Abstract
RecF pathway proteins play an important role in the restart of stalled replication and DNA repair in prokaryotes. Following DNA damage, RecF, RecR, and RecO initiate homologous recombination (HR) by loading of the RecA recombinase on single-stranded (ss) DNA, protected by ssDNA-binding protein. The specific role of RecF in this process is not well understood. Previous studies have proposed that RecF directs the RecOR complex to boundaries of damaged DNA regions by recognizing single-stranded/double-stranded (ss/ds) DNA junctions. RecF belongs to ABC-type ATPases, which function through an ATP-dependent dimerization. Here, we demonstrate that the RecF of Deinococcus radiodurans interacts with DNA as an ATP-dependent dimer, and that the DNA binding and ATPase activity of RecF depend on both the structure of DNA substrate, and the presence of RecR. We found that RecR interacts as a tetramer with the RecF dimer. RecR increases the RecF affinity to dsDNA without stimulating ATP hydrolysis but destabilizes RecF binding to ssDNA and dimerization, likely due to increasing the ATPase rate. The DNA-dependent binding of RecR to the RecF-DNA complex occurs through specific protein-protein interactions without significant contributions from RecR-DNA interactions. Finally, RecF neither alone nor in complex with RecR preferentially binds to the ss/dsDNA junction. Our data suggest that the specificity of the RecFOR complex toward the boundaries of DNA damaged regions may result from a network of protein-protein and DNA-protein interactions, rather than a simple recognition of the ss/dsDNA junction by RecF.
Homologous recombination (HR)2 is one of the primary mechanisms by which cells repair dsDNA breaks (DSBs) and ssDNA gaps (SSGs), and is important for restart of stalled DNA replication (1). HR is initiated when RecA-like recombinases bind to ssDNA forming an extended nucleoprotein filament, referred to as a presynaptic complex (2). The potential for genetic rearrangements dictates that HR initiation is tightly regulated at multiple levels (1). During replication, the ssDNA-binding protein (SSB) protects transiently unwound DNA chains, preventing interactions with recombinases. Following DNA damage, recombination mediator proteins (RMPs) initiate HR by facilitating the formation of the recombinase filaments with ssDNA, while removing SSB (3, 4). Mutations in human proteins involved in HR initiation are linked to cancer predisposition, chromosome instability, UV sensitivity, and premature aging diseases (4–8). To date, little is known about the mechanism by which RMPs regulate the formation of the recombinase filaments on the SSB-protected ssDNA.
In Escherichia coli, there are two major recombination pathways, RecBCD and RecF (9, 10). A helicase/nuclease RecBCD complex processes DSBs and recruits RecA on ssDNA in a sequence-specific manner (11–13). The principle players in the RecF pathway are the RecF, RecO, and RecR proteins, which form an epistatic group that is important for SSG repair, for restart of stalled DNA replication, and under specific conditions, can also process DSBs (14–20). Homologs of RecF, -O, and -R are present in the majority of known bacteria (21), including Deinococcus radiodurans, extremely radiation-resistant bacteria that lacks the RecBCD pathway, yet is capable of repairing thousands of DSBs (22, 23). In addition, the sequence or functional homologs of RecF pathway proteins are involved in similar pathways in eukaryotes that include among others WRN, BLM, RAD52, and BRCA2 proteins (4–8).
The involvement of all three RecF, -O, and -R proteins in HR initiation is well documented by genetic and cellular approaches (18, 24–30), yet their biochemical functions in the initiation process remain unclear, particularly with respect to RecF. RecO and RecR proteins are sufficient to promote formation of the RecA filament on SSB-bound ssDNA in vitro (27). The UV-sensitive phenotype of recF mutants can be suppressed by RecOR overexpression, suggesting that RecF may direct the RMP complex to DNA-damaged regions where HR initiation is required (31). In agreement with this hypothesis, RecF dramatically increases the efficiency of the RecA loading at ds/ssDNA junctions with a 3′ ssDNA extension under specific conditions (32). RecF and RecR proteins also prevent the RecA filaments from extending into dsDNA regions adjacent to SSGs (33). These data suggest that RecF may directly recognize an ss/dsDNA junction structure (34). However, DNA binding experiments have not provided clear evidence to support such a hypothesis (11).
The targeting promoted by RecF may also occur through more complex processes. RecF shares a high structural similarity with the head domain of Rad50, an ABC-type ATPase that recognizes DSBs and initiates repair in archaea and eukaryotes (35). All known ABC-type ATPases function as oligomeric complexes in which a sequence of inter- and intra-molecular interactions is triggered by the ATP-dependent dimerization and the dimer-dependent ATP hydrolysis (36–39). RecF is also an ATP-dependent DNA-binding protein and a weak DNA-dependent ATPase (11, 40). RecF forms an ATP-dependent dimer and all three conserved motifs (Walker A, Walker B, and “signature”) of RecF are important for ATP-dependent dimerization, ATP hydrolysis, and functional resistance to DNA damage (35). Thus, RecF may function in recombination initiation through a complex pathway of protein-protein and DNA-protein interactions regulated by ATP-dependent RecF dimerization.
In this report, we present a detailed characterization of the RecF dimerization, and its role in the RecF interaction with various DNA substrates, with RecR, and in ATP hydrolysis. Our data outline the following key findings. First, RecF interacts with DNA as a dimer. Second, neither RecF alone nor the RecFR complex preferentially binds the ss/dsDNA junction. Finally, RecR changes the ATPase activity and the DNA binding of RecF by destabilizing the interaction with ssDNA, and greatly enhancing the interaction with dsDNA. Our results suggest that the specificity of RecF for the boundaries of SSGs is likely to result from a sequence of protein-protein interaction events rather than a simple RecF ss/dsDNA binding, underlining a highly regulated mechanism of the HR initiation by the RecFOR proteins.
EXPERIMENTAL PROCEDURES
Expression and Purification of RecF, RecO, and RecR
RecF, RecR, and RecO proteins are from D. radiodurans, unless stated otherwise. All chemicals were purchased from Sigma and Fisher Scientific and were of the highest grade available. The recF, recR, and recO genes were amplified from D. radiodurans R1 genomic DNA (American Type Culture Collection), cloned into the pMCSG7 plasmid as described elsewhere (41), and expressed in the Escherichia coli strain BL21(DE3) pLysS (Novagen). Cells were harvested by centrifugation and resuspended in a buffer containing 1.0 m KCl, 10% glycerol (v/v), 20% sucrose (w/v), 20 mm HEPES sodium (pH 8.0), 0.2% Triton X-100 (v/v), 0.5 mm TCEP (tris(2-carboxyethyl)phosphine), 1 mm phenylmethanesulfonyl fluoride, and 1 mg ml–1 lysozyme. Cells were lysed by a freeze-thaw cycle, incubated at room temperature for 30 min, and then sonicated on ice. Insoluble cellular material was cleared by centrifugation at 14,000 rpm (Sorvall SS-40 rotor) for 40 min. The expressed His-tagged proteins were isolated using Ni-NTA (Qiagen) affinity resin. The N-terminal His tag was removed by incubation with tobacco etch virus protease. Uncleaved His-tagged proteins and other contaminants with elevated Ni-NTA affinity were removed by passing through the Ni-NTA resin again. The proteins were additionally purified with size-exclusion chromatography (Sephacryl 200, 26/60 column). Fractions corresponding to the monomeric protein were pooled, 10% DMSO added, concentrated again, and dialyzed against the storage buffer (40% glycerol (v/v), 10% DMSO (v/v), 1 m KCl, 20 mm HEPES sodium (pH 8.0), 0.5 mm TCEP). All protein stocks were stored at –80 °C.
