Abstract
Chemically modified antisense oligonucleotides (ASOs) are widely used as a tool to functionalize microRNAs (miRNAs). Reduction of miRNA level after ASO inhibition is commonly reported to show efficacy. Whether this is the most relevant endpoint for measuring miRNA inhibition has not been adequately addressed in the field although it has important implications for evaluating miRNA targeting studies. Using a novel approach to quantitate miRNA levels in the presence of excess ASO, we have discovered that the outcome of miRNA inhibition can vary depending on the chemical modification of the ASO. Although some miRNA inhibitors cause a decrease in mature miRNA levels, we have identified a novel 2′-fluoro/2′-methoxyethyl modified ASO motif with dramatically improved in vivo potency which does not. These studies show there are multiple mechanisms of miRNA inhibition by ASOs and that evaluation of secondary endpoints is crucial for interpreting miRNA inhibition studies.
INTRODUCTION
MicroRNAs (miRNAs) are a class of noncoding RNAs that regulate gene expression post-transcriptionally (1,2). In mammals, regulatory roles have been identified for miRNA in many areas of biology (3,4), highlighting miRNA as an exciting new class of therapeutic targets with broad applications. Antisense oligonucleotide (ASO) inhibition of miRNA has been a useful tool for their functionalization and shows promise as a therapeutic strategy. Chemical modification of ASOs to improve stability to nucleases and affinity for target RNA is generally necessary for robust activity. Effective targeting of miRNA by ASOs with various backbone and 2′ sugar modifications including 2′-O-methyl (2′-OMe), 2′-O-methoxyethyl (2′-MOE), 2′-fluoro (2′F) and locked nucleic acid (LNA) has been reported by several groups in cell culture (5–9) and after systemic delivery in vivo (10–13) and have been widely used for miRNA functionalization.
In the course of our own screening efforts to identify more effective anti-miRNA oligonucleotides, we recognized the importance of a reliable endpoint to evaluate miRNA inhibition. Measurement of miRNA levels by northern blotting or RT–PCR is commonly reported to assess miRNA inhibition. However, inhibition of miRNA activity, measured by modulation of miRNA target genes, without degradation of the miRNA has recently been reported (10). In addition, hybridization-based measurements of miRNA levels can be problematic, as they are potentially subject to interference by the complementary ASO present. If miRNA levels are not affected by ASO inhibition or cannot be measured reliably, a secondary endpoint—modulation of a miRNA target gene—is required to measure inhibition of miRNA activity.
We set out to develop a reliable method to measure miRNA levels in the presence of ASO, to better understand the outcome of miRNA inhibition and determine how best to measure ASO efficacy. We previously showed that a uniformly modified 2′-MOE phosphorothioate backbone ASO was an effective inhibitor of the abundant liver-expressed miR-122 in mice after systemic delivery in saline (12), and reduced miR-122 levels. Using our novel method to quantitate miRNA in the presence of ASO, we have confirmed this result. Our screening efforts have additionally identified a chimeric 2′-F/MOE ASO as a potent inhibitor of miRNA activity in vivo, when measured by derepression of miRNA target genes. Surprisingly, the mechanism of miRNA inhibition by 2′-F modified ASOs does not involve degradation of the miRNA. This suggests that there are multiple outcomes of miRNA inhibition by ASOs and that evaluation of secondary endpoints is crucial for interpreting miRNA inhibition studies.
METHODS
Animal care and treatments
All animal experiments were conducted according to the Institutional AAALAC Guidelines. Male C57BL/6 mice were housed four to five animals per cage with a 12 h light/dark cycle. Oligonucleotides were dissolved in saline and administered to mice based on body weight by intraperitoneal (i.p.) injection, twice weekly.
