zarivach et al. (2008) to speculate that a shortened C-terminal helix may bypass the dependence of the needle length control mechanism on a FliK homologue. In our structure of Spa40, we observe a C-terminal helix of equivalent length to that of EscU. As the length of the S. flexneri T3SS needle is regulated by the FliK homologue, Spa32, and Spa40 possesses the shorter form of the C-terminal helix, the length of this helix is not crucial for regulation of T3SS needle length.
The structures of EscUC, SpaSC and Spa40C all reveal that the N-terminal region of the cytoplasmic domain (CN), which links the folded domain to the inner membrane domain, is highly disordered (Fig. 5). Deletions or point mutations within this highly conserved region of EscU or FlhB abolish the secretion of both translocators and effectors (Fraser et al., 2003; Zarivach et al., 2008). Thus, mutations in this region affect the export capability of the apparatus rather than the substrate specificity. This observation suggests a distinct role for this flexible region in the secretion of substrates in general. The efficient secretion of substrates in both the pathogenic type III and flagellar systems is dependent on a soluble ATPase (FliI in flagella) (Woestyn et al., 1994; Fan and Macnab, 1996). In addition to their interactions with secretion substrates, the cytoplasmic domains of YscU and FlhB have been implicated in the recruitment of the ATPase regulatory complex [YscK–YscL–YscQ in Yersinia (Riordan and Schneewind, 2008), FliH and the general chaperone FliJ in flagella (Zhu et al., 2002)] and the ATPase itself [FliI in flagella, (Zhu et al., 2002)]. In this case, the changes within the CN domain may affect the correct positioning or movement of the Spa40C domain and so alter its ability to correctly recruit other components to the secretion apparatus.
Fig. 5.
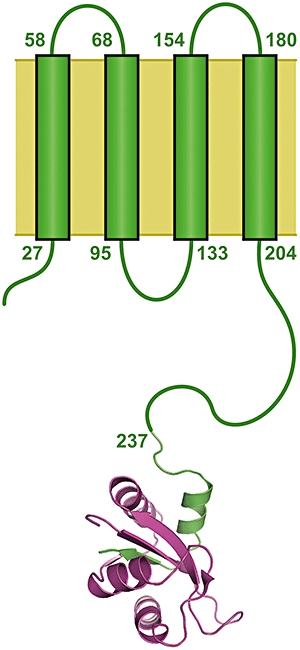
Relative size of the structured region of Spa40C within the context of the full-length membrane protein. Positions of transmembrane helices are labelled as predicted by the Phobius web server (Kall et al., 2007).
Several mutations have been identified within the first 30 residues of FlhB that can partially rescue the non-motile phenotype of a FliI (ATPase) knockout. This gain of function has been attributed to an increase in the probability of entry of flagellar proteins into the export gate (Minamino et al., 2003; Minamino and Namba, 2008). Thus, in addition to the proposed role of FlhB in recruitment of the ATPase complex to the secretion apparatus, these observations support a model whereby the FlhB family of proteins also plays a role in the blockage of secretion and identifies mutations that alleviate this block. It has been shown in both Yersinia and flagella that production of the cytoplasmic domain of YscU/FlhB exerts a dominant negative phenotype by blocking type III secretion (Fraser et al., 2003; Riordan and Schneewind, 2008). It remains uncertain whether the mechanism of this blockage involves sequestration of soluble components or localization of the cytoplasmic domain to the apparatus and direct blockage of the channel.
Following assembly of the extracellular components, the T3SS secretion apparatus must be negatively regulated in order to block secretion of late effectors prior to host-cell contact. This negative regulation is mediated, in part, by proteins belonging to the YopN family (Deng et al., 2005; Schubot et al., 2005). Functional knockouts of members of this family have no effect on needle formation, but result in enhanced effector secretion as well as altered secretion of translocators (Forsberg et al., 1991; Kubori and Galan, 2002; O'Connell et al., 2004). Interestingly, despite these proteins playing a role in the correct targeting of translocators and in the temporal regulation of effector secretion, the structures of the Yersinia and Shigella members of this family possess no structural homology with Spa40C (Schubot et al., 2005; Deane et al., 2008). Thus, it is likely that the mechanisms of secretion regulation by the T3SS will differ at distinct stages of regulation.
In summary, we have shown that the S. flexneri homologue of the T3SS substrate switching protein, Spa40, is cleaved on the N-terminal side of P258 within the highly conserved NPTH sequence to produce a stable complex consisting of the subdomains, Spa40CN and Spa40CC. The structure of this complex is similar to those of E. coli EscU and S. typhimurium SpaS, confirming that the CN region of these proteins forms part of the core of the fold. Cleavage at N257 creates new charged termini and results in the rearrangement of the surface-exposed PTH loop, altering the surface properties of Spa40C in the vicinity of a conserved highly charged patch. The change in surface properties upon cleavage is likely to be critical for the correct binding of additional T3SS components. The mapping of several functional mutations identified in different regions of the Yersinia and flagella homologues onto our structure suggests possible mechanisms of action for this class of proteins in substrate switching and secretion regulation.