For all assays, the proteins were dialyzed against Buffer A (10% glycerol (v/v), 10% DMSO (v/v), 50 mm KCl, 20 mm HEPES sodium (pH 8.0), 10 mm MgCl2, 0.5 mm TCEP) unless stated otherwise. After dialysis, the insoluble material was removed by centrifugation at 14,000 rpm (Spectrafuge M16, Labnet) for 60 min at 4 °C. The aggregation states of the obtained proteins were monitored using dynamic light scattering with the DynaPro Titan (Wyatt, Inc.). All measurements were performed at room temperature to minimize protein aggregation, observed to occur at higher temperatures in previous studies (11).
RecF Mutagenesis
Several mutations were introduced in RecF: signature motif, S279R; Walker A motif, K39R; and C-terminal flexible tail, A355C. Site-specific mutations were generated on the pMSCG7 plasmid using the Quick-change II site-directed mutagenesis kit (Stratagene). Reactions were performed according to the manufacturer's protocol, and mutations were confirmed by sequencing (Retrogene).
Labeling of RecF(A355C) Mutant with Fluorescein (FAM) and Cy3
Cysteine reactive dyes were purchased from Molecular Probes (Invitrogen). Conjugation of the dyes to RecF was carried out according to the manufacturer's protocol in Buffer A without reducing agent. The overnight reaction mixture was quenched by adding an excess of dithiothreitol to a final concentration of 10 mm and incubating on ice for 10 min. The labeled proteins were separated from free dye on a size-exclusion chromatography column, Sephacryl-200 16/60 (GE Healthcare).
Dimerization Assay
The Föster resonance energy transfer (FRET) technique was utilized to quantitatively characterize the dimerization of RecF under equilibrium conditions. RecF(A355C)-FAM and RecF(A355C)-Cy3 proteins were mixed in equimolar amounts, dialyzed overnight against Buffer A, and titrated with ATP or ADP. The absence of higher molecular weight aggregates of RecF was monitored with dynamic light scattering. The mixture was incubated for 10 min at room temperature and transferred to a Corning assay plate (384 well, low volume). FRET measurements were taken on a Tecan SPECTRAFluor Plus fluorescence reader using excitation wavelength 490 nm, with emitted intensities measured at 530 nm (IFAM) and 570 nm (ICy3), corresponding to the emissions of the FAM and Cy3 fluorophores, respectively. The FRET signal was calculated according to Equation 1,
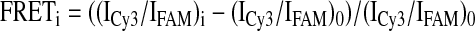 |
(Eq. 1) |
where (ICy3/IFAM)0 and (ICy3/IFAM)i are measurements at 0 and “i” ATP concentration points.
The FRET assay was conducted at six different RecF concentrations: 0.1, 0.2, 0.4, 1, 2, and 4 μm. Three independent measurements at each concentration were obtained to determine the experimental error. Data were treated by two different methods as follows.
Method 1—A plot of the estimated maximal FRET signal (Bmax) against the concentration of RecF was used to derive the dimerization constant. The FRET data were analyzed as a one-step reaction model as described in Reaction 1 using numerical nonlinear regression in the program SCIENTIST (Micromath Inc.).
 |
REACTION 1 |
where RecFATP is the RecF-ATP complex, and (RecFATP)2 is the dimer of the RecF-ATP complex. La is the association constant.
The data were fitted into an equilibrium equation (Equation 2),
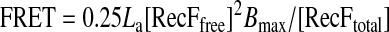 |
(Eq. 2) |
where [RecFfree] is the concentration of free RecF, [RecFtotal]is the concentration of total RecF, Bmax is the maximal FRET signal observed, and RecF is an equimolar mixture of FAM- and Cy3-labeled proteins.
Concentration of free RecF was calculated numerically for each point according to the corresponding mass conservation equations. Similar calculations were done for the data obtained in other experiments described below. More detailed descriptions of calculation schemes are provided in the supplemental materials.
Method 2—All six datasets corresponding to the six RecF concentrations were analyzed as a two-step reaction model described by Reaction 2 to estimate nucleotide binding and dimerization constants using numerical nonlinear regression with SCIENTIST program (Micromath Inc).
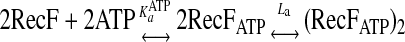 |
REACTION 2 |
The data were fitted into an equilibrium equation (Equation 3),
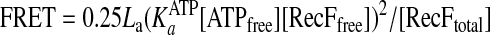 |
(Eq. 3) |
where [ATPfree] is the concentration of free ATP, KaATP is the association constant of ATP binding, [RecFfree] is the concentrations of free protein, and [RecFtotal] is the total RecF concentration.
DNA Binding Assay
All oligonucleotides were purchased from Integrated DNA Technologies. The D1 duplex DNA (20-mer) was prepared by annealing fluorescein-labeled S1 single-stranded oligonucleotide (20-mer) TATCCGCAGAGTTGGCTGGT with its complementary oligonucleotide. The J1 junction (15/15) was prepared by annealing of the fluorescein-labeled 30-mer TAT CCG CAG AGT TGG CTG GTA GTT CAG CCC with its complementary 15-mer oligonucleotide CCA ACT CTG CGG ATA. Oligonucleotide sequences were designed to avoid alternative secondary structures.
The reaction mixtures contained 20 nm of fluorescein-labeled oligonucleotides, various concentrations of RecF, and 30 μm of RecR, where indicated. All DNA concentrations are expressed in molecules. The total volume of reaction was 70 μl. After incubation at room temperature for 10 min, 20 μl of reaction mixture was transferred onto a Corning assay plate (384 well, low volume). Fluorescence polarization was measured on a Biosystems Analyst AD 96–384, and the normalized fluorescence polarization value (FP) in arbitrary units was calculated according to Equation 4,
 |
(Eq. 4) |
where Pi is the fluorescence polarization value of a given sample, P0 is the fluorescence polarization value at 0 μm concentration of RecF. The data were analyzed using SCIENTIST according to Equation 5,
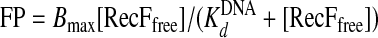 |
(Eq. 5) |
where Bmax is the maximal signal, [RecFfree] is the concentration of free RecF, and K DNAd is the dissociation constant of the RecF-DNA complex.
ATPase Assay
The oligonucleotides were prepared as described for the fluorescence polarization assay with non-labeled oligonucleotides. The J2 junction was prepared by annealing the 20-mer S2 oligonucleotide TTT TTT TTT TGC TGC CCA CA with 10-mer complementary strand TGT GGG CAG C. RecF and RecR were separately dialyzed overnight against Buffer A containing 2 mm ATP. The ATPase reactions were initiated by mixing the RecF solutions with DNA. RecR was included in the mixture where indicated. The final volume of the reaction mixture was 25 μl with 10 μm of RecF, and 50 μm of RecR, when present. Aliquots of 5 μl were taken at different time points and mixed with 200 μl of HCl (pH 2) to quench the ATP hydrolysis. The quenched reaction was mixed with Malachite Green reagent (BioAssay), incubated at 20 °C for 10 min, and optical density (A) was measured on a plate reader (Molecular Devices, Thermomax Microplate Reader, Softmax) at 650 nm wavelength. Three independent experiments were conducted for each data point. The standard curve of inorganic phosphate concentration versus A value was built according to the manufacturer's protocol. Initial trials with different incubation times ranging from 30 to 120 min yielded similar results, and the 120-min incubation was chosen to minimize overall error.
Experimental data were analyzed using one site saturation model with SCIENTIST and SigmaPlot8.0. Two values were obtained: K DNAd, which represents the apparent dissociation constants of the RecF-DNA complexes, and Bmax, which represents the maximal rate of ATP hydrolysis at the saturating concentration of a given DNA substrate. These values were calculated according to Equation 6,
 |
(Eq. 6) |
where ΔA is a difference between optical densities at 0 min and a given time point, K DNAa is the association constant of the corresponding RecF-DNA complex, [RecFtotal] is concentration of total RecF, and [DNAfree] and [RecFfree] are concentrations of free DNA and RecF, respectively. The ΔA was converted to the amount of hydrolyzed ATP according to the manufacturer's protocol.
