Abstract
Trichomonas vaginalis is one of a few eukaryotes that have been found to encode several homologues of flavodiiron proteins (FDPs). Widespread among anaerobic prokaryotes, these proteins are believed to function as oxygen and/or nitric oxide reductases to provide protection against oxidative/nitrosative stresses and host immune responses. One of the T. vaginalis FDP homologues is equipped with a hydrogenosomal targeting sequence and is expressed in the hydrogenosomes, oxygen-sensitive organelles that participate in carbohydrate metabolism and assemble iron-sulfur clusters. The bacterial homologues characterized thus far have been dimers or tetramers; the trichomonad protein is a dimer of identical 45-kDa subunits, each noncovalently binding one flavin mononucleotide. The protein reduces dioxygen to water but is unable to utilize nitric oxide as a substrate, similarly to its closest homologue from another human parasite Giardia intestinalis and related archaebacterial proteins. T. vaginalis FDP is able to accept electrons derived from pyruvate or NADH via ferredoxin and is proposed to play a role in the protection of hydrogenosomes against oxygen.
Flavodiiron proteins (FDPs) constitute a recently established superfamily of soluble enzymes, thus far exclusively found in anaerobic and facultative aerobic organisms (2, 19, 54). Originally, the function ascribed to these proteins was the reduction of molecular oxygen to water as reported for Desulfovibrio gigas rubredoxin:oxygen oxidoreductase, the first thoroughly characterized protein of this type. This protein was found to utilize electrons derived from glycolysis for safe, four-electron reduction of dioxygen, thus protecting the anaerobic bacterium from the deleterious effects of oxidative stress (19). Later, some of these proteins were also shown to be involved in the reduction of nitric oxide in addition to their oxygen-reducing activity, thereby probably protecting the microbial organism against NO released during the immune response of the higher eukaryote host. The ratio of FDP activity toward oxygen and NO may differ substantially in various organisms; in some cases, FDP is almost exclusively reactive with oxygen, in others it is reactive with NO (20, 21, 43).
FDPs are modular proteins, with flavodoxin-like and metallo-β-lactamase-like domains as their core modules. This two-domain structure is found in the simplest and most common members of the family, named class A FDPs. These proteins are the terminal elements of a multicomponent electron transporting chain that uses the reducing power of NAD(P)H to reduce and detoxify dioxygen and/or nitric oxide (41). Proximal electron donors to most class A FDPs are soluble electron transfer proteins. In the class A FDP rubredoxin:oxygen oxidoreductase from the sulfate-reducing bacterium Desulfovibrio gigas, the electron donor is a small protein, rubredoxin, that itself is reduced by an NADH:rubredoxin oxidoreductase (9, 10, 22). Besides rubredoxin, roles for other iron-sulfur flavoproteins in electron transport to FDPs have been suggested in several Archaea (41); coenzyme F420H2 is the electron donor for the FDP in the methanogenic archaeon Methanothermobacter marburgensis (44). The members of other FDP classes have additional domains fused to the C terminus that participate in electron transfer from the ultimate donor molecule [NAD(P)H] to the terminal electron acceptor (41).
While originally believed to be restricted solely to prokaryotes, recent progress in genome sequencing projects have revealed homologous protein sequences in the genomes of several “amitochondriate” anaerobic protists, mostly with parasitic lifestyles, such as Trichomonas, Giardia, Entamoeba, Spironucleus, and a free-living Mastigamoeba (1, 2, 33, 42). Giardia intestinalis is the only eukaryotic organism to have had data on its FDP published recently. In line with what is known for the prokaryotic homologues, the giardial protein was shown to possess high oxygen (but not NO)-reducing activity and was therefore proposed to participate in protection against oxidative stress (13).
Trichomonas vaginalis is an anaerobic (or microaerophilic) protozoan parasite causing human trichomoniasis, the most common nonviral sexually transmitted infection (38), for which oxygen concentrations higher than those encountered in situ in the vagina (i.e., concentrations above ∼60 μM) are toxic (17). The glucose metabolism of T. vaginalis is compartmentalized; while the reactions of classical glycolysis producing lactate, as well as the branch resulting in the formation of glycerol (8, 48) occur in the cytosol, a substantial portion of glycolytic carbon is diverted into the hydrogenosome, a mitochondrion-related organelle where the reactions of extended glycolysis produce additional ATP by oxidative decarboxylation of pyruvate (47, 48). Typical in the trichomonad hydrogenosome is the presence of the iron-sulfur (FeS) cluster-containing enzymes pyruvate:ferredoxin oxidoreductase (PFOR), hydrogenase, and the electron carrier ferredoxin, which are involved in the generation of molecular hydrogen using electrons released from pyruvate (36). PFOR and hydrogenase are highly oxygen-sensitive enzymes (29, 32), and it is likely that the sensitivity of trichomonads to oxygen could at least in part be due to the inactivation of these key hydrogenosomal proteins.
T. vaginalis must cope with low oxygen concentrations in its natural environment and, accordingly, possesses defense mechanisms to combat oxidative damage caused by oxygen itself or by reactive oxygen species that arise either enzymatically or when the reduced prosthetic groups of enzymes such as flavins and FeS clusters come into contact with oxygen. Most eukaryotes utilize glutathione as a key redox buffer and antioxidant, but trichomonads lack this and similar thiols (17). Cysteine has been suggested as a major reducing buffer and antioxidant (17), and it is believed that the organism relies upon cytosolic NADH oxidase (reducing oxygen to water) and NADPH oxidase (reducing oxygen to hydrogen peroxide) to prevent the permeation of oxygen into the hydrogenosomes (31). Proteins of the peroxiredoxin cascade (11) are also important for cytosolic peroxide detoxification. The identified defense mechanisms of hydrogenosomes include superoxide dismutase activity (17, 30) and recently found putative peroxidases that might provide protection against peroxides (39), but the protein that was suggested long ago to be responsible for oxygen uptake and detoxification has never been identified (6).
We describe here the properties of a class A FDP from T. vaginalis hydrogenosomes and suggest its role in the metabolism of oxygen and protection of the organelle.
