Abstract
We demonstrate the reversibility of RecA-promoted strand exchange reaction between short oligonucleotides in the presence of adenosine 5′-O-(thiotriphosphate). The reverse reaction proceeds without the dissociation of RecA from DNA. The reaction reaches equilibrium and its yield depends on the homology between the reaction substrates. We estimate the tolerance of the RecA-promoted strand exchange to individual base substitutions for a comprehensive set of possible base combinations in a selected position along oligonucleotide substrates for strand exchange and find, in agreement with previously reported estimations, that this tolerance is higher than in the case of free DNA. It is demonstrated that the short oligonucleotide-based approach can be applied to the human recombinases Rad51 and Dmc1 when strand exchange is performed in the presence of calcium ions and ATP. Remarkably, despite the commonly held belief that the eukaryotic recombinases have an inherently lower strand exchange activity, in our system their efficiencies in strand exchange are comparable with that of RecA. Under our experimental conditions, the human recombinases exhibit a significantly higher tolerance to interruptions of homology due to point base substitutions than RecA. Finding conditions where a chemical reaction is reversible and reaches equilibrium is critically important for its thermodynamically correct description. We believe that the experimental system described here will substantially facilitate further studies on different aspects of the mechanisms of homologous recombination.
Homologous recombination (HR)2 is a fundamental genetic process with a great variety of biological implications spanning from microbial evolution and the development of pathogens to genome instability and carcinogenesis in higher eukaryotes. It also is an example of an intricate macromolecular choreography. HR has been finding a widening scope of practical applications such as DNA recombineering, eukaryotic gene targeting etc. (for recent reviews see Refs. 1–3 and references therein).
The central event of HR is based on a peculiar type of recognition of homology between DNA molecules and until now the mechanism of this recognition remains enigmatic (for recent reviews see Refs. 4–9). This process is accompanied by drastic changes in DNA conformation (10, 11) (for recent reviews see Refs. 5, 9, and 12–14) and allosteric effects (15–18) in the proteins that promote DNA strand exchange, the central step of HR.
The main prerequisite for the thermodynamically correct description of a chemical reaction is finding conditions where the reaction is reversible and reaches equilibrium. However, enzymatic processes are often too complex to separate reversible phases and do not reach equilibrium. In the case of RecA-promoted DNA strand exchange the complexity is the result of the multistep nature of the reaction; the filamentous structure and high tendency to aggregate of the RecA protein and its complexes with long DNA molecules; the tendency of ssDNA to form extensive secondary structure, etc. Many of these complexities can be overcome by using short oligonucleotides as substrates for the strand exchange reaction.
The conventional pathway of the RecA-promoted DNA strand exchange reaction involves formation of a RecA protein co-filament with ssDNA that further interacts with the second substrate of the reaction, the dsDNA molecule, and displaces one of the DNA strands resulting in the formation of a dsDNA heteroduplex. The classic definition of enzymatic catalysis assumes the existence of a reverse reaction promoted to the same extent as the forward one and equilibration of the net reaction. For the RecA protein from Escherichia coli the question of whether there is a reverse reaction is unclear. Equilibration of the RecA-promoted strand exchange reaction in the presence of ATP was documented in a series of studies (19, 20) but most probably this equilibration was the result of the recycling of the RecA protein coupled with ATP hydrolysis (21, 22). In this case the apparently reverse reaction proceeds due to the release of RecA from the heteroduplex dsDNA formed in the course of the first round of strand exchange, its subsequent binding to displaced ssDNA with formation of a conventional presynaptic complex that further interacts with the heteroduplex dsDNA. Thus, the conventional “direct” pathway of strand exchange can result in this case in an apparent reversibility of the reaction, although the resulting steady state is not an equilibrium in the correct thermodynamics sense because it is supported by the permanent flow of free energy supplied by ATP hydrolysis.
A truly inverse process, that is, the interaction and strand exchange between a RecA protein complex with dsDNA and free ssDNA molecule, was documented earlier (23) but the possibility could not be excluded that the observed activity was independent and irrelevant to the canonical direct RecA-promoted strand exchange. It was suggested that true reversibility of the conventional reaction pathway might take place in reactions with ssDNA substrates not containing secondary structures and that another prerequisite to this activity is the formation of a high affinity DNA binding state of RecA (24). Shortening of oligonucleotide substrates may satisfy both conditions because shorter oligonucleotides exhibit a lower tendency to form secondary structures, and, at the same time, as we demonstrated earlier (25, 26), an oligonucleotide length as short as 18–20 nt is still sufficient for the formation of stable presynaptic complexes with RecA that are active in strand exchange.
RecA protein is the central component of HR in E. coli and a universal prototype of recombinases from all kingdoms of living organisms. This recombinase is the best characterized among all of the proteins of HR. A number of RecA protein analogues from different organisms exhibit a basically similar structure and activities, although, often differ in some important details of their mechanisms.
In humans, the main players of HR are the Rad51 and Dmc1 proteins. Rad51 protein is a universal recombinase and is found in all human cells where HR takes place. In contrast, Dmc1 is a meiosis-specific protein. The necessity for the concerted action of two different recombinases in meiosis remains unexplained and probably requires a more profound understanding of the mechanistic aspects of DNA strand exchange reactions promoted by these proteins.
The conventional reaction of strand exchange between long natural DNA molecules is a complex multistep process consuming free energy from ATP hydrolysis (27–32). There are two commonly used approaches to obtain possibly simpler experimental systems for the study of the important phases of strand exchange, the use of possibly shorter oligonucleotides as substrates for the strand exchange and slowly or non-hydrolyzable ATP analogues as cofactors for proteins that promote the strand exchange.
Earlier we elaborated an experimental system to study the RecA-promoted strand exchange reaction using very short fluorescent dye-tagged oligonucleotides and ATPγS as a cofactor (25, 26). It allowed us to follow the binding of substrates and products of the reaction in the recombinational nucleoprotein filaments and to reveal the phenomenon of RecA monomer phasing (33–35). In the present paper we describe the results of the further characterization of the RecA-promoted strand exchange between very short oligonucleotides and our first attempts to expand this approach to the reaction promoted by human recombinases. Here we demonstrate that the RecA-promoted strand exchange reaction under these conditions is reversible and attains equilibrium in the absence of ATP hydrolysis. This reaction is sensitive to point interruptions of homology between partners of strand exchange. The applicability of this approach to the characterization of human recombinases is also demonstrated.
MATERIALS AND METHODS
RecA protein from E. coli and human Rad51 and Dmc1 proteins purified according to published protocols (15, 36, 37) were kindly provided by Oleg Voloshin, Roberto Pezza, and Janet Donaldson.
Fluorescent dye-labeled 20-nt long oligonucleotide (*20-mer) was used as a substrate for the formation of recombinationally active complexes with RecA and human recombinases. We demonstrated earlier (25, 26) that fluorescent dye molecule stimulates binding of RecA protein to short oligonucleotides with formation of complexes active in strand exchange reactions. Fluorescence of the dye molecule was used to quantify the amounts of the *20-mer in ss and ds forms after their electrophoretic resolution and to follow the progress of the strand exchange under different conditions.