Stoichiometry of RecF-DNA-RecR Interaction
The DNA binding assay described above was implemented to estimate the stoichiometry of proteins in the RecFR-DNA complex. The FAM-labeled dsDNA substrate D1 was diluted in a 1:49 ratio with non-labeled D1, mixed with equimolar amount of RecF, and titrated with RecR. The fluorescence polarization signal was plotted against the RecR concentration for each RecF concentration, then analyzed with SigmaPlot 8.0 (SPSS, Inc.) using a single-site ligand-interaction model and Equation 7,
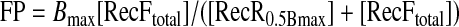 |
(Eq. 7) |
where Bmax is the maximal signal observed, [RecFtotal]isthe total RecF concentration, and [RecR0.5Bmax] is the RecR concentration at the half saturation signal.
Values of [RecR0.5Bmax] were plotted against [RecFtotal], and the best linear fit was calculated with SigmaPlot 8.0 according to Equation 8,
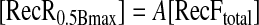 |
(Eq. 8) |
where the slope A corresponds to the stoichiometry for RecF-RecR interaction in the presence of dsDNA.
RESULTS
Experimental Design Rationale—In this report we characterized solution properties of RecF and RecR proteins from D. radiodurans, and we will refer to them as RecF and RecR unless stated otherwise. RecF pathway proteins are the only known RMPs in this extremely DNA damage-resistant bacteria, hence studies on these organisms are critical to better understand repair mechanisms. The primary amino acid sequence of RecF from this organism lacks cysteine residues, which was convenient for introduction of this residue at specific sites for modification with fluorescent dyes. This was important to study RecF interactions under equilibrium conditions.
RecF has a high propensity to form nonspecific aggregates, and such high molecular weight aggregates are capable of DNA binding (34, 42, 43). To avoid spontaneous aggregation, the concentration of RecF was limited to 15 μm, all protein solutions were clarified by centrifugation, and the absence of nonspecific aggregates was monitored using dynamic light scattering.
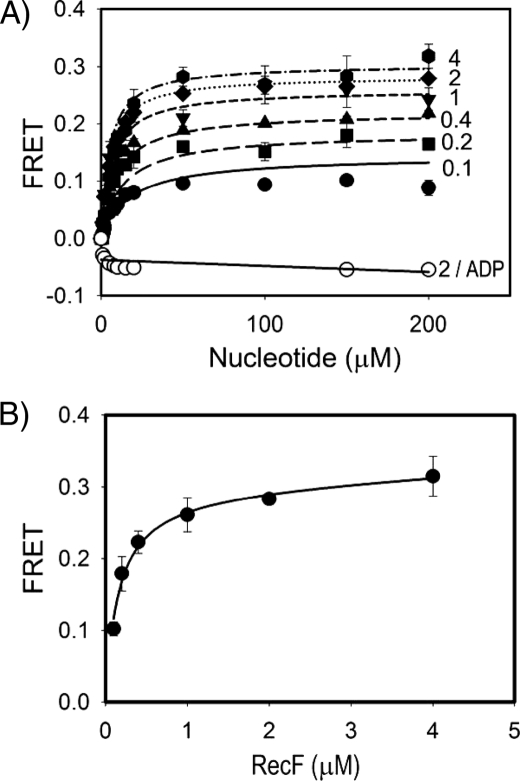
Characterization of ATP-dependent RecF Dimerization with FRET Assay—Previously we demonstrated that RecF forms a dimer in the presence of ATP using two methods. First, size-exclusion chromatography combined with static light scattering was utilized to measure the absolute molecule mass of protein complexes in solution, and, second, dynamic light scattering was used to monitor changes in oligomerization state of the wild-type protein and mutants under different buffer conditions (35). To quantitatively characterize RecF dimerization in solution and its role in interactions with DNA and other recombination proteins, we utilized the FRET technique. An equimolar mixture of the RecF cysteine mutants (supplemental Fig. S1) labeled either with fluorescein or with Cy3 was titrated with ATP or ADP. An increase in FRET signal due to the dimerization of RecF was measured at different protein concentrations (Fig. 1A). A secondary plot of the maximal FRET signals determined for each RecF concentration against the corresponding protein concentration was used to derive the dimerization constant (Fig. 1B). The data were fitted into Equation 1 according to Reaction 1, and a dimerization constant of Ld = 0.15 ± 0.02 μm was obtained. Alternatively, all six data sets were globally fitted into a two-step reaction model consisting of the ATP-binding and dimerization processes (Reaction 2 and Equation 2). This model resulted in a dimerization constant of Ld = 0.13 ± 0.02 μm, and an ATP-binding constant of K ATPd = 13 ± 2 μm. Additional tryptophan quenching experiments produced a similar estimation of the ATP-binding constant (data not shown).
FIGURE 1.
FRET measurements of ATP-dependent RecF dimerization. A, reactions were carried out as described under “Experimental Procedures.” Signals from FAM and Cy3 labels were measured upon titration of an equimolar mixture of FAM- and Cy3-labeled RecF proteins with ATP or ADP. Changes of FRET signal in arbitrary units were plotted against corresponding nucleotide concentrations. The lanes represent theoretical fit performed with Scientist software and plotted with SigmaPlot 8.0 (SPSS Inc.). Solid, long-dash, medium-dash, short-dash, dotted, and dash-dotted lines with filled symbols correspond to 0.1, 0.2, 0.4, 1, 2, and 4 μm concentrations of RecF, respectively. The solid line with open circles corresponds to titration of 2 μm RecF with ADP. B, a secondary plot of the Bmax values versus the RecF concentrations derived from the FRET measurements shown in A. The curve represents theoretical fitting of data with SigmaPlot.
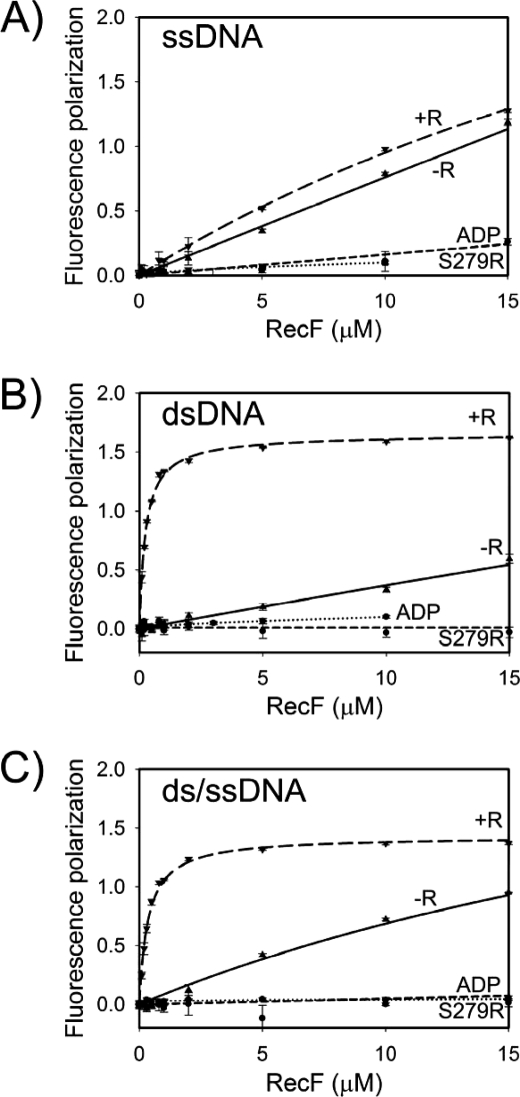
RecF-DNA Binding Requires the RecF Dimerization—Because ATP is required for DNA binding in the case of the E. coli RecF, and for dimerization in the case of the D. radiodurans RecF, it is reasonable to suggest that RecF interacts with DNA as a dimer. We compared DNA binding of the wild-type RecF and of the dimerization-deficient signature motif mutant S279R (35), and the DNA binding of the wild-type protein in the presence of ATP or ADP. The fluorescence polarization technique was utilized where different 20-mer oligonucleotides were modified with fluorescein (FAM) and titrated by RecF. The ssDNA (S1) (Fig. 2A), dsDNA (D1) (Fig. 2B), and junction DNA (J1) (Fig. 2C) substrates were tested. The wild-type RecF interacted with all DNA substrates in the presence of ATP. However, DNA binding was not observed for the wild-type RecF in the presence of ADP and for the RecF(S279R) mutant in the presence of ATP. Because all conditions that prevent dimerization were also unfavorable for DNA binding, the ATP-dependent dimerization of RecF must be essential for interactions with different DNA substrates.
