Abstract
Many proteins contain reiterated glutamine residues, but polyglutamine of excessive length may result in human disease by conferring new properties on the protein containing it. One established property of a glutamine residue, depending on the nature of the flanking residues, is its ability to act as an amine acceptor in a transglutaminase-catalyzed reaction and to make a glutamyl–lysine cross-link with a neighboring polypeptide. To learn whether glutamine repeats can act as amine acceptors, we have made peptides with variable lengths of polyglutamine flanked by the adjacent amino acid residues in the proteins associated with spinocerebellar ataxia type 1 (SCA1), Machado–Joseph disease (SCA3), or dentato-rubral pallido-luysian atrophy (DRPLA) or those residues adjacent to the preferred cross-linking site of involucrin, or solely by arginine residues. The polyglutamine was found to confer excellent substrate properties on any soluble peptide; under optimal conditions, virtually all the glutamine residues acted as amine acceptors in the reaction with glycine ethyl-ester, and lengthening the sequence of polyglutamine increased the reactivity of each glutamine residue. In the presence of transglutaminase, peptides containing polyglutamine formed insoluble aggregates with the proteins of brain extracts and these aggregates contained glutamyl–lysine cross-links. Repeated glutamine residues exposed on the surface of a neuronal protein should form cross-linked aggregates in the presence of any transglutaminase activated by the presence of Ca2+.
Keywords: polyglutamine as amine acceptor, neuronal proteins
Glutamine is the most common repeated amino acid in eukaryotic proteins (1–3), and it has been postulated that the introduction of such repeats is a general evolutionary mechanism for adding new amino acid sequence. Involucrin, a product of the terminally differentiating keratinocyte, a substrate of transglutaminase (4), and a precursor of the cross-linked envelope, probably began in evolution as polyglutamine and was subsequently modified by mutation (5, 6).
Five different proteins containing a sequence of polyglutamine are associated with disease of the human nervous system. These polyglutamine sequences have been evolving toward greater length within the human lineage, since they are much shorter in the corresponding proteins of nonhuman primates (7, 8); only when the polyglutamine in the human protein exceeds about 35 residues does the protein produce disease of the nervous system (for recent reviews, see refs. 9 and 10). The allele encoding an abnormally long segment of polyglutamine is dominant over the normal allele, but as disease does not occur when a single allele is disrupted or deleted (11, 12), the disease resulting from the longer sequence of polyglutamine must be due to a dominant gain of function. This gain-of-function results in cell lethality (9, 12).
Two of the numerous explanations suggested for the disease-producing dominant gain of function are (i) polyglutamine sequences are highly insoluble because they form hydrogen-bonded polar zippers (13) and a polypeptide containing such a sequence will form multimeric but not covalently bonded aggregates (14), and (ii) if polyglutamine is a transglutaminase substrate, it should, in the presence of an active enzyme, become cross-linked with polypeptides containing lysyl groups to form covalently bonded aggregates (15).
To participate in a transglutaminase-catalyzed reaction, glutamine residues must be part of a peptide or polypeptide. It has long been known that in certain small proteins, most or all scattered glutamine residues may act as amine acceptors, at least in the absence of secondary or tertiary structure preventing access of the enzyme (16, 17); but in native proteins, the nature of the neighboring residues has appreciable influence on the reactivity of a glutamine residue (18–21), and it has been shown that some glutamine residues are greatly preferred to others (22, 23). Among preferred glutamine residues are ones adjacent to a second glutamine residue.
We describe here a variety of synthetic polypeptides containing polyglutamine. As long as the polypeptides are rendered sufficiently soluble by the flanking residues, all are excellent substrates of transglutaminase.
MATERIALS AND METHODS
Peptide Synthesis.
Peptides were synthesized at the Biopolymers Facility of the Howard Hughes Medical Institute, Harvard Medical School, and purified by HPLC. The quality of synthesis was verified by mass spectrometry and amino acid analysis. Peptides were dissolved in a buffer containing 50 mM Tris·HCl (pH 7.5) and 1 mM EDTA, and stored at 4°C at concentrations estimated to be between 0.4 and 5.0 mM. The precise concentration was determined by amino acid analysis of an aliquot.
Transglutaminase-Catalyzed Incorporation of Labeled Glycine Ethyl-Ester into Synthetic Peptides.
The reaction volume (25 μl) contained 100 mM Tris·HCl (pH 8.2), 10 mM CaCl2, 10 mM DTT, 20% glycerol, 19.3 mM unlabeled glycine ethyl-ester, 0.73 mM 14C-labeled glycine ethyl-ester (1 μCi; 1 Ci = 37 GBq), and 0.144 milliunit/μl of purified guinea pig liver transglutaminase (Sigma) for each series of peptide, except for the dentato-rubral pallido-luysian atrophy (DRPLA) peptides, which we found to be more soluble when the Tris concentration was reduced to 50 mM. All peptides were used at a final concentration of 0.5 mM, except for R5QnR5, which was at 0.08 mM. Negative controls contained 20 mM EDTA (pH 8.0).
