Abstract
Use of highly potent small interfering RNAs (siRNAs) can substantially reduce dose-dependent cytotoxic and off-target effects. We developed a genetic forward approach by fusing the cytosine deaminase gene with targets for the robust identification of highly potent siRNAs from RNA interference (RNAi) libraries that were directly delivered into cells via bacterial invasion. We demonstrated that two simple drug selection cycles performed conveniently in a single container predominately enriched two siRNAs targeting the MVP gene (siMVP) and one siRNA targeting the egfp gene (siEGFP) in surviving cells and these proved to be the most effective siRNAs reported. Furthermore, the potent siRNAs isolated from the surviving cells possessed noncellular toxic characteristics. Interestingly, the length of highly potent siMVPs identified could be as short as 16-mer, and increasing the length of their native sequences dramatically reduced RNAi potency. These results suggest that the current approach can robustly discover the most potent and nontoxic siRNAs in the surviving cells, and thus has great potential in facilitating RNAi applications by minimizing the dose-dependent and sequence nonspecific side effects of siRNAs.
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved mechanism for silencing gene expression via the sequence-specific degradation of target RNA. In the past years, RNAi has become a very powerful genetic tool for functional genomics study, and its rapid advance has led to the development of RNAi-based therapeutics for treating human diseases. A prerequisite in the use of RNAi as a reverse genetic tool or as a therapeutic is the choice of potent and specific small interfering RNA (siRNA) sequences. It has been reported that the application of low doses of potent siRNAs/short hairpin RNAs (shRNAs) in vivo greatly reduced their off-target effects and cytotoxicities (1,2), which are the two major concerns in RNAi applications. Further studies also revealed that low-doses of potent siRNAs/shRNAs minimized the possibility of competition with endogenous miRNA machinery (2). At present, for RNAi studies most researchers use the reported algorithms simply to design several siRNA/shRNA sequences per target gene for synthesis (1,3,4). However, since the RNAi mechanism is currently not fully illustrated, it is acknowledged that the RNAi potency of such empirically designed siRNAs is uncertain until experimentally validated, and highly potent siRNAs, which would be particularly valuable for therapeutics, could not be predicted by the current design algorithms (1,5,6). Accordingly, alternative approaches have been made to define effective siRNAs/shRNAs for RNAi. The siRNAs as pools have been directly generated by the enzymatic digestion of double-stranded RNAs (dsRNAs), and have proven effective for silencing target genes (7,8). Furthermore, effective siRNAs/shRNAs could be identified from RNAi libraries through high-throughput screening (5,6), or by experimental defining, such as by systematically extending the duplex length of siRNAs beyond longer than 19–21 nt (9). Here we describe a forward approach for the robust selection of the most effective siRNAs from RNAi libraries that were delivered directly into an entire population of mammalian cells via bacterial invasion. We show that instead of the usual laborious cell-based assays of all wells in all plates, we performed the entire process of both delivering siRNA library and screening siRNAs in a single container (dish), and then immediately identified the most potent and nontoxic siRNAs from surviving cells upon drug selection. Thus we demonstrated the potential of this method for the robust discovery of the most suitable siRNAs for functional genomics studies and the development of RNAi-based therapeutics.
MATERIALS AND METHODS
The siRNA library construction
To construct a siRNA library, we first replaced human H1-U6 polymerase III promoter cassette in pHippy (10) (a gift of Dr Moon, University of Washington) with the double-promoter siRNA cassette of pFIV-H1/U6-Puro-siRNA (System Bioscience, CA) to produce pHippy-SBI. We digested the vector with BbsI and inserted two annealed oligos (Sense: 5′-AAAATATTAAGCTTAATATTC-3′ and Antisense: 5′-AAAAGAATATTAAGCTTAATA -3′) into the digested pHippy-SBI vector to position two SspI sites for blunt insertion of DNA fragments. The modified vector designated pHippy-SspI. To construct siRNA libraries, we PCR-amplified a 1.3 kb-cDNA fragment encoding the N-terminus of the major vault protein (MVP) gene or a 700 bp-cDNA fragment encoding the egfp gene with the two specific primers for the each gene. The amplified PCR products were partially digested by DNase I in the presence of 1 mM MnCl2. We isolated DNA fragments (∼20–30 pb) from gel, and ends-filled with dNTPs using T4 DNA polymerase to make blunt ends. The 5′ phosphates of the filled DNA fragments were removed with calf intestinal phosphatase. We ligated the DNA fragments with SspI digested pHippy-SspI vector at 16°C overnight with DNA ligase, and redigested the ligated products with SspI for 3 h at 37°C to destroy the self-ligated pHippy-SspI vector. We transformed the ligation reaction into Escherichia coli DH5α. After overnight cultivation on plates, the library plasmid DNA was purified and transformed into E. coli BT203 in which the asd gene was deleted and plasmid pGBΩinv-hly was transformed for delivery of siRNA libraries into mammalian cells via bacterial invasion (5).
