Abstract
Replication Protein A is a single-stranded (ss) DNA-binding protein that is highly conserved in eukaryotes and plays essential roles in many aspects of nucleic acid metabolism, including replication, recombination, DNA repair and telomere maintenance. It is a heterotrimeric complex consisting of three subunits: RPA1, RPA2 and RPA3. It possesses four DNA-binding domains (DBD), DBD-A, DBD-B and DBD-C in RPA1 and DBD-D in RPA2, and it binds ssDNA via a multistep pathway. Unlike the RPA1 and RPA2 subunits, no ssDNA-RPA3 interaction has as yet been observed although RPA3 contains a structural motif found in the other DBDs. We show here using 4-thiothymine residues as photoaffinity probe that RPA3 interacts directly with ssDNA on the 3′-side on a 31 nt ssDNA.
The replication protein A (RPA) is a single-stranded (ss) DNA-binding protein that is highly conserved in eukaryotes (1–3). RPA is one of the key players in various essential processes of DNA metabolism including replication, recombination, DNA repair and telomere maintenance (1,2,4–9). The functions of this protein are based on its DNA-binding activity and specific protein–protein interactions. Its ssDNA binding properties depend on DNA length and nucleotide sequence (6,10–13). RPA is a heterotrimeric protein, composed of 70-, 32- and 14-kDa subunits, commonly referred to as RPA1, RPA2 and RPA3, respectively. There are four DNA-binding domains (DBD) located in RPA1 (DBD A, DBD B, DBD C and DBD F), one located in RPA2 (DBD D) and one belongs to RPA3 (DBD E). RPA interacts with ssDNA via four DBD: DBD A, DBD B, DBD C and DBD D (14).
It is now accepted (11) that RPA binds to ssDNA in a sequential pathway with a defined polarity (15–17). RPA binds ssDNA with three different binding modes. First, binding initially involves an unstable recognition site of 8–10 nt with the high-affinity DBD A and DBD B domains on the 5′-side of the occluded ssDNA; it is designated ‘compact conformation’ or 8–10 nt binding mode. Second, this step is followed by the weaker binding of DBD C, on the 3′-side, leading to an intermediate or ‘elongated contracted’ (13–22 nt) binding mode (18–19). Finally binding of DBD D on the 3′-side forms a stable ‘elongated extended’ complex characterized by a 30 nt long occluded binding site (30 nt binding mode). Although RPA3 contains an Oligonucleotide-Binding (OB)-fold motif found in the other DBDs, there is presently no biochemical evidence that this subunit directly contacts DNA. Thus positioning of the RPA3 subunit relative to the other domains is still speculative (11,20). It has been clearly demonstrated that RPA3 is crucial for RPA function (1,2): RPA3 is involved in heterotrimer formation and is responsible for the polarity of binding to DNA (11,21,22). The scope of the data indicates that either RPA3 participates only in protein–protein interactions or that putative interaction of RPA3 with ssDNA is unstable and too transient to be detected by standard biochemical experiments. This latter possibility is likely if such interaction is provided by the 3′-side of the ssDNA, since it has been suggested that this region might be transiently accessible to the RPA DBD domains (23,24).
In the past few years, thionucleobases have been extensively used as intrinsic photolabels to probe the structure in solution of folded molecules and to identify transient contacts within nucleic acids and/or between nucleic acids and proteins, in nucleoprotein assemblies (25). Thio residues such as 4-thiothymine and 6-thioguanine absorb light at wavelengths longer than 320 nm, and thus can be selectively photo-activated. Owing to the high photo-reactivity of their triplet state, they exhibit high photo-cross-linking ability towards nucleic acid bases as well as towards amino acid residues. Here we used a combination of approaches including gel retardation assays, chemical cross-linking and cross-linking with photoreactive ssDNA probes containing 4-thiothymine, introduced at a defined site in the sequence of the ssDNA, to study interactions present in human RPA (hRPA): ssDNA complexes. These studies coupled with the identification of cross-linked targets using specific antibodies revealed that in the elongated extended hRPA:ssDNA complex RPA3 closely contacts the 3′-end positioned nucleotide and yields a covalent adduct with zero-length photolabel.
MATERIALS AND METHODS
Materials
BSA was from Roche, [γ-32P]ATP (6 μCi/pmol) was from Amersham and T4 polynucleotide kinase from BioLabs. The RNase free-DNase I was from Roche. The oligonucleotide 5′-d(GATCTCGGCGACGCACACGCGTCCTAACTCG)-3′ (Oligo 31) and thiolated oligonucleotides (Oligo 31S) were synthesized by MWG (The Genomic Company). Recombinant hRPA was expressed in the Escherichia coli BL21 (DE3) strain (the three entire subunits RPA1, RPA2 and RPA3 were coexpressed with plasmid pET11ahRPA generously provided by Dr Klaus Weisshart, IMB, Jena, Germany), and purified using an Affi-Gel Blue, HAP and Q-Sepharose chromatographic columns according to Gomes et al. (26). hRPA was quantified using the Bradford assay. Rabbit anti-RPA1 polyclonal antibodies were obtained from Chemicom International. Mouse anti-RPA2 and RPA3 monoclonal antibodies were obtained from Novus Biologicals. The gels were analyzed with a Phosphorimager STORM 860 instrument (Molecular Dynamics).