Northern blotting
Total RNA was purified using Trizol reagent (Invitrogen). 10 μg total RNA in 1× TBE-Urea loading buffer (Invitrogen) was separated on a 15% TBE-Urea polyacrylamide gel (Invitrogen) or a 14% TBE-Urea polyacrylamide gel with 20% formamide. Samples run on formamide containing gels were also loaded in 20% formamide. RNA was transferred to Hybond N + membranes (Amersham) at 30V for 2 hr and UV cross-linked. Blots were pre-hybridized in Rapid-Hyb Buffer (GE Healthcare) for 1 hr at 42°C and then hybridized with a 32P end-labeled DNA probe for 4 hr at 42°C. Blots were washed in 5× SSC buffer + 0.1% SDS twice for 5 min at room temperature and exposed overnight. Quantitation was done using a Storm 860 phosphoimager (Molecular Dynamics) and ImageQuant software.
Oligonucleotide synthesis and purification
2′-MOE, 2′-OMe, 2′-MOE/LNA and 2′-deoxy-2′-Fluoro/2′-MOE-modified oligonucleotides were synthesized on AKTA Oligopilot 10 (Amersham/GE Healthcare) oligonucleotide synthesizer. The 2′-MOE amidites were prepared as reported (14–16). The LNA, 2′-OMe and 2′-deoxy-2′-fluoro modified pyrimidine nucleoside phosphoramidites were purchased from commercial sources. The 2′-deoxy-2′-F purine nucleoside phosphoramidites were synthesized as described in the literature (16,17). Oligonucleotides were prepared as reported earlier (18) with minor modifications described here. The 3 min detritylation step was performed with 6% DCA/toluene. A 0.1 M solution of the phosphoramidites in anhydrous acetonitrile was used for the synthesis. For sulfurization, 0.2 M Phenylacetyl disulfide (PADS) in 1:1 3-picoline/CH3CN was used with 6 min contact time. After completion of the synthesis, solid support was treated with triethyl amine:acetonitrile (1:1) for 30 min. It was suspended in a mixture of aqueous ammonium hydroxide (30 wt.%): ethanol (3:1) and heated at 55°C for 9 h to complete the removal of all protecting groups.
The oligonucleotides were purified by ion exchange chromatography on an AKTA Explorer (GE healthcare) HPLC system on a strong anion exchange column (source 30 Q, GE Healthcare). Fractions containing full-length oligonucleotides were pooled together (assessed by LC MS analysis) and evaporated. The ASOs were desalted by reverse phase HPLC to furnish modified oligonucleotides in 30–40% yield based on the loading of the solid support. All the modified oligonucleotides were characterized by ion-pair-HPLC–MS analysis with Agilent 1100 MSD system.
Competitor PNA assay
Competitor PNA was obtained from Panagene. PNA was added to 10 μg total RNA in 1× TBE-Urea loading buffer (Invitrogen). Samples were mixed and heated at 95°C for 3 min prior to loading on 15% TBE-Urea polyacrylamide gel (Invitrogen). Prior to electroblotting, the top portion of the gel containing excess competitor PNA was cut and separated from lower miRNA-containing portion. Use of a 23-bp fluoroscein-labeled oligonucleotide aided in gel separation. After electroblotting, UV crosslinking, and pre-hybridization as described, the pre-hybridization buffer was exchanged for fresh buffer and blots were then hybridized for 4 h at 42°C. Blots were washed in 5× SSC buffer + 0.1% SDS (Invitrogen) twice for 5 min and exposed overnight.
RT–PCR analysis
qRT–PCR analysis was performed with a Prism 7700 Sequence Detector (Applied Biosystems). All RT-PCR reagents were obtained from Invitrogen. ALDOA primer/probe sequences: Forward primer, 5′-AGGCTCTTTCCCATCACTCTTG-3′; Reverse primer, 5′-GATGGCAGATTTAGCATTCACAGA-3′, Probe: 5′-FAM-TGTGCCCTCGTGTGCGGTG-3′-TAMRA. Real-time RT-PCR analysis of miR-122 levels performed with ABI Taqman miRNA assay kit.