Synthesis of siRNA duplex
We chemically synthesized siRNAs (Integrated DNA Technologies Inc.,) or synthesized them in vitro by T7 RNA polymerase using the Silencer siRNA construction kit (Ambion).
Evaluation of RNAi potency
For siMVPs, we constructed a pEGFP-MVP vector by directly inserting the 1.3-kb coding sequence of the MVP gene right after the stop codon of the egfp gene at the HindIII site for expression of egfp-MVP hybrid transcripts in cells. We transfected the synthesized siRNAs using Lipofectamine 2000 (Invitrogen) into mammalian cells at various concentrations in 96-well plates for RNAi assays. We monitored the siMVP-mediated knockdown of the expressed egfp-MVP hybrid transcript in 96-well plates (75 ng/150 μl) by fluorescence plate reading (Perkin Elmer Envision 2100 Multilabel Reader). We normalized RNAi potency with the activity of RLuc expressed by cotransfection of plasmid pRL-SV40 (5 ng). For siEGFPs, similarly we monitored the siEGFP-mediated knockdown of the transfected egfp gene in cells and normalized RNAi potency with the activity of RLuc as above. We also further examined the effectiveness of siMVP candidates with transfection of synthesized siMVPs into HeLa cells on 12-well plates, and monitored knockdown of the endogenous MVP genes by western blot analysis with anti-MVP antibody.
Expression of cytosine deaminase-target hybrid transcript in cells
To expressing the cytosine deaminase (CD)-target hybrid transcript in cells, we inserted the 1.3-kb MVP fragment coding the N-terminal sequence (or 700 bp of the egfp gene) into the HindIII site located right after the stop codon of the CD gene (11) (Invivogen). We stably transfected the constructs into HEK 293 cells for expression of the CD-MVP and CD-egfp hybrid transcripts, respectively, in the transfected cells.
Infection of mammalian cells with invasive bacteria
We seeded human embryonic kidney (HEK) 293 cells expressing the CD-target hybrid transcripts at a density of 3.0 × 106 cells per dish on 100 mm dish or 1.5 × 105 cells per well on 12-well plates in Dulbecco's; modified eagle's medium (DMEM) medium supplemented with 10% fetal calf serum at 37°C in 5% CO2. For delivery of siRNA libraries into cells via bacterial invasion, we grew the library-transformed E. coli BT203 (see above) in brain heart infusion (BHI) broth (Difco Laboratories) containing 0.5 mM DAP (Sigma), 40 μg/ml of streptomycin and 25 μg/ml of zeromycin at 30°C with shaking for 0.5 h. Then we further diluted overnight culture into BHI-DAP medium to make OD600 ≤ 0.1 and grew them for another 2 h at 30°C with shaking. We collected the bacteria by centrifugation, resuspended them at 1 × 107 cells/ml in DMEM with 0.5 mM DAP. We incubated the prepared bacteria with HEK 293 cells expressing the CD-target hybrid transcript at an m.o.i. of 20–30 for 2 h at 37°C. Under these conditions, the efficiency of bacterial internalization was 2–3 bacteria/cell. We washed cells three times with serum-free DMEM and refed cells with complete medium containing 25 μg/ml of gentamicin to kill extracellular bacteria, and added 5-fluorocytosine (FC) (Invivogen). At 48 h after addition of 5FC, we trypsinized the surviving cells in dish, and then collected and reseeded them in dish for 5 h to eliminate any contaminated dead cells. We extracted plasmid DNAs from the reseeded cells survived from the primary drug selection, and transformed them into E. coli DH5α by electroporation for amplification and preparation of the extracted plasmid DNAs. Then we transformed the prepared plasmid DNAs into E. coli BT203 for the next cycle of bacterial invasion and drug selection.