hRPA:Oligo 31 binding
Oligonucleotides were labeled with [γ-32P]ATP using T4 polynucleotide kinase (27). 32P-labeled oligonucleotides were purified using denaturing 15% polyacrylamide gel electrophoresis (PAGE). hRPA was diluted and pre-incubated (10 min at 4°C) in buffer containing 50 mM Tris–HCl pH 7.5, 100 mM KCl, 1 mM DTT, 10% glycerol, 0.2 mg/ml BSA and 0.1 mM EDTA. The radioactively labeled oligonucleotide (2 nM) was mixed with various amounts of protein in 10 ml reaction buffer [25 mM Tris–HCl pH 7.5, 1 mM EDTA, 2 mM MgCl2, 50 mM NaCl and 6% glycerol]. hRPA:Oligo 31 binding reactions were conducted at 10°C for 10 min. Longer incubation times (up to 1 h) did not affect the band patterns or intensities, indicating that the systems had reached thermodynamic equilibrium in 10 min. Individual reaction mixtures were analyzed by native 5 or 7% PAGE in 0.5 × TBE for 3 h at 7 V/cm and at 10°C.
Glutaraldehyde cross-linking experiments
The protein–DNA complexes obtained in the hRPA:Oligo 31 binding assays and prepared as described above, were cross-linked by the addition of 0.1% of glutaraldehyde at 10°C for 10 min. Individual reaction mixtures were analyzed by native 5% PAGE in the conditions as described above.
Photo-cross-linking experiments
Thiolated oligonucleotides (Oligo 31S) were labeled with [γ-32P]ATP using T4 polynucleotide kinase. Complete purification of the oligonucleotides was performed in two steps: first truncated oligonucleotides were separated from full-length oligonucleotides by gel electrophoresis on a denaturing 20% polyacrylamide gel; thiolated-oligonucleotides were then separated from non-thiolated oligonucleotides on an affinity gel slowing down the desired oligonucleotides (28).
32P-labeled Oligo 31S (2 nM) were mixed with the indicated amounts of protein using the protocol described above. Each sample (10 μl) was introduced in a siliconized glass capillary and placed 5 cm from the exit slit of a multichromator equipped with a Schott WG 345 filter. The samples thermostated at 15°C were irradiated for 45 min. Laemmli gels (10%) were used to separate multiple cross-linked species.
Immunoblotting procedure
The multiple cross-linked species were transferred from the Laemmli gel to a nitrocellulose membrane at 4°C. The membrane was then incubated in PBS (Phosphate Buffered Saline)— 0.1% Tween buffer containing 5% non-fat milk (at room temperature for 1 h in an orbital shaker) and rinsed three times with PBS-Tween. The membrane with the immobilized material was processed with RNase free-DNase I (40 U/ml) in 20 ml of DNase buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 0.1 mM DTT) to facilitate protein antigens-antibodies interactions. After 12 h of incubation with DNase I at room temperature using an orbital shaker the membrane was washed in PBS-Tween buffer for 1 h (4 ml/cm2). The standard ECL western blotting protocol from Amersham Biosciences served to reveal the identity of the cross-linked RPA subunit with antibodies specific of RPA1, RPA2 or RPA3 subunits.
RESULTS
Binding of hRPA to Oligo 31 forms 1:1, 2:1 and 3:1 complexes
hRPA binds ssDNA with three different binding modes but to date only the 30 nt binding mode has been observed by electrophoretic mobility shift assays (EMSA) (12,29–35). Yet, the 8–10 nt binding mode has been trapped after glutaraldehyde cross-linking showing that the 8–10 nt and probably the 13–22 nt binding mode are unstable (36). We wished to determine whether the three binding modes could be observed on native gels without trapping.
Folding program predicts a stem loop structure with Oligo 31. However, several hard points permit us to maintain that complexes described below are formed with an unstructured ssDNA. Indeed, even if the stability of this structure in our conditions is good (▵G = –7 kcal/mol, Tm = 68°C), this hairpin shows two long ssDNA strands at both extremities (9 nt at 5′-end and 8 nt at 3′-end). These ssDNA strands are very good targets for the ssDNA binding protein as RPA. In addition, RPA is well known to remove secondary and tertiary DNA structures by a simple destabilization process (37,38). Recently, we have shown that hRPA can unwind very stable compact G-quadruplex structures [▵G < –15 kcal/mol, Tm ≈ 70°C (39)] in < 2 min (6). Thus, we used incubation times of hRPA with Oligo 31 sufficiently long to remove theoretical hairpin structures.