FIGURE 2.
RecF-DNA interaction measured by the fluorescence polarization assay. The labeled DNA substrates (20 nm) were titrated with RecF in the presence of 2 mm nucleotide. RecR was included where indicated at final concentration of 30 μm. Normalized fluorescence polarization signal in arbitrary units was plotted against protein concentrations. The graphs were created with SigmaPlot 8.0 (SPSS Inc.). RecF interactions with ssDNA (S1) are shown in panel A, with dsDNA (D1) in panel B, and with ds/ssDNA (J1) junction in panel C. Dotted lines represent the interaction of the RecF(S279R) mutant with DNA in the presence of ATP; short-dashed lines, a wild-type RecF in the presence of ADP; solid lines, a wild-type RecF in the presence of ATP; long-dashed lines, a wild-type RecF in the presence of ATP and RecR.
The direct DNA binding assays should be interpreted with caution because RecF hydrolyzes ATP in the presence of DNA (11, 42), and ATP hydrolysis can potentially disrupt the dimer. Previously, it was shown that ATP hydrolysis interferes with DNA binding of E. coli RecF (11, 42). A slow decrease in RecF dimerization in the presence of DNA was also detected in FRET assay (supplemental Fig. S2). The decrease was significant only when ATP concentration was <50 μm, and after extended incubation time longer than 10 min, due to slow ATP hydrolysis by RecF. This supports the idea that DNA-dependent ATP hydrolysis may disrupt the RecF dimer and, consequently, DNA binding. At the same time, the DNA binding experiments conducted with 2 mm ATP and a relatively short period of incubation time served as a good approximation of the initial DNA binding by the RecF dimer when the effect of ATP hydrolysis was negligible.
Alternative approaches to prevent ATP hydrolysis were not implemented. First, non-hydrolyzable ATP analogs such as ATPγS, AMPPNP, and ADP·AlF4 did not support stable dimerization under the experimental conditions in the absence of DNA and RecR, even though the tryptophan-quenching measurements demonstrated that the binding of analogs to RecF was comparable to the binding of ATP (data not shown). Second, previously utilized mutation of the conserved lysine in the Walker A motif to arginine (11, 35, 44) to prevent ATP hydrolysis resulted in a protein with much higher propensity to aggregate and lower solubility. Therefore, all direct DNA binding experiments were performed with 2 mm ATP, and the measurements were taken within 10-min incubation time. Alternatively, ATP hydrolysis at an extended period of time was measured to quantify the ATPase activity of RecF, and to measure binding to different DNA substrates.
RecR Stabilizes the RecF Interaction with dsDNA, but Not with ssDNA—The E. coli RecR interacts with the E. coli RecF only in the presence of dsDNA and stabilizes the dsDNA-RecF interaction (11, 42). We tested the D. radiodurans counterparts for the same properties using different DNA substrates. RecR strongly increased the binding of RecF to dsDNA (Fig. 2B). In the presence of RecR, the apparent dissociation constant for the RecF-dsDNA interaction K DNA(RecR)d = 0.25 ± 0.02 μm, was at least an order of magnitude stronger than without RecR. In the case of ssDNA, the presence of RecR did not result in significant changes of RecF-ssDNA interaction (Fig. 2A). No interactions of RecR with any of the DNA substrates were observed under similar conditions (data not shown). Thus, RecR specifically stabilized only the RecF-dsDNA complex.
Interestingly, RecF alone did not show an obvious preferential binding to dsDNA. Similar binding experiments with the crowding agent polyethylene glycol 10,000 (PEG10K) showed a slight preference of RecF toward ssDNA in the absence of RecR, with the apparent dissociation constants K ssDNA(PEG10K)d = 0.33 ± 0.04 μm and K dsDNA(PEG10K)d = 0.77 ± 0.08 μm (supplemental Fig. S3).
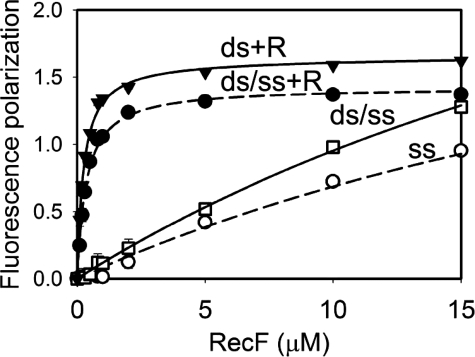
RecF Does Not Have Higher Affinity for the ss/dsDNA Junction—RecF was suggested to recognize the ss/dsDNA junction structure providing the RecFOR complex specificity toward boundaries of SSGs (11, 34). Previous experiments did not define whether RecF alone had elevated affinity toward the junction DNA due to the large DNA plasmids employed as substrates. We addressed this question using short oligonucleotides. RecF binding to junction DNA J1 was within the similar range of binding to single-stranded or double-stranded substrates (Figs. 2 and 3). Additional DNA binding experiments under conditions in the presence of crowding agent yielded similar results (supplemental Fig. S3). No preference for the junction DNA was detected in the presence of RecR either (Figs. 2 and 3). Similarly, a lack of specificity toward the junction DNA was observed at higher salt concentration of 200 mm KCl (supplemental Fig. S4). Our results demonstrate that RecF, neither alone nor in the presence of RecR, binds the ss/dsDNA junction with higher affinity.
FIGURE 3.
Lack of specificity of RecF to the junction DNA. Measurements were performed as described in Fig. 2. Interactions of RecF with ds/ssDNA (J1) and ssDNA (S1) in the absence of RecR are shown by open circles and open squares, respectively. Interactions of RecF with dsDNA (D1) and ds/ssDNA (J1) in the presence of RecR are shown by closed triangles and closed circles, respectively.
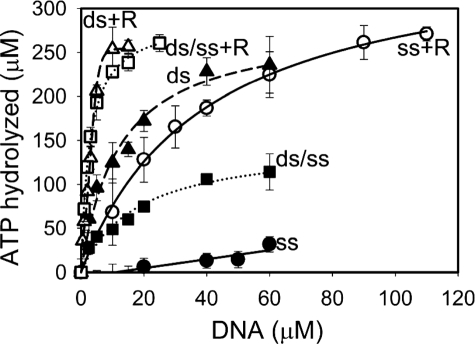
RecR Has an Opposite Effect on the RecF DNA Binding and ATP Hydrolysis in the Presence of ssDNA and dsDNA—Previously, we demonstrated that RecF hydrolyzes ATP under crowding conditions in the presence of 30-mer dsDNA twice faster than in the presence of 30-mer ssDNA (35). In this report, we measured the ATP hydrolysis in the presence of different 20-mer oligonucleotides under conditions identical to those employed in the dimerization and DNA binding experiments (Figs. 2 and 3) to compare maximal ATPase rates and DNA binding constants.