Evaluation of the cellular toxicity of siMVPs with cell viability assays
We isolated siMVP-containing plasmids either directly from the library (0-cycle) or from surviving cells in each selection cycle at 150 μM 5FC or from the dead cells that were suspended in the medium after the first bacterial invasion and drug selection cycle. We transformed the siMVP-containing plasmid DNAs by electroporation into E. coli DH5α. We randomly selected 30 colonies from each transformation, and reisolated the siMVP-containing plasmids from the cultures derived from individual colonies. We transfected the prepared siMVP-containing plasmid DNAs from individual colonies into HeLa cells in triplicate for cell viability assays. For this we seeded HeLa cells in antibiotic-free media at 0.5 × 104 cells per well in a 96-well plate for 24 h before conducting transfection. We transfected cells with siRNAs chemically synthesized or synthesized in vitro by T7 RNA polymerase using the Silencer siRNA construction kit (Ambion) (10 nM) or the plasmid DNAs prepared as above (0.2 μg/well). We determined cell viability using Alamar Blue (BioSource Int.) according to the manufacturer's instructions. Briefly, 72 h after transfection, 25 μl of Alamar Blue dye was added into each well containing cells in 100 μl of media. Cultures were then incubated for 0.5 h at 37°C in a humidified atmosphere with 5% CO2. For the purpose of this study, siRNAs were defined as toxic when the average values (mean ± SD) from three independent experiments each performed in triplicate showed cell viability below 75%.
RESULTS
A method for forward selection of effective siRNAs
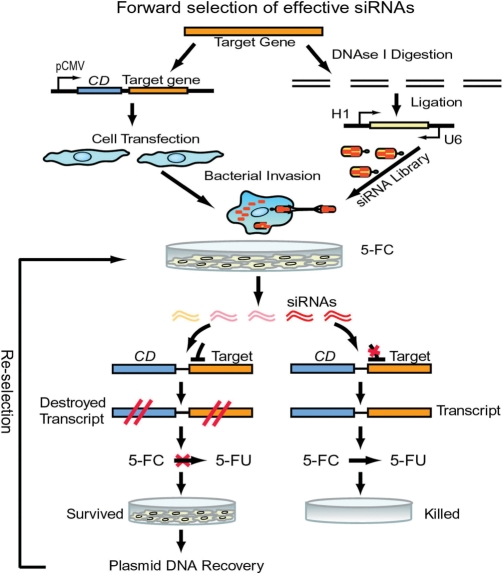
To develop a forward approach for the robust selection of effective siRNAs from an RNAi library, we fused a target gene 3′ to the gene encoding CD (11) (Figure 1), which converts 5FC to toxic 5-fluorouracil (5FU). We directly delivered siRNA-expression libraries into mammalian cells via integrin receptor-mediated bacterial invasion (12). Effective siRNAs in the library degrade the CD-target hybrid transcript to prevent the conversion of 5FC to toxic 5FU. Therefore, only those cells that contain highly potent siRNAs can survive in the presence of 5FC since toxic 5FU cannot accumulate in these cells (Figure 1). Furthermore, instead of the laborious RNAi assays in multiwell plates required by all reported other methods, we simply performed the entire forward selection process in a single container (dish), and then immediately identified the most effective and nontoxic siRNAs from surviving cells in the container.
Figure 1.
Schematic of protocol for forward and robust selection of effective siRNAs in a single container. Mammalian cells are transfected with pCMV-CD-taget gene constructs. Target genes are digested. siRNA libraries are constructed from DNase I-digested target genes and transformed into E. coli BT203, and delivered into cells via bacterial invasion. Upon drug (5FC) selection, potent and nontoxic siRNAs are immediately identified in surviving cells.
