Abstract
Cell proliferation and programmed cell death are closely controlled during animal development. Proliferative stimuli generally also induce apoptosis, and anti-apoptotic factors are required to allow net cell proliferation. Genetic studies in Drosophila have led to identification of a number of genes that control both processes, providing new insights into the mechanisms that coordinate cell growth, proliferation, and death during development and that fail to do so in diseases of cell proliferation. We present evidence that the Drosophila Sterile-20 kinase Slik promotes cell proliferation and controls cell survival. At normal levels, Slik provides survival cues that prevent apoptosis. Cells deprived of Slik activity can grow, divide, and differentiate, but have an intrinsic survival defect and undergo apoptosis even under conditions in which they are not competing with normal cells for survival cues. Like some oncogenes, excess Slik activity stimulates cell proliferation, but this is compensated for by increased cell death. Tumor-like tissue overgrowth results when apoptosis is prevented. We present evidence that Slik acts via Raf, but not via the canonical ERK pathway. Activation of Raf can compensate for the lack of Slik and support cell survival, but activation of ERK cannot. We suggest that Slik mediates growth and survival cues to promote cell proliferation and control cell survival during Drosophila development.
Identification and characterization of Slik as a new regulator of cell growth and survival based on loss-of-function and overexpression analysis
Introduction
Growth of tissues and organs during animal development involves careful coordination of the rates of cell proliferation, cell death, and differentiation (Neufeld et al. 1998; Conlon and Raff 1999). Cell proliferation depends on signals to stimulate cell growth and cell division. Cell survival is also dependent on intercellular signaling. A diverse array of long-range signals mediated by growth factors and cytokines as well as short-range signals mediated by cell surface proteins has been implicated in providing cells with growth and survival cues during development.
Systematic genetic screens for alterations in tissue growth in Drosophila imaginal discs have led to identification of a number of genes that promote cell proliferation and cell survival. ras, myc, TSC1/TSC2, and genes in the insulin pathway have been implicated primarily in control of cellular growth rates (Johnston et al. 1999; Prober and Edgar 2000; Saucedo and Edgar 2002 [reviewed in Oldham and Hafen 2003]). Cell growth and division rates are normally well coordinated, but excess activity of the insulin pathway, Ras, or Myc can cause cells to grow faster than they divide, leading to cellular overgrowth and concomitant tissue overgrowth. Other genes that cause tissue overgrowth when overexpressed do so without distorting cell size. Cyclin D and Cdk4 act together to promote coordinated cellular growth and cell division, leading to net cell proliferation (Datar et al. 2000; Meyer et al. 2000). The bantam gene encodes a microRNA that promotes net cell proliferation (Hipfner et al. 2002; Brennecke et al. 2003). bantam also prevents apoptosis by regulating translation of the apoptosis-inducing gene hid (Brennecke et al. 2003).
A different group of growth regulators have been identified in screens for loss-of-function mutations that cause tissue overgrowth. The warts/lats, salvador/shar-pei, and hippo genes promote cell cycle exit and also stimulate developmentally controlled apoptosis (Justice et al. 1995; Xu et al. 1995; Kango-Singh et al. 2002; Tapon et al. 2002; Harvey et al. 2003; Wu et al. 2003). Salvador serves as a scaffold protein to bind the Warts serine–threonine protein kinase and the recently identified Hippo Sterile-20 (Ste20) kinase. Cells mutant for any of the components of this complex exhibit elevated expression of cyclin E, which promotes cell proliferation and overrides developmentally controlled exit from proliferation, leading to tissue overgrowth. Mutant cells are also resistant to developmentally regulated apoptosis by virtue of increased expression of the apoptosis inhibitor DIAP1.
Here we present the identification of another member of the Ste20 kinase family that contributes to the control of cell proliferation and apoptosis during imaginal disc development. We have named the Drosophila gene slik (SLK- and LOK-like kinase) on the basis of its similarity to the human SLK and LOK Ste20 kinases. Slik activity is required to support cell survival. Survival of mutant cells is impaired when they are in competition with normal cells, but also occurs in the absence of cell competition, indicating that it is an intrinsic defect. Slik also promotes cell proliferation. Elevated Slik activity increases the rate of cell proliferation and also increases apoptosis. Tumor-like tissue overgrowth results when apoptosis is prevented. The phenotypes associated with slik mutants are essentially opposite to those of the Ste20 kinase Hippo, for which loss-of-function mutants cause tissue overgrowth and reduced apoptosis (Harvey et al. 2003; Wu et al. 2003). We suggest that Slik activity mediates growth and survival cues to promote cell proliferation and cell survival and present evidence that this depends on the activation of the MAP kinase kinase kinase (MAP3K) Raf, but not via the canonical extracellular signal-regulated kinase (ERK) MAP kinase (MAPK) pathway.
Results
Slik, a Ste20 Kinase Involved in Growth Control
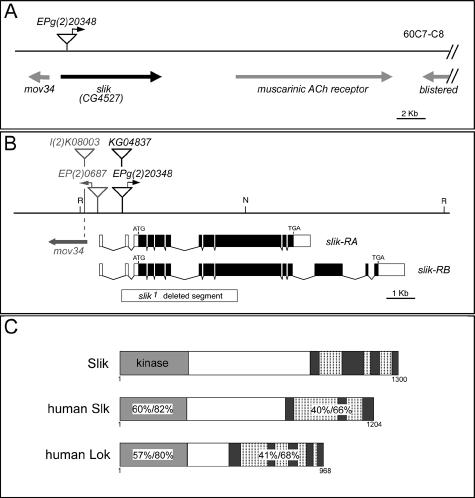
Genes involved in regulating tissue growth were identified in a systematic overexpression screen (Hipfner et al. 2002). EPg(2)20348 caused increased growth of the posterior compartment of the wing by 6% without inducing pattern abnormalities when expressed under control of the enGAL4 driver. Though small in magnitude, this difference was reproducible and statistically significant. As will be shown below, considerably stronger growth effects were obtained when compensating apoptosis was prevented. To identify the gene overexpressed by EPg(2)20348, DNA flanking the single P-element insertion was sequenced. The EPg element was inserted in the first intron of CG4527, which we now name slik (Figure 1A and 1B). According to GADFLY (release 3), there are likely to be two transcripts produced by alternative splicing at the 3′ end of slik. The shorter of these, slik-RA, encodes a 1300 amino acid protein and is identical in sequence to the cDNA we assembled from expressed sequence tags (ESTs) LD34405 (AY119617) and GH20991 (AY058322). The slik-RB transcript contains three additional 3′ exons and is based on gene prediction and incomplete EST data.
Figure 1. Molecular Characterization of the slik Locus.
(A) Schematic representation of the slik region. Predicted genes are indicated.
(B) Detailed view of the slik region. The insertion sites of EPg(2)23048 and KG04837 in the first intron and the extent of the slik1 deletion are indicated. l(2)K08003 is an allele of mov34. N, NotI; R, EcoRI.
(C) Comparison of Slik with human SLK and LOK proteins. Numbers show sequence identity/similarity within the indicated domains. Predicted coiled-coil regions in the C-terminal domain are indicated by hatching.
Both predicted slik transcripts encode Ste20 group kinases. The Ste20 group is a large and heterogeneous group of kinases, divided into two large families and ten distinct subfamilies (Dan et al. 2001). Members of different subfamilies show little sequence similarity outside of the kinase domain, reflecting the diversity of functions that have been attributed to these proteins. Drosophila contains one member of each of the ten subfamilies. The predicted Slik protein shows highest homology to two human proteins, SLK and LOK. Together these proteins form a subfamily of StE20 group kinases. The sequence similarity between them is largely restricted to an N-terminal kinase domain and a conserved C-terminal domain containing several coiled-coil motifs, separated by a nonconserved domain of variable length (Figure 1C). EPg(2)20348 is located upstream of the entire coding sequence of slik and is oriented to drive its expression (Figure 1B). We confirmed by antibody labeling that EPg(2)20348 directs GAL4-dependent Slik protein expression (data not shown).
slik Mutants Display a Larval Growth Defect
Relatively little is known about the functions of Slk and Lok, though they may influence cytoskeletal dynamics and cell adhesion (Endo et al. 2000; Sabourin et al. 2000; Wagner et al. 2002). To investigate the function of slik during Drosophila development, we screened for mutants generated by imprecise excision of EPg(2)20348. slik1 is a deletion that removes exons 2–8 and part of exon 9 of the slik transcript, including the translation start site and the entire kinase domain (Figure 1B). slik1 is expected to be a null allele. slik1 is not mutant for the adjacent mov34 gene and thus only affects the slik locus. The P-element KG04837 is inserted in the first intron of slik and causes a partial loss of slik function (see below).