Oligonucleotides (MWG Biotech Ltd.) were purified by PAGE. The oligonucleotide list is presented in Table 1. ds-16-mers were obtained by mixing of equal molar amounts of the corresponding complementary oligonucleotides followed by incubation for 10 min at 37 °C. To ensure that the preparations of ds-16-mers do not contain contamination of ss-16-mers (“–”-strand) that might distort our results of RecA-promoted strand exchange the ds-16-mers obtained were checked by mixing with the corresponding *ss-20-mers at 0 °C and subsequent electrophoresis under native conditions.
TABLE 1.
The list of oligonucleotides used in the present study
The asterisk designates the molecule of fluorescein. Variable nucleotides are boxed and given in bold.
| *20-mer (G) | * | A | T | G | T | A | A | A | A | C | G | A | C | G | G | C | C | A | G | T | C 3′ | |||
| *20-mer (T) | * | A | T | G | T | A | A | A | A | C | T | A | C | G | G | C | C | A | G | T | C 3′ | |||
| *20-mer (C) | * | A | T | G | T | A | A | A | A | C | C | A | C | G | G | C | C | A | G | T | C 3′ | |||
| *20-mer (A) | * | A | T | G | T | A | A | A | A | C | A | A | C | G | G | C | C | A | G | T | C 3′ | |||
| ds16-mer (G) | +16-mer (G) | 5′ G | T | A | A | A | A | C | G | A | C | G | G | C | C | A 3′ | ||||||||
| -16-mer (C) | 3′ C | A | T | T | T | T | G | C | T | G | C | C | G | G | T 5′ | |||||||||
| ds16-mer (T) | +16-mer (T) | 5′ G | T | A | A | A | A | C | T | A | C | G | G | C | C | A 3′ | ||||||||
| -16-mer (A) | 3′ C | A | T | T | T | T | G | A | T | G | C | C | G | G | T 5′ | |||||||||
| ds16-mer (C) | +16-mer (C) | 5′ G | T | A | A | A | A | C | C | A | C | G | G | C | C | A 3′ | ||||||||
| -16-mer (G) | 3′ C | A | T | T | T | T | G | G | T | G | C | C | G | G | T 5′ | |||||||||
| ds16-mer (A) | +16-mer (A) | 5′ G | T | A | A | A | A | C | A | A | C | G | G | C | C | A 3′ | ||||||||
| -16-mer (T) (GGT) 16 | 3′ C | A | T | T | T | T | G | T | T | G | C | C | G | G | T 5′ | |||||||||
| Hetero 21-mer | 5′ A | T | T | A | G | T | A | G | G | C | T | A | T | A | A | G | T | C | C | C |
The strand exchange reaction was performed by the conventional two-step procedure. At first, the complex of RecA protein with fluorescent dye-labeled ss-oligonucleotide (the presynaptic complex) was formed. Then the second reaction substrate, usually ds-oligonucleotide, was added and the time course of the transition of the dye-labeled oligonucleotide from the ss to the ds form was recorded.
Presynaptic complexes were formed by incubation of near stoichiometric amounts of RecA protein (8.6 μm) and dye-labeled 20-mer (22 μm of bases) in 20 mm Tris acetate buffer (20 mm, pH 7.3), 2 mm Mg(OAc)2, and 0.5 mm ATPγS at 37°C for 30 min. The mixture was then brought to the desired temperature for the second incubation, and the Mg(OAc)2 concentration was raised to 10 mm, followed by the addition of the second substrate of the reaction (ss- or ds-oligonucleotides usually in equimolar concentration). When it is not specially noted the RecA-promoted strand exchange reaction was performed at 32 °C.
To imitate the reverse strand exchange the complexes of RecA protein with ds-oligonucleotides corresponding to the product of the strand exchange reaction were obtained by the addition of equimolar amounts of “–”-strand 16-mers to preformed complexes of RecA with dye-labeled 20-mer and subsequent incubation for 5 min at 37 °C. The quick annealing of such oligonucleotides under these conditions was demonstrated earlier (26). Then “+”-strand 16-mer was added and the time course of the transition of dye-labeled oligonucleotide from the ds to the ss form was followed.
In control experiments the ds-oligonucleotides were initially formed by annealing of the dye-labeled *20-mer with the complementary strand and the preformed duplex was then co-incubated with RecA protein. Addition of equimolar amounts of +strand 16-mers to these complexes prepared in this manner resulted in the same kinetic curve of the reverse reaction (not shown).
The same procedure was used for the preparation of complexes with the human recombinases and their subsequent strand exchange reactions. The reactions were performed at 37 °C, Mg(OAc)2 was replaced by CaCl2 (3 mm) and ATPγSby ATP (1 mm). The reactions also contained 100 mm NaCl for Rad51 and 5 mm KCl for Dmc1 introduced from the protein storage buffers.
After appropriate incubation the reaction mixture was placed on ice, deproteinized by the addition of SDS to 0.2%, and electrophoresed in 16% polyacrylamide gel, TAE buffer (40 mm Tris acetate, pH 8.0, 1 mm EDTA) for 2 h at 16 V/cm performed in a cold room. Gels were scanned on a FluoroImager Storm 840, or photographed using the gel documentation system ChemiImager™ 5500 (Alpha Innotech) and the images obtained were analyzed using the ImageQuant 5.0 program.
RESULTS
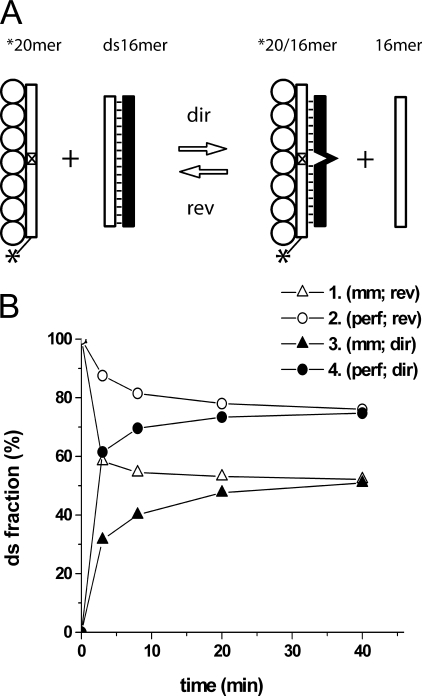
Reversibility of the RecA-promoted Strand Exchange—Fig. 1 presents a reaction scheme for the RecA-promoted strand exchange between short oligonucleotides (Fig. 1A). As seen in Fig. 1B, RecA protein efficiently promotes the direct reaction but the strand exchange does not reach completion and approaches a steady state at a level dependent on the extent of homology between the oligonucleotide partners of the strand exchange (curves 3 and 4). Under these conditions RecA protein also promotes an efficient strand exchange in the “reverse” direction (curves 1 and 2), that is, exchange of strands between ds-oligonucleotide in complex with RecA protein and free ss-oligonucleotide added in solution. The last reaction corresponds in the right to left direction of the reaction scheme depicted in Fig. 1A. The corresponding kinetic curves for the direct and reverse reactions converge and reach a steady state in a reasonable reaction time demonstrating that the most probable reason for the saturation of the reactions is that they reach equilibrium. The equilibrium level depends on the extent of homology between the reaction substrates and is higher in the case of perfect homology demonstrating the sensitivity of the reaction to an individual base substitution.
FIGURE 1.