MATERIALS AND METHODS
Organism.
The T. vaginalis strain T1 (J.-H. Tai, Institute of Biomedical Sciences, Taipei, Taiwan) was grown in Diamond's TYM medium without agar as previously described (35).
TvFDP constructs.
Several versions of recombinant TvFDP were prepared.
(i) His-FDP.
His-FDP is a recombinant TvFDP without the hydrogenosomal targeting sequence fused with a His6 tag at the C terminus. The PCR-amplified gene was cloned into the pQE 30 Escherichia coli expression system (Qiagen). The purified protein was used for TvFDP characterization, polyclonal antiserum preparation, spectroscopic analysis, and functional studies.
(ii) TvFDPHa.
TvFDP with the hydrogenosomal targeting sequence was fused with a C-terminal 2x hemagglutinin (Ha) tag. The complete TvFDP gene was cloned into the Master Neo (25) vector (kindly provided by Patricia J. Johnson, University of California, Los Angeles, CA) and used to transform trichomonad cells. This construct was used to immunolocalize the protein inside the cell.
(iii) TvFDPStrep.
TvFDP with the hydrogenosomal targeting sequence and fused with a streptavidin tag at the C terminus was expressed in the TagVag (12) vector (T. vaginalis expression system). TvFDPStrep protein was isolated from transformed trichomonads and used to identify the flavin cofactor.
(iv) TvFDPWT.
TvFDP protein with the hydrogenosomal targeting sequence was overexpressed in trichomonads (Master Neo vector) without any additional tags. Hydrogenosomes with TvFDPWT were used to determine the native molecular mass of TvFDP.
Isolation of hydrogenosomes and partial purification of TvFDP.
Hydrogenosomes were isolated from the T. vaginalis homogenate by differential centrifugation of sonicated cells, followed by isopycnic centrifugation of the hydrogenosome-enriched fraction on a self-forming gradient of 45% Percoll (Sigma) as previously described (49).
TvFDP was identified in the T. vaginalis hydrogenosomes during the search for an NADH:ferredoxin oxidoreductase (which was eventually found to be an extremely reduced homologue of mitochondrial respiratory complex I) (25). Briefly, hydrogenosomes from 2 to 4 liters of culture were resuspended in a morpholinepropanesulfonic acid-phosphate buffer (40 mM morpholinepropanesulfonic acid, 400 mM KH2PO4, 2 mM EDTA, 10% glycerol [pH 7.0]) with protease inhibitors (2 mg of leupeptin and N-α-tosyl-l-lysine chloromethyl ketone/ml) and then extracted by sonication (using a Vibra cell sonicator) three times for 1 min each time at an amplitude of 60, with 1-s pulses. The sonicate was centrifuged at 200,000 × g for 45 min to obtain a soluble hydrogenosomal matrix. This fraction was diluted 2.5 times with 20 mM Tris-HCl (pH 7.5) and loaded onto an SP-Sepharose FastFlow (GE Healthcare) column. Bound protein was eluted with 0 to 1 M gradient of NaCl in the same buffer. The fractions containing NADH:ferredoxin oxidoreductase activity (monitored as described in reference 25) were collected, diluted three times with 20 mM Tris-HCl (pH 9.0), and loaded onto a Reactive Blue 2 Sepharose (Sigma) column equilibrated with the same buffer. The protein was eluted with an NaCl gradient (0 to 1 M), and the active fractions were pooled and concentrated by ultrafiltration. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis revealed that the sample was particularly enriched in two barely separable polypeptides with molecular masses of ∼45 kDa. After transblotting onto a polyvinylidene difluoride membrane and Coomassie blue staining, the enriched bands were cut out and subjected to Edman degradation (performed at the Protein/DNA Technology Center, Rockefeller University, New York, NY) to determine the amino-terminal sequences of the polypeptides. The heavier polypeptide was a homologue of the 51-kDa subunit (also called NuoF in bacteria) of the electron-input module of respiratory complex I (25), while the lighter polypeptide was found to be a homologue of bacterial FDPs and named TvFDP. Edman degradation also allowed determination of the processing site where the hydrogenosomal targeting peptide is cleaved from the preprotein by a hydrogenosomal processing peptidase.
Expression and purification of TvFDP.
To express the His-tagged protein (His-FDP) in E. coli M15 cells, the bacteria were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown for 7 h at 28°C in LB medium supplemented with 400 μM ammonium ferrous sulfate and 200 μM flavin mononucleotide (FMN). The harvested cells were homogenized by passage through a French press at 18,000 lb/in2. The soluble fraction obtained by ultracentrifugation (250,000 × g, 1 h, 4°C) was applied to a Ni-NTA column (Qiagen) and eluted with a stepwise gradient of 20 mM imidazole (buffer A: 20 mM imidazole, 50 mM Tris-HCl, 300 mM NaCl, 10% glycerol [pH 7.6]) and 400 mM imidazole (buffer B: 400 mM imidazole, 50 mM Tris-HCl, 300 mM NaCl, 10% glycerol [pH 7.6]) at a flow rate of 1 ml/min using a BioLogic HR system (Bio-Rad). His-FDP-containing fractions were dialyzed overnight against 50 mM Tris-HCl with 10% glycerol (pH 7.6), concentrated (Amicon UltraConcentrator, 30 kDa; Millipore), and analyzed by SDS-PAGE.
Characterization of TvFDP.
The flavin cofactor was characterized by reversed-phase high-pressure liquid chromatography using a nucleosil 100-5 C18 column and a thin-layer chromatography (TLC) method (HPTLC-Alufolien; Merck). The TLC mobile phase consisted of n-butanol-acetic acid-water (6:2:4). The flavin was extracted from the protein with trichloroacetic acid at a final concentration of 10%, followed by centrifugation and supernatant neutralization with 1 M ammonium acetate (pH 7.0). The identity of the flavin was determined for recombinant His-FDP, as well as for TvFDPStrep purified from T. vaginalis hydrogenosomes according to the IBA Strep-tag protein purification protocol. FMN and FAD (Sigma) were used as standards. The amount of flavin in TvFDP was assessed after protein denaturation with 80% trichloroacetic acid, using an extinction coefficient of 12,500 M−1 cm−1 at λ = 450 nm (21).