Selection of effective siRNAs targeting the MVP gene
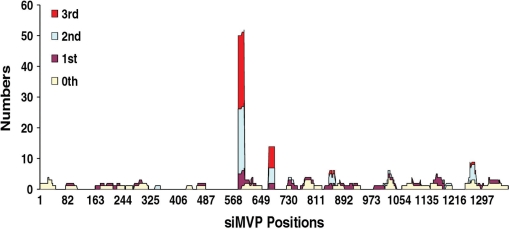
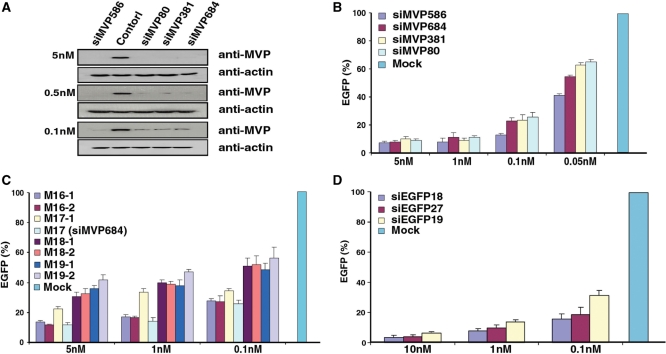
We first generated a stable cell line expressing a CD-MVP (13) hybrid transcript. Using this cell line, we found that without expressed siMVPs almost the entire cell population was killed when the concentration of drug 5FC reached >100 μM (Figure 2). To identify the most potent siMVPs, we performed the forward selection of siRNAs with 150 μM 5FC. After the second drug selection cycle we found that two siRNAs, namely siMVP-586 and siMVP-684 (17-mer), were significantly enriched in surviving cells (Table 1). Following the third drug selection cycle, these two siRNAs became further predominant as 24 and 7 of 34 siRNAs were siMVP-586 and siMVP-684, respectively (Table 1). In addition of a number of unenriched siMVPs observed inconsistently in each drug selection cycle (Supplementary Table 1), there were a very few other less-enriched siMVP sequences that were identified in surviving cells (Figure 3 and Table 1). We chemically synthesized siMVP-586 and siMVP-684 to evaluate their RNAi potencies in comparison with siMVP-80 and siMVP-381 both of which were the two most potent siMVPs at the time identified (5). Western blot analysis showed that siMVP-586, the mostly enriched siRNA in the drug selection (24 of 33), knocked down the endogenous cellular-MVP expression by ∼90% at a concentration as low as 0.1 nM (Figure 4A). The siMVP-684 also silenced the endogenous MVP gene by ∼80%, similar to siMVP-80 and siMVP-381 (Figure 4A). Using a more precise quantitative assay for silencing the egfp-MVP hybrid transcript (5), siMVP-586 consistently exhibited the highest inhibitory activity at all tested concentrations (Figure 4B), further confirming that siMVP-586 was the most potent siRNA among the identified siMVPs in the screenings.
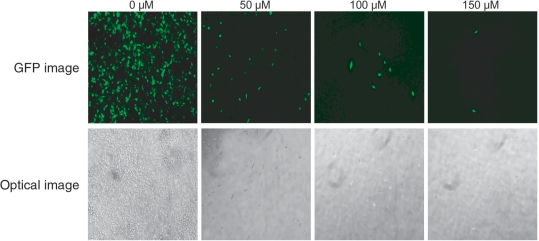
Figure 2.
Dose-responses of cells to 5FC. HEK 293 cells stably expressing the CD-MVP hybrid transcript were transfected with pEGFP-C1 for EGFP expression. After 24 h, the pEGFP-C1 transfected cells were incubated with E. coli BT-203 that had been transformed with vector pHippy-SspI (without siMVP library) at a bacteria-to-cell ratio (m.o.i.) of 20 at 37°C for 2 h. After washing, cells were re-fed with medium containing 25 µg/ml of gentamicin for overnight, and then incubated with 5FC at various concentrations. Photomicrographs of surviving cells showing EGFP-expressing were taken 48 h after addition of 5FC. Upper panel, GFP Images; lower panel, Optical Images.
Table 1.