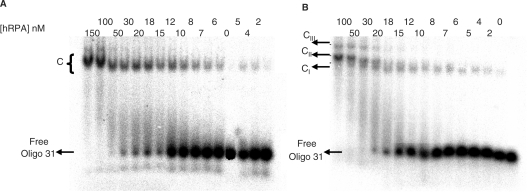
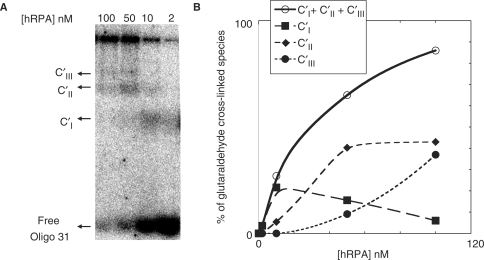
Oligo 31 incubated for 10 min at 10°C with a defined amount of hRPA was loaded onto a native 5 or 7% acrylamide/bisacrylamide gel; migration was performed at 10°C (Figure 1). Oligo 31 (2 nM) was titrated with hRPA (from 2to 150 nM). Figure 1A illustrates the results of EMSA experiments obtained when 5% non-denaturating PAGE was used to separate the binding products. In the presence of hRPA, a single major band with reduced mobility was detected. In accordance with previously reported data (12,29–35) the protein binds Oligo 31 and forms a non-covalent complex designated C. Higher concentrations of hRPA did not result in the detection of additional retarded bands. It should however be noted that at the two highest concentrations (100 and 150 nM) the single band was slightly more retarded and an additional band was discerned. When the same mixtures were separated by non-denaturating 7% PAGE three retarded complexes of different mobilities were detected (Figure 1B). At hRPA concentrations up to 10 nM, a single band designated CI appeared, that migrated like the C complex observed in Figure 1A; as the hRPA concentration increased, a second and then a third more slowly migrating bands, designated complex CII and CIII, respectively, appeared. To identify the nature of these retarded complexes, protein–DNA cross-linking experiments with 0.1% glutaraldehyde were performed (36) and the reaction mixtures loaded onto 5% native gels. As shown in Figure 2A, glutaraldehyde treatment revealed the existence of three bands designated C′I, C′II, C′III. These data indicate that C′I is the 1:1 (hRPA:Oligo 31) covalent complex, C′II is the 2:1 covalent complex and C′III is the 3:1 covalent complex, and suggest that the retarded species observed (Figure 1B) are respectively 1:1, 2:1 and 3:1 complexes. This is supported by our recent results that showed that a shorter oligonucleotide (21 nt) is able to bind at least two hRPA molecules (6). The 1:1 stoichiometry suggests that hRPA binds Oligo 31 in the stable 30 nt binding mode, while in the 2:1 complexes hRPA interacts in the 13–22 nt binding mode and finally formation of 3:1 complexes would be in accordance with a less stable 8–10 nt binding mode (Figure 1A at 100 and 150 nM hRPA).
Figure 1.
Titration of Oligo 31 as a function of hRPA concentration. 32P-Oligo 31 (2 nM) was incubated with the indicated amounts of hRPA and separated on a native 5% (A) or 7% (B) polyacrylamide gel. C represents non-covalent complexes. CI, CII and CIII represent 1:1, 2:1 and 3:1 complexes, respectively.
Figure 2.
Determination and quantification of complexes as a function of hRPA concentration. (A) 32P-Oligo 31 (2 nM) was incubated with the indicated amounts of hRPA and then cross-linked by the addition of 0.1% glutaraldehyde for 10 min. Individual reaction mixtures were analyzed on a native 5% polycarylamide gel. (B) The total amounts of complexes (CI + CII + CIII, solid line), 1:1 complex (CI, long dashed line), 2:1 complex (CII, medium dashed line) and 3:1 complex (CIII, short dashed line) were quantified for each hRPA concentration from hRPA:Oligo 31 EMSA data (Figure 2A). The relative error is ±20%.
Gel shift experiments with an 31 nt polydT named poly(dT)31 have not shown significant difference compared to Oligo 31: we have observed the same migration profil and the same glutaraldehyde cross-links (data not shown). However, hRPA has shown a higher affinity (∼10-fold) for poly(dT)31 compared to Oligo 31 in accordance with the preference of RPA for polypyrimidine sequences (1,2,12). Thus results obtained with Oligo 31 could be generalized to all ssDNA.
Figure 2B shows the quantification of complex formation upon increasing hRPA concentration. At low concentrations, 1:1 complexes are the major complexes whereas 2:1 complexes predominate at higher hRPA concentrations. At the highest hRPA concentrations, a mixture of 2:1 and 3:1 complexes appears. These results demonstrate that in solution the three types of complexes can coexist in equilibrium as previously suggested (40) and that this equilibrium can be shifted to higher order complexes at high protein concentrations.
hRPA-Oligo 31S photo-cross-linking demonstrates direct contact with RPA3
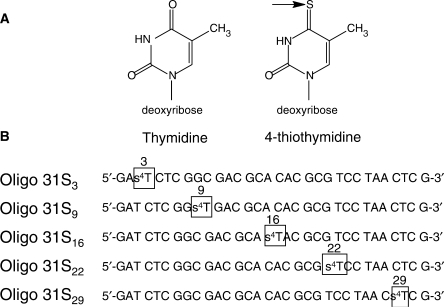
To reveal characteristic features of the complexes, a photo-cross-linking strategy was used to form the specific covalent adducts of hRPA and ssDNA. For this purpose a series of Oligo 31 analogues (Oligo 31S) were synthesized. Each Oligo 31S contains the photoactivable 4-thiothymine residue (s4T) at a single defined position (Figure 3A) in place of T. The following positions were selected: for s4T located in the 5′ region, positions 3 and 9 (Oligo 31S3 and Oligo 31S9, respectively); for centrally located s4T, position 16 (Oligo 31S16) and for s4T located in the 3′ region, positions 22 and 29 (Oligo 31S22 and Oligo 31S29, respectively) (Figure 3B).