Tm measurements
Absorbance versus temperature curves were measured at 260 nm using a Beckman DU 640 Spectrophotometer and High Performance Temperature Controller. The buffer contained 100 mM Na+, 10 mM phosphate and 0.01 mM EDTA, pH 7.0 with sufficient Cl− to achieve ionic neutrality. Oligonucleotide concentration was 4 μM each strand determined from the absorbance at 85°C and extinction coefficients calculated according to Puglisi and Tinoco (19). Tm's of duplex formation were obtained from fits of data to a two-state model with linear sloping baselines (20).
Data analysis
ED50 calculations were performed using GraphPad Prism software.
RESULTS
Novel chimeric 2′-F/MOE ASO potently inhibits miR-122 in vivo
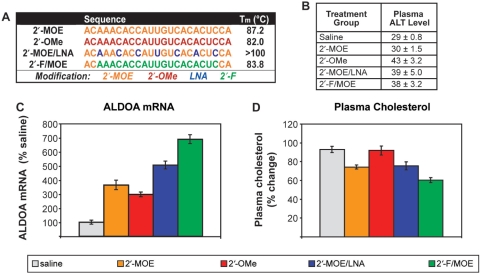
We have evaluated a panel of over 60 chemically-modified ASOs for their ability to inhibit the liver-expressed miR-122 in mice after systemic delivery. Initially, a panel of various 2′sugar, heterocycle and backbone modifications were evaluated (for an example, see Supplementary Figure 1), and modifications that supported anti-miR activity were then further explored to optimize their placement in the anti-miR ASO. In the course of our screening efforts, we were faced with the question of how best to evaluate miRNA inhibition. We and others had previously demonstrated a derepression of ALDOA and other miR-122 target genes at the mRNA level, reduced plasma cholesterol, and an apparent reduction of miR-122 levels measured by northern blotting after inhibition of miR-122 in vivo (12,13). MiRNA level may be the most direct measurement, but as outlined above, may not be the expected outcome of ASO inhibition. As there are well-validated target genes for miR-122, we compared ASO activity by measuring their derepression by RT-PCR. Several of the most active ASOs identified from our screening efforts were compared in a repeat dosing study. Normal mice were treated intraperitoneally (i.p.) with 25 mg/kg ASO twice weekly for three weeks. The 2′-MOE ASO previously described (12) was compared with 2′-OMe, a chimeric 2′-MOE/LNA and a 2′F modified ASO with two 2′-MOE modifications on either end (2′-F/MOE) for added stability in vivo (Figure 1A). All of the ASOs had fully modified phosphorothioate backbones, which aids delivery to tissues without formulation and provides stability to nuclease degradation (21). The 2′-F/MOE ASO was the most active as measured by a 7-fold increase in ALDOA mRNA in the liver compared to saline-treated mice, and nearly 40% reduction in plasma cholesterol (Figure 1C and D). The 2′-MOE/LNA compound was the next most active, resulting in a 5-fold increase in ALDOA mRNA and approximately 20% plasma cholesterol lowering. The activity of the 2′-MOE ASO was similar to that previously reported, with ALDOA mRNA levels increased 3.5-fold compared to saline and a 20% reduction in plasma cholesterol. However, treatment with the 2′-OMe ASO, while causing a 3-fold increase in ALDOA mRNA levels, did not result in plasma cholesterol lowering. Measuring several other miR-122 target genes gave similar results (data not shown). There were only mild increases in plasma transaminase levels in the mice treated with the 2′-OMe, 2′-MOE/LNA and 2′-F/MOE ASOs, and no significant change in the mice treated with the 2′-MOE ASO (Figure 1B), indicating that none of the ASOs caused liver toxicity.
Figure 1.
In vivo evaluation of miR-122 ASOs identifies 2′-F/MOE as potent miRNA inhibitor. Normal mice were treated i.p. with 25 mg/kg dose of miR-122 ASOs twice weekly for 3 weeks. Animals were sacrificed 48 hr after the last dose. n = 5. Error = SEM (A) Chemical modification of anti-miR ASOs evaluated for activity against miR-122 in mouse liver. All ASOs have completely modified phosphorothioate backbones. (B) Plasma transaminase levels. (C) Real-time RT–PCR measuring levels of miR-122 target gene ALDOA in liver RNA. (D) Change in total plasma cholesterol levels.