Enrichment of siMVPs in each drug selection cycle
| Name | Total sequenced clones | Selection cycle |
Potency (knockdown%) | |||
|---|---|---|---|---|---|---|
| 0th | 1st | 2nd | 3rd | |||
| 43 | 30 | 36 | 34 | |||
| siMVP586 | 5′-GGGGAAGAAUGGCUGGUCA-3′ | 0 | 5 | 21 | 24 | 87.3 |
| siMVP684 | 5′-GAAAAGACAGCCCUGCA-3′ | 0 | 2 | 5 | 7 | 77.3 |
| siMVP857 | 5′-UGCGUGAUUCUCGACCCUG-3′ | 0 | 0 | 2 | 1 | 72.1 |
| siMVP1270 | 5′-UGCCUCCCGGGUGGAGGAG-3′ | 0 | 0 | 3 | 2 | 75.8 |
The names of siMVPs were designated according to their positions in 1.3kb-MVP cDNA clone.
Only the sense strand is shown.
HeLa cells were transfected with 60 ng pEGFP, 1.2 ng pRLSV40 (Rluc) and 0.1nM siMVPs synthesized in vitro. Three independent assays each in triplicate were performed. The potency of siMVPs was calculated by the ratio of EGFP/Rluc against the control of unrelated siRNA (siHB1).
Figure 3.
The positions and numbers of siMVPs identified in each drug selection cycle. HeLa cells expressing the CD-MVP hybrid transcript were incubated with E. coli BT-203 containing the siMVP library for delivery of siMVPs into cells at an m.o.i. of 20 at 37°C for 2 h. After washing as before, 5FC was added to cells at 150 µM concentration. At 48 h after adding 5FC plasmid DNAs containing siMVPs were extracted from surviving cells and transformed into E. coli DH5α. About 30–40 colonies were randomly picked up from the transformants, and plasmid DNAs were reisolated from the cultures of individual colonies for PCR amplification of siMVPs using two primers located the H1 and U6 promoter regions, respectively. The sequences of siMVPs amplified from individual colonies were determined by DNA sequencing. At the meantime, plasmid DNA mixtures were reextracted from the transformed E. coli DH5α, and directly transformed into E. coli BT-203 for the second cycle of bacterial invasion and forward drug selection, and so repeatedly for the third selection cycle.
Figure 4.
Evaluation of RNAi potency. (A) HeLa cells were transfected with chemically synthesized siMVPs at indicated concentrations with similar transfection efficiency measured as coexpressed luciferase activities. Cell lysates were subjected to western blot analysis with anti-MVP after transfection for 48 h. (B) HeLa cells were transfected with 60 ng pEGFP-MVP, 1.2 ng pRLSV40 (Rluc) and chemically synthesized siMVPs at indicated concentrations. (C) As with (B), HeLa cells were transfected with indicated concentrations of siMVP-684 derivatives synthesized in vitro. (D) As with (B) and (C), HeLa cells were transfected with indicated concentrations of siEGFPs synthesized in vitro. Results in (B), (C) and (D) were obtained from three independent experiments each performed in triplicate. RNAi potency of siMVPs was normalized by Rluc activity for transfection efficiency against unrelated siRNA control (siHB1). Data are mean ± SD.
Since siMVP-684 was a 17-mer siRNA the length of which was much shorter than the most effective conventional 21-mer (14–16), we changed its native length to examine the relative potencies. Adding one or two nucleotides dramatically reduced RNAi potency (Figure 4C). In contrast, its derivative 16-mers with one nucleotide reduction (M16-1: 5′-GAAAAGACAGCCCUGC-3′; M16-2: 5′-AAAAGACAGCCCUGCA-3′) retained RNAi potency nearly as potent as siMVP-684 (M17). Interestingly, adding one native nucleotide to M16-1 also substantially reduced the RNAi potency of the resulting M17-1 (5′-GGAAAAGACAGCCCUGC-3′). To investigate whether the knocking down of MVP by the 16-mer siRNA (M16-1) is through the Argonaut protein Ago-2-mediated RNAi pathway(17), we examined the potency of the 16-mer siRNA (M16-1) in U87-MG cells that express no or very little Ago-2 (18). While M16-1 exerted no detectable silencing activity in inhibiting MVP in parental U87 cells even with a high dose (5 nM), transfected expression of Ago-2 in U87 completely restored its RNAi potency comparable to that observed in HeLa cells indicated in Figure 5 (Supplementary Figure 1). These results suggest that the length of exogenous potent siRNAs could be shorter than 17-mer.