It was known from earlier work that transglutaminase can itself stably incorporate amines (24). We have confirmed this autoreactivity and found that it leads to inactivation of the enzyme. A 1-h preincubation reduced subsequent activity to 40%, and a 4-h preincubation reduced it to 20%. No degradation of transglutaminase by Ca2+-activated proteases occurred, since the transglutaminase band remained of equal intensity by Coomassie blue staining. To retain enzymatic activity in long-term experiments on peptide substrates, additional transglutaminase was added repeatedly during the reaction. Experiments were carried out for as long as 10 h at 37°C in a reaction volume of 125 μl and 3.6 milliunits of transglutaminase per 25 μl of reaction volume were added every hour. Samples of 25 μl were taken at successive intervals, and EDTA was added to 20 mM to stop the reaction.
The reaction products were subjected to electrophoresis on 12.5% SDS/polyacrylamide gels by the system of Swank and Munkres (25) for oligopeptide analysis. Involucrin, fibronectin, and Machado–Joseph disease (SCA3) peptides were loaded on gels consisting of 1:10 bis/acrylamide, 0.1% SDS, 8 M urea, 0.075% N,N,N′,N′-tetramethylethylenediamine, 0.07% ammonium persulfate, and 0.1 M H3PO4. Trizma base was added in sufficient amount to bring the pH to 9.0 for the SCA3 peptide and 6.8 for involucrin and fibronectin peptides. The electrophoresis buffer consisted of 0.1 M H3PO4 and 0.1% SDS, to which trizma base was added to pH 6.8 (about 1.2 M Tris was required). The loading buffer contained 4 M urea, 0.5% SDS, 5% glycerol, 71.5 mM 2-mercaptoethanol, and 0.01% phenol red. The pH was brought to 6.8 or 9.0 with H3PO4 and Tris, as for the preparation of the gels.
For DRPLA, SCA1, and R5QnR5 peptides, we used 25 mM Tris·HCl in both the gel and the buffer because we found that a lower Tris concentration increased the solubility of the peptides. A small amount of H3PO4 was added to 25 mM trizma base to bring the pH to 6.8. We also omitted the urea, because gels lacking urea gave better resolution of the peptides and the intensity of the spots detected by direct autoradiography was increased. Electrophoresis was at 12.5 V/cm, for 1.5 h to resolve the spinocerebellar ataxia type 1 (SCA1) peptide, and for 2.5 h to resolve the other peptides. After electrophoresis, gels were exposed directly under SaranWrap to a Biomax-MR film (Kodak) overnight. Gel fragments (2 cm × 1 cm) corresponding to the spots on the autoradiographs were excised, dissolved in 10 ml of Biolute-S (EG & G, Salem, MA), and counted in 10 ml of Optiphase “HiSafe” (EG & G) scintillation liquid.
Preparation of Brain Extract and Incorporation of Glycine Ethyl-Ester by DRPLA Peptide Q12 and Brain Protein.
Brain extract was prepared according to ref. 26 with some modification. A rat brain was homogenized in two volumes of 50 mM Tris·HCl (pH 7.5), containing 1 mM EDTA and 0.1 mM leupeptin, using a motor-driven Teflon pestle (12 strokes at 900 rpm). The homogenate was centrifuged at 1000 × g for 10 min at 4°C. The supernatant was collected, clarified at 200,000 × g for 1 h, and concentrated about five times by centrifugation through an ultrafilter, using either a Millipore column (10,000 cut off) or a Sartorius column (centrisart 1, cut off 5000). The concentrate was used as source of both transglutaminase and substrate proteins. Protein concentration was determined with a Bio-Rad protein assay kit using IgG as protein standard. Incorporation of 14C glycine ethyl-ester into brain proteins was measured after trichloracetic acid precipitation of the proteins (4).
Incorporation of glycine ethyl-ester into DRPLA Q12 peptide by brain transglutaminase was measured by precipitating the peptide with 350 mM Tris·HCl (pH 7.5), washing the pellet with the same buffer five times, dissolving it in 100 μl of water, and counting in 10 ml of scintillation fluid.
Cross-Linking of Labeled R5Q18R5 with Polylysine or Brain Proteins.
Labeling of the peptide with glycine ethyl-ester was carried out in 50 μl of 100 mM Tris buffer identical to that used for other peptides, except that the concentration of R5Q18R5 was 0.2 mM, and the amount of 14C glycine ethyl-ester was 2 μCi. The tubes and pipette tips were siliconized. Transglutaminase was added every hour. At the end of 3 h, either polylysine or brain proteins were added; the final reaction volume was 80 μl. Reaction was stopped by addition of 20 mM EDTA, and the products were boiled for 5 min in the presence of either 4.0 M urea or 1% SDS and 20 mM 2-mercaptoethanol and centrifuged at 20,000 × g for 5 min. Pellets were washed twice with water and counted in 10 ml of scintillation fluid.
RESULTS
Reactivity of Model Fibronectin Peptide Containing Small Numbers of Glutamine Residues.
For our first experiments, we obtained a family of synthetic peptides consisting of the sequence pEA(Q)nIV, where n = 1, 2, or 6. This sequence, where n = 2, as in bovine fibronectin, had been used earlier as a model substrate for transglutaminase (27). Our experiments with purified liver transglutaminase showed that Q2 was much more reactive with 14C-labeled glycine ethyl-ester, a commonly used amine donor, than Q1 (Fig. 1). Q6 incorporated a considerable quantity of the amine, but most of the product remained at the top of the gel, in keeping with the tendency of the peptide to precipitate, even in pure solution, presumably because of the aggregating properties of polyglutamines. The effect of further increases in the length of polyglutamine on the reactivity catalyzed by transglutaminase could therefore not be examined in this family of peptides.