Reversible strand exchange reaction between short oligonucleotides in the presence of ATPγS. A, the reaction scheme. The presynaptic complex was formed by incubation of RecA protein with a fluorescent dye-tagged *20-mer. Position of the dye is designated by the asterisk. The interruption of homology is designated by the crossed rectangle in the dye-tagged oligonucleotide. Left to right direction corresponds to forward strand exchange reaction. For a more detailed description of the experimental procedure see “Materials and Methods” and Ref. 26. B, kinetic curves of forward (3 and 4) and reverse (1 and 2) strand exchange reactions. The single-stranded substrate of the reaction (fluorescently labeled oligonucleotide) contained no (curves 2 and 4) and one (curves 1 and 3) base substitution in comparison to the ds-16-mer (see supplemental Fig. S1 for the original data).
Essentially the same results were obtained for the temperature range from 24 to 37 °C. In this temperature interval RecA promotes efficient strand exchange between short oligonucleotides, which is reversible and reaches a steady state at a reasonable time, depending on the temperature. At the same time, the saturation levels of the reaction at equilibrium do not depend on the temperature (see below for the reverse reaction).
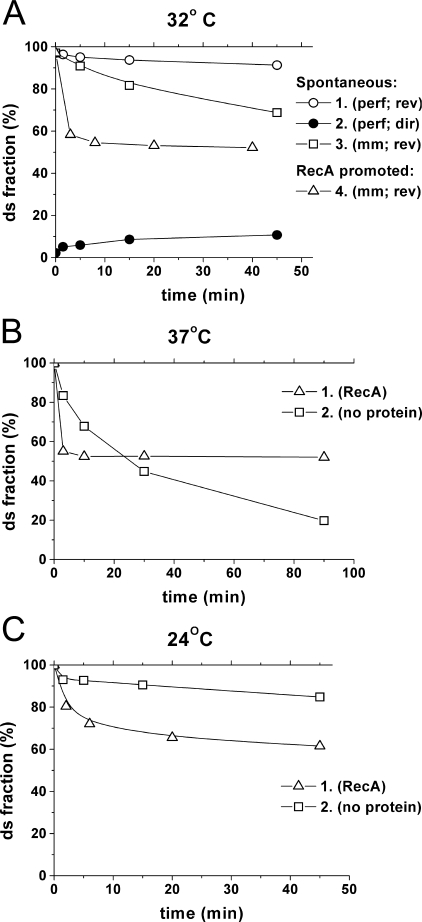
To estimate the stability of the free ds-oligonucleotides, corresponding to the reaction substrates and products under our experimental conditions, they were incubated in the absence of RecA protein. As seen in Fig. 2, the perfect ds-oligonucleotides were rather stable and the extent of spontaneous exchange between completely homologous substrates did not exceed 10%. Mismatch containing oligonucleotides exhibited a lower stability that was apparent from the significant level of spontaneous strand exchange in this case (curve 3 in Fig. 2A).
FIGURE 2.
Stability of the strand exchange reaction substrates and products. A, direct (2) and reverse (1 and 3) strand exchange reactions were performed in conditions identical to those used for RecA-promoted reactions but in the absence of the protein. Mismatch containing (3) and completely homologous (1 and 2) pairs of oligonucleotide substrates were used. For comparison purposes, the kinetic curve for reverse RecA-promoted strand exchange reaction is presented (4). B and C, comparison of spontaneous (2) and RecA-promoted (1) reverse strand exchange between mismatch containing pairs of oligonucleotide substrates at different temperatures.
Comparison of the time courses of the spontaneous and RecA-promoted reverse strand exchange reactions in the case of mismatch containing reaction products (curves 3 and 4 in Fig. 2A, correspondingly) demonstrates that RecA in these conditions accelerates the reverse reaction in the beginning of the reaction course but stabilizes it at a higher steady state level. This level does not depend on the reaction temperature, whereas the rate of the spontaneous reaction significantly varies with temperature (Fig. 2, B and C) demonstrating the capability of RecA protein to actively promote the reverse reaction and the efficient stabilization of the double-stranded reaction product by RecA protein (see also below).
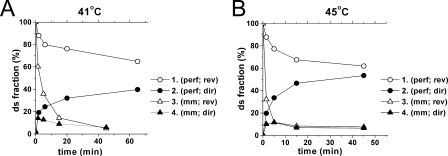
Theoretically, spontaneous strand exchange reaction in the case of perfectly homologous substrates must reach equilibrium at a yield of about 50%. To check if this is the case under our conditions and to drive the reactions closer to completion the spontaneous reactions were performed at elevated temperatures. The results are presented in Fig. 3 and demonstrate that the equilibrium level of spontaneous strand exchange is really about 50%, whereas, as expected, in the case of mismatch containing substrates the equilibrium is strongly shifted in the reverse direction. Less than 10% of mismatch containing product is formed in the last case. As demonstrated in Fig. 1B, the corresponding RecA-promoted reactions reach equilibrium at about 75% for the reaction with the perfectly homologous substrates and about 55% for the mismatch containing substrates. Thus, in both cases RecA shifts the equilibrium in the “forward” direction but in the case of mismatched substrates this shift is significantly higher and as a result the RecA-promoted exchange exhibits lower sensitivity to base substitution in comparison to spontaneous strand exchange. This result is in agreement with estimates of the free energy of mismatch formation in DNA in complexes with RecA (38).
FIGURE 3.
Spontaneous strand exchange at elevated temperatures. Direct and reverse strand exchange reactions were performed in conditions identical to those used for RecA-promoted reactions but in the absence of the protein and at elevated temperatures, 41 °C (A) and 45 °C (B). Kinetic curves for reverse (1 and 3) and direct (2 and 4) strand exchange are presented for the pair of completely homologous (1 and 2) and mismatch containing (3 and 4) substrates.
A necessary condition for the equilibration of any reaction is the existence of a pathway for the decay of the reaction product and returning the system to its initial state. When this decay proceeds via the same steps as the direct reaction but the reaction intermediates appear in the reverse order we consider the process as a true reverse reaction. In the RecA system the reverse reaction should involve the RecA promoted process of invasion of naked ssDNA in the dsDNA bound in the nucleoprotein filaments that results in displacement of a strand from this dsDNA and formation of a new dsDNA molecule corresponding to the initial double-stranded substrate of the direct (conventional) strand exchange reaction (see Fig. 1A).
In the present system, in addition to the genuine reverse process, there are two possible pathways for the decay of the strand exchange reaction product. The first is the melting of dsDNA (especially unstable in the case of a mismatched product) and release of the free–16-mer in solution followed by its annealing to the +16-mer. The second possibility is the dissociation of the RecA protein from the complex and its recycling in the strand exchange reaction. Release of RecA protein from the double-stranded reaction product has been well documented in the presence of ATP. In contrast, in the presence of ATPγS RecA remains bound with the reaction product and its complexes with dsDNA are very stable in the presence of this cofactor. In particular, we have demonstrated earlier the stability of RecA protein complexes with short dye-labeled oligonucleotides (25). To ascertain the absence of RecA recycling in the present system we performed experiments on the strand exchange with a different order of additions and characterized the influence of heterologous oligonucleotides on the exchange reaction.