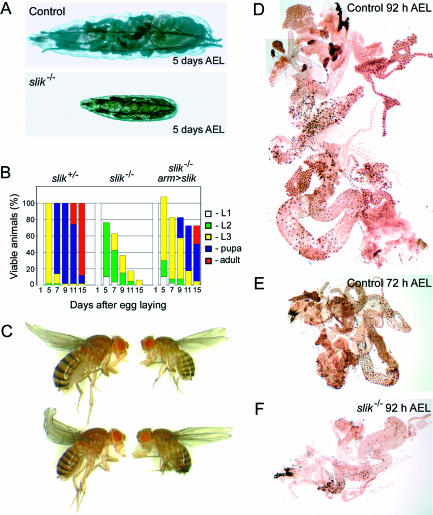
Homozygous slik1 mutant animals showed a striking larval growth defect (Figure 2A). To characterize this in more detail, we collected first-instar larvae shortly after hatching, cultured them at low density, and tracked their viability and developmental progress. slik1 homozygous mutant larvae were compared with similarly staged heterozygous control larvae. slik mutants were delayed with respect to growth and developmental timing (Figure 2B). After 5 d, the largest mutant larvae had grown to about one-third the size of controls (Figure 2A). Relatively few progressed as far as the third-larval instar. However, some larvae had an abnormally long lifespan. More than 5% of the mutant larvae remained alive for 15 d (three times longer than normal), and some reached a relatively normal third-larval instar size. To confirm that these defects are due to loss of slik function, we tested whether they could be reversed by expression of a slik transgene. Ubiquitous armadilloGAL4-driven slik expression rescued the larval growth defect and lethality (Figure 2B and 2C). Of 160 rescued mutant larvae examined, 115 survived to adulthood, albeit with a developmental delay of several days. These gave rise to adult flies of reduced body size, with mildly rough eyes, but of otherwise normal appearance (Figure 2C).
Figure 2. Growth Defects in slik Mutants.
(A) Heterozygous slik1/+ control and homozygous slik1 mutant larvae after 5 d of growth under uncrowded conditions.
(B) Growth and survival characteristics of slik1/+ control and homozygous slik1 mutant larvae. The percentage of animals at each developmental stage on the indicated days are shown by the colored bars. (Left) slik1/+ control animals. (Middle) Homozygous slik1 mutant animals. (Right) Percent expected homozygous slik1 mutant females rescued by expression of Slik under control of armadilloGAL4. Note that only females received both the armadilloGAL4 driver and the UAS-slik transgene. In competition with rescued females, male larvae die earlier than when all animals are homozygous mutant. The few surviving males are included in the107% recovery at 5 d.
(C) Comparison of slik1/+ control females (left) with homozygous slik1 mutant females rescued by expression of Slik under control of armadilloGAL4. Note the reduced body size of the rescued flies.
(D–F) Larval internal organs labeled by BrdU incorporation. (D and E) Control larvae fed BrdU from 76 to 92 h and 56 to 72 h AEL, respectively. All nuclei are brown, indicating BrdU incorporation during endoreplication. (F) slik1 mutant larva fed from 76 to 92 h. Few nuclei were labeled. Note that the size is comparable to the much younger wild-type control.
Most larval tissues grow by endoreplication, a process in which cells progress through a modified cell cycle consisting of rounds of DNA synthesis with no mitoses in between. The resulting polyploidy is necessary for growth of the larval tissues (Royzman et al. 1997; Britton and Edgar 1998). Mutations affecting the ability of larval cells to become polyploid by endoreplication can mimic the growth defects caused by starvation (Royzman et al. 1997; Galloni and Edgar 1999). We confirmed in control experiments using dye-stained food that over 90% of slik mutant larvae were feeding during the period from hatching until 72 h after egg laying (AEL), ruling out altered feeding behavior as a major cause of their growth defect. To ask whether slik regulates the endoreplication cycle, we cultured staged control and slik1 larvae on food containing bromodeoxyuridine (BrdU) to label cells that had undergone DNA synthesis. Control larvae fed BrdU from 56 to 72 h or from 76 to 92 h showed extensive BrdU incorporation in larval tissues including the gut, fat body, and salivary glands (Figure 2D and 2E). BrdU incorporation in slik mutant larvae was more variable. Most animals showed a marked decrease in the number of labeled cells (Figure 2F), suggesting that the proportion of cells undergoing endoreplication during the labeling period was decreased in the mutants. These observations indicate that slik is important for normal larval growth. Although not strictly necessary for endoreplication, slik may play a role in regulating the rate of endocycle progression in larval cells.
slik Promotes Cell Survival
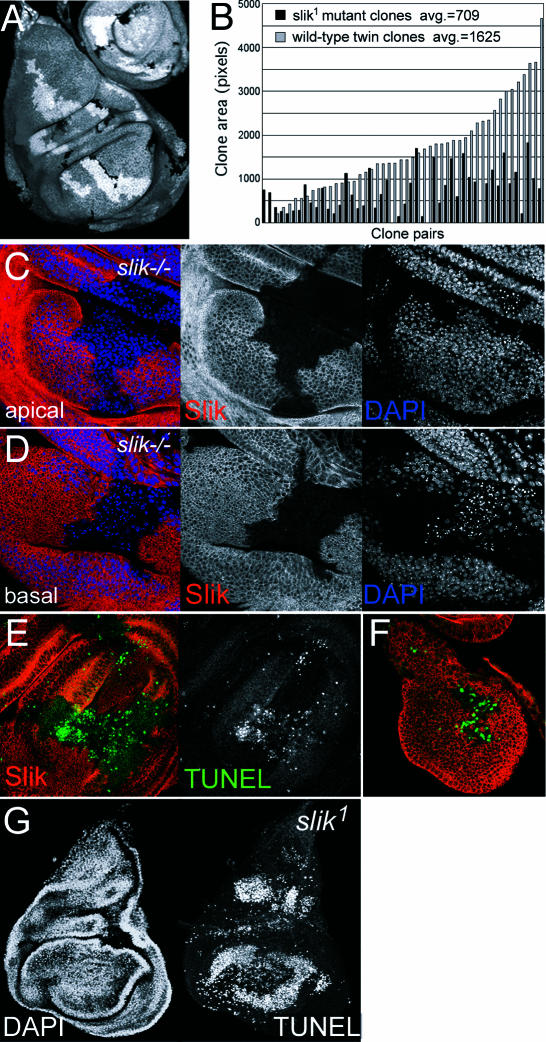
To evaluate the cellular basis for the larval growth defects, we examined the requirement for slik in diploid cells of the imaginal discs. Antibody labeling showed that Slik protein is expressed at a uniform level in the discs. We generated mosaic animals bearing slik mutant clones using FLP/FRT-mediated recombination. slik mutant clones were smaller than their simultaneously generated wild-type twin clones in the wing disc (Figure 3A). Mutant clones generated at 48 ± 2 h covered on average only 44% the area of the corresponding wild-type twin clones (Figure 3B) and rarely reached a large size. When clones were induced earlier, many discs were found to contain wild-type twinspots with no mutant clones, indicating that the slik mutant cells were eliminated. We did not observe any position-dependent effects on clonal growth, suggesting that slik function is required in all wing disc cells.
Figure 3. Growth and Survival Defects in slik Mutant Clones.
(A) Wing imaginal disc with several homozygous slik1 mutant clones and homozygous wild-type twin clones (part of a leg disc is visible at upper right). The homozygous wild-type and mutant cells are produced in the same cell division, so differences in size reflect differences in growth or cell survival after clone induction. Homozygous slik1 mutant cells lack the βGAL marker protein and are unlabeled (black). Homozygous wild-type cells have two copies of the marker and appear brighter than heterozygous slik1/+ cells.
(B) Area measurements of 48 pairs of homozygous slik1 mutant and wild-type twin clones.
(C and D) Wing disc with a large homozygous Minute+ slik1 mutant clone produced in a Minute heterozygous background. Slik protein is shown in red. Blue shows DAPI-labeled nuclei. (C) and (D) are different optical sections of the same disc. (D) shows the pyknotic nuclei below the epithelial layer.
(E and F) Wing discs with large homozygous Minute+ slik1 mutant clones. Red shows a single optical section showing Slik protein. Green shows a projection of several optical sections showing TUNEL labeling to visualize apoptotic cells. (F) Mid-third instar disc.
(G) DAPI and TUNEL labeling of a slik1 homozygous mutant wing disc.