Figure 1.
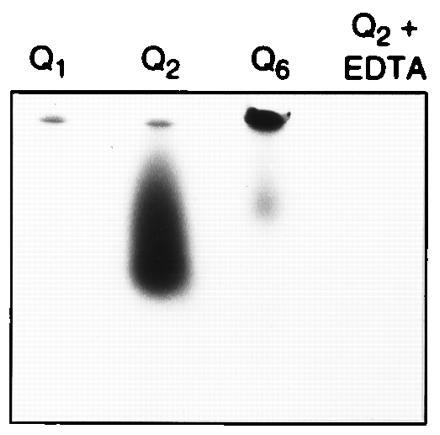
Activity of fibronectin peptides with small numbers of glutamine residues as amine acceptors. The different peptides were incubated with 5 μCi of 14C-labeled glycine ethyl-ester for 1 h. Reaction was stopped by addition of EDTA, and the products were resolved on a gel prepared according to Swank and Munkres (25). The gel was fixed and dried, and the radioactivity incorporated in the peptide was revealed by fluorography after a 24-h exposure. Q2 is much more active than Q1. Q6 is active, but most of the product did not enter the gel. The control incubated in the presence of EDTA shows no incorporation.
Choice of Peptides Containing Polyglutamine Flanked by Charged Residues to Increase Solubility.
Families of synthetic peptides were prepared so as to contain polyglutamine flanked by residues that normally flank a polyglutamine sequence in four proteins (Fig. 2). The first example was human involucrin, a demonstrated transglutaminase substrate (4, 29). The other three were polyglutamine-containing proteins, none of which has yet been shown to be a transglutaminase substrate, but all of which produce disease of the nervous system when the polyglutamine is of excessive length: SCA1, DRPLA, or SCA3.
Figure 2.

Peptide substrates containing polyglutamine. Residues with a charged side chain are indicated by an asterisk. The first 10 amino acids of the involucrin sequence (n = 2) constitute a B-type repeat (28), which contains, at site Qn, the most reactive glutamine in the entire protein (29). The residues K H L E are the first four of the succeeding A repeat. The flanking amino acids for the other peptides were chosen from the sequences of the proteins associated with SCA1 (30), SCA3 (31), and DRPLA (32).
In human involucrin (where n = 2), Qn is the site preferred by transglutaminase, and it is the second of the two glutamines that is most reactive (29). In the regions flanking the Qn, the principal charged residue is glutamic acid, but there are also a histidine and a lysine. The flanking residues in the SCA1 and SCA3 peptides also include mixed positively and negatively charged residues, whereas the only charged flanking residues of the DRPLA peptide are histidines. In the case of the involucrin and SCA3 peptides, the N-terminal glutamic acid was cyclized, as in the fibronectin model substrate (27). The sequences flanking the polyglutamine in Huntingtin and the androgen receptor protein were not used to prepare peptides, because they seemed to lack charged residues in sufficient number to promote the solubility of the peptide.
The Reactivity of Polyglutamine in the Involucrin Peptide.
Involucrin peptides containing different lengths of polyglutamine were incubated with 1 μCi of glycine ethyl-ester (20 mM) and purified liver transglutaminase for 1 h. The reaction was stopped by the addition of EDTA, the products were resolved on urea gels and their radioactivity was measured. It can be seen that Q0 and Q1 had no detectable reactivity (Fig. 3), but the addition of increasing numbers of Q to this site made the peptide progressively more active. Reactivity was completely dependent on Ca2+.
Figure 3.
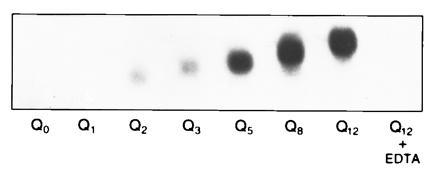
Reactivity of involucrin peptides containing Qn of different lengths. Autoradiography of the resolved products of transglutaminase action on peptides Q0–Q12 in the presence of glycine ethyl-ester, 20 mM, at a specific activity of 2 μCi/μmol. The larger the value of Qn, the greater the reactivity and the slower the mobility.
With rare exception, earlier studies of transglutaminase action have been carried out over an incubation period of 1–2 h, but as the involucrin peptide continued to react over long periods of time, quantitative studies of reactivity were carried out over a period of 10 h (Fig. 4). Because of the instability of the enzyme (see Materials and Methods), fresh enzyme was added every hour. Although it possessed a glutamine residue downstream of the site of Qn, Q0 showed no reactivity with glycine ethyl-ester even after 10 h (Fig. 4A). Q1 also incorporated practically no glycine ethyl-ester in a 1-h period, but incorporated small amounts over succeeding hours. Q3 was a better substrate than Q1, but Q5, Q8, and Q12 were better still. Maximum incorporation per mol of peptide at 10 h, obtained with Q12, was 6 mol of amine (Fig. 4B). Since the larger peptides had, per mol of peptide, more glutamine residues that could act as amine acceptors, their quality as substrates could also be expressed as mol of amine incorporated per mol of peptide glutamine. The presence of higher numbers of glutamine residues resulted in increasing reactivity per glutamine residue, up to Q5, which was 80% saturated with amine (Fig. 4C). At n > 5, the reactivity per glutamine residue declined; but in contrast to the peptide Q5, which was freely soluble, Q8 and Q12 were close to maximum solubility and were likely to have been partly aggregated.