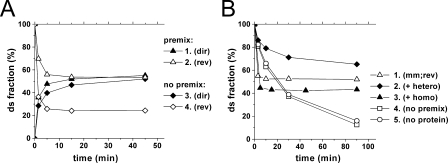
The recycling of RecA protein implies its rapid dissociation from dye-labeled substrate and interaction with other reactants. In this case there must be no effect of preincubating RecA protein with the dye-labeled oligonucleotides on the strand exchange kinetics. The results of control experiments with a different order of addition of RecA protein are presented in Fig. 4A. Addition of RecA protein to the mixture of the dye-labeled ss-20-mer and ds-16-mer resulted in strand exchange in the forward direction that attains equilibrium with the same yield as the reaction performed in the conventional manner, that is, with preincubation of RecA with the dye-labeled ss-20-mer (Fig. 4A, curves 3 and 1, respectively). In contrast, when RecA protein was added to oligonucleotide reactants corresponded to the reverse reaction, namely, a mixture of +16-mer with a mismatch containing *20-mer/–16-mer ds-oligonucleotide, the reaction is saturated at a significantly lower yield of the dye-labeled ds-product (curve 4). Therefore, in this design, when RecA protein is added after all oligonucleotide substrates direct and reverse reactions do not converge (compare curves 3 and 4).
FIGURE 4.
The influence of the order of protein addition and an excess of single-stranded oligonucleotides. A, different order of addition of RecA protein to the reaction substrates. Direct (1) and reverse (2) strand exchange were performed in the conventional manner after preincubation of RecA protein with the dye-labeled *20-mer. 3 and 4 RecA protein was added after mixing the dye-labeled *20-mer with the ds-16-mer (3) and of the +16-mer with the mismatch containing *20-mer/–16-mer (4) (see supplemental Fig. S2 for the original data). B, the kinetic experiments were performed at 37 °C. A RecA-promoted reverse strand exchange reaction with the mismatch containing *20-mer/–16-mer oligonucleotides were performed under conventional conditions (1); in a 4-fold excess of (GGT)16 oligomer (2); and in a 4-fold excess of +16-mer (3). The reverse reactions with an equimolar ratio of oligonucleotide substrates in 4-fold excess of (GGT)16 oligomer with addition of RecA protein after mixing of all DNA substrates (4) and with no RecA and (GGT)16 oligomer addition (5) are also presented. The addition of the 4-fold excess of (GGT)16 oligomer did not influence the kinetic of the spontaneous strand exchange (not shown) (see supplemental Fig. S3 for the original data).
The most straightforward explanation of these results is that the RecA protein binds irreversibly in this time scale to the added single strand substrate and the further reaction proceeds via interactions of other reactants with this complex. Kinetic curves for the direct reaction are very similar when the same ss-oligonucleotide was used in experiments with (Fig. 4A, curve 1) and without (curve 3) preincubation with RecA protein. That the last reaction is slightly but reproducibly slower may reflect some time delay needed for the formation of the RecA protein-*20-mer complex. In contrast, the order of addition of RecA protein substantially influences the yield of the reverse reaction (curves 2 and 4). Apparently, in the absence of preincubation of RecA protein with *20-mer, the protein following the conventional reaction scheme initially forms the presynaptic complex with single-stranded +16-mer that further interacts with the double-stranded mismatch-containing *20-mer/–16-mer substrate and efficiently displaces *20-mer from this substrate. Therefore, in both cases when RecA protein is added to oligonucleotides with no preincubation the strand exchange reactions proceed by the pathway corresponding to the conventional reaction scheme and depending on what oligonucleotides were introduced in the reaction in the single-stranded form the reaction outcome is dramatically different.
These results confirm that RecA shifts the equilibrium in the direction of the formation of the heteroduplex ds product and that RecA remains bound with the oligonucleotide that was introduced in the reaction in the single-stranded form and was coated by RecA protein at the stage of the formation of the presynaptic complex. This course of the reaction has been well documented for the RecA-promoted strand exchange between long DNA molecules and the present results demonstrate that RecA protein complexes with short oligonucleotides are sufficiently stable to recapitulate this reaction pattern.
The results of another control experiment are presented in Fig. 4B. Addition of a 4-fold excess of heterologous ss-oligomer (GGT)16 significantly retards the reverse reaction. In the first few minutes of the reaction, when in the absence of heterologous oligonucleotides the reverse reaction reaches saturation (kinetic curve 1), the progress of the reverse reaction in the presence of heterologous oligonucleotides is much slower (curve 2). The reverse reaction in the last case is also slower than spontaneous strand exchange in these conditions (curve 5) demonstrating the stability of the RecA protein complex with dsDNA. These results were obtained with complexes formed in the conventional manner, by preincubating RecA protein with the dye-labeled 20-mer before addition of the other reaction substrates. If RecA protein was added immediately after the mixing of the substrates in the presence of excess heterologous oligonucleotides (as in the experiments described above, see Fig. 4A) the kinetic curve (curve 4) of the strand exchange coincided with that for spontaneous exchange (curve 5) demonstrating that heterologous oligonucleotides efficiently inhibit formation of active complexes of RecA protein with the dyelabeled substrate. Therefore, stabilization of the ds-oligonucleotide in the presence of the heterologous oligomer is the result of the irreversible binding or the very slow dissociation of RecA protein from the complex with the ds-oligonucleotide corresponding to the strand exchange reaction products.
The inhibition of the reverse reaction by the heterologous DNA demonstrates also that the reverse reaction is not a consequence of melting ds-oligonucleotides in the complex with RecA protein with release of –16-mer in solution and its subsequent spontaneous annealing with the added +16-mer. Such annealing would not be inhibited by heterologous oligonucleotides (compare curves 4 and 5).
One may suggest that excess heterologous DNA stabilizes the complexes with RecA and/or retards the melting and release of the –16-mer in solution in some nonspecific (not related to homology or complementarity) manner. This possibility is disproved by the finding that the reverse reaction is accelerated by an excess of +16-mers (Fig. 4B, curve 3) in contrast to the retardation observed in the presence of a heterologous competitor (curve 2). That this excess of the reaction product (+16-mer) also shifts the steady state level in the direction of the reaction substrates (compare curves 1 and 3) confirms that the observed steady state corresponds to the reaction equilibrium. The addition of excess heterologous oligonucleotides with a random or scrambled sequence instead of (GGT)16 oligomer resulted in essentially the same results (not shown).
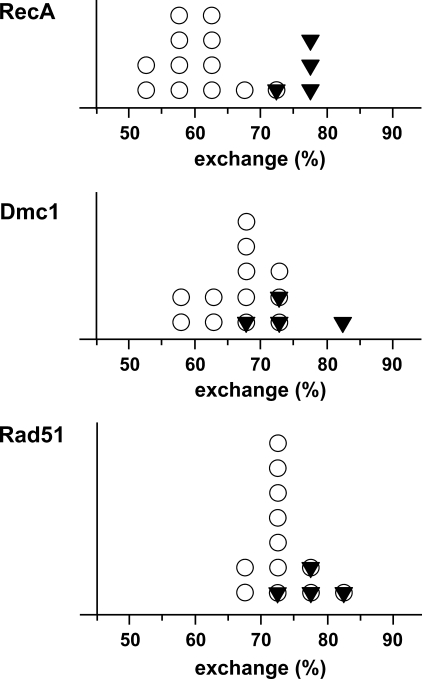
Tolerance of the RecA-promoted Strand Exchange to Different Types of Base Substitutions—An important feature of the RecA protein is its ability to discriminate DNA substrates that are not perfectly homologous. As seen in Fig. 1B the strand exchange between short oligonucleotides in our experimental conditions is sensitive to an interruption of homology. To systematically estimate the tolerance of the RecA-promoted strand exchange to different types of individual base substitutions the strand exchange reactions were performed with substrates containing all possible combinations of base substitutions at one position in the central part of the oligonucleotides (see Fig. 1A and Table 1). The results are presented in Table 2 and demonstrate a significant dependence on the nature of the base substitution in the reactions between oligonucleotide substrates with perfect homology and different heterologies (see also below, the histogram in Fig. 7A).