Experimental procedures
Protein production and purification
Constructs were made encoding the full-length protein (Spa40FL) and the cytoplasmic domain (residues 207–342, Spa40C). DNA fragments were produced by PCR (FLfwd 5′-AGGAGATATACCATGGCAAATAAAACAGAAAAGCCGACAC-3′, FLrev 5′-GTGATGGTGATGTTTATGAGTGTTTTCAACCTGCTCAAGC-3′, Cfwd 5′-TACCATGGATATGATGATGGATAAACAGGAG-3′ and Crev 5′-TGCTCGAGATGAGTGTTTTCAACCTGC-3′) and cloned into pOPINE (Berrow et al., 2007) for the Spa40FL construct and pET28b (Novagen) for the Spa40C construct to produce C-terminally His6-tagged proteins. Spa40FL and Spa40C were expressed in E. coli B834 (DE3) cells grown in LB media at 37°C until an A600nm of ∼0.6 was reached, whereupon the cultures were cooled to 30°C and protein production was induced by the addition of 0.5 mM IPTG. After 4 h, cells were harvested by centrifugation (4000 g, 15 min, 4°C). The cell pellet was re-suspended in lysis buffer (20 mM Tris, pH 8.0, 500 mM NaCl and Complete EDTA-free protease inhibitor cocktail from Roche) and lysed using an Emulsiflex-C5 Homogeniser (Glen Creston, UK). Following centrifugation (20 000 g, 20 min, 4°C) the supernatant fraction was flowed over a 5 ml Ni-NTA Superflow column (Qiagen) at 1 ml min−1. Protein was eluted using a step gradient, with elution of Spa40C at 300 mM imidazole. Fractions containing Spa40C were further purified by size-exclusion chromatography using a HiLoad 26/60 Superdex 75 column (Amersham Biosciences) equilibrated with 20 mM Tris, pH 8.0 and 500 mM NaCl. Spa40C was also purified under denaturing conditions by re-suspending the pellet following cell lysis in 20 mM Tris pH 8.0, 500 mM NaCl, 2 M Urea and 2% (v/v) Triton X-100. This suspension was centrifuged (15 000 g, 30 min, 4°C) and the pellet re-suspended in the same buffer without Triton X-100 and centrifuged again. This pellet was re-suspended in 20 mM Tris pH 8.0, 500 mM NaCl and 6 M GnCl and stirred overnight at 4°C. This suspension was centrifuged as before and the supernatant was applied to a 5 ml Ni-NTA Superflow column pre-equilibrated in 20 mM Tris pH 8.0, 500 mM NaCl and 8 M Urea and eluted using a step gradient as for the native purification except for the presence of 8 M Urea. Cells expressing Spa40FL were grown as for Spa40C. To fractionate cells, the supernatant following cell lysis and removal of cell debris (20 000 g, 20 min, 4°C) was centrifuged to isolate the membrane fraction (94 000 g, 1.5 h, 10°C). Western blots were carried out using an HRP-conjugated antibody to the His6 tag (Qiagen).
Crystallization
The purified complex of Spa40CN with Spa40CC was concentrated using Millipore Ultra-15 5k MWCO centrifugal filtration devices to 3.3 mg ml−1 and immediately put into crystallization trials. Initial crystallization conditions were obtained by sparse-matrix screening using the sitting drop vapour diffusion technique. Drops were prepared using an OryxNano crystallization robot (Douglas Instruments) by mixing 0.2 μl of protein (3.3 mg ml−1, 20 mM Tris pH 8.0, 500 mM NaCl) with 0.2 μl of reservoir solution and were equilibrated against 100 μl of reservoir solution at 20°C. Diffraction quality crystals in crystal form 1 grew in 3 weeks in condition P2-14 (0.1 M bisTris propane pH 6.5, 0.2 M NaBr and 20% w/v PEG 3350) of the PACT Premier screen (Molecular Dimensions). Crystals that diffracted to higher resolution in a different P1 crystal form were grown in condition P1-32 (0.1 M Hepes pH 7.0, 0.2 M NH4Cl and 20% PEG w/v 6000) of the PACT Premier screen. Crystals were cryoprotected in reservoir solution supplemented with 25% (v/v) glycerol for 15 s and flash-cryocooled by plunging into liquid nitrogen.
Data collection
Initial diffraction data were collected on small plate-like crystals (50 μm × 20 μm × 5 μm) of form 1 of the Spa40C complex at the microfocus beamline ID23-2 at the European Synchrotron Radiation Facility (ESRF), France (Table 1). Data were collected in three batches, each of 180° with 1.0° oscillations, from different regions of the same crystal using a helical-collection strategy, whereby the crystal is moved linearly between two points on the crystal as it is rotated, in order to minimize radiation damage effects (D. Flot et al., in preparation). This method was necessary because of the small size of the crystal and the number of images required for a complete data set in the space group P1. This data set was used for initial structure determination (below). Higher-resolution data were collected from optimized crystals in the second P1 crystal form at beamline ID23-1 (Nurizzo et al., 2006) at the ESRF and used for structure refinement. All data collection was performed at 100 K. Data were indexed and integrated in Mosflm (Leslie, 1992) and scaled with Scala (Evans, 1997) within the CCP4 program suite (CCP4, 1994).