Figure 4.
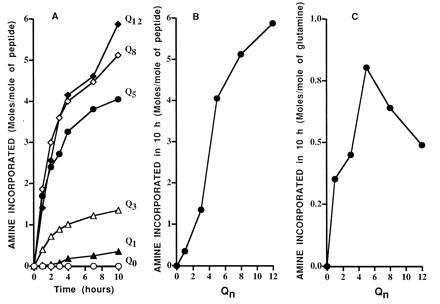
Quantitation of reactivity of involucrin peptides. (A) Time course of reaction of peptides containing variable length of polyglutamine. (B) Stoichiometry of glycine ethyl-ester incorporated after 10-h incubation, expressed per mol of peptide. (C) Stoichiometry of same incorporation, expressed per mol of glutamine (complete reaction = 1 mol/mol of glutamine).
Reactivity of Polyglutamine in DRPLA, SCA1, and SCA3 Peptides.
DRPLA peptide containing a single glutamine residue was not appreciably reactive in a 3-h incubation (Fig. 5A). Q2 and Q5 were increasingly better substrates, not only when incorporation is expressed per mol of peptide (Fig. 5A), but also when it is expressed per mol of glutamine (Fig. 5B). By increasing the number of glutamine residues from 2 to 5, the reactivity of each glutamine residue increased over 3-fold. Q5 was nearly saturated with amine (0.95 mol of amine per mol of glutamine).
Figure 5.
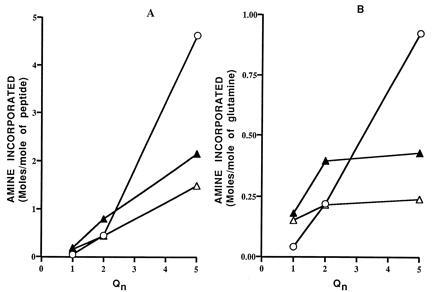
Reactivity of DRPLA, SCA1, and SCA3 peptides. Incorporation of glycine ethyl-ester during a 3-h period per mol of peptide (A) and per mol of glutamine (B). ○, DRPLA peptides; ▴, SCA1 peptides; ▵, SCA3 peptides.
SCA1 peptides also improved markedly as substrates when n increased from 3 to 5, though not as greatly as the DRPLA peptide; the reactivity per mol of glutamine reached only 0.43 for SCA1 and 0.24 for SCA3.
The SCA1 peptide Q12 could not be dissolved in aqueous solution at all and the SCA3 peptide Q12 dissolved to form a very viscous solution that resembled a gel; both peptides behaved poorly as substrates. The DRPLA peptide Q12, being more soluble, incorporated 1.5 mol of amine/mol of peptide in a 3-h reaction. Although this peptide was not as satisfactory a substrate as the involucrin Q12, we used it as a substrate for experiments on brain extracts (see below).
Polyglutamine Flanked by Arginine Residues.
To see whether the flanking residues found in proteins are necessary for the amine-accepting function of the glutamine residues, and in the hope that more strongly charged flanking residues would be more effective in maintaining solubility of the peptides with longer polyglutamine stretches, we prepared a family of peptides of sequence R5QnR5. At all values of n up to 18, the peptides were soluble, and increasing the value of n improved incorporation of glycine ethyl-ester (Fig. 6). By the end of a 10-h incubation period, when incorporation was still continuing, Q18 had incorporated over 8 mol of glycine ethyl-ester per mol of peptide, equivalent to 45% saturation of the glutamine residues. Over the same period, Q3 incorporated a maximum of about 1 mol of amine per mol of peptide, equivalent to 33% saturation of the glutamine residues. Peptides with intermediate values of n incubated for a single period of 3 h confirmed the increasing reactivity with increasing values of n. The arginine-flanked family of polyglutamines was the best of the peptide substrates tested in the sense that the reactivity per mol of glutamine increased up to Q12.
Figure 6.
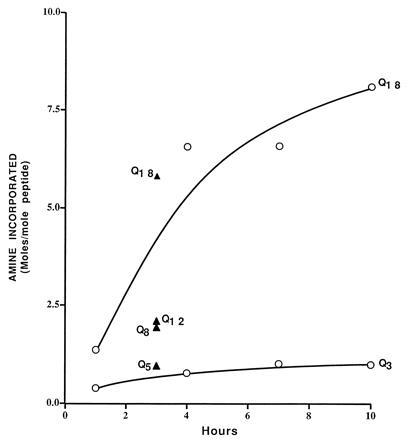
Reactivity of R5QnR5. For Q3 and Q18, the incorporation was measured after different incubation times (○). For intermediate values of Qn, values are plotted for a single incubation time of 3 h (▴).
DRPLA Peptide Q12 and Brain Proteins as Substrates for Transglutaminase.