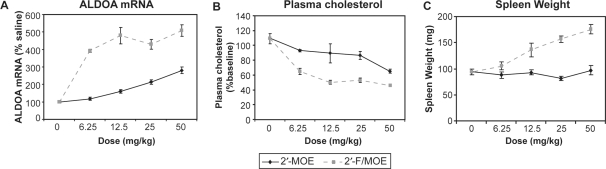
The improved in vivo activity of the 2′-F/MOE ASO over the higher affinity 2′-MOE and 2′-MOE/LNA ASOs was surprising. We next compared its potency to the 2′-MOE ASO by performing a dose response. Normal mice were treated i.p. with 6–50 mg/kg 2′-F/MOE or 2′-MOE ASOs twice weekly for 3 weeks. Treatment with the 2′-MOE ASO resulted in a dose-responsive increase in ALDOA mRNA levels in the liver, reaching a maximum 3-fold increase at the highest dose (Figure 2A). By contrast, all of the 2′-F/MOE treatment groups showed a 4–5-fold increase in ALDOA mRNA levels, suggesting that the maximal effect was achieved by the 12.5 mg/kg dose level (Figure 2A). The 2′-F/MOE ASO was more efficacious and at least 8-fold more potent than the 2′-MOE ASO, since the lowest dose of 2′-F/MOE was already more effective than an 8-fold higher dose of 2′-MOE ASO. A similar result was observed for plasma cholesterol lowering (Figure 2B). Evaluation of plasma transaminase levels showed no significant elevations in ALT from any of the treated mice. However, a dose-dependent increase in spleen weight at sacrifice was observed after treatment with the 2′-F/MOE ASO which was not observed after 2′-MOE treatment (Figure 2C). This suggested that the 2′-F/MOE ASO may have a mild immunostimulatory effect in the mice. The introduction of three additional 2′-MOE modifications into the 2′-F/MOE ASO was able to blunt this immune stimulation without affecting anti-miR-122 activity (Supplementary Figure 2), an effect which has previously been observed for phosphorothioate oligodeoxynucleotides (22). Interestingly, placement of 2′-MOE modifications was only tolerated in the 5′ half of the ASO. Additional 2′-MOE modifications in the 3′ end of the ASO negatively affected anti-miR activity.
Figure 2.
Improved potency and efficacy of 2′-F/MOE compared to 2′-MOE ASO. Normal mice were treated i.p. with indicated doses of miR-122 ASOs twice weekly for 3 weeks, and animals were sacrificed 48 hr after the last dose. n = 5 Error = SEM (A) Real-time RT–PCR measuring levels of miR-122 target gene ALDOA in liver RNA. (B) Change in total plasma cholesterol levels. (C) Spleen weight.
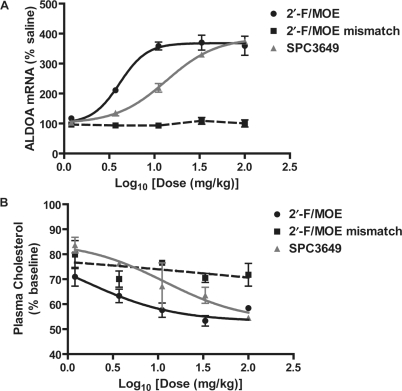
We next compared the activity of the 2′-F/MOE ASO with a 15 mer LNA/DNA chimeric ASO (SPC3649), which was reported to inhibit miR-122 activity in mice (11). Additional studies had revealed that robust anti-miR-122 activity could be measured after a single administration of the 2′-F/MOE ASO, so the activity of the ASOs was compared four days after a single treatment of 1.2–100 mg/kg. A six-base mismatch 2′-F/MOE ASO was also included. Levels of the miR-122 target gene ALDOA were increased almost 4-fold after treatment with the highest, 100 mg/kg dose of the 2′-F/MOE miR-122 ASO, with near-maximal activity already observed at the 11 mg/kg dosing level (Figure 3A). The mismatched 2′-F/MOE ASO had no effect on ALDOA mRNA levels. Treatment with the LNA/DNA ASO resulted in a dose-dependent increase in ALDOA mRNA levels, but was approximately 3.5-fold less potent than the 2′-F/MOE ASO (calculated ED50 14 mg/kg compared to 4 mg/kg). Plasma cholesterol lowering followed a similar trend for the three ASOs (Figure 3B). As measured by miRNA target gene modulation, the 2′-F/MOE ASO is the most potent miRNA inhibitor reported to date.