The iron content of TvFDP (His-FDP was used for analysis) was determined by using the 2,4,6-tripyridyl-1,3,5-triazine method (18).
The native molecular mass of TvFDP was determined by gel filtration chromatography using a BioLogic HR system (Bio-Rad). The purified recombinant His-FDP, as well as the hydrogenosomal extract (prepared as described above) from FDP-overexpressing trichomonads (TvFDPWT), were run on a Superdex 75 (XK 16 column; GE Healthcare) column equilibrated with 400 mM imidazole-50 mM sodium phosphate buffer (pH 8.0) using a flow rate of 1 ml/min. The native molecular mass of the recombinant enzyme was calculated from the calibration curve determined by running the standards under the same conditions. TvFDPWT contained in the hydrogenosomal extract was determined in the elution profile by Western blotting, followed by immunodetection of TvFDP using a specific rabbit polyclonal antiserum raised against His-FDP purified on an Ni-NTA column. The rabbit antiserum was prepared at the Institute of Parasitology, Academy of Sciences of the Czech Republic, České Budějovice, according to a published protocol (53).
Protein concentration was determined by the Lowry assay (34) using bovine serum albumin as a standard.
Spectroscopic analysis of TvFDP.
The redox titrations were performed anaerobically at 25°C under an argon atmosphere by the stepwise addition of buffered sodium dithionite (250 mM Tris-HCl [pH 9.0]), in the presence of suitable redox mediators, in 50 mM Tris-HCl-18% glycerol (pH 7.5) as previously described (51). The samples were analyzed by visible spectroscopy (Shimadzu UV-1603 spectrophotometer) or by electron paramagnetic resonance (EPR) spectroscopy using an EMX Bruker spectrometer equipped with an Oxford Instruments ESR900 continuous flow cryostat. EPR spectra were recorded at 10 K, 9.39 GHz, 2.0 mW, and 1 mT of modulation amplitude. The electrodes (a silver/silver chloride combined electrode, or silver chloride and platinum electrodes) were calibrated against a saturated quinhydrone solution at pH 7.0, and the recorded potentials were normalized against that of the standard hydrogen electrode.
Protein localization.
T. vaginalis cells overexpressing TvFDPHa were used for immunodetection of TvFDP. The cells were placed on glass slides coated with 3-aminopropyltriethoxysilane (Sigma), fixed with methanol (5 min), permeabilized with acetone (5 min) (both steps at −18°C), preincubated for 1 h in phosphate-buffered saline-0.25% bovine serum albumin-0.25% gelatin, and treated with antibodies as described previously (49). Anti-Ha tag monoclonal antibody (kindly provided by Patricia Johnson, University of California at Los Angeles) and hydrogenosomal malic enzyme polyclonal antiserum (15) were used as the primary antibodies. Anti-mouse immunoglobulin G (IgG) labeled with Alexa Fluor 488 (catalog no. A21202; Molecular Probes) and anti-rabbit IgG labeled with Alexa Fluor 549 (catalog no. A21207; Molecular Probes) were used as the secondary antibodies for fluorescent immunolocalization. Anti-TvFDP rabbit polyclonal serum (see above) and anti-rabbit IgG antibody conjugated with alkaline phosphatase (ICN/Cappel) were used for Western blot analysis to visualize TvFDP in T. vaginalis subcellular fractions obtained from untransformed, wild-type trichomonads and TvFDPWT-overexpressing trichomonads.
Enzymatic reduction of TvFDP.
TvFDP spectra were recorded on a Shimadzu UV-1601 spectrophotometer. The enzymatic reduction of TvFDP (His-FDP construct, 30 to 150 nmol) was monitored in stoppered cuvettes with a silicone septum in 860 μl of phosphate buffer (100 mM KH2PO4/KOH, 150 mM NaCl, 10% glycerol [pH 7.4]) using 44 mM pyruvate, 0.25 mM coenzyme A (CoA), ∼40 μg of T. vaginalis ferredoxin 1 (52), and ∼10 μg of T. vaginalis PFOR as elements of the reduction pathway. Alternatively, another enzymatic system consisting of ∼10 μg of T. vaginalis complex I homologue purified from hydrogenosomes (25), ∼40 μg of T. vaginalis ferredoxin 1 (52), and 220 μM NADH in the same buffer as described above was used, but the spectra obtained with this system were less informative due to partially overlapping absorptions of NADH and TvFDP. Anaerobiosis of the system was achieved by degassing the buffer followed by flushing with N2 and the addition of glucose oxidase (28 U), catalase (104 U), and glucose (3 mM) into the reaction mixture (50). This enzymatic oxygen- and hydrogen peroxide-scavenging system was omitted from experiments aimed to identify the product of oxygen reduction by TvFDP.
H2O2 was determined by the FOX method using xylenol orange as an indicator in a spectrophotometric assay at 560 nm. This assay is able to reliably detect 100 pmol of hydrogen peroxide in a 50-μl sample (55). Approximately 150 nmol of TvFDP was reduced by the enzymatic system (see above) and briefly exposed to air to oxidize TvFDP; 50 μl of the reaction mixture was mixed with 950 μl of FOX1 reagent (100 μM xylenol orange, 250 μM ammonium ferrous sulfate, 100 mM sorbitol, 25 mM H2SO4) (55). Hydrogen peroxide (0 to 250 pmol) was used to construct the calibration curve and as a positive control.
DPTA-NONOate (Alexis Biochemicals, Switzerland) was used as a nitric oxide donor to prepare solutions of 12 μM to 1 mM of nitric oxide in the reaction mixture (37). The presence of chemically generated NO in the reaction mixture was verified by using a membrane-type NO-sensitive electrode (ISO-NOP, World Precision Instruments) connected to an ISO-NO Mark II meter (World Precision Instruments). Possible inactivation of the enzymatic reduction system by NO was excluded by checking for persistent TvFDP-reducing activity after anaerobic exposure to NO followed by reoxidation of TvFDP with air and resealing of the cuvette.