In all the experiments described above, we used a commercial preparation of tissue type transglutaminase (type II) purified from guinea pig liver. The brain contains the same enzyme (33) but also an uncharacterized transglutaminase associated with synaptic vesicles (34, 35). Since the two brain transglutaminases may be different, we determined whether either could act on the DRPLA Q12 peptide. When a crude rat brain extract was incubated in the presence of the DRPLA Q12 under optimal conditions for catalysis by transglutaminase, we observed significant incorporation of labeled glycine ethyl-ester into the peptide. A crude rat liver extract incorporated about 15 times more amine into the DRPLA Q12 than the brain extract, in keeping with the higher concentration of transglutaminase in liver than in brain (36, 37).
We then compared the DRPLA Q12 peptide and the proteins of a crude brain extract as amine acceptors (Table 1). Brain extract was incubated in the presence of labeled glycine ethyl-ester, either with or without added DRPLA Q12. The DRPLA peptide incorporated about 16-fold more glycine ethyl-ester than did the same amount of brain protein. When purified liver transglutaminase was used as enzyme with either the brain protein or the DRPLA Q12 as substrate, the incorporation of glycine ethyl-ester was about 50-fold higher by the DRPLA peptide than by the average brain protein (Table 1).
Table 1.
Incorporation of glycine ethyl-ester by DRPLA peptide O12 and brain protein
| dpm × 10−3 incorporated by
|
||||
|---|---|---|---|---|
| Brain transglutaminase into
|
Purified
liver transglutaminase into
|
|||
| Brain proteins | DRPLA peptide | Brain proteins | DRPLA peptide | |
| + Ca2+ | 1.3 | 22.2 | 11.4 | 548 |
| + EDTA | 0.24 | 6.3 | 0.60 | 5.3 |
Results are expressed as dpm of amine incorporated into 100 μg of protein or peptide. For incorporation by brain transglutaminase, 1.4 mg of brain extract was incubated with or without 100 μg of DRPLA peptide. For incorporation by purified liver transglutaminase, 100 μg of brain extract or of DRPLA peptide were incubated with 11.5 milliunits of the transglutaminase. Reactions were for 2 h at 37°C.
Experiments carried out with R5Q12R5 as amine acceptor yielded very similar results. In this case, the labeled product was separated from the proteins by trichloracetic acid precipitation of the latter, prior to electrophoresis.
Cross-Linking of R5Q18R5 to Polylysine and the Formation of Aggregates.
In all the experiments described above, we measured the cross-linking of glycine ethyl-ester into a monomeric peptide. To determine whether a polyglutamine-containing peptide acting as amine acceptor could be cross-linked to another polypeptide acting as amine donor and form polymeric aggregates, we incubated 36 μg of R5Q18R5 in the presence of 100 μg of polylysine (molecular weight range 10–30 kDa) together with purified transglutaminase. After 3 h, we observed the development of aggregates. No aggregates were visible in the controls lacking R5Q18R5, transglutaminase, or polylysine, or containing excess EDTA. Aggregates were not dissolved by boiling for 5 min in a solution containing 4 M urea and 20 mM 2-mercaptoethanol. (In this experiment, urea was used as denaturant rather than SDS since the latter caused polylysine itself to precipitate.)
R5Q18R5 was labeled for 3 h with 0.2 mM dansylcadaverine (38). At the end of this period, polylysine was added and incubation was continued for an additional 3 h. The aggregates that formed after addition of polylysine were boiled in the presence of urea and 2-mercaptoethanol. Examination under UV light showed that the aggregates were brightly fluorescent. Since polylysine cannot incorporate free dansylcadaverine, we concluded that the R5Q18R5 labeled with dansylcadaverine was subsequently polymerized with the polylysine.
To quantitate this process, we labeled R5Q18R5 with 2 μCi of 14C glycine ethyl-ester in the presence of 11.5 milliunits of purified liver transglutaminase for 3 h, and removed an aliquot for subsequent electrophoresis and counting of the reaction product. Meanwhile, we added polylysine to the rest of the reaction mixture and continued the incubation overnight, adding three successive aliquots of the transglutaminase. Aggregates that formed during this incubation were heated in the presence of urea and 2-mercaptoethanol, centrifuged for 5 min at 20,000 × g, washed twice with water and counted by liquid scintillation. The aggregates contained from 0.62 to 1.06 × 105 dpm, values corresponding to 10–20% of the labeled R5Q18R5 (Table 2).
Table 2.