In the case of D1 substrate, the calculated maximal ATPase activity of RecF was similar (0.23 ± 0.01 min–1) in the absence or presence of RecR (Fig. 4). Saturation of ATPase activity in the presence of RecR was achieved at lower DNA concentrations than in the absence of RecR due to a stronger RecF-DNA interaction with K dsDNA(RecR)d = 0.5 ± 0.21 μm versus K dsDNAd = 8.3 ± 1.6 μm (Fig. 4). Estimation of the DNA binding constants yielded by the ATPase assay were in an excellent agreement with the direct binding experiments described earlier for dsDNA (Fig. 2B). Under slightly different conditions, with a longer DNA substrate and in the presence of 5% PEG10K, the ATPase activity of the RecFR complex was lower than that of RecF alone in the presence of dsDNA (supplemental Table S1, lane ds(30)) (35). The data suggest that RecR may stabilize interaction between RecF and dsDNA via two mechanisms: first, by increasing the affinity of RecF to dsDNA, and, second, by decreasing the ATPase rate of RecF in the presence of dsDNA. The lack of dsDNA-dependent stimulation of ATPase rate by RecR contrasts with the results for the E. coli proteins (11). The apparent contradiction may be due to the fact that RecF hydrolyzes ATP only when bound to DNA and ATPase rates must be normalized by the amount of the RecF-DNA complex rather than total protein. The apparent ATP hydrolysis by RecF in the presence of RecR at low protein concentration will be greater than that by RecF alone due to considerably higher amount of protein-DNA complex formed in the presence of RecR, whereas the estimated Bmax values are the same (Fig. 4).
FIGURE 4.
RecF ATPase activity in the presence of different DNA substrates and RecR. Reactions were carried out as described under “Experimental Procedures.” RecF at 10 μm concentration was titrated with different DNA substrates in the presence of 2 mm ATP. RecR at 50 μm was included where indicated. Amount of hydrolyzed ATP was calculated based on the amount of released inorganic phosphate and was plotted against DNA concentrations. The ATPase activity in the absence of RecR is shown with closed circles for ssDNA (S1), with closed squares for ds/ssDNA junction (J2), and with closed triangles for dsDNA (D1). The activity in the presence of RecR is shown by open circles, squares, and triangles for S1, J2, and D1 substrates, correspondingly. The lines represent theoretical fitting using the Scientist software and Equation 5.
In the case of ssDNA substrate S1, the ATPase measurements clearly demonstrated interaction between RecF and RecR (Fig. 4), not evident from the direct DNA binding experiments (Fig. 2A). RecR significantly increased the ssDNA-stimulated maximal ATPase activity of RecF from 0.01 ± 0.01 min–1 to 0.31 ± 0.01 min–1 (Fig. 4). This effect also allowed us to estimate the RecF-ssDNA dissociation constant in the presence of RecR with K ssDNA(RecR)d = 35 ± 2.3 μm. Fluorescence polarization assay clearly showed the RecF-ssDNA interaction (Fig. 2A) in contrast to almost no ATPase activity (Fig. 4), meaning that the RecF-ssDNA complex is a poor ATPase. In previous studies with lower DMSO concentration (5%) and longer ssDNA substrate (30-mer), the ATPase rate of RecF was 0.08 ± 0.01 min–1 (35). An apparent lack of the effect of RecR on ssDNA binding observed in fluorescence polarization experiment may be explained by strong increase of ATPase rate by RecFR complex and faster dissociation of the ssDNA-protein complex. This will result in overall small contribution of RecFR complex into RecF interaction with ssDNA.
Similarly to the direct DNA binding measurements (Figs. 2 and 3), the ATPase assays did not show any preferential binding of RecF to the DNA junction J1 in the absence of RecR (K ds/ssDNAd = 8 ± 1.4 μm, maximal ATPase rate 0.1 ± 0.003 min–1) or in the presence of RecR (K ds/ssDNA(RecR)d = 0.32 ± 0.25 μm, maximal ATPase rate 0.21 ± 0.01 min–1) (Fig. 4).
RecR Stabilizes the RecF Dimerization and DNA Binding with ATP Analogs—Because RecR enhances interaction between the RecF dimer and dsDNA, it should stabilize weak RecF dimers formed in the presence of ATP analogs not observed for RecF alone under employed conditions. The fluorescence polarization DNA binding assay was utilized as a measurement of both the RecF-DNA interaction and the RecF dimerization, because the latter is required for DNA binding. Interaction of DNA with RecF and RecR was observed in the case of all ATP analogs, although ATP was much more efficient (Fig. 5). At the highest protein concentration a weak DNA binding was detected even with ADP, suggesting that RecR may stabilize the RecF dimer bound to dsDNA even after ATP hydrolysis.
FIGURE 5.
RecFR-dsDNA interaction in the presence of ATP analogs measured with the fluorescence polarization assay. The assay was performed similarly to Fig. 2. 20 nm dsDNA was titrated with different RecF concentrations in the presence of 36 μm RecR and 2 mm of the following nucleotides: ATP (solid line), ATPγS (long dashed line), AMPPNP (short dashed line), and ADP (dotted line).
The FRET-based dimerization measurements demonstrated similar results. The RecF dimerization can be stimulated by ATPγS in the presence of both dsDNA and RecR (supplemental Fig. S5). The E. coli RecF binds plasmid DNA in the presence of ATPγS (11, 40), while in our experiments ATPγS did not support stable interaction between the D. radiodurans RecF and short oligonucleotides in the absence of RecR. Such a difference may arise from slightly shifted equilibrium of RecF dimerization caused by either different experimental conditions, different substrates (short oligonucleotides versus plasmid DNA where binding of multiple dimers to the same DNA molecule may play a stabilizing effect), or minor differences in dimerization constants of E. coli and D. radiodurans RecF proteins.
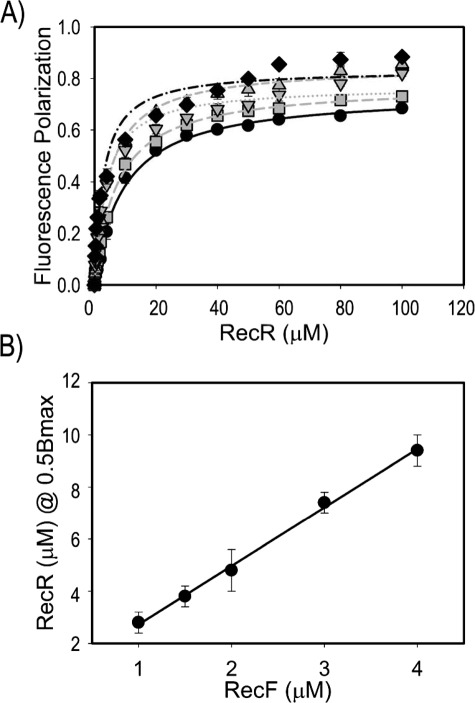
RecF Interacts with RecR with 2:4 Stoichiometry—The structural studies revealed a DNA-clamp shape of a tetrameric RecR, and a dimer-to-tetramer transition has been suggested as a mechanism of loading a putative RecR clamp on DNA (45). RecR interacts with RecF only when the RecF dimer is bound to DNA. This leads to a hypothesis that the RecF dimerization may serve as a clamp-loading mechanism where the RecF dimer promotes tetramerization of RecR on DNA. RecF and RecR from Thermus thermophilus form a stable complex in solution even without DNA, with 2:4 stoichiometry (46). We measured the stoichiometry of more transient and DNA-dependent complexes of the D. radiodurans proteins. The property of RecR to stimulate the RecF-DNA interaction was utilized to measure the stoichiometry of the RecF-RecR interaction by means of the DNA binding assay. Five different RecF-dsDNA concentrations were titrated with RecR (Fig. 6A). The analysis of RecR concentrations required for half-saturation of binding versus the RecF concentration resulted in a stoichiometry of 1:2.06 ± 0.08 corresponding to a 1:2 ratio of RecF to RecR (Fig. 6B). RecF binds DNA as a dimer, meaning that the stoichiometry of the RecF-RecR complex should be 2:4.