Figure 5.
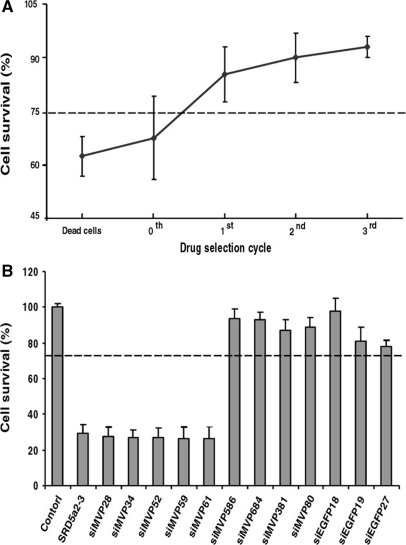
Evaluation of siMVP cytotoxicity. (A) siMVP-containing plasmids were isolated either directly from the library (selection cycle 0), or from cells that survived in each selection cycle with 150 μM 5FC, or from dead cells resulted in the first cycle of bacterial invasion and drug selection and transformed into E. coli. For toxicity evaluation, 30 colonies were randomly selected from each transformation for reisolation of siMVP-containing plasmids. HeLa cells in 96-well plate transfected in triplicate with siMVP-containing plasmids (0.2 μg) prepared from individual colonies. (B) HeLa cells transfected with 10 nM of siRNAs synthesized in vitro. Cell survival values represent the mean of three independent transfections each performed in triplicate. Five highly toxic siMVPs, siMVP-28, siMVP-34, siMVP-52, siMVP-59 and siMVP-61, were identified from dead cells (A) after screening the transformed100 colonies. Their toxicity was compared with most toxic SRD5a2-3 reported. Cellular viability assays in both (A) and (B) were performed at 72-h after transfection. The dotted line represents the 75% cell viability threshold. Data in (A) and (B) are mean ± SD.
We performed the second independent forward selection, and found that siMVP-586 and siMVP-684 again predominately enriched. However, reducing the concentration of 5FC to 100 μM enriched several additional siMVPs (data not shown), suggesting that this approach could also define more effective siRNAs, but not the most potent ones under conditions of low stringency.
Identification of the most potent siRNAs from enhanced green fluorescence protein library
The gene encoding enhanced green fluorescence protein (EGFP) has been widely used as a target for RNAi studies, and a highly potent siRNA sequence was reported previously (9). To demonstrate the general usefulness of this approach, we used the egfp gene as a target. As before, we delivered siEGFP library via bacterial invasion into mammalian cells, which stably express a CD-EGFP hybrid transcript. After the second drug selection cycle, siEGFP-18 (5-AAGCUGACCCUGAAGUUC-3′) was predominately enriched because 24 of 28 siRNAs were siEGFP-18. There were a number of other unenriched siEGFPs that were inconsistently observed in each drug selection cycle (Supplementary Table 2). We compared its RNAi potency with that of the two other siEGFPs. One was a 27-nt siRNA designated as siEGFP-27, the most effective siEGFP identified at the time as it was >100-fold more potent than the corresponding conventional 21-mer siEGFP (9). The other was siEGFP-19 (5′-GGCACAAGCUGGAGUACAA-3′), which rated the highest score (9 out of 10) according to the algorithm (1,19). We in vitro synthesized the correspondent siEGFP sequences and evaluated their potencies in knockdown of the transfected EGFP reporter gene. Silencing assays revealed that siEGFP-18 was significantly more potent than either siEGFP-27 or siEGFP-19 (Figure 4D).
Together, these results showed that our approach indeed identified the most potent siRNAs from the libraries.