Mutations that slow the rate of cell proliferation can cause cells to be out-competed by faster-growing wild-type cells. Cell competition can be reduced if mutant cells are provided with a growth advantage relative to neighboring cells. Minute genes encode ribosomal proteins and mutations impair growth by reducing the biosynthetic capacity of cells. We generated slik−/− Minute+/+ clones in a slik+/− Minute+/− background. When given a growth advantage, slik mutant clones grew to relatively large sizes. The appearance and density of nuclei were normal in apical optical sections through Minute+ slik1 mutant clones; however, many mutant cells with pyknotic nuclei were extruded beneath the epithelium (Figure 3C and 3D), a feature typical of wing disc cells undergoing apoptosis. Apoptosis was verified using TUNEL staining to visualize DNA cleavage products in Minute+ slik1 mutant cells (Figure 3E). Outside the clones, a few dispersed clusters of TUNEL-positive cells appeared sporadically, as in normal discs (Milán et al. 1997). Many TUNEL-positive cells were observed in late-second/early-third instar Minute+ slik1 mutant clones (Figure 3F) and in wing discs taken from 13-d-old homozygous slik1 mutant larvae, in which all cells are mutant (Figure 3G). These observations indicate that survival of slik mutant cells is impaired even when they are not in competition with wild-type cells. This suggests an intrinsic survival deficit in the mutant cells.
Amplification of Apoptosis in slik Mutant Cells by the JNK Pathway
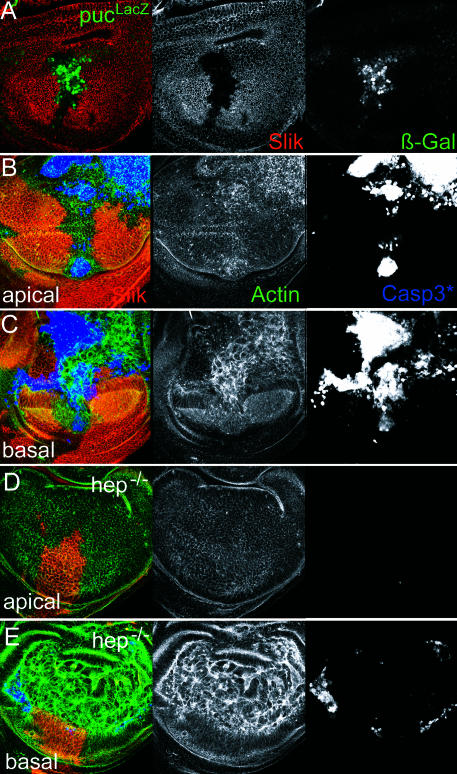
The c-Jun N-terminal kinase (JNK) pathway is an effector of apoptotic cell death during imaginal disc development (Adachi-Yamada et al. 1999; Moreno et al. 2002a). JNK pathway activity can be monitored in the discs by transcriptional activation of puckered (puc), a dual-specificity phosphatase that acts in a negative-feedback loop to regulate the JNK pathway (Martin-Blanco et al. 1998). Transcription of a puc–lacZ reporter gene is normally low or absent in the wing disc. However, in response to apoptotic stimuli, puc–lacZ is induced in a JNK-dependent manner (Adachi-Yamada et al. 1999). We observed that puc–lacZ was induced in Minute+ slik1 mutant clones (Figure 4A), indicating apoptosis of slik mutant cells involves recruitment of the JNK pathway to amplify the apoptotic trigger. To confirm this, we compared the level of apoptosis in slik mutant clones induced in flies lacking the hemipterous (hep) gene, which encodes the kinase that activates JNK (JNKK). Removing Hep activity has been shown to suppress JNK-induced apoptosis (Adachi-Yamada et al. 1999). Apoptosis was visualized using an antibody to the activated form of caspase 3. Activated caspase was readily detected throughout the Minute+ slik1 mutant clones in a wild-type background (Figure 4B and 4C). The staining was most prominent in basal optical sections of the disc. Using phalloidin to label cortical actin, we observed that dying Slik-negative cells were extruded out the basal surface of the epithelium. Large Minute+ slik1 mutant clones in a hep-null background caused much less distortion of the discs and looked essentially normal in apical optical sections (Figure 4D). Levels of activated caspase were strongly reduced in these clones compared to slik mutant clones in the JNKK+ background, even though many mutant cells were extruded on the basal surface of the epithelium (Figure 4D and 4E). These observations indicate that activation of the JNK pathway contributes to apoptosis in slik mutant clones.
Figure 4. JNK Activity and slik-Dependent Apoptosis.
(A) Wing disc with a Minute+ slik1 mutant clone. Red shows Slik protein. Green shows puc–lacZ reporter gene expression visualized by anti-βGAL. Increased βGAL staining in the clone indicates puc transcription in response to JNK pathway activation.
(B and C) Wing disc with large Minute+ slik1 mutant clones in an otherwise wild-type background. Blue shows activated caspase 3. Green shows actin visualized by phalloidin to show cell outlines. (B) and (C) are different optical sections of the same disc. Genotype: +/Y; FRT42D P(πmyc) M(2)531/FRT42D slik1; hsFLP388/+.
(D and E) Wing disc with large Minute+ slik1 mutant clones in a hemipterous mutant background. (D) and (E) are different optical sections of the same disc. Genotype: hepr75/Y; FRT42D P(πmyc) M(2)531/FRT42D slik1; hsFLP388/+. Note the dramatic increase in clone size, relatively normal apical appearance, and reduction of apoptosis in the clones (detected by activated caspase 3 staining) when JNK pathway activity is reduced in the absence of the hep JNKK.
slik Mutant Cells Have the Capacity to Differentiate Normally
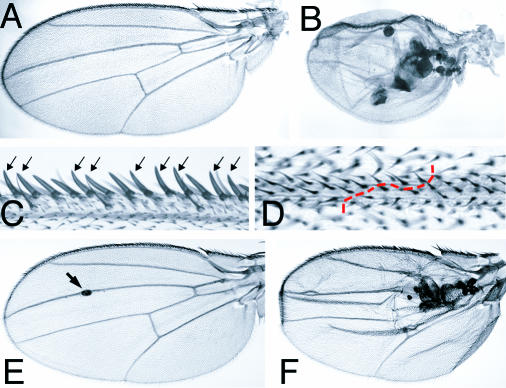
Despite the elevated rate of apoptosis observed in mutant clones and discs, many slik mutant cells survived and were integrated normally in the disc epithelium. To assess the developmental capacity of these cells, we examined adult wings bearing large Minute+ slik1 mutant clones generated at 60 ± 12 h. Most of these wings curved upward or downward to varying degrees, suggesting that there were differences in the sizes of the dorsal and ventral surfaces of the wing blade. In more severe cases, the wings were small and contained vesicles of blackened tissue (Figure 5A and 5B). These vesicles may derive from cells extruded on the basal side of the epithelium, which come to lie between the two layers of the wing blade. slik mutant cells that remained in the epithelium differentiated into morphologically normal wing blade cells, margin bristles, and wing veins (Figure 5C and 5D). Mutant cells were the same size as wild-type cells.
Figure 5. slik Mutant Clones in Adult Wings.
(A and B) Cuticle preparations of adult wings from w f36a hs-FLP1/Y; FRT42D P(f+) P(f+) M(2)l2/FRT42D slik1 larvae.
(A) Adult wing from a larva not subjected to heat shock to induce clones.
(B) Wing with large homozygous Minute+ slik1 mutant clones. Note the small size of the wing and the vesicles of black necrotic tissue between the layers of the wing.
(C) Detail of a clone in the wing margin. Mutant cells, marked by forked, differentiate as normal wing margin bristles (arrows).
(D) Detail of a clone in the wing blade. Mutant cells differentiate as normal wing blade and wing vein cells. The boundary of the clone in the vein is indicated by the dashed red line.
(E and F) slikKG04837/slik1 wings. In (E), the arrow indicates a small vesicle in a mildly affected wing.
The slikKG04837 mutant produced a similar, but milder, phenotype than slik1. slikKG04837/slik1 flies showed a 13% decrease in viability. Nearly 40% of the wings in the surviving flies had phenotypes similar to those caused by slik1 mutant clones. Most showed curvature of the wing blade surface or small isolated vesicles (Figure 5E, arrow). However, 30% of affected wings showed a stronger phenotype characterized by accumulation of vesicles and reduction in wing size (Figure 5F). These defects correlated with an increased level of apoptosis in wing discs (see Figure 10D).