Figure 3.
Improved potency of 2′-F/MOE compared to 15 mer LNA/DNA ASO (SPC3649) after single administration. Normal mice were treated i.p. with a single administration of the indicated dose of miR-122 ASO, and anti-miR-122 activity was measured four days later. n = 2–4 Error = SEM. (A) Real-time RT–PCR measuring miR-122 target gene mRNA ALDOA in liver RNA. (B) Change in total plasma cholesterol levels from baseline.
Measuring miRNA levels in the presence of ASO
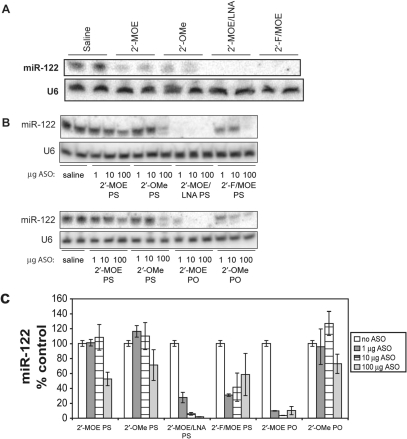
We next measured by northern blotting the miR-122 levels in the livers of mice from the experiment described in Figure 1. Krutzfeldt et al. (13) had reported that interference of miRNA detection by ASO could be eliminated by running the RNA samples in a denaturing gel containing 20% formamide, so northern blotting was performed under these conditions (Figure 4A). Treatment with all of the ASOs led to significant reduction in the levels of miR-122 detected, with no measurable signal from the 2′-MOE/LNA or 2′-F/MOE ASO-treated samples. However, the complete absence of miR-122 after treatment with the high-affinity 2′-MOE/LNA led us to suspect interference with miR-122 detection, in spite of the presence of formamide, similar to what has been previously reported (23). Control experiments confirmed that in the case of the high-affinity 2′-MOE/LNA, as well as the 2′-F/MOE ASO, significant interference with miR-122 detection was observed when the ASO was added to liver tissue lysates from untreated mice (Figure 4B). Although phosphorothioate backbone-modified 2′-OMe and 2′-MOE ASOs only minimally interfered with miR-122 detection, the higher affinity phosphodiester backbone 2′-OMe and 2′-MOE ASO interfered substantially. A similar pattern of interference by the ASOs with Taqman RT-PCR detection of miR-122 was also seen (Figure 4C).
Figure 4.
Some anti-miRNA ASOs interfere with miRNA detection. (A) Northern blotting for miR-122 in liver RNA from mice treated as described in Figure 1. (B) Inhibition of miR-122 detection by northern blotting after spiking the indicated amount of ASO (μg) into Trizol lysates containing 50 mg liver tissue before RNA purification. PS, phosphorothioate backbone; PO, unmodified phosphodiester backbone. RNA is separated on polyacrylamide gel containing 8 M urea and 20% formamide. (C) Real-time RT–PCR detection of miR-122 with TaqMan miRNA Assay in same samples shown in (B).