Transglutaminase-catalyzed formation of labeled insoluble aggregates from labeled R5O18R5 and polypeptide amine donors
|
14C incorporation into aggregates,
dpm × 10−5
|
|||
|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 3 | |
| Polylysine + purified transglutaminase | |||
| + Prelabeled R5Q18R5 | 0.62 | 1.06 | 0.78 |
| + Unlabeled R5Q18R5 + 14C - GEE | 0.003 | 0.001 | |
| + Prelabeled R5Q18R5 + EDTA | 0.08 | 0.12 | 0.06 |
| − R5Q18R5 + 14C - GEE | 0.006 | 0.002 | |
| − Polylysine | 0.06 | 0.15 | |
| Brain extract + purified transglutaminase | |||
| + Prelabeled R5Q18R5 | 2.5 | 3.4 | |
| + Unlabeled R5Q18R5 + 14C - GEE | 0.18 | ||
| + Prelabeled R5Q18R5 + EDTA | 0.12 | 0.07 | |
| − R5Q18R5 + 14C - GEE | 0.007 | ||
| − Brain extract | 0.12 | ||
| Brain extract + any residual purified transglutaminase from preincubation | |||
| + Prelabeled R5Q18R5 | 0.21 | 0.21 | |
| + Unlabeled R5Q18R5 + 14C - GEE | 0.001 | 0.0009 | |
| + Prelabeled R5Q18R5 + EDTA | 0.014 | 0.033 | |
| − R5Q18R5 + 14C - GEE | 0.0024 | 0.0014 | |
R5Q18R5 was labeled by preincubation with 2 μCi of 14C glycine ethyl-ester (GEE) for 3 h in the presence of purified transglutaminase. Subsequent incubation with amine donors (brain proteins or polylysine) resulted in the formation of aggregates insoluble in the presence of 20 mM 2-mercaptoethanol and either 4 M urea or 1% SDS at 100°C for 5 min. The aggregates were centrifuged, washed, and counted. Included among the negative controls was one in which the R5Q18R5 was not prelabeled with 14C glycine ethyl-ester, but the latter was incubated together with the unlabeled R5Q18R5 during the enzymatic coupling to polylysine or brain protein.
Cross-Linking of R5Q18R5 to Brain Proteins and the Formation of Aggregates.
An aliquot of brain extract containing 200 μg of protein was incubated in the presence of R5Q18R5 and purified transglutaminase, but without polylysine. Aggregation of the brain proteins was visible within 5 min, but with time the aggregate increased in amount and settled more rapidly to the bottom of the tube. No aggregate was apparent in controls lacking R5Q18R5 or transglutaminase or containing an excess of EDTA. The aggregate remained insoluble after boiling in the presence of 1% SDS and 20 mM 2-mercaptoethanol (Fig. 7).
Figure 7.

Insoluble aggregates formed by reaction of R5Q18R5 with brain proteins. Brain proteins (200 μg) incubated overnight with 11.5 milliunits of purified transglutaminase in the presence of 36 μg of R5Q18R5 resulted in slow formation of a dense aggregate that settled to the bottom of the tube. Addition of SDS to 1% and 2-mercaptoethanol to 20 mM, followed by heating to 100°C, did not affect the appearance of the aggregate (Center). The aggregate contained 70% of the total initial protein and peptide. A control lacking the peptide (Right) remained completely clear. A second control (Left) containing 100 mM EDTA immediately formed a faint opalescence (not reaction-dependent), but no aggregate at the bottom of the tube. An additional control without brain proteins (not shown) yielded no aggregate.
We then repeated this experiment with R5Q18R5 previously labeled with glycine ethyl-ester and purified transglutaminase, as described above. In two separate experiments, a 3-h incubation with transglutaminase added every hour, resulted in incorporation of 2.5 × 105 dpm and 3.4 × 105 dpm into the aggregate (Table 2). These values corresponded to 30–60% of the labeled R5Q18R5.
Finally, we determined whether brain transglutaminase alone would suffice to form aggregates in the presence of R5Q18R5 as amine acceptor and brain proteins as amine donors. After a 3-h incubation of brain extract containing 1 mg of protein in the presence of R5Q18R5, an aggregate was evident, although it was not as voluminous as when additional enzyme was added. No aggregate appeared in the presence of EDTA or in the absence of R5Q18R5. The polymerization of the brain proteins must have been due solely to the endogenous transglutaminase activity in the extract. Enzymatically pre-labeled R5Q18R5, when incubated with brain extract, was incorporated into aggregates, but not as extensively as when purified transglutaminase was added to the reaction mixture (Table 2).
In experiments combining the DRPLA peptide Q12 with brain extract in the absence of any additional transglutaminase, incubation for 24 h led to the formation of insoluble aggregates similar to those produced by R5Q18R5.
To demonstrate that isopeptide bonds were formed during the aggregation, the reaction was carried out in the presence of R5Q18R5, brain proteins and purified transglutaminase for 24 h, and the resulting aggregates were centrifuged, boiled in the presence of SDS and 2-mercaptoethanol, and subjected to terminal enzymatic hydrolysis (39). The enzymes were precipitated with trichloracetic acid and the supernatant was analyzed by high pressure liquid chromatography at the Harvard Microchemistry Facility. A peak at the correct position for ɛ(γ glutamyl) lysine equaled 2.7% of the total free lysine. When the enzymatic hydrolysate was subjected to acid hydrolysis before the chromatography, there was no peak at the position of ɛ(γ glutamyl) lysine.
DISCUSSION
The nervous system diseases resulting from polyglutamine in proteins require years to develop, whereas the properties of polyglutamine as substrates for transglutaminase are necessarily studied in short-term experiments. Short polyglutamine stretches in native proteins should be exposed to water, and the only crystallographic study on such material shows that Q5 in a small protein is located in a loop protruding on the surface (40). In a larger protein, access of transglutaminase to such a loop could be hindered by the tertiary structure of the protein; for this reason, the peptide facsimiles of the disease-producing proteins, lacking steric hindrance, may reveal with greater sensitivity the substrate properties conferred by polyglutamine than would the intact proteins. This conjecture is supported by the recent demonstration that expanded polyglutamine itself is more cell-lethal when it is expressed in the brain or in cultured cells than the same polyglutamine expressed as part of the protein of SCA3 (41). In any of the family of intact proteins, a polyglutamine sequence of more than 35 residues is usually required to produce disease; this suggests that a longer sequence of polyglutamine forces some of the residues into a configuration favoring reactivity with transglutaminase. Such an effect could explain the behavior of the monoclonal antibody 1C2, which recognizes polyglutamine in intact proteins only if the sequence contains more than 32 residues (42).