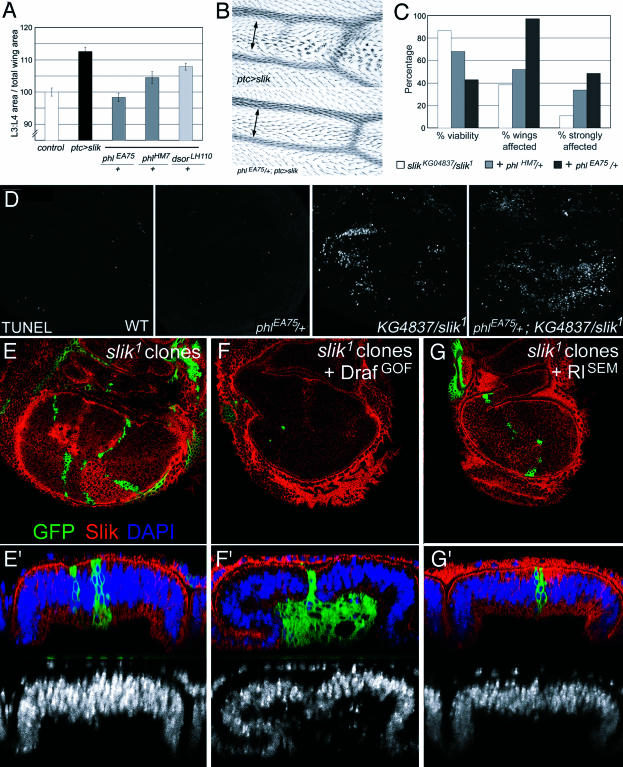
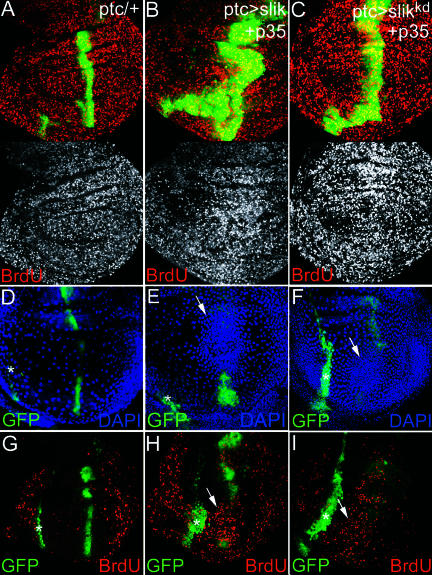
Figure 10. Slik Acts via Raf to Control Cell Survival and Cell Proliferation.
(A and B) ptcGAL4 UAS-slik phenotype in different genetic backgrounds.
(A) Quantification of the area between veins 3 and 4 as a function of total wing area. Control: wild-type flies. Other genotypes as indicated.
(B) Detail of the vein 3–4 region in ptcGAL4 UAS-slik and Raf heterozygous phlEA75/+; ptcGAL4 UAS-slik wings. The multiple wing hair phenotype characteristic of Slik overexpression was suppressed when one copy of Raf was removed and the spacing between the veins was reduced.
(C) Survival rates and assessment of the severity of the wing phenotypes in flies of the indicated genotypes.
(D) TUNEL labeling of wing discs to visualize apoptotic cells. Genotypes as indicated. (E–G) slik1 mutant clones labeled by expression of GFP (green) and by the absence of Slik protein (red). Larval genotypes: UAS-CD8-GFP hsFlp; FRT42 Gal80/FRT42 slik1; Tub-Gal4/+. (F) Plus UAS-RafGOF. (G) Plus UAS-RlSEM. Lower panels show optical sections perpendicular to the plane of the epithelium. DAPI (blue) shown alone below.
slik Promotes Cell Proliferation and Tissue Overgrowth
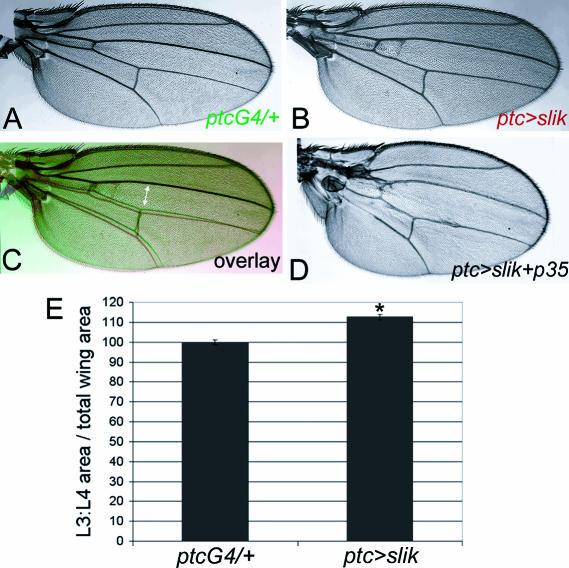
We identified slik by its ability to promote overgrowth of the wing when expressed under GAL4 control. Overexpression of slik in the region between the third and fourth wing veins using ptcGAL4 caused overgrowth, increasing the distance between these veins (Figure 6A–6C). In larvae raised at 18°C, transgene-driven slik expression resulted in a 13% increase in the area bounded by veins 3 and 4 as a proportion of total wing area (p < 0.001; Figure 6E). When larvae were raised at 25°C, which normally results in higher levels of transgene activation, we saw only a 5% increase in area, suggesting that slik-driven overgrowth is offset by a counteracting process. This was examined in more detail in imaginal discs using green fluorescent protein (GFP) to mark cells overexpressing slik. We noted abnormal apoptotic cell death in the domain of slik and GFP expression, even when the larvae were raised at 18°C. Cellular and nuclear morphology were normal within the plane of the epithelium, but pyknotic nuclei were visible in GFP-expressing cells extruded below the epithelium (Figure 7A and 7B). High levels of activated caspase were also detected in this region (data not shown). These observations suggested that slik, like many oncogenes, promotes cell proliferation and apoptosis in parallel.
Figure 6. Slik-Induced Overgrowth in the Wing.
(A) Cuticle preparation of a ptcGAL4 adult wing.
(B) Cuticle preparation of a ptcGAL4 UAS-slik adult wing.
(C) Overlay of (A) (green) and (B) (red) aligned in the anterior margin. Note the increased separation of veins 3 and 4 in the center of the wing (arrow).
(D) Cuticle preparation of a ptcGAL4 UAS-slik UAS-p35 adult wing. The separation of veins 3 and 4 was larger than in (C).
(E) Measurement of the area enclosed by veins 3 and 4 in ptcGAL4 and ptcGAL4 UAS-slik wings. Error bars indicate standard deviation (*: p < 0.001 using a Student's t-test).
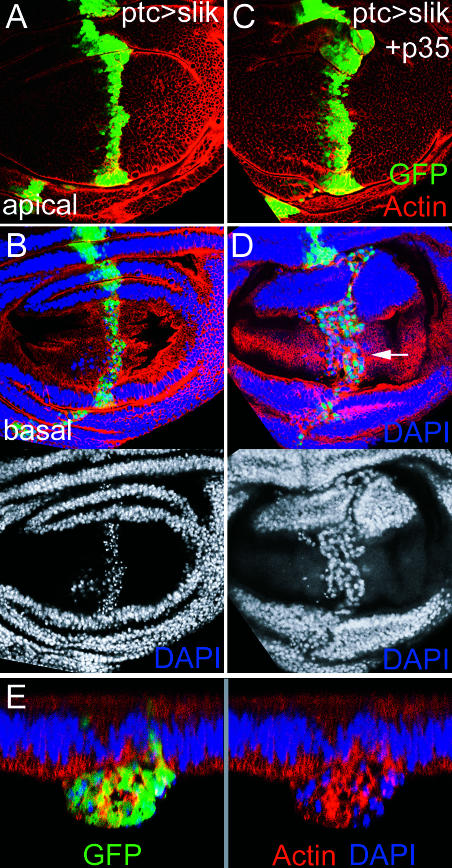
Figure 7. Slik Overexpression Induces Apoptosis.
(A and B) Two views of one ptcGAL4 UAS-slik UAS-GFP wing disc.
(A) Optical section in the columnar epithelial layer of the wing pouch. Red shows actin staining. Green shows ptcGAL4-expressing cells visualized by GFP. Blue shows DAPI staining.
(B) Basal optical section to show extruded GFP+ cells with pyknotic nuclei. DAPI is shown below.
(C–E) ptcGAL4 UAS-slik UAS-GFP UAS-p35 wing disc. The GFP-expressing stripe is wider. Basally extruded cells have morphologically normal nuclei (arrow). In (E), optical cross-section show a large accumulation of ptcGAL4 UAS-slik UAS-GFP UAS-p35 cells below the epithelial layer.