TABLE 2.
The tolerance of the RecA protein promoted strand exchange reaction to substitutions of individual nucleotides
The reaction yields (in percent of ds-product of strand exchange) for all possible combinations of individual base substitutions are presented. For each substrate configuration direct strand exchange reaction was performed (with a 1-h incubation with ds-substrtae). The results of at least three independent experiments for each point were averaged. Data for exchanges between completely homologous substrates are given in bold.
|
RecA
|
Incoming strand
|
|||
|---|---|---|---|---|
| G | T | C | A | |
| Outgoing strand | ||||
| G | 77.4 ± 1.8 | 56.6 ± 2.8 | 58.7 ± 3.2 | 52.1 ± 0.8 |
| T | 71.2 ± 1.4 | 76.2 ± 1.8 | 64.2 ± 3.1 | 58.6 ± 0.55 |
| C | 63.8 ± 0.8 | 56.8 ± 3.1 | 73.5 ± 1.3 | 51.8 ± 2.3 |
| A | 69.2 ± 0.1 | 60.5 ± 4.3 | 62.1 ± 2.1 | 76.5 ± 0.32 |
FIGURE 7.
Distribution of the yields of strand exchange reactions promoted by RecA protein and human recombinases. Each symbol corresponds to one entry in Table 2 for RecA and Table 3 for human recombinases and presents one of the 16 combinations of the reaction substrates with a reaction yield in a particular 5% interval. Open circles present exchanges between heterologous substrates (off the diagonal entries in Table 2 and 3); closed triangles, between completely homologous substrates (diagonal entries in Table 2 and 3).
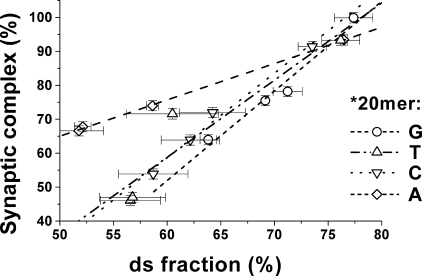
In Fig. 5 the yields of RecA-promoted strand exchange reaction between short oligonucleotides are plotted against previously reported yields of RecA protein assisted synaptic complexes of ss-oligonucleotides with targets in plasmid dsDNA (38). The plots for four different base substitutions at the same site on the *20-mers are presented in Fig. 5. As seen, there is a rather good correlation between the present and previously reported data in the series with the same ss-oligonucleotide substrate. Three plots (for bases C, G, and T in the ss-*20-mer) practically coincide, whereas one (for A base) is slightly different. Given the rather strong difference in the present and previously reported experimental systems these two sets of data may be considered as well correlated.
FIGURE 5.
The tolerance of RecA-promoted strand exchange to individual base substitutions compared with previously reported data. Previously reported data on the formation of D-loop in plasmid DNA by RecA-coated oligonucleotides (Table 1 in Ref. 38) are plotted against yields of the strand exchange reaction between oligonucleotides presented in Table 2. Each line corresponds to one of the four possible base substitutions in the *20-mer.
Strand Exchange Promoted by Human Recombinases—The eukaryotic recombinases on their own usually exhibit rather low strand exchange activity in comparison with the RecA protein from E. coli (39–42). Recently it has been demonstrated that activities of human Rad51 and Dmc1 proteins are significantly higher in the presence of calcium ions (42–44). These conditions result in a substantial drop in ATP hydrolysis and deprive the strand exchange reaction of an external source of free energy making these conditions similar to those for the RecA protein-promoted reaction in the presence of ATPγS. Like RecA-promoted strand exchange, the first stages of the Rad51-promoted reaction do not require ATP hydrolysis (39, 45). This opens the possibility to apply the same approaches described to the characterization of these human recombinases and to directly compare the different aspects of the strand exchange reactions promoted by RecA protein and the human recombinases.
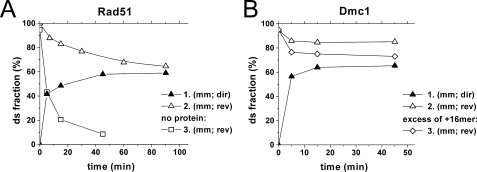
In our preliminary experiments we found that under these conditions human recombinases efficiently bind short fluorescent dye-tagged oligonucleotides (not shown). Both human recombinases promote efficient strand exchange at 37 °C and in contrast to RecA protein their activities sharply drop with decreasing temperature (not shown). Therefore, the reactions of strand exchange promoted by human recombinases were performed at 37 °C. An extent of exchange comparable with those promoted by RecA protein was achieved at a stoichiometric (3 nucleotides per Rad51 monomer) DNA/protein ratio in the case of Rad51 protein and required a 1.5 excess (2 nucleotides per monomer) of Dmc1 protein. The results are presented in Fig. 6. Like RecA protein both recombinases stabilize mismatched reaction products against spontaneous decay. The Rad51-promoted reaction is reversible and equilibrates, although the reverse reaction is slower than that promoted by RecA and the spontaneous exchange under similar conditions. Dmc1 exhibits a lower efficiency in promoting the reverse reaction and the direct and reverse reaction do not converge completely. The reverse reaction proceeds with higher efficiency with an excess of +16-mer. Therefore, there are some obstacles to the reverse reaction that can be overcome with an excess of +16-mer.
FIGURE 6.
Strand exchange between short oligonucleotides promoted by human recombinases. A, kinetic curves for Rad51 promoted (1 and 2) and spontaneous (3), direct (1) and reverse (2 and 3) strand exchange reactions. All oligonucleotide substrates are in equimolar amount. B, kinetic curves for Dmc1 promoted direct (1) and reverse (2 and 3) strand exchange at an equimolar ratio of oligonucleotide substrates (1 and 2) and with a 3-fold excess of the +16-mer (3).
The results of the experiments on sensitivity of Rad51 and Dmc1 proteins promoted strand exchange reactions to individual base substitutions are presented in Table 3. The set of oligonucleotides was the same as used in the experiments with RecA described above (Table 1). The data for RecA and the human proteins are summarized in Fig. 7. Both human proteins exhibit significantly higher tolerance to base substitutions in comparison to RecA protein as can be estimated from the distributions of the reaction yields for the reactions with perfectly homologous and containing base substitution substrates. In the case of RecA-promoted strand exchange these distributions are well resolved, whereas Dmc1 and Rad51 promoted reactions with base-substituted substrates exhibit only a slight downward shift of the reaction yields distributions in comparison with exchanges between substrates with perfect homology.
TABLE 3.