FIGURE 6.
Estimation of RecF-RecR stoichiometry using DNA binding fluorescence polarization assay. A, solutions with five different RecF and dsDNA concentrations were titrated with RecR. Data for 1μm RecF and dsDNA are shown by black diamonds, for 1.5 μm, by gray downward triangles, for 2 μm, by gray upright triangles, for 3μm, by gray squares, for 4μm, by black circles. The curves represent theoretical fitting calculated with SigmaPlot. B, concentrations of RecR necessary for half saturation of the RecF-dsDNA complex were plotted against the RecF concentrations and fitted by linear approximation.
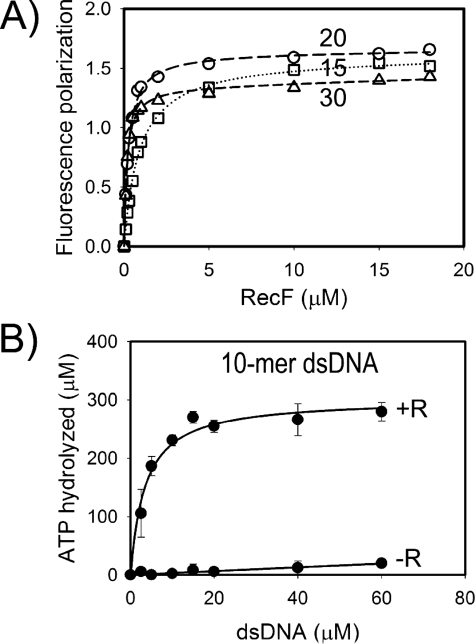
The RecR-DNA Interaction Is Not Essential for RecR Binding to the RecF-DNA Complex—The stoichiometry analysis supported the functional importance of RecR tetramer. Yet, the role of RecR DNA binding in the process of recombination initiation is not clear. Moreover, it is not clear if RecR binds DNA as a clamp. The E. coli RecR protein has never been reported to interact with DNA without RecF. The binding of D. radiodurans RecR to plasmid DNA in the presence of 40 mm Mg2+ (45) and weak interactions of T. thermophilus RecR with DNA have been demonstrated (46). We tested whether a potential weak RecR affinity to DNA contributes to formation and stability of the RecF-DNA-RecR complex. Structural modeling suggested that the RecF dimer can completely occlude from 10 to 15 bp between the claw-shaped subdomains 2 of the dimer (supplemental Fig. S1) (35). Thus, longer oligonucleotides would be more preferable substrates for the triple complex to accommodate the RecF dimer and RecR clamp. The RecR-dependent DNA binding of RecF was assayed with 10-, 15-, 20-, and 30-mer dsDNAs to address this question (Fig. 7). RecR stabilized the RecF-dsDNA binding for all substrates with 15 bp or longer in the fluorescent polarization DNA binding assay (Fig. 7A). Interpretation of data for the 10-mer was complicated due to the interaction between DNA-conjugated fluorophore and RecF (data not shown), and the ATPase assay was utilized in this case (Fig. 7B). The ATPase activity of RecF alone was very weak, likely due to a weak binding of RecF to the 10-mer dsDNA. RecR greatly stimulated the ATPase rate, indicating that RecR interacts with RecF bound to the 10-mer dsDNA. RecR is unlikely to interact with DNA directly in this case, because the 10-mer dsDNA should be mostly buried within the RecF dimer. Therefore, we concluded that RecR recognizes RecF in a DNA-bound form, likely due to specific DNA-dependent conformational changes of the RecF structure, and the RecR-DNA interaction does not contribute significantly in RecR binding to the RecF-DNA complex.
FIGURE 7.
Dependence of RecR - RecF interaction on the length of dsDNA substrate. A, the fluorescence polarization assay was used to measure the RecF binding to 15-mer (squares), 20-mer (circles), and 30-mer (triangles) dsDNA in the presence of RecR. The reactions were performed with labeled DNA similarly to Fig. 2. B, the ATPase assay was used to show the stimulation of the ATPase activity of RecF by RecR in the presence of the 10-mer dsDNA substrate. The ATPase activity of RecF was measured similarly as described in Fig. 5.
DISCUSSION
The specific role of RecF in recombination remains much less understood than the functions of other members of the RecF pathway, despite its early discovery (1, 15). The DNA binding properties of RecF together with its ability to promote initiation of presynaptic complex formation at ds/ssDNA junctions suggest that RecF provides specificity for the detection of a DNA damage site by RecFOR proteins. The specific mechanism of binding to the boundaries of SSGs and the role of ATP hydrolysis by RecF are unknown. Our previous structural and functional studies suggested that the model of ATP-dependent RecF dimerization was similar to dimerization of other SMC proteins, where the signature motif plays a critical role in dimer formation (35). The results presented here demonstrate that dimerization plays a central role in RecF interactions with DNA and RecR, and, likely, in recognition of SSG boundaries.
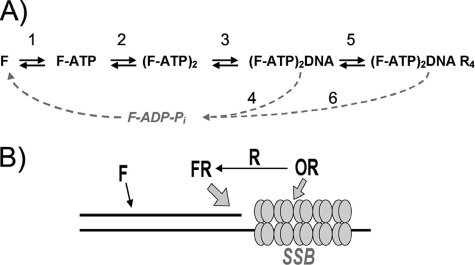
Initial steps of RecF function may be described in first approximation by the scheme shown in Fig. 8A. RecF interacts with DNA and RecR, only as an ATP-dependent dimer. In turn, a slow DNA-stimulated ATP hydrolysis leads to dimer dissociation. Equilibrium between different complexes and ATPase rate also depend on the structure of DNA substrate. The scheme is based on studies of D. radiodurans RecF. Known properties of E. coli RecF fit this pathway as well. A complex nature of the described RecF interactions indicates that RecF dimerization plays a regulatory role in the recombination initiation and that this is a highly regulated process that depends on multiple intermolecular interactions.
FIGURE 8.
A, scheme of initial steps of RecF dimerization, DNA recognition, and interaction with RecR. RecF binds to ATP (Step 1) and undergoes dimerization (Step 2), which enables RecF to bind to DNA. The RecF-DNA interaction (Step 3) activates the ATPase activity of RecF (Step 4) and dissociates the RecF dimer. The DNA-bound RecF binds to RecR (Step 5), which alters the ATPase activity of RecF (Step 6) also in a DNA-dependent manner. B, a hypothetical mechanism of SSG boundary recognition. A boundary of SSG is represented by a ds/ssDNA junction (drawn as black lines), with an ssDNA region occupied by SSBs (tetrameric clusters of four circles). RecF alone weakly interacts with dsDNA, but the interaction is greatly stabilized by RecR attracted to the ssDNA region since RecR binds to RecO, and the latter binds to SSB that is bound to ssDNA.
A simple model of ds/ssDNA junction recognition by RecF was previously suggested (34) but was not confirmed (11). We did not observe preferential binding of RecF to ds/ssDNA junctions in any of the employed assays and solution conditions. Based on structural modeling, we previously proposed that asymmetrical interaction of two ATP-binding sites of RecF dimer with ss- and dsDNA structures may trigger specific conformational changes important for interaction with other protein partners (35). Solution studies presented in this report do not show any preference for the junction DNA in the case of RecF binding to RecR. Therefore, the role of potential specific conformational changes of the complex bound to the junction DNA should be addressed at the next level of RecFR interactions with other protein partners.