Figure 3.
Thiolated Oligo 31S molecules used to probe hRPA—DNA interactions (A) Comparison of the structure of thymidine and 4-thiothymidine. (B) 4-thiothymidine was introduced at different strategic positions on Oligo 31S near the 5′-side (positions 3 and 9), in a central position (position 16) and near the 3′-side (positions 22 and 29).
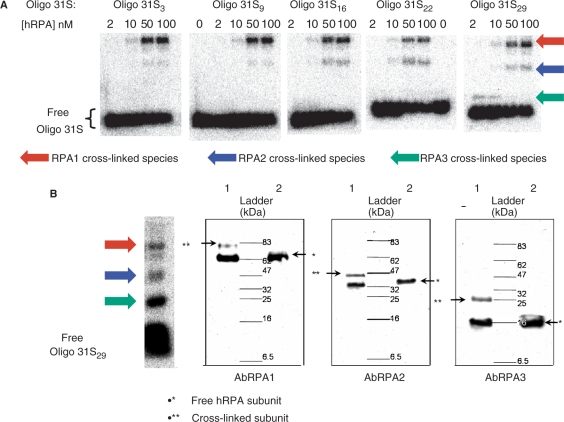
Samples containing one of the Oligo 31S were irradiated in the presence of increasing hRPA concentrations and analyzed by SDS gel electrophoresis As seen in Figure 4A, the modification patterns obtained are specific for each Oligo 31S. Three distinct retarded bands were detected for each DNA probe used in the assay. To identify the nature of the retarded bands, the appropriate control experiments were performed. No crosslink formed without UV-irradiation or upon irradiation of a mixture of hRPA and Oligo 31. Addition of proteinase K to the irradiated mixture before loading onto the gel led to the disappearance of retarded bands yielding only free DNA (not shown). These results clearly demonstrate that the retarded bands correspond to adducts of ssDNA with hRPA subunits. Cross-linking efficiencies are presented in Table 1. As expected, the cross-linking efficiency varies with the hRPA concentration. At low protein concentrations, no cross-links were observed for Oligo 31S3-22. However a protein–DNA adduct was observed with Oligo 31S29 (Figure 4A). Thus at low protein concentrations there is a favorable contact between the 3′-side of ssDNA and hRPA. The apparent molecular weight of each cross-linked species has been estimated to be close to 25, 42 and 83 kDa. Cross-linking of individual subunits to DNA is expected to increase their apparent masses by 10 kDa suggesting that the 25, 42 and 83 kDa species indeed correspond to the cross-linked RPA3, RPA2 and RPA1, respectively. Nevertheless, from this experiment it cannot be excluded that the fastest migrating bands (25 and 42 kDa species) correspond to cross-linked products of proteolyzed forms, generated from the larger subunits (RPA1 and RPA2), retaining their ability to bind ssDNA.
Figure 4.
Photo-cross-linking reactions between hRPA and Oligo 31S and identification of complexes by immunoblotting. (A) 32P-labeled Oligo 31S (2 nM) were mixed with the indicated amounts of hRPA then irradiated at 15°C for 45 min. SDS-PAGE (10%) were used to separate multiple cross-linked species. Cross-linked species (arrows) were detected and quantified by the phosphorimager—Molecular Dynamic. (B) After electrophoresis and transfer onto a nitrocellulose membrane, DNA was removed by incubation of the membrane with DNase I. ECL western blotting used anti-RPA1 (AbRPA1), anti-RPA2 (AbRPA2) and anti-RPA3 (AbRPA3) antibodies to reveal the cross-linked RPA1, RPA2 and RPA3 subunits, respectively. The immunoblot shown here identifies each subunit implicated in photo-cross-linking with Oligo 31S29 and 10 nM of hRPA (lanes 1). For comparison, free hRPA (10 nM) was loaded (lanes 2).
Table 1.
Quantification of photo-cross-linking yield
| Oligo 31S | Oligo 31S3 | Oligo 31S9 | Oligo 31S16 | Oligo 31S22 | Oligo 31S29 |
|---|---|---|---|---|---|
| [hRPA] nM | 2 10 50 100 | 2 10 50 100 | 2 10 50 100 | 2 10 50 100 | 2 10 50 100 |
| Total cross-linking yield | 0 8 23 24 | 0 5 15 23 | 0 9 25 27 | 0 12 19 21 | 7 14 27 31 |
| Error (±) | 4 8 10 | 3 6 7 | 5 7 3 | 7 8 8 | 4 8 10 8 |
For each Oligo 31S, the overall yield of cross-linking is reported as a function of hRPA concentration. Several irradiation experiments with different hRPA preparations allowed us to determine error bars.