The approach eliminates potential toxic siRNAs during screening
A substantial number of siRNAs can induce sequence unspecific cytotoxicity that also greatly correlates with off-target effects (20). Since the forward approach described here is based on the selection of effective siRNAs from surviving cells, and toxic siRNAs can reduce cell viability and enhance or can expedite cell death during bacterial invasion and drug selection, it is expected that toxic siRNAs should be preeliminated from surviving cells during the drug selection. In this regard, we compared the cellular toxicity of the siRNAs isolated at each drug selection cycle with those that were unselected or isolated from dead cells. Figure 5A shows that while the viability of cells transfected with siMVPs from the library (0-selection cycle) was only ∼68%, that of cells transfected with siMVPs isolated from the first-drug selection cycle reached ∼85%. The viability of cells transfected with siMVPs isolated from the second or third selection cycles further increased (∼90%). Notably, siMVPs derived from the dead cells caused further decreases in cell viability (∼62%). Consistently, increasing 5FC concentration also significantly reduced the cytotoxicity of siMVPs isolated from the surviving cells in the drug selection (Supplementary Figure 2). Moreover, we readily identified siMVPs from the dead cells with toxicity equal to or more than that of the most toxic siRNA (SRD5a2-3) reported (20). Further, the most potent identified siRNAs, siM586, siM684 and siEGFP18, consistently displayed noncytotoxic characteristics in cell survival assays (Figure 5B). These results suggest that this approach is able to eliminate toxic siRNAs from the selected siRNAs.
DISCUSSION
The development of robust methods for the rapid identification of highly potent and nontoxic siRNAs to silence any gene of interest has great potential not only in facilitating functional genomics studies, but also in applications for RNAi-based therapeutics. Use of the most effective siRNAs can reduce the possibility of competition with endogenous miRNA machinery and the required dose, resulting in minimizing the dose-dependent off-target effects (20,21). Thus, highly potent and nontoxic siRNA offer considerable advantages to the development of RNAi-based therapeutics through eliminating or decreasing the side effects of siRNAs. Accordingly, we applied a forward approach to selection the most effective and nontoxic siRNAs using the CD suicide gene in the presence of 5FU.
Using the current approach, we could rapidly identify the most effective and nontoxic siRNAs from surviving cells in a forward and robust selection manner. Instead of the laboriously assaying RNAi potency of all wells in plates as required by all other methods reported, we simply performed the entire selection process in a single container, and then immediately identified the most effective and nontoxic siRNAs from surviving cells upon drug selection. Prior assay of the RNAi potency of individual siRNAs in the screening was not required. Any contaminating or less potent siRNAs in the primary drug selection could be eliminated in a second or third drug selection cycle if necessary. We examined the power of this method for selecting the most effective siRNAs using both the MVP and the egfp genes as targets. Using the MVP gene as a target, we identified siMVP-586 as the most potent siMVP among siMVPs tested after the second drug selection cycle. Using the egfp gene as a target, we similarly found that only one siEGFP, siEGFP-18, was predominantly enriched after the second drug selection cycle, and that this was the most potent among the tested siEGFPs. These results showed that our approach indeed identified the most potent siRNAs from the libraries. However, since the entire RNAi library was generated by digesting the target transcript and then isolating the digested fragments for library construction, we could not completely exclude other highly potent siRNAs against the target due to the incomplete inclusion of all potential potent siRNAs in the libraries. In addition, the specificity of the identified siRNAs might be further examined by comparing the results obtained from a library made from a nontarget transcript though it is very unlikely that predominant sequences could be consistently isolated from the nontarget library in the surviving cells.
Recent studies have shown that both siRNAs and shRNAs exhibited dose-dependent cytotoxicity (2) that is probably caused by competing with endogenous RNAi machinery (2,22). In the current approach, all highly potent siRNAs were isolated from the cells that survived in drug selection. Thus, toxic siRNAs in the library should be eliminated from surviving cells during screening because these toxic siRNAs can reduce cell viability and enhance and/or expedite cell death during bacterial invasion and drug selection. Cellular toxicity assays showed that cell viability was greatly increased following drug selection (Figure 5A), and that all of the identified potent siRNAs, such as siMVP-586, siMVP-684 and siEGFP-18, were nontoxic (Figure 5B). In addition, highly toxic siRNAs were readily identified from the dead cells during the drug selection (Figure 5B). Hence, the current approach can eliminate toxic siRNAs from those selected, resulting in robust enrichment of the most potent and nontoxic siRNAs.