We assumed that the degree of interference observed on the northern blot corresponded to the affinity of the ASO for the miRNA, although the data were not entirely consistent with this (e.g. 2′-F/MOE ASO). One possible explanation is that recovery of the 2′-F/MOE ASO is greater than the 2′-MOE ASO during the RNA purification process, leading to greater interference with miR-122 detection during northern blotting. To investigate this, we measured the recovery of radiolabeled ASOs spiked into Trizol lysates of mouse liver tissue (Supplementary Figure 3A). Recovery of all the modified ASOs was similar, however, making it unlikely that differential ASO recovery accounted for the different degrees of interference by the ASOs.
We observed in these experiments that while a large portion of the ASOs persist through the RNA preparation process, the majority of ASO remains in the organic phase during the first step of the Trizol isolation protocol. Some ASOs may remain bound to the miRNA during the Trizol purification, partitioning the miRNA into the organic phase during the phase separation step of the RNA isolation. To evaluate this, we measured recovery of a radiolabeled miR-122 RNA through the Trizol purification process in the presence of the ASOs (Supplementary Figure 3B). Overall recovery of the miRNA in the presence of the high affinity 2′-MOE/LNA ASO was 4-fold reduced compared to the other ASOs, none of which had any significant effect on miR-122 recovery. The recovery of miR-122 during the precipitation step was similar for all the ASOs, indicating that the miRNA is primarily lost during the initial phase separation. We also observed other high affinity ASOs, including the antagomir described by Krutzfeldt et al. (13), similarly retaining the miRNA in the organic phase (data not shown).
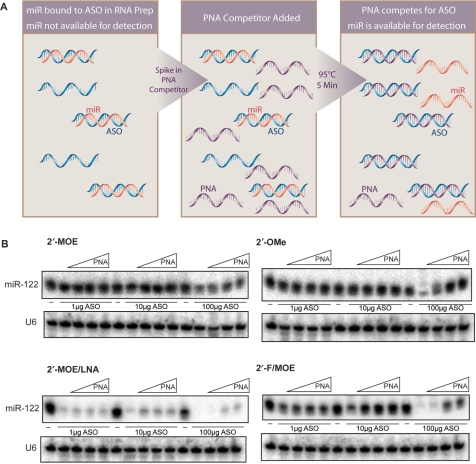
To more accurately quantitate miRNA levels in the presence of ASO, we devised a strategy to free the miRNA from the ASO after RNA purification by adding an excess of high affinity peptide nucleic acid (PNA) complementary to the ASO to compete for its binding, thereby making it available for northern hybridization (Figure 5A). When the RNA sample is separated on a polyacrylamide gel by electrophoresis, the PNA-bound ASO is shifted on the gel and migration of free PNA will also be retarded compared to the miRNA due to its neutral charge. This is important, as the PNA is the same sequence as the miRNA and will be detected by the northern probe.
Figure 5.
Competitor PNA strategy recovers detection of miRNAs in the presence of anti-miRNA ASOs. (A) Competitor PNA strategy to recover detection of miRNAs in the presence of anti-miR ASOs. (B) Recovery of miR-122 detection in total liver RNA in presence of ASO by addition of competitor PNA.
The feasibility of this approach was shown by its ability to free radiolabeled miR-122 from a miRNA-ASO duplex (Supplementary Figure 4). We next evaluated the competitor PNA's ability to restore northern detection of miR-122 in total liver RNA in the presence of ASO (Figure 5B). Increasing amounts of ASO were added to Trizol liver lysate, and RNA was isolated. Competitor PNA was added to the purified RNA samples, which were separated on a denaturing gel and northern blotting for miR-122 was performed. Under conditions in which the 2′-MOE and 2′-OMe ASOs interfered with miRNA detection, the competitor PNA restored miR-122 detection, although in the case of the 2′-MOE ASO the recovery was not complete. The competitor PNA also restored the miR-122 signal in the presence of 1 and 10 μg of the 2′-F/MOE ASO, but only partially in the samples containing 100 μg of the ASO. The inability of the PNA competitor to restore the miR-122 signal in the presence of the 2′-MOE/LNA ASO was not surprising, since miR-122 is retained by this ASO in the organic phase during RNA purification and is therefore not present to be recovered.