The tolerance of Rad51- and Dmc1-promoted strand exchange reactions to substitutions of individual nucleotides
See legend for Table II.
|
Incoming strand
|
||||
|---|---|---|---|---|
| G | T | C | A | |
| Rad51 | ||||
| Outgoing strand | ||||
| G | 71.5 ± 3.0 | 67.9 ± 1.4 | 67.7 ± 3.8 | 70.8 ± 2.6 |
| T | 70.8 ± 0.46 | 78.5 ± 0.57 | 81.4 ± 3.6 | 74.4 ± 3.0 |
| C | 71.1 ± 2.1 | 70.1 ± 2.24 | 83.9 ± 2.3 | 71.0 ± 2.1 |
| A | 75.5 ± 2.7 | 71.7 ± 2.46 | 79.5 ± 1.8 | 79.5 ± 2.7 |
| Dmc1 | ||||
| Outgoing strand | ||||
| G | 72.7 ± 1.3 | 68.7 ± 2.3 | 64.4 ± 1.0 | 72.8 ± 0.9 |
| T | 64.9 ± 1.1 | 72.7 ± 1.9 | 65.3 ± 3.5 | 73.6 ± 1.7 |
| C | 59.2 ± 1.7 | 66.7 ± 1.8 | 67.8 ± 0.6 | 74.6 ± 1.4 |
| A | 58.3 ± 2.6 | 69.8 ± 0.4 | 65.7 ± 2.4 | 79.6 ± 1.7 |
DISCUSSION
The alignment of homologous DNA strands in the course of HR is one of the most intricate examples of molecular recognition. The RecA-promoted DNA strand exchange reaction that underlies HR is a complex multistep process (20, 21, 46, 47). Thus, the elucidation of the strand exchange mechanisms requires characterization of the different phases of the reaction, identification of the reaction intermediates and pathways, the influence of different conditions, the effect of substrate configurations, etc.
Shortening the Substrates of the Strand Exchange Reaction—The use of short oligonucleotides substrates avoids many of the complications related to the intricacy of the RecA-promoted strand exchange reaction between long molecules of natural DNAs.
Mechanistically the RecA-promoted strand exchange reaction may be divided in two main stages. First is the recognition of homology between the DNA molecules involved, their initial alignment, and the nucleation of the strand invasion. The second stage is the directed expansion of the synaptic complex that ultimately can span over thousands of base pairs of DNA. The mechanisms of these events are most probably different and to separate the first stage shorter oligonucleotides should be useful as substrates for RecA binding and strand exchange.
Commonly used DNA substrates for physicochemical studies of RecA-promoted strand exchange reactions are oligonucleotides 30–50 nt long. Yet even these lengths are apparently too large to separate the stages of strand exchange. Even at a length of homology of 27-nt substitutions of nucleotides located at different ends of the region of homology resulted in different patterns of mismatch tolerance implying that the length of DNA regions involved in the initial step of strand exchange is shorter (38, 46). Experimentally the synaptic complexes were observed between DNA molecules that shared as little as eight bases of homology (50, 51); theoretical estimations predict even less bases of DNA for the nucleation of homology recognition (4, 52). Therefore, to approach these limits for the length of the reaction substrates shorter oligonucleotides should be used. But diminishing the oligonucleotide length is limited by two factors, the stability of ds-oligonucleotides at temperatures suitable for RecA protein activity and the cooperativity of RecA-DNA interaction that makes unstable the complexes of this protein with oligonucleotides that are too short.
As we demonstrated earlier, the efficiency of RecA protein binding to short oligonucleotides can be increased by using fluorescent dye-tagged oligonucleotides as the substrates for RecA protein binding. This approach allowed us to decrease the acceptable oligonucleotide length down to 18 nucleotides and obtain complexes with RecA that are active in strand exchange (25, 26). In the present work we expand this approach to study the reversibility of RecA-promoted strand exchange and the sensitivity of this reaction to heterologous base substitutions. The present data as well as our previously published results demonstrate that a significant decrease in the length of the oligonucleotides can be used for the formation of active recombinational nucleoprotein filaments, to clearly identify RecA-promoted processes, and to reveal a new reaction (the reverse reaction) in RecA-promoted strand exchange.
The Use of Slow or Non-hydrolyzable Analogues of ATP—Another common approach to simplify the reaction of strand exchange is using non-hydrolysable analogues of the ATP, the natural cofactor of RecA. Although the initial phases of the reaction do not require ATP hydrolysis (27, 28, 30–32, 54, 55) ATP hydrolysis supplies the free energy only for the dissociation of RecA from the ds-reaction product and allows the recycling of RecA protein in the strand exchange reaction. In the absence of ATP hydrolysis the cycle of RecA protein turnover is interrupted at the stage of the formation of the heteroduplex dsDNA that remains bound to RecA protein potentially allowing us to identify reaction phases associated with the recognition of homology and the nucleation of strand displacement.
As we demonstrated earlier (25) in the presence of ATPγS RecA protein formed stable complexes with short double-stranded fluorescent dye-labeled oligonucleotides that closely resemble the products of the reaction described in the present study. The stabilization of the strand exchange ds-product documented in the present paper demonstrates the effective binding of this product by RecA protein in agreement with our previous data (25) and a number of other studies on the stability of RecA-dsDNA complexes (27, 46, 54).
Reversibility of the Strand Exchange Reaction—It is generally accepted that the main pathway of RecA-promoted strand exchange reaction in E. coli is the formation of a RecA protein cofilament with ssDNA (presynaptic complex) and subsequent interaction of this complex with dsDNA. The stringent definition of the reverse reaction implies an invasion of free ssDNA in dsDNA bound with RecA protein. Such a pathway has been documented for RecA from Deinococcus radiodurans (57). For RecA protein from E. coli a similar process has been described earlier (23) but the results were interpreted by the authors in favor of the idea that the observed activity is independent and irrelevant to the canonical direct RecA-promoted strand exchange reaction. Besides, the effect could be observed only in the presence of ATP but not ATPγS, an additional complication to the interpretation of the results.
Due to difference in the experimental design and conditions it is difficult to directly compare our results with those previously published (23). We believe that the activity we demonstrate in the present work is a true reverse process to the conventional strand exchange because the two processes attain a steady state at the same level regardless of the extent of the reaction with the different configuration of the substrates. This equilibration does not require dissociation and recycling of RecA and, therefore, the RecA complex with the dye-labeled oligonucleotides may be considered as a distinct catalytic unit that interacts with the other partners of strand exchange, in the present case two complementary 16-mers. The extent of the strand exchange in this complex is determined only by the free energy of binding to the DNA reactants in different centers of the nucleoprotein filament and their interaction between each other (see scheme in Fig. 1A). The final product of the strand exchange reaction in our conditions is a complex of RecA protein with the heteroduplex reaction product. In the case of perfect homology between the reaction substrates, where there is no gain or loss of free energy, the bound RecA protein shifts the equilibrium toward the formation of the heteroduplex reaction product.
There is considerable evidence that in the ternary complex of RecA protein with DNA formed at the last stage of the strand exchange reaction the displaced DNA strand is still bound to the nucleoprotein filament more weakly than to the heteroduplex dsDNA, as evidenced by the fact that the displaced strand is located at some distance from the heteroduplex product and its accessibility to chemical modification resembles that of free DNA (58–61). The reversibility of the strand exchange demonstrated in the present work implies that this location of the outgoing strand does not prevent its transient interaction with the heteroduplex product of the strand exchange and that there is a dynamic equilibrium of the ternary complexes between the states in which the complementary strand pairs to the incoming or outgoing strand.