To examine the effects of slik overexpression when cell death was blocked, we coexpressed slik and GFP with the viral caspase inhibitor p35. The combination of slik expression and suppression of apoptosis resulted in a strong further increase in tissue growth (Figure 7C). In addition to an increase in the number of slik-, p35-, and GFP-expressing cells in the epithelium, many GFP-positive cells with normal nuclear appearance were found in an abnormal outgrowth below the epithelial layer (Figure 7D and 7E). Co-expression of slik with p35 was lethal during pupal stages with most GAL4 drivers tested. However, we found some escapers at 18°C that showed additional overgrowth in the ptcGAL4 expression domain (see Figure 6D). Abnormal outgrowths of tissue were also often found proximally in these wings.
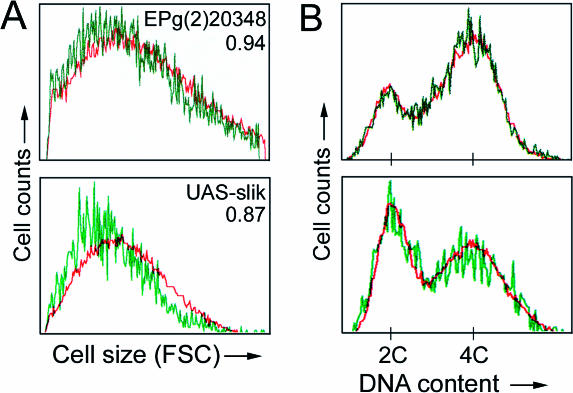
To ask whether slik-driven tissue growth resulted from increased cell proliferation rather than from increased cell size (as results from activation of the insulin signaling pathway; Stocker and Hafen 2000), we performed flow cytometry on cells from dissociated discs. Slik-overexpressing cells showed a modest decrease in size (6% smaller than control cells using EPg(2)20348; 13% smaller using a stronger UAS-slik transgene; Figure 8A). Slik also had little effect on cell cycle profile (Figure 8B).
Figure 8. Analysis of Slik-Expressing Cells.
Clones of cells expressing GFP and Slik expressed from EPg(2)20348 (top) or UAS-slik transgene (bottom) were induced at 48 ± 2 h AEL. Discs were dissected at 112 h AEL (EPg(2)20348) or 98 h AEL (UAS-slik), and cells were dissociated and analyzed by flow cytometry. Data for GFP and EP or transgene-expressing clonal cells are in green and nonexpressing control cells from the same discs are in red.
(A) Cell sizes estimated by forward scatter values. Numbers represent the ratio of forward scatter values for GFP+/GFP− cells.
(B) Distribution of cells in G1 (2C DNA content), S (2C–4C), and G2 (4C).
To verify that slik-induced tissue overgrowth resulted from increased cell proliferation, we used BrdU incorporation to label cells that had undergone DNA replication. During a 1-h labeling period cells in wing discs from control ptcGAL4, UAS-GFP larvae incorporated BrdU in a uniformly random pattern, typical of normal wing discs (Figure 9A). In contrast, there was a considerable increase in the number of cells that incorporated BrdU in the center of discs overexpressing slik under ptcGAL4 control (Figure 9B). We noted that the region of increased proliferation was centered on the stripe of cells expressing the GAL4 driver, but was not limited to it.
Figure 9. Nonautonomous Stimulation of Cell Proliferation by Slik-Expressing Cells.
(A, D, G) ptcGAL4 UAS-GFP wing discs. (B, E, H) ptcGAL4 UAS-slik UAS-GFP UAS-p35 wing discs. (C, F, I) ptcGAL4 UAS-slikkd UAS-GFP UAS-p35 wing discs. (A–C, G–I) BrdU incorporation (red). (A–C) Projections of several optical sections. (G–I) Sections of the overlying peripodial layer. (D–F) Peripodial cell nuclei visualized by DAPI. Arrows show high nuclear density above the ptcGAL4 UAS-GFP stripe in the columnar epithelium in (E) and (F). Asterisks indicate the peripodial extension of the ptcGAL4 stripe. (H and I) Cells in this region have incorporated more BrdU than control disc in (G).
To test the requirement for the kinase activity of Slik, we constructed a kinase-inactive version of the protein (Slikkd) by mutating aspartate176 in the nearly invariant aspartate–phenylalanine–glycine triplet of kinase subdomain VII to asparagine. Expression of Slikkd resulted in significantly more lethality than wild-type Slik with most GAL4 drivers tested (data not shown). However, the effects of the two proteins on proliferation of wing disc cells were remarkably similar. When coexpressed with p35 and GFP using ptcGAL4, Slikkd caused an increase in the number of GFP-expressing cells and an increase in the number of cells that incorporated BrdU (Figure 9C). This suggests that Slik can drive cell proliferation by a kinase-independent mechanism.
In addition to the increased proliferation of the disc epithelium caused by Slik or Slikkd overexpression, we noted a striking increase in cell proliferation in the overlying peripodial cell layer, reflected by increased nuclear density and BrdU incorporation (Figure 9D–9I). The region of elevated proliferation in the peripodial layer was situated directly above the ptcGAL4 stripe in the columnar epithelial layer and was clearly separated from the ptcGAL4 stripe in the peripodial layer. Note that the overproliferating cells were normal peripodial cells that were not expressing p35. This suggests that slik overexpression in the columnar epithelium led to production of a signal that was able to stimulate proliferation of cells in the peripodial layer. Communication between these cell layers has previously been implicated in growth regulation, but the signals have been suggested to flow in the opposite direction (Gibson and Schubiger 2000; Gibson et al. 2002).
Slik Acts via Raf to Control Proliferation and Cell Survival
A few Ste20 group kinases have been shown to act as upstream activators of MAPK-type pathways by regulating MAP3K activity (Dan et al. 2001). To determine whether slik function involves activation of MAPK signaling, we tested the ability of mutants in two Drosophila MAPK-type pathways (those mediated by JNK and ERK) to suppress slik-driven tissue growth. We observed limited modulation of slik-driven overgrowth in animals heterozygous for mutations in the JNKK hep, the JNK basket, the transcription factor jun, or the JNK phosphatase puc (20% or less). In contrast, slik showed strong genetic interactions with upstream components of the ERK pathway. Removing one copy of the Drosophila MAP3K Raf using the null allele phlEA75 completely suppressed the effects of slik-driven overgrowth, restoring the wings to a wild-type appearance (Figure 10A and 10B). A weaker allele of Raf, phlHM7 produced a milder suppression of overgrowth (64%, p < 0.001). Overgrowth was reduced by 37% in flies heterozygous for the MAPKK Sor, which acts downstream of Raf (p < 0.005). A deficiency removing the Drosophila ERK Rolled had only a modest effect (15%). These observations raise the possibility that ERK may not be a major effector of Slik in promoting cell proliferation, although Raf appears to be one (see below).
If Raf acts as a downstream effector of Slik, we would expect removing one copy of the raf gene to enhance the severity of slik mutant phenotypes. The combination of the hypomorphic allele slikKG04837 and slik1 produced viable flies at approximately 90% of the expected frequency (Figure 10C). Removing one copy of raf reduced the viability of the slik hypomorphs to approximately 40% and increased the penetrance of their wing phenotype from 39% to nearly 100%. The frequency of strongly affected wings increased from 11% to nearly 50% of the wings examined. The weaker phlHM7 allele caused a milder enhancement of these phenotypes.
slikKG04837/slik1 wing discs showed a considerable amount of apoptosis by TUNEL labeling (Figure 10D). The amount of apoptosis in the slikKG04837/slik1 background increased when one copy of raf was removed (a 1.75-fold increase in the average number of TUNEL-positive cells, 2.0-fold increase in median; p < 0.05). We did not observe an increase in TUNEL-positive cells in phlEA75/+ heterozygotes compared to wild-type. Thus, reduction of Raf levels specifically increased the amount of apoptosis and the severity of the subsequent phenotypic effects when Slik activity was compromised. These results, together with the ability of Raf to suppress Slik gain-of-function phenotypes, suggest that Raf is an important downstream effector of Slik in providing cell survival and cell proliferation cues.
To test this, we asked whether expression of an activated form of Raf could compensate for the absence of Slik. Clones of slik1 mutant cells were produced using the Marcm system (Lee and Luo 1999) to allow GAL4-dependent expression of activated Raf in the slik1 mutant cells. Clones of GFP-expressing slik1 mutant cells were much smaller than their wild-type twin clones. In contrast, large slik1 mutant clones were recovered that expressed activated Raf (Figure 10E and 10F). These clones were located on the basal surface of the epithelium, grew to quite large sizes, and had few pyknotic nuclei (and little evidence of apoptosis by TUNEL labeling; data not shown). This contrasts with Minute+ slik1 mutant clones, which showed very strongly elevated cell death inside the clones (see Figure 3E). Expression of p35 produced similar effects on survival of slik1 mutant clones (data not shown). Comparable experiments expressing an activated form of ERK (RlSEM) did not suppress the survival defect of slik1 mutant clones (Figure 10G). These clones are considerably smaller than their twins and contained many pyknotic nuclei, indicative of apoptosis (Figure 10G). The level of ERK activity generated by this transgene was sufficient to elicit the high threshold ERK response of vein formation. Thus, the failure to suppress cell death in slik mutant clones cannot be attributed to insufficient ERK activity (discussed below). These observations indicate that activation of Raf can suppress the survival defect and promote proliferation of slik1 mutant cells and suggest that Raf does not act via the canonical ERK MAPK pathway to do so. These differences may explain the strong genetic interactions between Slik and Raf mutants and the very limited genetic interaction between Slik and ERK (Rolled) mutants.