In our synthetic peptides, all polyglutamine sequences from Q2 to Q18 were substrates for transglutaminase. The most soluble family of peptides was R5QnR5, followed by the involucrin family and the DRPLA family. In contrast to the transglutaminase substrates studied earlier, the nature of the sequences flanking the reactive glutamines becomes of little consequence as n increases beyond 2, apart from the necessity for charged residues to maintain solubility. The addition of increasing numbers of glutamine residues not only increases the number of cross-links per peptide molecule, but also makes each residue more reactive as long as the peptides remain soluble: in the involucrin family, reactivity per glutamine residue continued to increase at least up to Q5, and in the case of R5QnR5, up to Q12; per mol of peptide, the largest number of cross-links formed when n was further increased, at least up to 12 for involucrin and 18 for R5QnR5.
Involucrin is a substrate for transglutaminase during terminal differentiation of the keratinocyte. The evolutionary introduction of polyglutamine in this protein, although mostly ancient, is still occurring in some lineages in a way similar to that of the neuronal proteins. Whereas human involucrin possesses only two glutamines at the preferred site, mouse involucrin has a sequence of 19 consecutive glutamines, most of which were added recently in evolution and are located in a region not present in the human protein (43). It would be interesting if the site preferred by the keratinocyte transglutaminase has shifted from its known location in the human protein to the recently introduced polyglutamine sequence of the mouse protein.
In our experiments, the formation of cross-linked protein aggregates by transglutaminase in vitro depended on the marked amine-accepting properties of the polyglutamine. It occurred when the transglutaminase was of brain type and when the amine donors were normal brain proteins. The mechanism of aggregation differs from that proposed by Perutz et al. (13) in that it occurs when the polypeptide containing the polyglutamine is itself soluble, as seems to be the case for far larger proteins. However, it does require the participation of a Ca2+-dependent enzyme. It is very likely that if the Ca2+ concentration rises, even transiently, in cells containing a protein with a long polyglutamine sequence, there will be action on that sequence by transglutaminase. This is particularly so for neurons, which are subjected to Ca2+ transients as part of their function in impulse-conduction. Evidence that the cross-linking occurs in neurons would be supported by detection of glutamyl-lysine in brain or cerebrospinal fluid.
Acknowledgments
We thank Mr. Neil Wright and Dr. John Rush of the Biopolymers Facility at Harvard Medical School for their valuable advice and their preparation of all peptides, and Dr. William Lane of the Harvard Microchemistry Facility for the quantitation of the isopeptide glutamyl-lysine. We are indebted for research support to the Centre National de la Recherche Scientifique, the Association Française contre les Myopathies, the Fondation pour la Recherche Médicale, and the National Cancer Institute.
Footnotes
Abbreviations: SCA1, spinocerebellar ataxia type 1; DRPLA, dentato-rubral pallido-luysian atrophy; SCA3, Machado–Joseph disease.
References
- 1.Stallings R L. Genomics. 1994;21:116–121. doi: 10.1006/geno.1994.1232. [DOI] [PubMed] [Google Scholar]
- 2.Green H, Wang N. Proc Natl Acad Sci USA. 1994;91:4298–4302. doi: 10.1073/pnas.91.10.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlin S, Burge C. Proc Natl Acad Sci USA. 1996;93:1560–1565. doi: 10.1073/pnas.93.4.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice R H, Green H. Cell. 1979;18:681–694. doi: 10.1016/0092-8674(79)90123-5. [DOI] [PubMed] [Google Scholar]
- 5.Eckert R L, Green H. Cell. 1986;46:583–589. doi: 10.1016/0092-8674(86)90884-6. [DOI] [PubMed] [Google Scholar]
- 6.Green H, Djian P. Mol Biol Evol. 1992;9:977–1017. doi: 10.1093/oxfordjournals.molbev.a040775. [DOI] [PubMed] [Google Scholar]
- 7.Rubinsztein D C, Amos W, Leggo J, Goodburn S, Ramesar R S, Old J, Bontrop R, McMahon R, Barton D E, Ferguson-Smith M A. Nat Genet. 1994;7:525–530. doi: 10.1038/ng0894-525. [DOI] [PubMed] [Google Scholar]
- 8.Djian P, Hancock J M, Chana H S. Proc Natl Acad Sci USA. 1996;93:417–421. doi: 10.1073/pnas.93.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross C A. Neuron. 1995;15:493–496. doi: 10.1016/0896-6273(95)90138-8. [DOI] [PubMed] [Google Scholar]