miR-122 levels after in vivo ASO inhibition
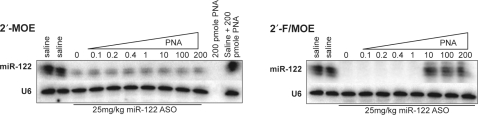
We next used this assay to evaluate the effect of in vivo ASO inhibition on miR-122 levels. Competitor PNA was added to mouse liver RNA from the study described in Figure 1, and northern blotting for miR-122 was performed (Figure 6). The tissue level of the 2′-MOE ASO after this 3 weeks dosing regimen is not expected to be in a range, where interference with miRNA detection is likely. Without any competitor PNA, a 5-fold reduction in miRNA levels was measured and addition of competitor PNA did not recover any further signal. While miR-122 detection in the RNA from 2′-F/MOE ASO-treated mice was completely inhibited, 80% of the miRNA signal could be restored by addition of the competitor PNA. Surprisingly, the most potent compound caused only a modest change in miRNA levels.
Figure 6.
Most potent 2′-F/MOE ASO does not cause substantial miRNA reduction. Northern blotting for miR-122 after separation of RNA by denaturing PAGE in presence of competitor PNA. Normal mice were treated i.p. with 25 mg/kg dose of miR-122 ASOs twice weekly for 3 weeks.
Consequences of miR-122 inhibition on global miRNA levels in the liver
Our data suggest that the 2′-MOE and 2′-F/MOE ASOs inhibit miRNA activity by different mechanisms. While the 2′-F/MOE apparently acts by sequestering the miRNA, the mechanism of miRNA reduction by the 2′-MOE or 2′-OMe ASOs is currently unknown. The ASOs may be accelerating turnover of the mature miRNA or acting on the precursor to prevent miRNA processing. Any of these mechanisms may have consequences for the miRNA pathway, if it results in sequestration, accelerated turnover or increased availability of RISC. We therefore evaluated the levels of other miRNAs in the liver after inhibition of miR-122 with the 2′-MOE and the 2′-F/MOE ASOs. Profiling of miRNA expression after 4 weeks of miR-122 inhibition with the 2′-MOE ASO or 6 weeks of inhibition with the 2′-F/MOE ASO, revealed that few miRNAs were modulated as a result of ASO treatment (Supplementary Table 1). The changes in miRNA levels were small (<2-fold) and not consistently up or downregulated, showing that in vivo inhibition of miR-122 by either ASO did not result in substantial dysregulation of miRNA levels in the liver.
DISCUSSION
The technical difficulty of measuring miRNA levels in the presence of abundant ASO has made the interpretation of many published miRNA inhibition studies challenging. The level of anti-miR-122 ASO accumulated in the liver after in vivo dosing may be as much as hundreds of micrograms per gram of liver tissue, with the majority of the ASO not bound to miRNA. The ratio of transfected ASO to miRNA in cell culture studies will also be high, leading to detection artifacts. While interference with miRNA detection or detection of a miRNA–ASO duplex is an indication that ASO is associated with the cell or tissue, it is not a measure of inhibition of miRNA activity. Furthermore, if miRNA degradation is not the primary outcome of ASO inhibition, measuring miRNA levels, even if done reliably, will not correlate with miRNA activity. Here we have described a novel strategy to measure miRNA levels in the presence of ASO, involving the addition of a PNA complementary to the ASO to compete it away from the miRNA, freeing it for northern blot detection. The results show that although treatment with the 2′-MOE ASO does result in reduced miRNA levels, the 2′-F/MOE ASO potently inhibits miRNA activity in vivo without substantial reduction in miR-122 levels. This suggests that for the most effective anti-miRNA ASO, measuring miRNA levels on a northern blot or by RT–PCR is not informative, making the identification of a secondary endpoint, a miRNA target gene, absolutely critical for interpretation of miRNA inhibition studies. The availability of well-validated miR-122 target genes that are robustly and dose-dependently modulated after miRNA inhibition has allowed us to usefully compare the potency of chemically modified miRNA inhibitors in vivo. However, in most miRNA inhibition studies, target genes are not known or only modestly modulated, making it difficult to assess miRNA inhibition. Consequently, many phenotypic outcomes of miRNA inhibition studies are difficult to interpret.