Purification of PFOR.
In order to obtain a homologous enzymatic system capable of reducing TvFDP, T. vaginalis PFOR was purified as follows: hydrogenosomes from 4 liters of culture were treated with 2% octylglucoside (MP Biomedicals) in 10 mM KH2PO4 (pH 6.8) on ice for 60 min and then centrifuged at 120,000 × g for 30 min. The resulting membrane pellet containing PFOR was extracted with 5 ml of degassed and nitrogen-equilibrated 1% deoxycholate (Sigma) in 10 mM KH2PO4-2 mM dithiothreitol (pH 7.4) for 60 min, followed by centrifugation at 120,000 × g for 30 min. The supernatant fraction was mixed with 1 ml of Ni-NTA agarose (Qiagen), followed by incubation for 60 min at 4°C. The mixture was loaded onto a disposable column and the flowthrough fraction containing PFOR plus minor contaminants was collected. This step was found to selectively remove the malic enzyme, the dominant hydrogenosomal protein with a native molecular mass close to that of PFOR (240,000 Da), due to specific binding of the malic enzyme to Ni-NTA agarose. The PFOR-enriched eluate was concentrated and loaded at a rate of 0.5 ml/min onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with degassed 10 mM KH2PO4-2 mM dithiothreitol-1% deoxycholate (pH 7.4). Fractions (750 μl) were collected, analyzed by SDS-PAGE, and checked for PFOR activity (24). The fraction with the strongest activity was used to reduce TvFDP in combination with T. vaginalis ferredoxin 1 that was expressed in E. coli BL21 cells and purified as described earlier (49).
RESULTS
Purification and properties of TvFDP.
Recombinant TvFDP with a His6 tag and without the hydrogenosomal targeting sequence (His-FDP) produced in M15 E. coli cells was purified by affinity chromatography on Ni-NTA agarose under native conditions. This procedure produced an almost homogeneous protein, as determined by SDS-PAGE (Fig. 1), with an approximate yield of 10 mg of His-FDP per liter of bacterial culture. The His-FDP polypeptide migrated on SDS-PAGE as a band with a molecular mass of ∼45 kDa, a finding in agreement with the molecular mass calculated from the amino acid sequence without the hydrogenosomal targeting peptide.
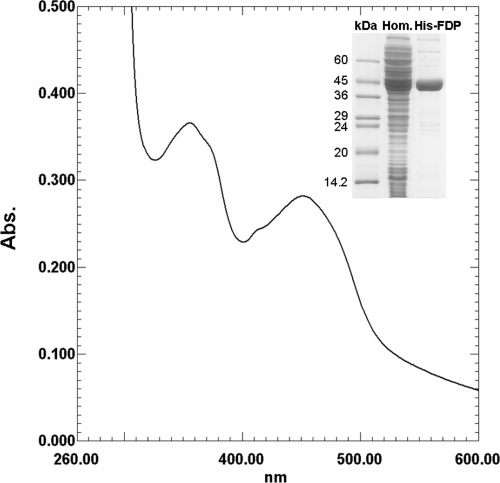
FIG. 1.
Spectroscopic analysis of recombinant T. vaginalis His-FDP. UV/VIS spectrum of pure His-FDP exhibits the typical features of flavoproteins. The inset shows the results of an SDS-PAGE analysis of purified recombinant His-FDP. Hom., bacterial lysate; FDP, purified His-FDP.
To determine the native molecular mass of TvFDP, the hydrogenosomal extract from transformed T. vaginalis cells overexpressing native, nontagged TvFDPWT, as well as the recombinant His-FDP isolated from E. coli, were analyzed by gel filtration chromatography. The native molecular mass of His-FDP was ∼92 kDa. The TvFDPWT in the elution profile was determined by SDS-PAGE and Western blot analysis with a specific antibody. TvFDPWT from the hydrogenosomal extract was eluted in the same elution volume as the purified recombinant protein (results not shown), suggesting that both the recombinant and the plasmid-encoded TvFDPWT expressed in trichomonads exist as dimers in vivo.
The cofactor contained in the flavodoxin domain was identified by TLC and high-pressure liquid chromatography as noncovalently bound FMN (data not shown). This cofactor was established for both the recombinant protein and the protein containing the Strep tag (TvFDPStrep) isolated by affinity chromatography from T. vaginalis hydrogenosomes. Freshly isolated recombinant protein contained ∼0.5 FMN and ∼1.5 iron atoms per monomer. This stoichiometry indicates that a fully occupied protein contains one molecule of FMN and two iron atoms per monomer; the incomplete occupancy is probably due to cofactor loss during purification. Pure, concentrated His-FDP was dark yellow and displayed a typical FDP UV/VIS spectrum (Fig. 1) dominated by the features of the flavin moiety, as the diiron center has a very low absorption.
Sequence analysis.
TvFDP displayed homology with other described FDPs from both prokaryotic and eukaryotic sources. The residues implicated in binding the binuclear iron center (His94, His161, His240, Glu96, Asp98, and Asp180), as well as the conservative flavodoxin-like signature motif starting at position 271 (Val271) (Fig. 2) (41, 54), were all conserved in the T. vaginalis protein.
FIG. 2.
Amino acid sequence of TvFDP (XP001583562, TVAG_036010). The residues binding the diiron ligands are marked with an asterisk (*), and the conserved flavodoxin signature motif is shaded. The position of the hydrogenosomal processing peptidase cleavage site is marked with an arrow.
Soluble hydrogenosomal proteins are synthesized in the cytoplasm and typically contain a short amino acid presequence with a targeting function on their amino termini (3, 14). These targeting signals are cleaved by a specific hydrogenosomal processing peptidase (4) upon translocation of the protein into the organelle. TvFDP also possesses such a signal. The processing peptidase cleavage site was determined by amino acid sequencing via Edman degradation of mature TvFDP partially purified from hydrogenosomes; the cleavage site was found to be located between serine 11 and alanine 12 (Fig. 2). The conserved arginine residue typically located in the −2 position relative to the cleavage site in hydrogenosomal presequences was in an unusual −4 position in this case.