Structure determination
The structure of Spa40C was solved by molecular replacement using the program phaser (McCoy et al., 2007) with a poly-serine search model generated using Chainsaw (CCP4, 1994; Schwarzenbacher et al., 2004) from the structure of the E. coli homologue EscUC (PDB code 3BZL). Iterative cycles of refinement with REFMAC5 (Murshudov et al., 1997) and model building in coot (Emsley and Cowtan, 2004) were performed in consultation with the validation tools present in coot and with the MolProbity web server (Davis et al., 2007). Molecular figures were generated using PyMol (DeLano, 2002).
The atomic co-ordinates and structure factors have been deposited at the RCSB Protein Data Bank with Accession Code 2VT1.
Acknowledgments
We thank Natalie Strynadka and Raz Zarivach for providing the atomic co-ordinates of EscU prior to publication, Andreas Veenendaal for original cloning of the Spa40C construct, Tony Willis for N-terminal sequencing, Joanne Nettleship for collection of mass spectrometry data and staff at beamlines ID23-1 and ID23-2 at the ESRF. The pOPINE vector was a gift from Ray Owens (Oxford Protein Production Facility). J.E.D. is funded by a Wellcome Trust Programme Grant (WT083599MA) to S.M.L. S.C.G. is a Nuffield Medical Fellow. S.J. is funded by a grant to S.M.L. from the Medical Research Council of the UK (G0400389).
References
- Allaoui A, Woestyn S, Sluiters C, Cornelis GR. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. What's the point of the type III secretion system needle. Proc Natl Acad Sci USA. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. Collaborative Computational Project, 4. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nature Rev. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, et al. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc Natl Acad Sci USA. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JE, Roversi P, King C, Johnson S, Lea SM. Structures of the Shigella flexneri type 3 secretion system protein MxiC reveal conformational variability amongst homologues. J Mol Biol. 2008;377:985–992. doi: 10.1016/j.jmb.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 [WWW document]. URL http://www.pymol.org.
- Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA, Lee S, et al. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun. 2005;73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, Wolf-Watz H, Lloyd SA. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–2266. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans PR. Scala. 1997. Joint Ccp4+ESF-EAMCB Newsletter on Protein Crystallography, No. 33.
- Fan F, Macnab RM. Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Minamino T. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 2006;14:519–526. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- Forsberg A, Viitanen AM, Skurnik M, Wolf-Watz H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, et al. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction – the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Galan JE. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J Bacteriol. 2002;184:4699–4708. doi: 10.1128/JB.184.17.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol. 2002;184:4500–4509. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW. Mosflm – a programme for processing X-ray diffraction data. 1992. Joint Ccp4+ESF-EAMCB Newsletter on Protein Crystallography, No. 26.
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol. 2000a;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, MacNab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000b;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- Minamino T, Gonzalez-Pedrajo B, Kihara M, Namba K, Macnab RM. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J Bacteriol. 2003;185:3983–3988. doi: 10.1128/JB.185.13.3983-3988.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nurizzo D, Mairs T, Guijarro M, Rey V, Meyer J, Fajardo P, et al. The ID23-1 structural biology beamline at the ESRF. J Synchrotron Radiat. 2006;13:227–238. doi: 10.1107/S0909049506004341. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ, Luo W, et al. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol. 2004;52:1613–1625. doi: 10.1111/j.1365-2958.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- Riordan KE, Schneewind O. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06247.x. (in press). doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubot FD, Jackson MW, Penrose KJ, Cherry S, Tropea JE, Plano GV, Waugh DS. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J Mol Biol. 2005;346:1147–1161. doi: 10.1016/j.jmb.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr. 2004;D60:1229–1236. doi: 10.1107/S0907444904010145. [DOI] [PubMed] [Google Scholar]
- Sorg I, Wagner S, Amstutz M, Muller SA, Broz P, Lussi Y, et al. YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J. 2007;26:3015–3024. doi: 10.1038/sj.emboj.7601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamano K, Katayama E, Toyotome T, Sasakawa C. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J Bacteriol. 2002;184:1244–1252. doi: 10.1128/JB.184.5.1244-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woestyn S, Allaoui A, Wattiau P, Cornelis GR. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S, Jin J, Lloyd SA. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J Bacteriol. 2008 doi: 10.1128/JB.00328-08. (in press). doi: 10.1128/JB.00328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, et al. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature. 2008;453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- Zenk SF, Stabat D, Hodgkinson JL, Veenendaal AK, Johnson S, Blocker AJ. Identification of minor inner-membrane components of the Shigella type III secretion system ‘needle complex’. Microbiology. 2007;153:2405–2415. doi: 10.1099/mic.0.2007/007781-0. [DOI] [PubMed] [Google Scholar]
- Zhu K, Gonzalez-Pedrajo B, Macnab RM. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry. 2002;41:9516–9524. doi: 10.1021/bi0203280. [DOI] [PubMed] [Google Scholar]