We also demonstrate that human Rad51 protein (and to some extent Dmc1) possess the capability to promote the reverse strand exchange. Therefore, the reversibility of the strand exchange may be a fundamental property of the proteins of HR and any organism may “choose” a particular direction of this reaction by modulating the recombinational proteins and possibly by loading onto different DNA substrates.
Tolerance of the Strand Exchange to Interruptions of Homology—One of the most important properties of a recombinase-promoted strand exchange reaction is its tolerance to a limited extent of heterology between the DNA substrates. It determines the efficiency of strand exchange between diverged DNA sequences and ultimately the fidelity of HR.
As expected, under our conditions formation of a mismatched product is thermodynamically unfavorable and results in a decrease of the reaction yield. In agreement with previously reported data (38, 62) this shift for complexes with RecA protein is lower than for free DNA.
The data on the sensitivity of RecA-promoted strand exchange reaction to individual base substitutions reported in the present paper are in reasonable agreement with previously reported observations (38). That the steady state in the formation of RecA-promoted synaptic complexes (38) is to some extent similar to the thermodynamically equilibrium state of the reaction between short oligonucleotide substrates reiterates the usefulness of our model system.
Human Recombinases—The eukaryotic recombinases usually exhibit significantly lower activity in traditional in vitro strand exchange and strand invasion assays than the E. coli RecA protein (39–42). At present, it is not clear if this is an intrinsic property of eukaryotic systems of HR, a consequence of non-optimal experimental conditions, or the higher requirement for additional control of the activity of these eukaryotic recombinases by additional cofactors or partner proteins. HR in eukaryotes is a much more complex process than in prokaryotes and proceeds as the concerted action of protein groups where partner proteins extensively interact physically and functionally. For example, recently, a strong stimulation of Rad51 and Dmc1 pairing activities by a Hop2-Mnd1 heterodimer was demonstrated (56, 63–65). Some of these factors may also influence the sensitivity to heterology of the human recombinase-promoted reactions. On the other hand, the increased tolerance to heterology may reflect intrinsic features of HR systems adapted to the size and complexity of the eukaryotic genomes and the roles that HR plays in these organisms. Earlier, reasoning from some features of the kinetic mechanisms of Rad51 promoted strand exchange (20, 40) indicated a lower fidelity for human Rad51. Probably, the enhanced tolerance of eukaryotic recombinases to heterology might explain the lower fidelity of modern approaches to gene correction based on HR in eukaryotes (see for review Refs. 49 and 53).
Crystal Structure of the RecA-DNA Complexes—While this paper was under revision the crystal structure of the RecA DNA complexes was published (48). There are several important implications of the published structure to our experimental system and the results obtained in our present and previous studies. In the crystals the structure of RecA protein complexes with very short (15 nt) oligonucleotides recapitulates the structure of the active complex of RecA protein with long DNA molecules and validates our experimental system based on the use of short oligonucleotides.
The published structure suggests a possible rationale for the lowered fidelity of RecA-assisted recognition of homology in comparison with that for free DNA observed in this study and earlier (38). In the triads of nucleotides in the RecA-DNA co-filament two bases out of three are stabilized by stacking only from one side and, therefore, must exhibit enhanced conformational flexibility and lowered fidelity of homology recognition.
Conclusion—In the present paper we describe a new reaction for the strand exchange among RecA protein-promoted reaction pathways. This reaction is reverse to the conventional RecA-promoted strand exchange and allows the strand exchange reaction between short oligonucleotides to reach thermodynamic equilibrium in the absence of ATP hydrolysis. We demonstrate that the capability to promote the reverse strand exchange is to some extent inherent to human recombinases as well. We directly demonstrate an increased tolerance to individual interruptions of homology between the substrates of strand exchange in the RecA-promoted reaction in comparison with spontaneous strand exchange under conditions of thermodynamic equilibrium. Finally, we show that human recombinases exhibit significantly higher tolerances to individual interruptions of homology than RecA.
Supplementary Material
Acknowledgments
We are grateful to Oleg Voloshin, Roberto Pezza, and Janet Donaldson for the proteins used in this study. We thank Paul Khil for useful discussions, Peggy Hsieh and Oleg Voloshin for a critical reading of the manuscript, and Linda Robinson for assistance.
This work was authored, in whole or in part, by National Institutes of Health staff. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
Footnotes
The abbreviations used are: HR, homologous recombination; ss, single-stranded; ds, double-stranded; ATPγS, adenosine 5′-O-(3-thiotriphosphate); nt, nucleotide.
References
- 1.Volodin, A. A., Voloshin, O. N., and Camerini-Otero, R. D. (2005) Trends Biotechnol. 23 97–102 [DOI] [PubMed] [Google Scholar]
- 2.West, S. C. (2003) Nat. Rev. Mol. Cell. Biol. 4 435–445 [DOI] [PubMed] [Google Scholar]
- 3.Dutreix, M., Fulconis, R., and Viovy, J.-L. (2003) ComPlexUs. 1 89–99 [Google Scholar]
- 4.Dorfman, K. D., Fulconis, R., Dutreix, M., and Viovy, J. L. (2004) Phys. Rev. Lett. 93 268102, 1–4 [DOI] [PubMed] [Google Scholar]
- 5.Prevost, C., and Takahashi, M. (2003) Q. Rev. Biophys. 36 429–453 [DOI] [PubMed] [Google Scholar]
- 6.Folta-Stogniew, E., O'Malley, S., Gupta, R., Anderson, K. S., and Radding, C. M. (2004) Mol. Cell 15 965–975 [DOI] [PubMed] [Google Scholar]
- 7.Voloshin, O. N., and Camerini-Otero, R. D. (2004) Mol. Cell 15 846–847 [DOI] [PubMed] [Google Scholar]
- 8.Lee, A. M., Xiao, J., and Singleton, S. F. (2006) J. Mol. Biol. 360 343–359 [DOI] [PubMed] [Google Scholar]
- 9.Egel, R. (2007) DNA Repair 6 669–675 [DOI] [PubMed] [Google Scholar]
- 10.Stasiak, A., Di Capua, E., and Koller, T. (1981) J. Mol. Biol. 151 557–564 [DOI] [PubMed] [Google Scholar]
- 11.Stasiak, A., and Di Capua, E. (1982) Nature 299 185–186 [DOI] [PubMed] [Google Scholar]
- 12.Egelman, E. (2000) Trends Biochem. Sci. 25 183–184 [DOI] [PubMed] [Google Scholar]