Subcellular localization.
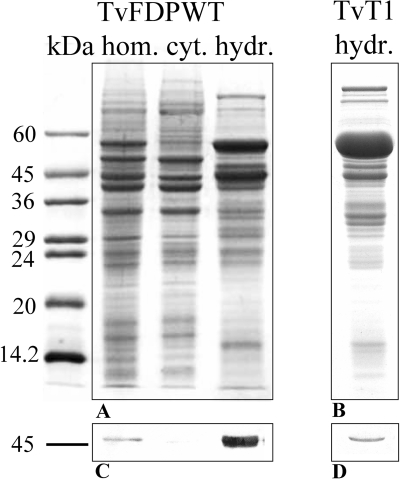
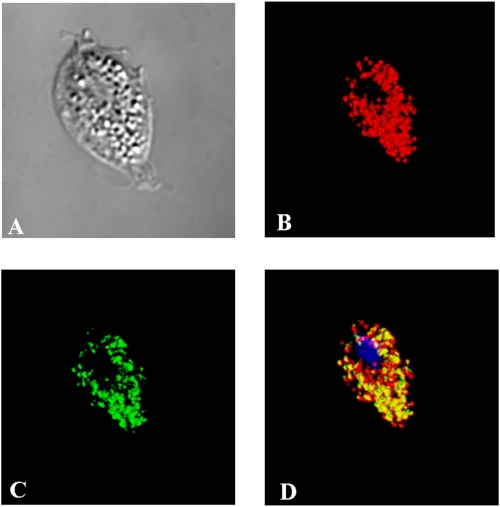
His-FDP was used to raise a specific antiserum that was subsequently utilized to determine the localization of native, nuclear-encoded TvFDP, as well as overexpressed TvFDPWT within trichomonad cells. Western blot analysis of T. vaginalis subcellular fractions showed that the TvFDP was expressed and specifically localized to the hydrogenosomal fraction (Fig. 3). To further verify the localization of TvFDP protein by immunofluorescence microscopy, T. vaginalis cells overexpressing TvFDPHa with an Ha tag at the C terminus were used. The protein was colocalized with the malic enzyme (hydrogenosomal marker enzyme) in the hydrogenosomal compartment (Fig. 4).
FIG. 3.
Subcellular localization of TvFDP. (A) SDS-PAGE of subcellular fractions of transformed T. vaginalis line overexpressing TvFDP (TvFDPWT); (B) SDS-PAGE of hydrogenosomes of untransformed T. vaginalis (TvT1); (C and D) Western blots probed with anti-TvFDP polyclonal antiserum. Molecular mass standards are indicated in kilodaltons. hom., homogenate; cyt., cytosol; hydr., hydrogenosomes.
FIG. 4.
Immunodetection of TvFDP in T. vaginalis cells. (A) Nomarski differential contrast; (B) visualization of malic enzyme, hydrogenosomal marker; (C) TvFDP labeling; (D) merge of color channels showing the presence of TvFDP in the hydrogenosomes with DAPI (4′,6′-diamidino-2-phenylindole) staining for nuclei.
Spectroscopic studies.
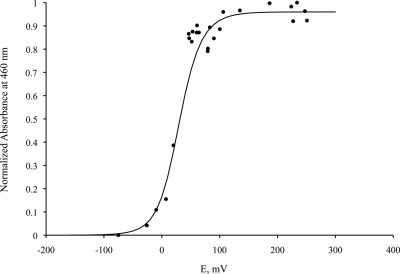
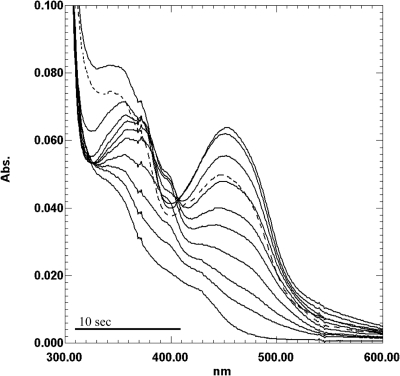
The visible spectrum of TvFDP was characteristic of a flavoprotein, with the main absorption bands at about 455 and 350 nm. Upon chemical reduction with sodium dithionite, there was no clear indication of the formation of a semiquinone radical, either of the blue or the red type, suggesting that the semiquinone form is not stabilized. Indeed, redox titration monitored by visible spectroscopy yielded a curve that could be fitted with two identical reduction potentials of approximately +25 mV for the FMNox/FMNSQ and the FMNSQ/FMNred forms (Fig. 5). However, during the course of the enzymatic reduction by PFOR, a small band appeared at 398 nm during the early stages of the reduction (Fig. 6). Such a peak has not been observed in any of the other FDPs studied thus far, and its origin remains to be clarified.
FIG. 5.
Redox titration of the flavin cofactor monitored by visible spectroscopy at 460 nm. The full line was calculated for two consecutive one-electron redox processes, using molar absorptivities for the oxidized and semiquinone forms of 14,400 and 4,500, respectively, and with E1 = E2 = 25 mV.
FIG. 6.
Enzymatic reduction of TvFDP. Spectra of gradual reduction of His-FDP were obtained in an anaerobic system consisting of 44 mM pyruvate, 0.25 mM CoA, T.vaginalis PFOR (10 μg), and recombinant T. vaginalis ferredoxin (40 μg) in a phosphate buffer (100 mM KH2PO4/KOH, 150 mM NaCl, 10% glycerol [pH 7.4]). The dashed spectrum indicates reoxidized His-FDP after introduction of air into the cuvette. The recording time for one spectrum was 38 s.