- 10.Ashley C T, Jr, Warren S T. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- 11.Ambrose C M, Duyao M P, Barnes G, Bates G P, Lin C S, Srinidhi J, et al. Somat Cell Mol Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- 12.Albin R L, Tagle D A. Trends Neurosci. 1995;18:11–14. doi: 10.1016/0166-2236(95)93943-r. [DOI] [PubMed] [Google Scholar]
- 13.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stott K, Blackburn J M, Butler P J G, Perutz M. Proc Natl Acad Sci USA. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green H. Cell. 1993;74:955–956. doi: 10.1016/0092-8674(93)90718-6. [DOI] [PubMed] [Google Scholar]
- 16.Pincus J H, Waelsch H. Arch Biochem Biophys. 1968;126:34–43. doi: 10.1016/0003-9861(68)90556-0. [DOI] [PubMed] [Google Scholar]
- 17.Toda H, Folk J E. Biochim Biophys Acta. 1969;175:427–430. doi: 10.1016/0005-2795(69)90022-1. [DOI] [PubMed] [Google Scholar]
- 18.Folk J E. In: Mechanism and Basis for Specificity of Transglutaminase-Catalyzed ɛ-(γ Glutamyl) Lysine Bond Formation. Meister A, editor. Vol. 54. New York: Wiley; 1983. pp. 1–56. [DOI] [PubMed] [Google Scholar]
- 19.Gorman J J, Folk J E. J Biol Chem. 1981;256:2712–2715. [PubMed] [Google Scholar]
- 20.Gorman J J, Folk J E. J Biol Chem. 1984;259:9007–9010. [PubMed] [Google Scholar]
- 21.Coussons P J, Price N C, Kelly S M, Smith B, Sawyer L. Biochem J. 1992;282:929–930. doi: 10.1042/bj2820929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aeschlimann D, Paulsson M, Mann K. J Biol Chem. 1992;267:11316–11321. [PubMed] [Google Scholar]
- 23.Aeschlimann D, Paulsson M. Thromb Haemostasis. 1994;71:402–415. [PubMed] [Google Scholar]
- 24.Birckbichler P J, Orr G R, Carter H A, Patterson M K., Jr Biochem Biophys Res Commun. 1977;78:1–7. doi: 10.1016/0006-291x(77)91213-x. [DOI] [PubMed] [Google Scholar]
- 25.Swank R T, Munkres K D. Anal Biochem. 1971;39:462–477. doi: 10.1016/0003-2697(71)90436-2. [DOI] [PubMed] [Google Scholar]
- 26.Selkoe D J, Abraham C, Ihara Y. Proc Natl Acad Sci USA. 1982;79:6070–6074. doi: 10.1073/pnas.79.19.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parameswaran K N, Velasco P T, Wilson J, Lorand L. Proc Natl Acad Sci USA. 1990;87:8472–8475. doi: 10.1073/pnas.87.21.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djian P, Green H. Proc Natl Acad Sci USA. 1989;86:8447–8451. doi: 10.1073/pnas.86.21.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon M, Green H. J Biol Chem. 1988;263:18093–18098. [PubMed] [Google Scholar]
- 30.Banfi S, Servadio A, Chung M-y, Kwiatkowski Jr T J, McCall A E, Duvick L A, Shen Y, Roth E J, Orr H T, Zoghbi H Y. Nat Genet. 1994;7:513–520. doi: 10.1038/ng0894-513. [DOI] [PubMed] [Google Scholar]
- 31.Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, Kimura J, Narumiya S, Kakizuka A. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 32.Nagafuchi S, Yanagisawa H, Ohsaki E, Shirayama T, Tadokoro K, Inoue T, Yamada M. Nat Genet. 1994;8:177–182. doi: 10.1038/ng1094-177. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg C S, Birckbichler P J, Rice R H. FASEB J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 34.Pastuszko A, Wilson D F, Erecinska M. J Neurochem. 1986;46:499–508. doi: 10.1111/j.1471-4159.1986.tb12996.x. [DOI] [PubMed] [Google Scholar]
- 35.Facchiano F, Benfenati F, Valtorta F, Luini A. J Biol Chem. 1993;268:4588–4591. [PubMed] [Google Scholar]
- 36.Clarke D D, Mycek M J, Neidle A, Waelsch H. Arch Biochem Biophys. 1959;79:338–354. doi: 10.1016/0003-9861(59)90613-7. [DOI] [PubMed] [Google Scholar]
- 37.Chung S I. Ann NY Acad Sci. 1972;202:240–255. doi: 10.1111/j.1749-6632.1972.tb16338.x. [DOI] [PubMed] [Google Scholar]
- 38.Lorand L, Conrad S M. Mol Cell Biochem. 1984;58:9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- 39.Rice R H, Green H. Cell. 1977;11:417–422. doi: 10.1016/0092-8674(77)90059-9. [DOI] [PubMed] [Google Scholar]
- 40.Ko T-P, Ng J D, McPherson A. Plant Physiol. 1993;101:729–744. doi: 10.1104/pp.101.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A. Nat Genet. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- 42.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, Agid Y, Brice A, Mandel J-L. Nature (London) 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 43.Djian P, Phillips M, Easley K, Huang E, Simon M, Rice R H, Green H. Mol Biol Evol. 1993;10:1136–1149. doi: 10.1093/oxfordjournals.molbev.a040069. [DOI] [PubMed] [Google Scholar]