- 13.Egelman, E. (2000) Trends Biochem. Sci. 25 179–182 [DOI] [PubMed] [Google Scholar]
- 14.Yu, X., VanLoock, M. S., Yang, S., Reese, J. T., and Egelman, E. H. (2004) Curr. Protein Pept. Sci. 5 73–79 [DOI] [PubMed] [Google Scholar]
- 15.Voloshin, O. N., Wang, L., and Camerini-Otero, R. D. (2000) J. Mol. Biol. 303 709–720 [DOI] [PubMed] [Google Scholar]
- 16.Yu, X., Jacobs, S. A., West, S. C., Ogawa, T., and Egelman, E. H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanLoock, M. S., Yu, X., Yang, S., Lai, A. L., Low, C., Campbell, M. J., and Egelman, E. H. (2003) Structure 11 187–196 [DOI] [PubMed] [Google Scholar]
- 18.McGrew, D. A., and Knight, K. L. (2003) Crit. Rev. Biochem. Mol. Biol. 38 385–432 [DOI] [PubMed] [Google Scholar]
- 19.Chow, S. A., Rao, B. J., and Radding, C. M. (1988) J. Biol. Chem. 263 200–209 [PubMed] [Google Scholar]
- 20.Bazemore, L. R., Takahashi, M., and Radding, C. M. (1997) J. Biol. Chem. 272 14672–14682 [DOI] [PubMed] [Google Scholar]
- 21.Bazemore, L. R., Folta-Stogniew, E., Takahashi, M., and Radding, C. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11863–11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugh, B. F., and Cox, M. M. (1987) J. Biol. Chem. 262 1337–1343 [PubMed] [Google Scholar]
- 23.Zaitsev, E. N., and Kowalczykowski, S. C. (2000) Genes Dev. 14 740–749 [PMC free article] [PubMed] [Google Scholar]
- 24.Zaitsev, E. N., and Kowalczykowski, S. C. (1998) Nucleic Acids Res. 26 650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volodin, A. A., Smirnova, H. A., and Bocharova, T. N. (1994) FEBS Lett. 349 65–68 [DOI] [PubMed] [Google Scholar]
- 26.Volodin, A. A., Smirnova, H. A., and Bocharova, T. N. (2000) FEBS Lett. 473 53–57 [DOI] [PubMed] [Google Scholar]
- 27.Rosselli, W., and Stasiak, A. (1990) J. Mol. Biol. 216 335–352 [DOI] [PubMed] [Google Scholar]
- 28.Ullsperger, C. J., and Cox, M. M. (1995) Biochemistry 34 10859–10866 [DOI] [PubMed] [Google Scholar]
- 29.Makino, O., Ikawa, S., Shibata, Y., Maeda, H., Ando, T., and Shibata, T. (1987) J. Biol. Chem. 262 12237–12246 [PubMed] [Google Scholar]
- 30.Cox, M. M., and Lehman, I. R. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riddles, P. W., and Lehman, I. R. (1985) J. Biol. Chem. 260 170–173 [PubMed] [Google Scholar]
- 32.Honigberg, S. M., Gonda, D. K., Flory, J., and Radding, C. M. (1985) J. Biol. Chem. 260 11845–11851 [PubMed] [Google Scholar]
- 33.Volodin, A. A., Smirnova, H. A., and Bocharova, T. N. (1997) FEBS Lett. 407 325–328 [DOI] [PubMed] [Google Scholar]
- 34.Volodin, A. A., and Camerini-Otero, R. D. (2002) J. Biol. Chem. 277 1614–1618 [DOI] [PubMed] [Google Scholar]
- 35.Volodin, A. A., Smirnova, E. A., Bocharova, T. N., and Camerini-Otero, R. D. (2003) FEBS Lett. 546 203–208 [DOI] [PubMed] [Google Scholar]
- 36.Baumann, P., Benson, F. E., Hajibagheri, N., and West, S. C. (1997) Mutat. Res. 384 65–72 [DOI] [PubMed] [Google Scholar]
- 37.Masson, J. Y., Davies, A. A., Hajibagheri, N., Van Dyck, E., Benson, F. E., Stasiak, A. Z., Stasiak, A., and West, S. C. (1999) EMBO J. 18 6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malkov, V. A., and Camerini-Otero, R. D. (1998) J. Mol. Biol. 278 317–330 [DOI] [PubMed] [Google Scholar]
- 39.Morrison, C., Shinohara, A., Sonoda, E., Yamaguchi-Iwai, Y., Takata, M., Weichselbaum, R. R., and Takeda, S. (1999) Mol. Cell. Biol. 19 6891–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta, R. C., Bazemore, L. R., Golub, E. I., and Radding, C. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, Z., Golub, E. I., Gupta, R., and Radding, C. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, K. P., Eggler, A. L., Sung, P., and Cox, M. M. (2001) J. Biol. Chem. 276 38570–38581 [DOI] [PubMed] [Google Scholar]
- 43.Bugreev, D. V., Golub, E. I., Stasiak, A. Z., Stasiak, A., and Mazin, A. V. (2005) J. Biol. Chem. 280 26886–26895 [DOI] [PubMed] [Google Scholar]
- 44.Bugreev, D. V., and Mazin, A. V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chi, P., Van Komen, S., Sehorn, M. G., Sigurdsson, S., and Sung, P. (2006) DNA Repair 5 381–391 [DOI] [PubMed] [Google Scholar]
- 46.Gumbs, O. H., and Shaner, S. L. (1998) Biochemistry 37 11692–11706 [DOI] [PubMed] [Google Scholar]
- 47.Xiao, J., Lee, A. M., and Singleton, S. F. (2006) Biopolymers 81 473–496 [DOI] [PubMed] [Google Scholar]
- 48.Chen, Z., Yang, H., and Pavletich, N. P. (2008) Nature 453 484–489 [DOI] [PubMed] [Google Scholar]
- 49.Sangiuolo, F., and Novelli, G. (2004) Cytogenet. Genome Res. 105 435–441 [DOI] [PubMed] [Google Scholar]
- 50.Hsieh, P., Camerini-Otero, C. S., and Camerini-Otero, R. D. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6492–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golub, E. I., Radding, C. M., and Ward, D. C. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7186–7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao, J., Lee, A. M., and Singleton, S. F. (2006) ChemBioChem 7 1265–1278 [DOI] [PubMed] [Google Scholar]
- 53.Igoucheva, O., Alexeev, V., and Yoon, K. (2004) Curr. Mol. Med. 4 445–463 [DOI] [PubMed] [Google Scholar]
- 54.Kowalczykowski, S. C., and Krupp, R. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3478–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menetski, J. P., Bear, D. G., and Kowalczykowski, S. C. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pezza, R. J., Voloshin, O. N., Vanevski, F., and Camerini-Otero, R. D. (2007) Genes Dev. 21 1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim, J. I., and Cox, M. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7917–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adzuma, K. (1992) Genes Dev. 6 1679–1694 [DOI] [PubMed] [Google Scholar]
- 59.Xiao, J., and Singleton, S. F. (2002) J. Mol. Biol. 320 529–558 [DOI] [PubMed] [Google Scholar]
- 60.Malkov, V. A., Panyutin, I. G., Neumann, R. D., Zhurkin, V. B., and Camerini-Otero, R. D. (2000) J. Mol. Biol. 299 629–640 [DOI] [PubMed] [Google Scholar]
- 61.Voloshin, O. N., and Camerini-Otero, R. D. (1997) Genes Cells 2 303–314 [DOI] [PubMed] [Google Scholar]
- 62.Malkov, V. A., Sastry, L., and Camerini-Otero, R. D. (1997) J. Mol. Biol. 271 168–177 [DOI] [PubMed] [Google Scholar]
- 63.Chi, P., San Filippo, J., Sehorn, M. G., Petukhova, G. V., and Sung, P. (2007) Genes Dev. 21 1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petukhova, G. V., Pezza, R. J., Vanevski, F., Ploquin, M., Masson, J. Y., and Camerini-Otero, R. D. (2005) Nat. Struct. Mol. Biol. 12 449–453 [DOI] [PubMed] [Google Scholar]
- 65.Pezza, R. J., Petukhova, G. V., Ghirlando, R., and Camerini-Otero, R. D. (2006) J. Biol. Chem. 281 18426–18434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.