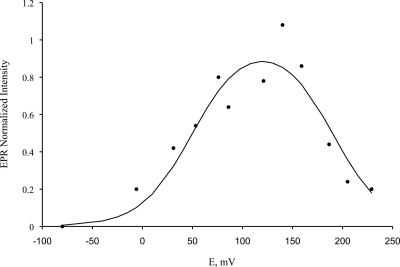
Redox titration, followed by EPR spectroscopy, allowed us to both observe the signature of the diiron center and to determine its reduction potentials (Fig. 7 and Fig. 8). The diiron center had resonances at g = 1.955, 1.805, and 1.57, which is characteristic of the mixed valence [Fe(III)-Fe(II)] state of a diiron site. By measuring the change in amplitude of the resonance at g = 1.8, the complete redox titration was constructed (Fig. 8) and analyzed with a Nernstian system of two consecutive one-electron transitions, yielding reduction potentials for the Fe(III)-Fe(III)/Fe(III)-Fe(II) and Fe(III)-Fe(II)/Fe(II)-Fe(II) transitions of 50 ± 20 and 190 ± 20 mV.
FIG. 7.
EPR spectrum of as-purified TvFDP at 10 K. Microwave frequency, 9.39 GHz; microwave power, 2.0 mW; modulation amplitude, 1 mT.
FIG. 8.
Redox titration of the diiron center monitored by EPR spectroscopy, following the changes in EPR intensity at the gmed value of the diiron center. The full line was also calculated for two one-electron reduction processes and corresponds to the formation and disappearance of the EPR resonance of the Fe(III)-Fe(II) mixed valence state, with reduction potentials of 190 and 50 mV.
Function.
In order to establish the physiological function of TvFDP, we studied the recombinant, His-tagged protein (His-FDP), which could be obtained in large quantities and displayed properties (i.e., FMN and iron cofactor, UV/VIS spectrum, and dimeric structure) similar to those of known FDPs. Studies of flavoproteins often utilize sodium dithionite as a reductant of the flavin moiety. This approach, however, is of little use when studying the physiological function of a protein whose presumed electron acceptor is oxygen. To reduce His-FDP enzymatically, we used a system consisting of pyruvate, CoA, PFOR purified from T. vaginalis hydrogenosomes, and purified recombinant T. vaginalis ferredoxin 1 (52). The primary electron source was pyruvate in a CoA-dependent PFOR reaction that oxidatively decarboxylates pyruvate, transfers electrons to ferredoxin, and releases acetyl-CoA. The reduced ferredoxin then served as a reductant for His-FDP in an anaerobic spectrophotometric assay. Catalytic amounts of PFOR and ferredoxin 1 fully reduced His-FDP in minutes. After complete reduction of His-FDP, the absorbance peak at 455 nm was totally bleached (Fig. 6). Upon introduction of air into the reaction mixture by opening the cuvette, His-FDP was immediately reoxidized (as documented by the regression of its spectral features to those of the native protein [Fig. 6]), indicating that an electron acceptor of TvFDP is indeed oxygen that is reduced with high affinity. When the cuvette was resealed again, the reduction of His-FDP resumed, indicating that the enzymatic system was not damaged by the short exposure to oxygen. To determine the product of the oxygen reduction, approximately 150 nmol of His-FDP was first fully enzymatically reduced as described above and then reoxidized with air. The mixture was subsequently analyzed by using the FOX method (55) for the presence of hydrogen peroxide. No hydrogen peroxide was detected, indicating that the product of oxygen reduction by TvFDP is water.
Since the ability of certain bacterial FDPs to reduce nitric oxide has been well documented (21, 40, 45, 46), we tested the NO-reducing activity of His-FDP; NO was chemically generated by using DPTA-NONOate. The presence of NO in the reaction mixture was verified by a NO-specific electrode connected to an ISO-NO Mark II meter. Approximately 30 nmol of His-FDP was first enzymatically reduced and then combined with DPTA-NONOate to obtain 12 μM to 1 mM NO in an anaerobic assay mixture. No change in the spectrum of the reduced protein was observed with any concentration of NO tested over a period of 15 min (data not shown), indicating that TvFDP has negligible, if any, reactivity toward nitric oxide.
DISCUSSION
T. vaginalis hydrogenosomes harbor oxygen-sensitive proteins and are accordingly equipped with defensive enzymes to neutralize oxidative stress. These protective proteins include superoxide dismutase (17) and putative peroxidases such as thiol-dependent peroxidase and rubrerythrin (39). However, the enzyme that is logically first in the line of defense, oxygen reductase, has remained elusive since its presence was first proposed in the 1970s (6). In the present study, we describe the properties of an FDP from T. vaginalis (TvFDP) that functions as a true oxygen reductase. This protein, first identified by proteomic methods (see Materials and Methods) during our search for other hydrogenosomal activities, localized to hydrogenosomes as demonstrated by immunoblotting with specific antibodies. The subcellular localization of TvFDP was further confirmed by immunofluorescence microscopy of transformed trichomonads expressing a tagged version of the enzyme. The organellar localization is consistent with the presence of a cleavable hydrogenosomal targeting peptide with a characteristic amino acid composition on the amino terminus of the conceptually translated protein (Fig. 2).
Analysis of cofactor content suggests that TvFDP binds two iron atoms and one FMN per monomer as in other homologues. Like most of its counterparts that have already been studied in this respect, native TvFDP is an ∼92-kDa homodimer of identical 45-kDa subunits. This dimeric quaternary structure is essential for efficient electron transfer from the flavin of one monomer to the diiron center of the other monomer; these regions come into close contact due to the head-to-tail organization of each monomer, a feature common to all FDPs whose structures have been determined thus far. Within a single monomer, the redox centers are too far apart to allow electron transfer at a physiologically relevant rate (50). The reduction potentials determined here for the FMN and the diiron center are higher than those observed for the E. coli enzyme, the only FDP extensively characterized in this respect thus far (50, 51). Another remarkable characteristic was observed in the reduction potentials of the flavin transitions, which were almost identical; this has not been reported for any other FDP thus far. This precludes the formation of a stable flavin semiquinone form as observed in other FDPs and favors an almost two-electron oxidation of the flavin. Nevertheless, the measured reduction potentials are suitable for either NO or O2 reduction, and further studies in other FDPs are needed to determine whether these differences are enzymatically relevant. Due to the high reduction potentials of either substrate, the specificity toward these small molecules cannot be related to their redox parameters. The UV-visible and EPR spectroscopic data are similar to those of other FDPs characterized thus far. The data for the diiron center showed that the iron ions were antiferromagnetically coupled in the half-reduced state, yielding a total spin of S = 1/2. For the oxidized and reduced forms, it was not possible to obtain any EPR spectral signature; in the oxidized state the total spin state is most probably S = 0, while in the reduced form it could be either zero or S = 4 as observed for the E. coli enzyme.