To identify the RPA subunits involved in the formation of each protein–DNA adduct, immunoblotting experiments were performed using antibodies specific of RPA1 (AbRPA1), RPA2 (AbRPA2) and RPA3 (AbRPA3) subunits. After treatment of the membrane-immobilized protein–DNA adducts by DNase I, the subunits were identified using Amersham ECL detection kits (Figure 4B). Two bands were detected after incubation with each type of Ab (lanes 1): one corresponds to non cross-linked hRPA subunit and the retarded band corresponds to the cross-linked subunit. The difference in migration for each couple of bands corresponds to the mass increment of the Oligo 31S (≈10 kDa). Thus incubation with antibodies specific of RPA1, RPA2 and RPA3 demonstrated that the 25, 42 and 83 kDa products are covalent adducts of ssDNA with the RPA3, RPA2 and RPA1, respectively.
The 1:1, 2:1 and 3:1 complexes reveal 30, 13–22 and 8–10 nt binding modes, respectively
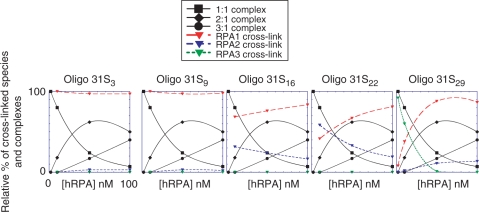
To illustrate the correlation of hRPA photo-cross-linking patterns with the presence of different types of complexes, the relative yields of each cross-linked species and each complex represented in the Figure 5 were evaluated. The data reveal that RPA1 cross-links at positions 3 and 9 whatever the nature of the complex. In the case of the centrally located s4T (Oligo 31S16) both RPA1 and RPA2 crosslink (with a higher efficiency for RPA1). In addition, formation of 2:1 and 3:1 complexes is followed by a decrease in RPA2 cross-linking and an increase in RPA1 cross-linking. However, the relative yield of RPA2 cross-linking slightly decreases upon increase in hRPA concentration suggesting that RPA2 remains in contact with ssDNA around this position in the 2:1 complex. When the photoactivable nucleotide is placed at position 22 and at low hRPA concentrations, where the 1:1 complex predominates, RPA2 is the major cross-linking target. In addition, this adduct disappears upon increasing hRPA concentrations showing a good correlation between the presence of the 1:1 complex and contact of RPA2 at position 22 of Oligo 31S. Finally, at low hRPA concentrations the DNA probe bearing photoactivable nucleotide at the position 29 (close to the 3′-side) forms single adduct, which corresponds to cross-linked RPA3. Positioning of RPA3 at the 3′-side is correlated with the existence of the 1:1 complex since disappearance of the RPA3 contact with DNA follows disappearance of the 1:1 complex. The appearance of the 2:1 and 3:1 complexes favors the RPA1 cross-link. Interestingly, even if the yield is low, a cross-link occurs between RPA2 and ssDNA when the 2:1 complex is formed indicating contact between RPA2 and the 3′-side of the ssDNA. This contact is still present at the highest hRPA concentration since the 2:1 complex remains the major complex. However, we cannot exclude that RPA2 is present in this region in the 3:1 complex.
Figure 5.
Quantification of cross-linked species. The relative amounts of cross-linked species involving RPA1 (red, long dashed line), RPA2 (blue, medium dashed line) and RPA3 (green, short dashed line) subunits were quantified for each hRPA concentration from the hRPA-Oligo 31S gel (Figure 4A). Error bar is ±20%. The results obtained for 2 nM of hRPA were not reported since no cross-links were observed except for Oligo 31S29. To correlate the nature of the complexes with the presence of the subunit on the DNA, the relative yield of each complex was taken from data in Figure 2B. The same scales are used for each graph, but to simplify the figure only scales for the Oligo 31S3 graph are shown.
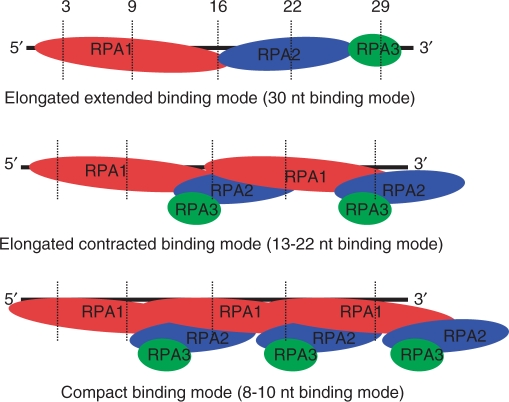
From all the results presented here and the hRPA binding modes demonstrated previously (12,15–19), a schematic model of hRPA subunits binding with 31 nt ssDNA is proposed for each type of complex (Figure 6). (i) In the 1:1 complex, only one molecule of the heterotrimer protein binds ssDNA in the 30-nt binding mode (elongated extended form). Thus RPA1 is localized at the 5′-side of ssDNA and occludes ∼14–16 nt. The following subunit bound to ssDNA is RPA2 and then RPA3 is positioned near the 3′-end of the ssDNA. This model is in accordance with the defined 5′- to 3′-binding polarity of hRPA (15–17) and clearly localizes the RPA3 subunit on the 3′-side of the ssDNA. (ii) In the 2:1 complex, two hRPA molecules bind ssDNA in the 13–22 nt binding mode. In this elongated contracted form, RPA3 subunits do not contact ssDNA while two RPA1 and two RPA2 subunits interact with ssDNA. The RPA2 subunits of two heterotrimers are placed around positions 16 and 29, respectively. (iii) In the 3:1 complex hRPA in mainly contacts ssDNA via RPA1 in accordance with 8–10 nt binding mode (41). In this compact conformation of hRPA RPA2 does not contact ssDNA. However a contact of one of the three RPA2 subunits at the 3′-side of the ssDNA cannot be excluded.