Our data suggest an alternative mechanism of DNA junction recognition by RecF and RecR proteins. Direct DNA binding experiments with ATP and its analogs and ATPase assay evidence significant stimulation of RecF binding to dsDNA by RecR. RecR is also known to bind RecO, which in turn interacts with SSB bound to ssDNA (32, 43, 48, 49). Thus, it is attractive to suggest a dynamic model for the recognition of SSGs boundaries through a network of DNA-protein and protein-protein interactions as illustrated in the Fig. 8B. Here, the ssDNA part of the ds/ssDNA junction is covered by SSB. The RecR-RecO-SSB interactions will result in an elevated local concentration of RecR, which may stimulate RecF binding to dsDNA adjacent to an SSB-coated ssDNA region. Alternatively, the additive effect of RecR binding to dsDNA through RecF, and to ssDNA through RecO-SSB, may result in overall increased affinity of RecR or of all three proteins to the junction. Such a scenario would be beneficial for both initiation of RecA loading at the boundaries of a DNA damage site, and, for protecting the RecA filament at the 3′ ssDNA region from disassembly (50). In earlier electron microscopy experiments, the E. coli RecFR complex was localized at the ss/dsDNA junction on a DNA plasmid, with ssDNA gaps occupied by RecA (33). In this situation, potential RecR interaction with RecA may stabilize the RecR-RecF-dsDNA binding next to the RecA-ssDNA region. Overall, the recognition of SSG boundaries would be a result of RecR-mediated interaction between ssDNA binding proteins, RecA or SSB-RecO, and dsDNA binding RecF. Thus, the recognition of the SSGs boundaries and the overall efficiency of the recombination mediation reaction in vivo and in vitro would depend on a precise stoichiometry of all protein components, including RecF, -O, -R, and -A and SSB, as well as the ds- and ssDNA substrates.
Earlier functional studies suggested that RecR is involved in protein-protein interactions in recombination mediation reaction, because it forms complexes either with RecO or with RecF, and potentially with RecA (32, 33, 51). A DNA clamp function of RecR was suggested by structural studies of D. radiodurans RecR (45, 46). A similar model of RecR-DNA interaction within the RecFR complex was proposed based on small angle x-ray scattering measurements of T. thermophilus proteins (52). The drawback of the latter studies was modeling of two independent RecF monomers into a complex with a RecR tetramer, rather than using a conserved Rad50-like model of the ATP-dependent RecF dimer (35). Our results also support the tetrameric functional form of RecR when binding to RecF-DNA. On the other hand, we found that RecR-mediated stabilization of the RecF-dsDNA complex does not depend on the length of the DNA substrate. Therefore, potential interaction of RecR with DNA does not significantly contribute to the recognition of DNA-bound RecF dimer. This does not rule out the possibility of the RecR clamp encircling longer DNA substrates. Accordingly, a complex of T. thermophilus RecF with RecR mutant with the reversed surface charge inside the clamp binds to DNA considerably weaker than in case of the wild-type RecR, indirectly supporting the model of the RecR being a DNA clamp (52).
Different role of the RecR clamp within the RecOR complex was suggested by structural and mutagenesis studies of the D. radiodurans RecOR complex (48). The purified complex binds to plasmid DNA, despite the fact that RecO partially occupies the inner space of the RecR clamp. The complex was also shown to have higher affinity to the ss/dsDNA substrate and was speculated to recognize the boundaries of SSGs during recombination initiation. Nevertheless, this model does not agree with other studies, in which the presence of junction DNA had no effect on the efficiency of E. coli RecOR proteins in presynaptic complex formation (32, 53).
The alternative model where RecR tetramer interacts only with RecF should be considered as well. For T. thermophilus proteins, RecF and RecO binding sites on RecR overlap (46). Lack of structural similarity between RecF and RecO makes it difficult to predict which part of RecF binds RecR. One potential region is a tip or an apical part of the subdomain 2, which is an arm embracing DNA (supplemental Fig. S1). Judging from the model of a RecF dimer, additional conformational changes have to occur to bring the tips of the arms closer together to fit inside the RecR clamp, at least partially. This model will be similar to the model of Rad50-Mre11 complex forming a potential heterotetrameric clamp around DNA (54) and will explain the stabilization of RecF-dsDNA binding by RecR even with short dsDNA fragments.
Our results demonstrate a complex nature of the interactions between RecF, DNA, and RecR, where the ATP-dependent dimerization and the ATPase activities of RecF play key regulatory role. Interactions of RecF with other proteins may be important in HR initiation as well. RecF interacts with the RecX protein antagonizing the RecA-ssDNA filament dissociation (33, 55). RecF is involved in an earlier step of the UV-damage response by limiting RecQ- and RecJ-mediated degradation of the lagging nascent DNA strand (56). With its diverse properties, RecF may be a key player in the integration of the protein networks of DNA replication and repair machineries.
Multistep reactions of the RecF ATP-dependent dimerization, ATP hydrolysis, and interactions with DNA and RecR resemble intricate activities of the Rad50 protein, which detects DSBs and initiates different pathways of the DSBs repair in eukaryotes (36, 54, 57). Understanding the mechanism of the RecF-mediated processes will be useful in studies of the much more complex Rad50 protein, and other ABC-type ATPases such as the structural maintenance of chromosome proteins cohesin and condensin (37, 47, 54, 58–61).
Supplementary Material
Acknowledgments
We thank Dr. T. Heyduk for help with the design and interpretation of fluorescence polarization experiments, and for helpful discussions, Dr. S. Patel for constructive suggestions, and Drs. J. Courcelle, I. Denisov, and D. Grandgenett for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM073837. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S5, and equations.
Footnotes
The abbreviations used are: HR, homologous recombination; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; ss/dsDNA, single-stranded double-stranded DNA junction; RMP, recombination mediator protein; SSB, ssDNA-binding protein; RecFOR, RecF with RecR and RecO proteins; RecFR, RecF and RecR proteins; RecOR, RecO and RecR proteins; RecBCD, RecB, RecC, and RecD protein complex; ABC, ATP-binding cassette; SSG, single-stranded gapped DNA; DSB, double-stranded DNA break; TCEP, tris(2-carboxyethyl)phosphine; FAM, fluorescein; ATPγS, adenosine 5′-O-(thiotriphosphate); AMPPNP, 5′-adenylyl-β,γ-imidodiphosphate; FRET, Föster resonance energy transfer; Ni-NTA, nickel-nitrilotriacetic acid.