In order to determine the activity and substrate specificity of TvFDP, we needed to reconstruct in vitro the electron transfer chain from an electron donor to the terminal acceptor, oxygen and/or nitric oxide. This was a nontrivial task, since T. vaginalis does not possess rubredoxin, the physiological reductant of some FDPs, and analysis of the T. vaginalis genome did not provide any clue about potential redox partners for TvFDP. Nevertheless, T. vaginalis encodes a large number of [2Fe-2S] ferredoxins, small electron carrier proteins linked to PFOR, hydrogenase, and a remnant of complex I (28). In addition, the genome also encodes several predicted hydrogenosomal iron-sulfur flavoproteins with unknown function but presumably involved in electron transfer (5). We have found that the enzymatic system consisting of PFOR purified from T. vaginalis hydrogenosomes and a homogeneous preparation of recombinant T. vaginalis ferredoxin 1 could effectively reduce TvFDP at the expense of electrons derived from pyruvate by the activity of PFOR. Strictly anaerobic conditions had to be used to follow the reduction of the flavin cofactor of TvFDP and also to prevent autooxidation of ferredoxin in the spectrophotometric assay. Alternatively, another redox system could be used to reduce TvFDP; this system consists of a purified hydrogenosomal remnant of complex I and recombinant T. vaginalis ferredoxin 1. In this case, the electron donor is NADH, but since its absorbance partially interfered with the absorbance of oxidized FMN of TvFDP in a spectrophotometric assay, the PFOR-based system was preferentially used. Under these experimental conditions, full reduction of TvFDP, monitored as disappearance of the absorbance of FMN, was achieved within a few minutes. The reaction time depended on the concentration of PFOR and ferredoxin in the assay (Fig. 6). Upon introduction of air into the cuvette, TvFDP immediately reoxidized as documented by the regression of the spectral curve to the original, oxidized pattern (Fig. 6). The immediate reoxidation of the flavin cofactor indicates a fast reaction with a high affinity for oxygen; however, the kinetic parameters of the electron transfer could not be determined due to the autooxidative nature of the trichomonad ferredoxin, which precluded a continuous aerobic spectrophotometric assay.
To function as a protective, O2 scavenging enzyme, TvFDP should reduce oxygen by four electrons to water. The product of oxygen reduction was determined by the FOX assay with enzymatically reduced His-TvFDP that was reoxidized by short exposure to air. No traces of hydrogen peroxide were detected, indicating that TvFDP indeed reduces dioxygen to water using four molecules of the one-electron carrier ferredoxin per cycle. It should be noted that FDPs do not reduce hydrogen peroxide (M. Teixeira, unpublished data). Experiments designed to verify the potential reactivity of enzymatically reduced TvFDP with nitric oxide, the known substrate of several members of the flavodiiron superfamily, did not show any such activity for the trichomonad protein, despite the broad range of NO concentrations and prolonged reaction times tested. This is similar to what was found for the only other eukaryotic FDP characterized, the protein from G. intestinalis, which is also virtually unreactive with NO (13).
Analysis of the primary structure of TvFDP shows that the enzyme belongs to the class A family of FDPs. Members of this family are the simplest representatives of the superfamily, consisting of only the core flavodiiron module, which is formed by a metallo β-lactamase-like domain in the amino-terminal half of the protein and by a flavodoxin-like domain forming the rest of the polypeptide chain (41). The mature (without the hydrogenosomal targeting peptide) trichomonad protein is colinear over its entire length with its homologues, with conservation of all residues implicated in binding the binuclear iron center and a well-conserved flavodoxin signature motif at the start of the flavodoxin-like domain (Fig. 2).
Attempts have been made to attribute the selective specificity of some FDPs for oxygen (and not NO) to particular amino acid residues (44). This issue has been discussed in the light of data recently obtained for Giardia FDP (13), and we can only conclude that the results concerning both the Giardia (13) and the Trichomonas FDPs (the present study) invalidate the original hypothesis (44) and leave the question of the substrate specificity of FDPs unsettled. In addition, tryptophan 361 (W347 in reference 44), which is missing in the FDP from M. marburgensis but conserved in rubredoxin-specific FDPs, has been proposed to play a role in electron shuttling between rubredoxin and the FMN of rubredoxin-specific FDPs (44). However, neither Trichomonas nor Giardia FDPs, which have W361 conserved, have rubredoxins as electron donors, since there are no genes encoding these proteins (http://www.trichdb.org/trichdb/; http://www.giardiadb.org/giardiadb/). The absence of this tryptophan in Methanothermobacter FDP might be related to the nature of the proposed electron donor to FDPs from methanogens, enabling easy access to F420H2.
FDPs have already been subjected to a phylogenetic study, as particularly good candidates for genes that have been acquired by certain unicellular eukaryotes via lateral gene transfer from prokaryotic sources (1). Eukaryotic FDP homologues have thus far been invariably found in the genomes of anaerobic fermentative protists, most with parasitic lifestyles. This suggests that the role of FDPs in these eukaryotes is similar to that in prokarytes, i.e., protection against oxidative/nitrosative stress. However, the only eukaryotic homologue studied to date is the FDP from G. intestinalis (13). Giardia (a member of Diplomonadida) is believed to share a common ancestor with trichomonads (23), and the T. vaginalis protein described in the present study is the closest relative to the giardial homologue (which, however, is natively a tetramer) (1), suggesting that this particular gene entered this eukaryotic lineage before the divergence of diplomonads and trichomonads. Notably, the sister group to diplomonad/trichomonad FDPs consists of the branch of archaebacterial FDPs of methanogens (1), some of which have been shown to utilize only oxygen and not nitric oxide (44), like the homologues from both human parasites.