Figure 6.
Schematic models of the 30, 13–22 and 8–10 nt binding modes. The s4T substituted positions are represented by dashed lines. RPA1 is represented by a red bubble, RPA2 by a blue bubble and RPA3 by a green bubble.
DISCUSSION
Human replication protein A is indispensable for multitude of DNA metabolism pathways including DNA replication, DNA recombination, DNA repair and substantially contributes to the coordination of these processes (1,2,4,9). This protein tightly binds ssDNA via a multi-step mechanism with a defined 5′→3′ polarity and very high affinity and takes part in resolution of complicated DNA structures (6,42,43).
The data presented demonstrate different binding modes of hRPA:ssDNA using EMSA without glutaraldehyde trapping. Several factors linked to the experimental conditions can explain the observation of these three binding modes. The experiments were performed at 10°C, instead of the widely used 25°C. These conditions stabilize hRPA:ssDNA complexes. Non-denaturating 7% PAGE provides better separation of different hRPA:ssDNA complexes than the widely used 5% acrylamide or 1% agarose gels. The same observations were made by Jiang et al. (44) using a 7.5% acrylamide gel. Thus, these data obtained in the optimized conditions demonstrate existence of three species of hRPA:ssDNA complexes at varying [hRPA]/[ssDNA] ratio. Observation of high order complexes at low NaCl concentration and at high RPA concentration is correlated with the recently demonstrated binding of four hRPA molecules to a 48 nt ssDNA in saturation conditions at equilibrium (45) and two Saccharomices cerevisiae RPA (scRPA) molecules to a 30 nt ssDNA at low NaCl concentration (20 mM) and at high [scRPA]/[ssDNA] ratio (46).
There are four DBD located in the RPA1 (DBD A, DBD B, DBD C and DBD F), one each in RPA2 (DBD D) and in RPA3 (DBD E). RPA1 has the highest ssDNA binding affinity of the three subunits (DBD A and DBD B) (41); the DBD C domain has a lower affinity for ssDNA (47) while the major function of the DBD F domain is to control specific interactions of RPA with other proteins (48). The DBD D domain of RPA2 has moderate ssDNA binding ability (49). Although RPA3 contains a similar OB-fold motif as found in DBDs, direct contacts of DNA with the small hRPA subunit have not been demonstrated in humans (1,2,50). However, recent studies suggest that in the 30 nt binding mode, the four DBDs (DBD A, DBD B, DBD C and DBD D) occlude only ∼26 nt of ssDNA implying that other parts or domains of hRPA could also be partially or indirectly involved, so that 30 nt of ssDNA would be occluded by hRPA (51). Using photo-cross-linking experiments based on photoreactivity of 4-thiothymine, and the product identification procedure based on the used specific antibodies we have demonstrated the existence of direct contact RPA3-ssDNA, bearing a photoreactive nucleotide near the 3′-end. Thus, even if this contact is low or transient, it is clear that RPA3 (DBD E) is positioned near the 3′-side of ssDNA in conditions of elongated extended hRPA:ssDNA complex formation. Transient or low contact could be explained by a very weak affinity of DBD E for ssDNA due to the significant structural differences observed in the OB-fold motif in DBD E (50). In addition, a comparison of the ssDNA binding surfaces between RPA1 and the RPA3/RPA2 dimer showed that the lower affinity of RPA3/RPA2 can be contributed to a shallower binding crevice with reduced positive electrostatic charge (52). This close contact observed here significantly advances our understanding of the 30 nt binding mode of RPA. The entire contact data presented in Figure 5A allow us to propose a schematic model of the 30 nt binding mode where RPA3 is clearly localized at the 3′-side of the ssDNA. This model agrees with the well-characterized polarity of RPA in which RPA1 is located at the 5′-side and RPA2 towards the 3′-side (15,17). Localization of RPA2 around position 22 agrees with recent data that place RPA2 around position 24 (51) with a straight 8-nt inserted into the ssDNA between the DBD C and DBD D domains. However, significant cross-linking with RPA2 around position 16 suggests that this subunit could move along the ssDNA and be placed more or less near RPA1. From the data obtained in conditions where the 2:1 complex predominates, we propose a 13–22 nt binding mode, RPA1 overall occluding the length of the ssDNA. On the basis of the known structures of DBD domains of RPA and of further recent data (51), we propose that RPA1 interacts with ssDNA by sequential loading, from the 5′- to 3′-end of ssDNA, of DBD A, DBD B and DBD C domains. DBD D of RPA2 is also placed along this length of the DNA strand. At saturating RPA concentrations, when 3:1 complexes are predominantly formed, only RPA1 directly contacts with DNA in accordance with an 8–10 nt binding mode of RPA in which only DBD A and DBD B interact with ssDNA.