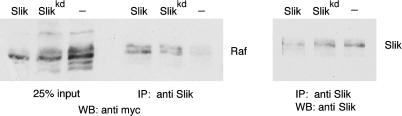
The strong genetic interactions between Slik and Raf prompted us to ask whether Slik can bind directly to Raf. Co-immunoprecipitation experiments were performed using S2 cells transfected to express Myc-tagged Raf. Immunoprecipitation of endogenous Slik protein from S2 cells was able to coprecipitate Raf (Figure 11, lane 6; the loading control represents 25% of input). Cotransfection of S2 cells to express Slik or the kinase inactive Slikkd increased the recovery of Raf in the coimmunoprecipitation (Figure 11, lanes 4 and 5). The relationship between Slik and Raf resembles that reported for the Ste20 kinase germinal center kinase (GCK) and the MAP3K MEKK1 in that GCK binds to MEKK1 and activates it in a kinase-independent manner (Chadee et al. 2002). Our results suggest that activation of Raf is mediated by binding to Slik, rather than by phosphorylation of Raf by Slik. We examined the level of ERK phosphorylation in S2 cells transfected to overexpress Slik or Slikkd. No significant difference was observed (data not shown). Taken together with the finding that expression of activated ERK cannot rescue the slik mutant survival defect in vivo, these findings suggest that Raf does not act via the canonical ERK MAPK pathway to mediate Slik's activity in supporting cell survival and promoting cell proliferation.
Figure 11. Slik Binds to Raf.
(Right) Endogenous Slik protein, or transfected Slik or Slikkd proteins were immunoprecipitated from S2 cells with anti-Slik. (Left) The blot was probed with anti-Myc to visualize co-precipitation of Myc-tagged Raf.
Discussion
In this report we have identified the Ste20 group kinase Slik as a mediator of cell survival and cell proliferation signaling in Drosophila. slik is an essential gene. Loss of slik function results in a larval growth defect characterized by slow growth and a decreased rate of endoreplicative cell cycling in polyploid larval cells, developmental delay, and eventually death. The behavior of slik mutant clones suggests that diploid cells lacking slik proliferate more slowly than normal and are eliminated by apoptosis. Even when removed from cell competition, slik mutant cells exhibit an intrinsic survival defect. Our findings suggest that this is due to insufficient activation of Raf. Indeed, activation of Raf can compensate for the absence of Slik in supporting cell survival. Whereas reduced Slik activity limits growth due to increased apoptosis, increased Slik activity promotes growth by increasing the rate of cell proliferation. Slik-induced proliferation is counteracted by increased apoptosis. The increase in proliferation and apoptosis is due to overactivation of Raf, as both proliferation and apoptosis can be suppressed by reducing Raf activity. These observations suggest that Raf may be the principal mediator of Slik activity in promoting cell survival and cell proliferation.
At present, the upstream signals that control Slik activity are not known. Activation of MAPK pathways by Ste20 kinases such as NCK and GCK has been linked to the interleukin and TNF receptor effector proteins TRAF2 and TRAF6 (Baud et al. 1999; Chadee et al. 2002). A Drosophila TNF ligand and receptor have been identified and shown to act via Apaf1 and DRONC to induce apoptosis (Igaki et al. 2002; Kanda et al. 2002; Moreno et al. 2002b). Cell survival and proliferation also depend on multiple inputs. Information about the nutritional state of the animal is transmitted by the insulin/PI3K pathway (e.g., Britton et al. 2002). In addition to the EGFR/ERK pathway, the Dpp, Wg, and Notch pathways provide growth and survival cues (Go et al. 1998; Miller and Cagan 1998; Baonza and Garcia-Bellido 1999; Milán et al. 2002; Moreno et al. 2002a). We do not know whether these or other signals act via Slik to control cell survival and cell proliferation during development.
Slik Can Activate Raf in a Kinase-Independent Manner
The proliferative and cell-survival signaling effects of Slik appear to be mediated through Raf. Signals from receptor tyrosine kinases (RTKs) act via the small GTPase Ras to activate Raf (Schlessinger 2000). In its GTP-bound active state, Ras recruits Raf to the cell membrane, where it is activated by a mechanism involving protein–protein and protein–lipid interactions as well as phosphorylation and dephosphorylation of key residues (reviewed in Dhillon and Kolch 2002). Some members of the Ste20 kinase family act as MAP4Ks to activate Raf and other MAP3Ks, whereas others act in a kinase-independent manner (reviewed in Dan et al. 2001). HPK1 and PAK2 act as MAP4Ks to phosphorylate MEKK1 and Raf. The Ste20 protein GCK activates MEKK1 in a kinase-independent manner to transduce signals from TNF family receptors via the JNK pathway. Oligomerization of MEKK1 by GCK1 is thought to lead to its activation by autophosphporylation (Chadee et al. 2002). Our finding that the proliferative effects of Slik and its ability to bind Raf are independent of its kinase activity suggests that Slik activates Raf as a consequence of binding, perhaps by oligomerization, as has been suggested for GCK and MEKK1.
Although the kinase-inactive form Slikkd can produce the same effects as the unmodified form of Slik in overexpression assays, low-level ubiquitous expression of Slikkd was not able to rescue slik1 mutant larvae. A wild-type slik transgene rescued the mutant to viability under comparable conditions. In fact, Slikkd expression enhanced the severity of the slik1 survival defect. The slik1 mutant lacks the kinase domain and should express no protein, so Slikkd is unlikely to act as a dominant negative for endogenous Slik. Indeed, low-level ubiquitous expression of Slikkd had no effect in wild-type animals. Thus, Slikkd may compete for the activity of a different kinase that phosphorylates Slik targets in the absence of endogenous Slik
One interpretation of our findings is that the kinase activity of Slik is required to activate Raf at normal Slik levels, but that this requirement can be circumvented under conditions of overexpression. If there is a difference in the ability of the two forms of Slik to activate Raf, it does not appear to be reflected in their ability to bind Raf as assayed by coimmunoprecipitation, but many other possibilities exist. For example, Slik kinase activity may not be required for Raf activation, but may be required to allow Slik to perform other functions during development.
Does Slik Have Other Functions?
Preliminary evidence suggests that Slik may also regulate the actin cytoskeleton. For example, rescue of the slik mutant cell-survival defect by expression of p35 or activated Raf did not prevent cells from exhibiting an abnormal arrangement of actin and dropping out of the disc epithelium. This suggests a Raf-independent role for Slik that is required to allow normal organization of the actin cytoskeleton and for normal cell interactions within the epithelium. In this context, it is interesting that human SLK and LOK are thought to influence cytoskeletal dynamics and cell adhesion (Endo et al. 2000; Sabourin et al. 2000; Wagner et al. 2002). Future work will be directed toward determining whether this reflects an independent function of Slik that requires it to function as a kinase.
Distinct Modes of ERK Activation for Cell Survival and Proliferation versus Cell Fate Specification
The Ras–Raf–MEK–ERK signaling cassette regulates cell fate specification, proliferation, and cell survival downstream of the EGFR in the imaginal discs. Clones of cells mutant for components of this pathway proliferate poorly and are frequently lost even when given a growth advantage (Xu and Rubin 1993; Diaz-Benjumea and Hafen 1994; Prober and Edgar 2000). In the eye, activity of this pathway is required for cell survival and cell cycle progression posterior to the morphogenetic furrow (Dominguez et al. 1998; Baker and Yu 2001; Halfar et al. 2001). Activated forms of Ras and Raf promote tissue growth, and high levels of activated Ras can induce apoptosis (Karim and Rubin 1998; Prober and Edgar 2000, 2002).