Previously studies of Drosophila melanogaster suggested that 21-bp RNA duplexes with two-base 3′ overhangs were the most effective siRNAs for gene silencing in vitro (14,15). Reducing the RNA duplexes to 20 or increasing the RNA duplexes to 23 bp, resulted in reduce potent (14). Interestingly, we found during screening for effective siMVPs that siMVP-684, one of the two most potent siRNAs identified, was a 17-mer, which was much shorter than the most effective conventional 21-mer. We further examined its RNAi potency relative to its length by systematic lengthening or shortening. Adding one or two nucleotides dramatically reduced its RNAi potency (Figure 4B). In contrast, reducing the 17-mer by one nucleotide to a 16-mer did not apparently affect its potency. Notably, adding back one nucleotide to the 16-mer (M16-1) also resulted in a substantial reduction of its RNAi potency. These results suggest that, unlike Dicer-produced endogenous siRNAs, the length of exogenous potent siRNAs can very greatly, and the 16-mers described here were the shortest and most sequence-specific potent siMVPs so far identified, although the functional mechanism of these short siRNAs in the RNA-induced silencing complex (RISC) remains elusive. The identification of the highly potent siMVP-684 and its two short M-16 derivatives (16-mers) further demonstrates the power of the current forward approach for identifying the most effective and nontoxic siRNAs from libraries though the applications of such short siRNAs could cause more off-target effects than that of conventional longer siRNAs. Since few highly potent siRNAs were identified by the current approach, these siRNA sequences can be readily modified chemically for stabilization in vivo and to eliminate their potential off-target effects (23). Hence, the current approach can eliminate toxic siRNAs from those selected to allow the robust enrichment of the most potent and nontoxic siRNAs, thus facilitating RNAi applications for studies of functional genomics and for the development of RNAi-based therapeutics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institute of Health Research (MOP82807); Natural Science and Engineering Research Council of Canada (GP0183691). Funding to pay the Open Access charge: Canadian Institute of Health Research (MOP82807).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Zhenbao Yu for valuable discussion and advice, Linhua Zhang, Meiqun Wu and Denis L’Abbe for technical assistances.
REFERENCES
- 1.Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 2.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 3.Clark J, Ding S. Generation of RNAi Libraries for High-Throughput Screens. J. Biomed. Biotechnol. 2006;2006:45716. doi: 10.1155/JBB/2006/45716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patzel V, Rutz S, Dietrich I, Koberle C, Scheffold A, Kaufmann SH. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat. Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 5.Zhao HF, L’Abbe D, Jolicoeur N, Wu M, Li Z, Yu Z, Shen SH. High-throughput screening of effective siRNAs from RNAi libraries delivered via bacterial invasion. Nat. methods. 2005;2:967–973. doi: 10.1038/nmeth812. [DOI] [PubMed] [Google Scholar]
- 6.Kumar R, Conklin DS, Mittal V. High-throughput selection of effective RNAi probes for gene silencing. Genome Res. 2003;13:2333–2340. doi: 10.1101/gr.1575003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 10.Kaykas A, Moon RT. A plasmid-based system for expressing small interfering RNA libraries in mammalian cells. BMC Cell Biol. 2004;5:16. doi: 10.1186/1471-2121-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erbs P, Regulier E, Kintz J, Leroy P, Poitevin Y, Exinger F, Jund R, Mehtali M. In vivo cancer gene therapy by adenovirus-mediated transfer of a bifunctional yeast cytosine deaminase/uracil phosphoribosyltransferase fusion gene. Cancer Res. 2000;60:3813–3822. [PubMed] [Google Scholar]
- 12.Grillot-Courvalin C, Goussard S, Huetz F, Ojcius DM, Courvalin P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 1998;16:862–866. doi: 10.1038/nbt0998-862. [DOI] [PubMed] [Google Scholar]
- 13.Mossink MH, van Zon A, Scheper RJ, Sonneveld P, Wiemer EA. Vaults: a ribonucleoprotein particle involved in drug resistance. Oncogene. 2003;22:7458–7467. doi: 10.1038/sj.onc.1206947. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Vickers TA, Lima WF, Nichols JG, Crooke ST. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res. 2007;35:6598–6610. doi: 10.1093/nar/gkm663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 20.Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquez RT, McCaffrey AP. Advances in MicroRNAs: implications for gene therapists. Hum. Gene. Ther. 2008;19:27–37. doi: 10.1089/hum.2007.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.