Recent data obtained using similar photoreactive ssDNA with a different non monotonous sequence also demonstrated formation of hRPA:ssDNA complexes of various stoichiometries and existence of photo-cross-linking product, which can be conceivably attributed as RPA3-DNA adduct (53). Several distinctions in non-covalent complexes formation and in photo-cross-linking patterns, as compare to the data presented here, can be explained by the differences of experimental conditions used. This also emphasized, that interaction of RPA with ssDNA is a dynamic process, which is sensitive to many factors (19).
At higher RPA concentrations, RPA molecules compete with each other to bind to short DNA. As DBD A and DBD B of RPA bind DNA with high affinity, each RPA molecule occludes 8–10 nt producing DNA saturated by several RPA molecules. This polymerization phenomenon is in accordance with recent data showing polymerization of RPA along long ssDNA by AFM microscopy (54). When the protein concentration decreases, more sites are available for each RPA molecule allowing binding of other DBD domains. Thus binding of DBD A and DBD B can be followed by the weakly binding DBD C on the 3′-side, resulting in the elongated contracted (13–22 nt) binding mode as well as by DBD D. Binding of DBD E is possible at lower protein concentrations leading to the formation of elongated extended RPA complexes characterized by a 30 nt long occluded binding site. It is interesting that RPA3 was never detected as cross-linking target of photoaffinity labeling of RPA using recessed template primers with various photoreactive groups introduced in 3′-end of the primer; 3′-protruding 30 nt ssDNA templates were used in this study (18). The major target of photo-cross-linking with an [RPA]/[ssDNA] ratio of 1/1 was RPA2. When a recessed template primer with a template length of 13–14 nt or less was used, the main target of cross-linking was RPA1. These data are in agreement with the model proposed here. The absence of RPA3 at the template primer—overhang might be due to the difference in mode of RPA interaction with ssDNA and recessed template-primer DNA structure as well as by the absence of direct contact of RPA3 with template-primer overhang (21).
In conclusion, in the present study RPA3 was identified as the subunit of the heterotrimer directly contacting the 3′-side of ssDNA when one molecule of hRPA in an elongated conformation binds a 31 nt ssDNA molecule. The proposed scheme of elongated complex is in good agreement with sequential 5′→3′ binding of RPA where the first DBDs A and B of RPA1 located to the 5′-side start to interact with the DNA (8–10 nt binding mode), followed along the DNA by DBD C (from RPA1), DBD D (from RPA2) and finally DBD E (from RPA3) binds to the DNA progressing to the 30 nt high-affinity binding mode. To identify the details of the interaction of each RPA subunit and ssDNA, isolation of protein–ssDNA adducts and analysis by mass spectrometry methods will be employed.
FUNDING
GEFLUC (to C.S.), INTAS-SBRAS grant and Program RAS Molecular and Cellular Biology (to O.L. and I.P.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Anne-Lise Haenni (IJM, Paris, France) for careful reading of the manuscript. We are grateful to CONACYT and SEP Mexico scholarships (to T.R.S.).
REFERENCES
- 1.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 3.Millership JJ, Cai X, Zhu G. Functional characterization of replication protein A2 (RPA2) from Cryptosporidium parvum. Microbiology. 2004;150:1197–1205. doi: 10.1099/mic.0.26833-0. [DOI] [PubMed] [Google Scholar]
- 4.Smith J, Zou H, Rothstein R. Characterization of genetic interactions with RFA1: the role of RPA in DNA replication and telomere maintenance. Biochimie. 2000;82:71–78. doi: 10.1016/s0300-9084(00)00183-8. [DOI] [PubMed] [Google Scholar]
- 5.Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Wilson E, Géli V. RPA regulates telomerase action by providing Est 1p access to chromosome ends. Nature Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- 6.Romero Salas T, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Fave A, Saintomé C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kibe T, Ono Y, Sato K, Ueno M. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol. Biol. Cell. 2007;18:2378–2387. doi: 10.1091/mbc.E06-12-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grudic A, Jul.Larsen A, Haring SJ, Wold MS, Lonning PE, Bjerkvig R, Boe SO. Replication protein A prevents accumulation of single-stranded telomeric DNA in cells that use alternative lengthening of telomeres. Nucleic Acids Res. 2007;35:7267–7278. doi: 10.1093/nar/gkm738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 10.Lao Y, Gomes XV, Ren Y, Taylor JS, Wold MS. Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry. 2000;39:850–859. doi: 10.1021/bi991704s. [DOI] [PubMed] [Google Scholar]
- 11.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyka IM, Dhar K, Binz SK, Wold MS. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003;42:12909–12918. doi: 10.1021/bi034930h. [DOI] [PubMed] [Google Scholar]
- 13.Mou TC, Shen M, Abdalla S, Delamora D, Bochkareva E, Bochkarev A, Gray DM. Effects of ssDNA sequences on non-sequence-specific protein binding. Chirality. 2006;18:370–382. doi: 10.1002/chir.20262. [DOI] [PubMed] [Google Scholar]
- 14.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 15.De Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers N.GJ, Hoeijmakers J. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iftode C, Borowiec JA. 5’-> 3’ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry. 2000;39:11970–11981. doi: 10.1021/bi0005761. [DOI] [PubMed] [Google Scholar]