Although Slik is not a core component of the ERK pathway, our findings suggest that Slik activates Raf to control cell survival and cell proliferation. What are the differences between RTK-mediated and Slik-mediated activation of Raf? The growth and survival functions of the ERK pathway are thought to require low-intensity signaling (Baker and Yu 2001; Halfar et al. 2001; Yang and Baker 2003). Stronger Raf-dependent ERK activation is required for cell fate specification in response to high-threshold EGFR signaling. Repeated cycles of strong ERK activation are responsible for sequential cell fate specification events in the eye imaginal disc (Freeman 1996). High-level activation of ERK specifies vein cell fate in the wing disc (as visualized by antibody to doubly phosphorylated ERK; Gabay et al. 1997). Activation of ERK in proliferating wing cells is not detectable by this antibody. Clones of cells lacking Slik activity do not cause defects in vein differentiation or in cell fate specification in the eye (data not shown). Elevated Slik expression stimulates cell proliferation, but does not cause a detectable increase in the level of ERK phosphorylation in S2 cells (data not shown). This suggests that Slik-mediated activation of Raf is not required for high-threshold ERK responses.
Both Raf and Sor (MAPKK) act downstream of Slik based on the ability of mutations in either kinase to dominantly suppress Slik-induced tissue growth. However, a deficiency removing ERK had little effect in the same assay. It may be that that ERK levels are not limiting because Slik is involved only in low-level ERK activation. Alternatively, Slik may regulate Raf- and MEK-dependent survival and proliferation independently of ERK. This view is supported by our finding that activation of Raf is able to replace the requirement for Slik and support survival of slik mutant cells, whereas activation of ERK is not able to do so. There is increasing evidence supporting an ERK-independent function of Raf proteins, particularly in the regulation of cell survival/apoptosis (reviewed in Hindley and Kolch 2002). Furthermore, Raf activity may be connected to cell survival signaling by other kinase-independent mechanisms (Chen et al. 2002).
Are Mammalian Homologues of slik Oncogenes?
The observation that increased slik expression promotes proliferation and tissue growth while at the same time increasing the rate of apoptosis is reminiscent of the effects of oncogenes. In mammalian systems, it has long been known that driving cell proliferation by expression of oncogenes such as MYC, E2F, and RAS or by removal of tumor suppressors, such as RB, concomitantly promotes apoptotic cell death. Similar observations have been made in flies, for example, by overexpression of ras or E2F (Du et al. 1996; Karim and Rubin 1998; Neufeld et al. 1998) and in cells lacking the tumor suppressor RBF (Datar et al. 2000). It has been suggested that the signal to proliferate inherently sensitizes cells to apoptosis. In order to respond to such a signal by dividing rather than dying, a cell must receive sufficient survival cues at the same time. This dual signal model provides a means for constraining cellular proliferation in the context of a multicellular organism by making any individual cell dependent upon survival cues from its neighbors in order to grow and divide productively (Evan et al. 1994; Evan and Littlewood 1998).
One prediction of this model is that a cancer cell must activate both growth-promoting genes and anti-apoptotic genes in order to continuously proliferate inappropriately in vivo (Pelengaris et al. 2000). Regulated activation of c-MYC in pancreatic β-cells induced β-cell proliferation accompanied by massive apoptosis, resulting in involution of the islets (Pelengaris et al. 2002). Co-expression of the anti-apoptotic protein Bcl-XL suppressed the MYC-induced apoptosis and resulted instead in rapid tumor formation. In flies, the bantam microRNA has been shown to promote cell proliferation and to simultaneously suppress apoptosis by translational repression of the proapoptotic gene hid (Brennecke et al. 2003). Like Ras and E2F, Slik activity simultaneously promotes cell proliferation and apoptosis. Substantial increases in cell number and tissue size are observed only when slik-induced apoptosis is blocked by coexpression of p35. In some cases, this resulted in tumor-like outgrowths in the wing. It will be of interest to learn whether the mammalian SLK and LOK proteins play a similar role in the control of growth and apoptosis.
Materials and Methods
Fly strains
The 2300 EP lines (Rorth et al. 1998) and 8500 EPg strains (Mata et al. 2000) were screened as described (Hipfner et al. 2002). KG04837 was kindly provided by Hugo Bellen. Other strains are described in FlyBase. A full-length UAS-slik transgene identical to the slik-RA transcript (GADFLY release 3) was prepared using ESTs LD34405 and GH20991. An EcoRI–NotI fragment from LD34405 (containing the 5′ UTR and 2502 nucleotides of coding sequence) and a NotI–XhoI fragment (containing 1401 nucleotides of coding sequence and the 3′ UTR) were cloned into the EcoRI–XhoI sites of pUAST. Several independent transformants produced similar phenotypes of varying strengths when crossed to a series of GAL4-driver lines. The transgene used in this report is an insert on the X-chromosome that gave the mildest phenotypes.
Generation and characterization of the slik1 allele
EPg(2)20348 is viable and causes no phenotype. Excisions of EPg(2)20348 were tested for complementation of the deletion Df(2R)Px2. Those failing to complement Df(2R)Px2 were analyzed by Southern blotting. slik1 was a deletion of approximately 4 kb. The adjacent NotI–EcoRI fragment containing the 3′ portion of the slik gene was unchanged. slik1 complemented l(2)K08003, an allele of the immediately upstream mov34 gene, suggesting that the deletion only mutates the slik locus.
Genotypes of larvae for generation of mosaic clones
Genotypes were w hs-FLP1; FRT42D P(armadillo-lacZ)/FRT42D slik1 and w hs-FLP1; FRT42D P(πmyc) M(2)531/FRT42D slik1 and w f36a hs-FLP1/Y; FRT42D P(f+) P(f+) M(2)l2/FRT42D slik1 and UAS-CD8-GFP hsFlp; FRT42 Gal80/FRT42 slik1; Tub-Gal4/+ and UAS-CD8-GFP hsFlp; FRT42 Gal80/FRT42 slik1; Tub-Gal4/UAS-RafGOFand hepr75/Y; FRT42D P(πmyc) M(2)531/FRT42D slik1; hsFLP3.
Analysis of larval and adult phenotypes
Crosses were carried out at 25°C unless otherwise indicated. For analysis of slik larval phenotypes, w; armGAL4 FRT42D slik1/CyO KrGAL4 UAS-GFP females were crossed to FRT42D slik1/CyO KrGAL4 UAS-GFP males. Embryos were collected for approximately 6 h on apple juice agar plates. After 24 h, newly hatched GFP-negative (slik homozygous mutant) and GFP-positive (heterozygous control) larvae were sorted, and 40–50 animals were transferred to vials containing softened food. Beginning 4 d later, larvae were collected every 2 d by floating them in 20% sucrose. After counting and assessment of the larval stage of each animal based on size and appearance of mouth hooks, they were returned to fresh vials. The progress to pupal stages and eclosion was also scored. To rescue the larval growth defect by transgene expression, females of the same genotype were crossed to UAS-slik/Y; FRT42D slik1/CyO KrGAL4 UAS-GFP males. In this case, only female GFP-negative offspring received the UAS-slik transgene. We therefore expected to see a rescue in only half of the animals. For BrdU labeling, larvae were transferred onto fly food containing 0.1 mg/ml BrdU. Larvae were dissected 16 h later and fixed in 8% formaldehyde/PBS, and BrdU incorporation was detected by standard techniques. Wing disc BrdU labeling was performed by incubating dissected larval anterior halves in serum-free medium containing 0.2 mg/ml BrdU for 1 h. To measure slik mutant clone areas, w hs-FLP1; FRT42D armadillo-lacZ females were crossed to FRT42D slik1/Cyo KrGAL4 UAS-GFP males. Embryos were collected for 4 h. After 24 h, freshly hatched larvae (50 per vial) were transferred onto fresh food. Larvae were heat-shocked at 48 ± 2 h for 1 h at 37°C and dissected at 112 ± 2 h. Mutant clone and twinspot areas were measured from confocal images using the histogram function of Adobe Photoshop (Adobe Systems Incorporated, San Jose, California, United States). TUNEL labeling was performed as described (Milαn et al. 1997). Levels of TUNEL staining were measured from confocal images of slikKG04837/slik1 (n = 13) and phlEA75/+;slikKG04837/slik1 (n = 16) wing discs using functions of Adobe Photoshop. For the analysis of Slik overexpression phenotypes, w; ptcGAL4 UAS-GFP females were crossed to either w1118, UAS-slik, UAS-slik;UAS-p35, UAS-slikkd, or UAS-slikkd; UAS-p35 males. Embryos were collected for approximately 6 h and transferred to 18°C. After 24 h, newly hatched first-instar larvae (40 per vial) were transferred onto fresh food and returned to 18°C. Larvae were dissected at the wandering third-instar stage. Co-expression of Slik and p35 caused a developmental delay of approximately 2 d. Some vials were left at 18°C and adult flies were collected. Wings were mounted and imaged, and areas were measured using NIH Image (National Institute of Mental Health, Bethesda, Maryland, United States).