References
- 1.Cox, M. M. (2007) Crit. Rev. Biochem. Mol. Biol. 42 41–63 [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski, S. C. (2000) Trends Biochem. Sci. 25 156–165 [DOI] [PubMed] [Google Scholar]
- 3.Beernink, H. T., and Morrical, S. W. (1999) Trends Biochem. Sci. 24 385–389 [DOI] [PubMed] [Google Scholar]
- 4.Kowalczykowski, S. C. (2005) Nature 433 591–592 [DOI] [PubMed] [Google Scholar]
- 5.Kantake, N., Madiraju, M. V., Sugiyama, T., and Kowalczykowski, S. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohaghegh, P., and Hickson, I. D. (2001) Hum. Mol. Genet. 10 741–746 [DOI] [PubMed] [Google Scholar]
- 7.Karow, J. K., Wu, L., and Hickson, I. D. (2000) Curr. Opin. Genet. Dev. 10 32–38 [DOI] [PubMed] [Google Scholar]
- 8.Yang, H., Li, Q., Fan, J., Holloman, W. K., and Pavletich, N. P. (2005) Nature 433 653–657 [DOI] [PubMed] [Google Scholar]
- 9.Kowalczykowski, S. C., Dixon, D. A., Eggleston, A. K., Lauder, S. D., and Rehrauer, W. M. (1994) Microbiol. Rev. 58 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzminov, A., and Stahl, F. W. (1999) Genes Dev. 13 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webb, B. L., Cox, M. M., and Inman, R. B. (1999) J. Biol. Chem. 274 15367–15374 [DOI] [PubMed] [Google Scholar]
- 12.Spies, M., Bianco, P. R., Dillingham, M. S., Handa, N., Baskin, R. J., and Kowalczykowski, S. C. (2003) Cell 114 647–654 [DOI] [PubMed] [Google Scholar]
- 13.Taylor, A. F., and Smith, G. R. (2003) Nature 423 889–893 [DOI] [PubMed] [Google Scholar]
- 14.Wang, T. V., and Smith, K. C. (1984) J. Bacteriol. 158 727–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horii, Z., and Clark, A. J. (1973) J. Mol. Biol. 80 327–344 [DOI] [PubMed] [Google Scholar]
- 16.Kolodner, R., Fishel, R. A., and Howard, M. (1985) J. Bacteriol. 163 1060–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asai, T., and Kogoma, T. (1994) J. Bacteriol. 176 7113–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courcelle, J., Carswell-Crumpton, C., and Hanawalt, P. C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courcelle, J., and Hanawalt, P. C. (2003) Annu. Rev. Genet. 37 611–646 [DOI] [PubMed] [Google Scholar]
- 20.Bidnenko, V., Seigneur, M., Penel-Colin, M., Bouton, M. F., Dusko Ehrlich, S., and Michel, B. (1999) Mol. Microbiol. 33 846–857 [DOI] [PubMed] [Google Scholar]
- 21.Rocha, E. P., Cornet, E., and Michel, B. (2005) PLoS Genet. 1 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V., and Daly, M. J. (2001) Microbiol. Mol. Biol. Rev. 65 44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, G., Wang, L., Chen, H., Lu, H., Ying, N., Tian, B., and Hua, Y. (2008) J. Bacteriol. 190 2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, G. R. (1988) Microbiol. Rev. 52 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdi, A. A., and Lloyd, R. G. (1989) Mol. Gen. Genet. 216 503–510 [DOI] [PubMed] [Google Scholar]
- 26.Wang, T. C., Chang, H. Y., and Hung, J. L. (1993) Mutat. Res. 294 157–166 [DOI] [PubMed] [Google Scholar]
- 27.Umezu, K., Chi, N. W., and Kolodner, R. D. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 3875–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Y. H., Cheng, A. J., and Wang, T. C. (1998) J. Bacteriol. 180 1766–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow, K. H., and Courcelle, J. (2004) J. Biol. Chem. 279 3492–3496 [DOI] [PubMed] [Google Scholar]
- 30.Ivancic-Bace, I., Peharec, P., Moslavac, S., Skrobot, N., Salaj-Smic, E., and Brcic-Kostic, K. (2003) Genetics 163 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandler, S. J., and Clark, A. J. (1994) J. Bacteriol. 176 3661–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimatsu, K., and Kowalczykowski, S. C. (2003) Mol. Cell 11 1337–1347 [DOI] [PubMed] [Google Scholar]
- 33.Webb, B. L., Cox, M. M., and Inman, R. B. (1997) Cell 91 347–356 [DOI] [PubMed] [Google Scholar]
- 34.Hegde, S. P., Rajagopalan, M., and Madiraju, M. V. (1996) J. Bacteriol. 178 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koroleva, O., Makharashvili, N., Courcelle, C. T., Courcelle, J., and Korolev, S. (2007) EMBO J. 26 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hopfner, K. P., and Tainer, J. A. (2003) Curr. Opin. Struct. Biol. 13 249–255 [DOI] [PubMed] [Google Scholar]
- 37.Junop, M. S., Obmolova, G., Rausch, K., Hsieh, P., and Yang, W. (2001) Mol. Cell 7 1–12 [DOI] [PubMed] [Google Scholar]
- 38.Smith, P. C., Karpowich, N., Millen, L., Moody, J. E., Rosen, J., Thomas, P. J., and Hunt, J. F. (2002) Mol. Cell 10 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moncalian, G., Lengsfeld, B., Bhaskara, V., Hopfner, K. P., Karcher, A., Alden, E., Tainer, J. A., and Paull, T. T. (2004) J. Mol. Biol. 335 937–951 [DOI] [PubMed] [Google Scholar]
- 40.Madiraju, M. V., and Clark, A. J. (1992) J. Bacteriol. 174 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R., and Donnelly, M. I. (2002) Protein Expr. Purif. 25 8–15 [DOI] [PubMed] [Google Scholar]
- 42.Webb, B. L., Cox, M. M., and Inman, R. B. (1995) J. Biol. Chem. 270 31397–31404 [DOI] [PubMed] [Google Scholar]
- 43.Hegde, S. P., Qin, M. H., Li, X. H., Atkinson, M. A., Clark, A. J., Rajagopalan, M., and Madiraju, M. V. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14468–14473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandler, S. J., Chackerian, B., Li, J. T., and Clark, A. J. (1992) Nucleic Acids Res. 20 839–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, B. I., Kim, K. H., Park, S. J., Eom, S. H., Song, H. K., and Suh, S. W. (2004) EMBO J. 23 2029–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honda, M., Inoue, J., Yoshimasu, M., Ito, Y., Shibata, T., and Mikawa, T. (2006) J. Biol. Chem. 281 18549–18559 [DOI] [PubMed] [Google Scholar]
- 47.Haering, C. H., Schoffnegger, D., Nishino, T., Helmhart, W., Nasmyth, K., and Lowe, J. (2004) Mol. Cell 15 951–964 [DOI] [PubMed] [Google Scholar]
- 48.Timmins, J., Leiros, I., and McSweeney, S. (2007) EMBO J. 26 3260–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hobbs, M. D., Sakai, A., and Cox, M. M. (2007) J. Biol. Chem. 282 11058–11067 [DOI] [PubMed] [Google Scholar]
- 50.Shan, Q., Bork, J. M., Webb, B. L., Inman, R. B., and Cox, M. M. (1997) J. Mol. Biol. 265 519–540 [DOI] [PubMed] [Google Scholar]
- 51.Bork, J. M., Cox, M. M., and Inman, R. B. (2001) EMBO J. 20 7313–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honda, M., Fujisawa, T., Shibata, T., and Mikawa, T. (2008) Nucleic Acids Res. 36 5013–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakai, A., and Cox, M. M. (November 4, 2008) J. Biol. Chem. 10.1074/jbc.M807220200 [DOI] [PMC free article] [PubMed]
- 54.Hopfner, K. P., Karcher, A., Craig, L., Woo, T. T., Carney, J. P., and Tainer, J. A. (2001) Cell 105 473–485 [DOI] [PubMed] [Google Scholar]
- 55.Lusetti, S. L., Hobbs, M. D., Stohl, E. A., Chitteni-Pattu, S., Inman, R. B., Seifert, H. S., and Cox, M. M. (2006) Mol. Cell 21 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Courcelle, J., and Hanawalt, P. C. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8196–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Jager, M., van Noort, J., van Gent, D. C., Dekker, C., Kanaar, R., and Wyman, C. (2001) Mol. Cell 8 1129–1135 [DOI] [PubMed] [Google Scholar]
- 58.Lowe, J., Cordell, S. C., and van den Ent, F. (2001) J. Mol. Biol. 306 25–35 [DOI] [PubMed] [Google Scholar]
- 59.Yuan, Y. R., Blecker, S., Martsinkevich, O., Millen, L., Thomas, P. J., and Hunt, J. F. (2001) J. Biol. Chem. 276 32313–32321 [DOI] [PubMed] [Google Scholar]
- 60.Harvey, S. H., Krien, M. J., and O'Connell, M. J. (2002) Genome Biology 3 REVIEWS3003.1–3003.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirano, T. (2002) Genes Dev. 16 399–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.