Several studies have evaluated the effects of sugar and backbone oligonucleotide modifications on anti-miRNA activity in cell culture, but this is the first study directly comparing the effectiveness of a panel of modifications in vivo. Our own in vitro studies evaluating anti-miRNA ASOs in Hela cells had highlighted the 2′-F modification as a surprisingly potent modification, and those findings have translated well to the in vivo setting. The reasons for the dramatic improvement in potency by the 2′-F/MOE ASO are not clear. It is at least partly independent of its affinity to the target miRNA, as it was more potent than the full-length 2′-MOE/LNA ASO with higher measured Tm. Accumulation of the 2′-F/MOE and 2′-MOE ASOs in the mouse liver are similar, suggesting that improved tissue delivery is not responsible for the improved potency. The 2′-F modification may allow more favorable interactions with the miRNA in RISC. The effect of the 2′-F/MOE ASO was sequence-specific, as a six-base pair mismatch of the same chemistry and 2′-F/MOE ASOs targeting several other miRNAs (data not shown), did not affect miR-122 target genes or plasma cholesterol. This argues against the possibility that the anti-miR-122 activity is an indirect result of the mild pro-inflammatory effects we observed with the parent 2′-F/MOE ASO. Further dissection of the contributions of length, affinity and pharmacokinetic properties to the anti-miR activity of the 2′F/MOE and LNA/DNA compounds should improve our understanding of the structure-activity relationship. Clarification of how inhibition with the 2′-MOE ASO leads to miRNA degradation, while the more potent 2′-F/MOE ASO does not, may give additional insight into the mechanisms of anti-miR inhibition and suggest new strategies for miRNA targeting.
The growing body of miRNA research is confirming that miRNA are an important class of regulators in many areas of biology. The application of antisense technology for functionalization and therapeutic targeting of miRNAs shows great promise. Twenty years of investment in the technology have advanced understanding of how oligonucleotide chemical modifications affect stability, affinity to RNA and uptake into tissues. This experience should accelerate the development of ASOs targeting miRNAs. However, our understanding of the optimal chemical modifications for anti-miRNA ASOs is only at the beginning, and targeting of miRNAs with ASOs presents challenges for evaluating target engagement which must be addressed in order to interpret ASO inhibition studies. Identification of the 2′-F/MOE ASO as a particularly effective in vivo inhibitor of miR-122, and clarification of the outcome of miRNA inhibition with ASO represents significant progress in both of these areas.
FUNDING
Funding for open access charge: Regulus Therapeutics.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health [R43 AI072802].
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 11.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 14.Martin P. Ein neuer Zugang zu 2-O-Alkylribonucleosiden und Eigenschaften deren Oligonucleotide. Helv. Chim. Acta. 1995;78:486–504. [Google Scholar]
- 15.Ross B, Song Q. 2004. US patent no. 20040082775. [Google Scholar]
- 16.Ross BS, Springer RH, Sprankle KG, Vasquez G. An efficient and scalable synthesis of arabinosylguanine and 2′-deoxy-2′-fluoroguanosine. Nucleosides Nucleotides. 1997;16:1645–1647. [Google Scholar]
- 17.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 18.Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP, Bennett CF. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 20.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 21.Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB, Sasmor H, Manoharan M, Levin AA. Pharmacokinetic properties of 2'-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 2001;296:890–897. [PubMed] [Google Scholar]
- 22.Henry S, Stecker K, Brooks D, Monteith D, Conklin B, Bennett CF. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J. Pharmacol. Exp. Ther. 2000;292:468–479. [PubMed] [Google Scholar]
- 23.Fabani MM, Gait MJ. miR-122 targeting with LNA/2'-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.