Antibodies
Mouse anti-BrdU was from PharMingen (San Diego, California, United States). Rabbit anti-GFP was from Torrey Pines Biolabs (Houston, Texas, United States). Anti-DP-ERK was from Sigma (St. Louis, Missouri, United States). Rabbit anticleaved human caspase 3 antibody was from Cell Signaling Technology (Beverly, Massachusetts, United States). An antiserum raised against the same epitope has been shown to react with the cleaved form of the Drosophila caspase Drice and to label apoptotic cells in situ (Yu et al. 2002).
Preparation of anti-Slik antibody
Sequences encoding the central nonconserved domain of Slik were PCR amplified (5′ primer: ATAGAATTCGACCTCGACGATGACTCTGC; 3′ primer ACGTAAGCTTTGATCACCACCTCCTCTTCCTC) and cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, California, United States). An EcoRI–HindIII insert fragment was cloned into pET-23a (Novagen, Madison, Wisconsin, United States). The resulting fusion protein consisted of amino acids 335–863 of Slik fused to a C-terminal His6 tag. The bacterially expressed fusion protein was solubilized in 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris (pH 8) and purified on a Ni2+ column under denaturing conditions. Guinea pigs were injected with 50 μg of purified fusion protein in RIBI adjuvant at 3-wk intervals.
Immunoprecipitation
Myc epitope-tagged Raf, Slik, and Slikkd were cloned into pUAST. S2 cells were cotransfected with 3 μg each of pRmHa3-Gal4, pUAST-myc-RAF, and either empty pUAST, pUAST-Slik, or pUAST-Slikkd using CellFectin (Invitrogen). Transfected cells were induced with 0.7 mM CuSO4 for 2 d. Cells were lysed in 200 μl of 50 mM Tris (pH 8), 150 mM NaCl, 0.5% CHAPS, protease inhibitors, and 75% of the cell lysate was diluted in 200 μl of lysis buffer and immunoprecipitated with 2 μl of anti-Slik. Western blots were probed with anti-Myc and then with anti-Slik.
Supporting Information
Accession Numbers
The accession numbers for the sequences reported in this paper are NM 138064 (slik-RA) and NM 166669 (slik-RB).
Acknowledgments
We thank Lidia Perez for excellent technical support and B. Starling Emerald for help in isolating the slik1 mutant. Marco Milαn and Julius Brennecke made helpful comments on the manuscript. This work was supported by the European Molecular Biology Laboratory. DRH was a recipient of a European Molecular Biology Organization Long-Term Fellowship and a Human Frontiers Science Program Fellowship.
Abbreviations
- AEL
after egg laying
- BrdU
bromodeoxyuridine
- ERK
extracellular signal-regulated kinase
- EST
expressed sequence tag
- FLP
yeast Flp recombinase
- FRT
Flp recombinase target site
- GCK
germinal center kinase
- GFP
green fluorescent protein
- lacZ
E. coli β-galactosidease gene
- MAP3K
MAP kinase kinase kinase
- MEK
mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- ptc
patched
- puc
puckered
- RTK
receptor tyrosine kinase
- slik
SLK- and LOK-like kinase
- Ste20
Sterile-20
Conflicts of Interest. The authors have declared that no conflicts of interest exist.
Author Contributions. DRH and SMC conceived and designed the experiments. DRH performed the experiments. DRH and SMC analyzed the data. DRH and SMC wrote the paper. SMC discussed the experiments, design, and interpretation with DRH.
Academic Editor: Matthew P. Scott, Stanford University
References
- Adachi-Yamada T, Fujimura-Kamada K, Nishida Y, Matsumoto K. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature. 1999;400:166–169. doi: 10.1038/22112. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Baonza A, Garcia-Bellido A. Notch signaling directly controls cell proliferation in the Drosophila wing disc. Proc Natl Acad Sci U S A. 1999;97:2609–2614. doi: 10.1073/pnas.040576497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, et al. Signaling by proinflammatory cytokines: Oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated micro-RNA that controls cell proliferation and regulates the pro-apoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: Nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Chadee DN, Yuasa T, Kyriakis JM. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol Cell Biol. 2002;22:737–749. doi: 10.1128/MCB.22.3.737-749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Fujii K, Zhang L, Roberts T, Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci U S A. 2002;98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Datar SA, Jacobs HW, de la Cruz AF, Lehner CF, Edgar BA. The Drosophila cyclin D–cdk4 complex promotes cellular growth. EMBO J. 2000;19:4543–4554. doi: 10.1093/emboj/19.17.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AS, Kolch W. Untying the regulation of the Raf-1 kinase. Arch Biochem Biophys. 2002;404:3–9. doi: 10.1016/s0003-9861(02)00244-8. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Hafen E. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Du W, Xie JE, Dyson N. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J. 1996;15:3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Endo J, Toyama-Sorimachi N, Taya C, Kuramochi-Miyagawa S, Nagata K, et al. Deficiency of a STE20/PAK family kinase LOK leads to the acceleration of LFA-1 clustering and cell adhesion of activated lymphocytes. FEBS Lett. 2000;468:234–238. doi: 10.1016/s0014-5793(00)01219-9. [DOI] [PubMed] [Google Scholar]
- Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- Evan G, Harrington E, Fanidi A, Land H, Amati B, et al. Integrated control of cell proliferation and cell death by the c-myc oncogene. Philos Trans R Soc Lond B Biol Sci. 1994;345:269–275. doi: 10.1098/rstb.1994.0105. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Galloni M, Edgar BA. Cell-autonomous and nonautonomous growth-defective mutants of Drosophila melanogaster. Development. 1999;126:2365–2375. doi: 10.1242/dev.126.11.2365. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Schubiger G. Peripodial cells regulate proliferation and patterning of Drosophila imaginal discs. Cell. 2000;103:343–350. doi: 10.1016/s0092-8674(00)00125-2. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Lehman DA, Schubiger G. Lumenal transmission of decapentaplegic in Drosophila imaginal discs. Dev Cell. 2002;3:451–460. doi: 10.1016/s1534-5807(02)00264-2. [DOI] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development. 2001;128:1687–1696. doi: 10.1242/dev.128.9.1687. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hindley A, Kolch W. Extracellular signal-regulated kinase (ERK)/mitogen activated protein kinase (MAPK)-independent functions of Raf kinases. J Cell Sci. 2002;115:1575–1581. doi: 10.1242/jcs.115.8.1575. [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranagam E, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kanda H, Igaki T, Kanuka H, Yagi T, Miura M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J Biol Chem. 2002;277:28372–28375. doi: 10.1074/jbc.C200324200. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rorth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, et al. Drosophila cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 2000;19:4533–4542. doi: 10.1093/emboj/19.17.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M, Campuzano S, Garcia-Bellido A. Developmental parameters of cell death in the wing disc of Drosophila. Proc Natl Acad Sci U S A. 1997;94:5691–5696. doi: 10.1073/pnas.94.11.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M, Pιrez L, Cohen SM. Short-range cell interactions and cell survival in the Drosophila wing. Dev Cell. 2002;2:797–805. doi: 10.1016/s1534-5807(02)00169-7. [DOI] [PubMed] [Google Scholar]
- Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002a;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002b;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: A TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Rudolph B, Littlewood T. Action of Myc in vivo: Proliferation and apoptosis. Curr Opin Genet Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Ras1 promotes cellular growth in the Drosophila wing. Cell. 2000;100:435–446. doi: 10.1016/s0092-8674(00)80679-0. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1–S progression from an associated transcriptional program. Genes Dev. 1997;15:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin LA, Tamai K, Seale P, Wagner J, Rudnicki MA. Caspase 3 cleavage of the Ste20-related kinase SLK releases and activates an apoptosis-inducing kinase domain and an actin-disassembling region. Mol Cell Biol. 2000;20:684–696. doi: 10.1128/mcb.20.2.684-696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA. Why size matters: Altering cell size. Curr Opin Genet Dev. 2002;12:565–571. doi: 10.1016/s0959-437x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–215. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Stocker H, Hafen E. Genetic control of cell size. Curr Opin Genet Dev. 2000;10:529–535. doi: 10.1016/s0959-437x(00)00123-4. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey K, Bell D, Wahrer D, Schiripo T, et al. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Wagner S, Flood T, O'Reilly P, Hume K, Sabourin LA. Association of the Ste20-like kinase SLK with the microtubule: Role in Rac1-mediated regulation of actin dynamics during cell adhesion and spreading. J Biol Chem. 2002;277:37685–37692. doi: 10.1074/jbc.M205899200. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan DJ. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, inivasan A, et al. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]