- 17.Kolpashchikov DM, Khodyreva SN, Khlimankov DY, Wold MS, Favre A, Lavrik OI. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 2001;29:373–379. doi: 10.1093/nar/29.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavrik OI, Kolpashchikov DM, Weisshart K, Nasheuer HP, Khodyreva SN, Favre A. RPA subunit arrangement near the 3’-end of the primer is modulated by the length of the template strand and cooperative protein interactions. Nucleic Acids Res. 1999;27:4235–4240. doi: 10.1093/nar/27.21.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alani E, Thressher R, Griffith JD, Kolodner RD. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 21.Pestryakov PE, Weisshart K, Schlott B, Khodyreva SN, Kremmer E, Grosse F, Lavrik OI, Nasheuer H.-P. Human replication protein A. The C-terminal RPA70 and the central RPA32 domains are involved in the interactions with the 3’-end of a primer-template DNA. J. Biol. Chem. 2003;278:17515–17524. doi: 10.1074/jbc.M301265200. [DOI] [PubMed] [Google Scholar]
- 22.Pestryakov PE, Khlimankov DY, Bochkareva E, Bochkarev A, Lavrik OI. Human replication protein A (RPA) binds a primer-template junction in the absence of its major ssDNA-binding domains. Nucleic Acids Res. 2004;32:1894–1903. doi: 10.1093/nar/gkh346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arunkumar AI, Klimovich V, Jiang X, Ott RD, Mizoue L, Fanning E, Chazin WJ. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005;12:332–339. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Favre A. Morrison H. Bioorganic Photochemistry. Vol. 1. New York: Willey & Sons; 1990. 4-thiouridine as an intrinsic photoaffinity probe of nucleic acid structure and interactions. pp. 379–425. [Google Scholar]
- 26.Gomes XV, Henricksen LA, Wold MS. Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry. 1996;35:5586–5595. doi: 10.1021/bi9526995. [DOI] [PubMed] [Google Scholar]
- 27.Dos Santos DV, Vianna A.-L, Fourrey JL, Favre A. Folding of DNA substrate-hairpin ribozyme domains: use of deoxy 4-thiouridine as an intrinsic photolabel. Nucleic Acids Res. 1993;21:201–207. doi: 10.1093/nar/21.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igloï GL. Interaction of tRNAs and phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry. 1988;27:3842–3849. doi: 10.1021/bi00410a048. [DOI] [PubMed] [Google Scholar]
- 29.Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 30.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 31.Gomes XV, Wold MS. Structural analysis of human replication protein A. J. Biol. Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 32.Sibenaller ZA, Sorensen BR, Wold MS. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- 33.Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry. 1999;38:3974–3984. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 34.Walther AP, Gomes XV, Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70 kDa subunit. Biochemistry. 1999;38:3963–3973. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- 35.Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackwell LJ, Borowiec JA, Mastrangelo IA. Single-stranded DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J. Mol. Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 38.Iftode C, Borowiec JA. Unwinding of origin-specific structures by human replication protein A occurs in a two-step process. Nucleic Acids Res. 1998;26:5636–5643. doi: 10.1093/nar/26.24.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Wu P, Ohmichi T, Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 40.Blackwell LJ, Borowiec JA. Human replication protein A binds single-stranded DNA in two distinct complexes. Mol. Cell. Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Crystal structure of the DNA binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 42.Misura M, Buterin T, Hindges R, Hüscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X, Klimovich V, Arunkumar A, Hysinger EB, Wang Y, Ott RD, Guler GD, Weiner B, Chazin WJ, Fanning E. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 2006;25:5516–5526. doi: 10.1038/sj.emboj.7601432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petruseva IO, Tikhanovich IS, Chelobanov BP, Lavrik OI. RPA repair recognition of DNA containing pyrimidines bearing bulky adducts. J. Mol. Res. 2008;21:154–162. doi: 10.1002/jmr.877. [DOI] [PubMed] [Google Scholar]
- 46.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single–stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brill SJ, Bastin-Shanower S. Identification and characterization of the four single-stranded-DNA binding domain of replication protein A. Mol. Cell. Biol. 1998;18:7225–7234. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daughdrill GW, Ackerman J, Isern NG, Botuyan MV, Arrowsmith C, Wold MS, Lowry DF. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29:3270–3276. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philipova D, Mullen JR, Maniar HS, Lu J, Gu C, Brill SJ. A hierarchy of SSB promoters in replication protein A. Genes Dev. 1996;10:2222–2223. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 50.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai L, Roginskaya M, Qu Y, Yang Z, Xu Y, Zou Y. Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007;46:8226–8233. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng X, Habel JE, Kabaleeswaran V, Snell EH, Wold MS, Borgstahl G.EO. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J. Mol. Biol. 2007;374:865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- 53.Pestryakov PE, Krasikova YS, Petruseva IO, Khodyreva SN, Lavrik OI. The role of p14 subunit of replication protein A in binding to single-stranded DNA. Doklady Biochem. Biophys. 2006;412:4–7. doi: 10.1134/s1607672907010024. [DOI] [PubMed] [Google Scholar]
- 54.Hamon L, Pastré D, Dupaigne P, Le Breton C, Le cam E, Piétrement O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein-DNA complexes. Nucleic Acids Res. 2007;35:e58. doi: 10.1093/nar/gkm147. [DOI] [PMC free article] [PubMed] [Google Scholar]