Class A FDP is not the only FDP present in T. vaginalis. In fact, four genes encoding FDP homologues could be identified in the genome of this parasite (5) (http://www.tigr.org/tdb/e2k1/tvg; locus numbers TVAG_036010, TVAG_129610, TVAG_049830, and TVAG_263800). One of these proteins is the subject of the present study; the remaining three are highly similar homologues that belong to a strongly separated clade and form a group with other eukaryotic sequences from Entamoeba and Mastigamoeba and with eubacterial sequences from Clostridium sp. (1). Unlike hydrogenosomal TvFDP, the genes of these three trichomonad proteins do not encode a clear amino-terminal extension with an organellar targeting function (and were not detected in the proteome of hydrogenosomes, unpublished data); thus, these putative proteins are likely localized to the cytoplasm. Because of their novel primary structure, these trichomonad homologues were proposed to constitute a fourth, D class of FDPs, together with clostridial proteins, that have an NADH:rubredoxin oxidoreductase and rubredoxin module fused to the flavodiiron core and could probably directly utilize NAD(P)H as an electron donor (50). In this respect, it should be mentioned that nitric oxide reducing activity has been detected in intact as well as lysed T. vaginalis cells; this activity has been ascribed to FDP partly because a cross-reactive band was visualized in a T. vaginalis cell lysate using a heterologous antibody against E. coli flavorubredoxin (42). Since the molecular size of the denatured candidate protein (∼60 kDa) was far from the predicted size of a class D FDP monomer (∼95 kDa), its identity is doubtful. Nevertheless, if these class D FDP homologues are indeed expressed, the observed nitric oxide reducing activity (42) might be due to their presence in the T. vaginalis cytosol, where they could provide protection against the host immune response as well as against oxygen.
Finally, it should be stated that despite the evidence collected in vitro using a functional electron transport chain consisting of the T. vaginalis hydrogenosomal proteins PFOR, ferredoxin 1, and TvFDP (or complex I, ferredoxin 1, and TvFDP), the identity of the actual physiological components of the presumed pathway remains uncertain. This is largely due to the complexity of the predicted electron transporting pathways of hydrogenosomes. The core electron-generating reactions utilizing malate and pyruvate as the carbon and electron source, as coined by Müller (36), are still valid, but research over the last 15 years, as well as the recently completed annotation of the T. vaginalis genome (5), has shown that most already identified hydrogenosomal proteins involved in electron generation and transport are coded by multiple and clearly distinct genes. These include seven genes encoding PFOR homologues, up to nine malic enzymes, seven ferredoxins, seven iron-sulfur flavoproteins, and up to nine putative hydrogenases, of which four are equipped with a predicted hydrogenosomal targeting peptide; many of these proteins are transcribed, also as indicated by complex EPR spectroscopic data (7, 16, 26-28; unpublished proteomic data). It is possible that certain PFOR homologues preferentially use particular ferredoxins, while others could function as electron acceptors of complex I or in iron-sulfur cluster assembly machinery (49). The involvement of iron-sulfur flavoproteins even in these known reactions also cannot be excluded, as indicated by preliminary experiments (unpublished data). Other possible sources of electrons besides pyruvate and NADH (generated by the malic enzyme), such as amino acids or α-glycerophosphate (6), could also play a role. Some combinations of substrate, electron-generating enzyme, and electron-transporting protein would likely be more efficient than others in shuttling electrons toward TvFDP.
In summary, by fusing a hydrogenosomal targeting peptide to a class A FDP, most likely obtained from an anaerobic bacterium by lateral gene transfer, T. vaginalis equipped the hydrogenosomes with a new enzyme. Functional data suggest that this acquisition helped the parasite to cope with oxidative stress in these oxygen-sensitive organelles.
Acknowledgments
This study was supported by the Czech Science Foundation grant 204/06/0944 to I.H. and by Ministry of Education, Youth, and Sports of the Czech Republic grant MSM0021620858. Further support was provided by the Fundação para a Ciência e Tecnologia-Portugal, projects PTDC/BIA-PRO/67263/2006, and Structure, Dynamics and Functions of Proteins (REEQ/336/BIO/2005). V.L.G. is the recipient of a Ph.D. grant from Fundação para a Ciência e Tecnologia-Portugal (SFRH/BD/29428/2006).
We acknowledge the help of J. Vicente (Instituto de Tecnologia Química e Biológica) with the EPR-monitored redox titration. We thank Míša Marcinčiková (Charles University) for excellent technical assistance, Petr Jedelsky (Charles University) for mass spectrometry analyses, and Jan Mach (Charles University) for help with the figures.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Andersson, J. O., R. P. Hirt, P. G. Foster, and A. J. Roger. 2006. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol. Biol. 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, J. O., A. M. Sjogren, L. A. M. Davis, T. M. Embley, and A. J. Roger. 2003. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 1394-104. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, P. J., C. J. Lahti, E. Plümper, and P. J. Johnson. 1997. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 163484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. T., H. M. H. Goldstone, F. Bastida-Corcuera, M. G. Gadillo-Correa, A. G. McArthur, and P. J. Johnson. 2007. A functionally divergent hydrogenosomal peptidase with protomitochondrial ancestry. Mol. Microbiol. 641154-1163. [DOI] [PubMed] [Google Scholar]
- 5.Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, J. R. Wortman, S. L. Bidwell, U. C. M. Alsmark, S. Besteiro, T. Sicheritz-Ponten, C. J. Noel, J. B. Dacks, P. G. Foster, C. Simillion, Y. Van de Peer, D. Miranda-Saavedra, G. J. Barton, G. D. Westrop, S. Müller, D. Dessi, P. L. Fiori, Q. H. Ren, I. Paulsen, H. B. Zhang, F. D. Bastida-Corcuera, A. Simoes-Barbosa, M. T. Brown, R. D. Hayes, M. Mukherjee, C. Y. Okumura, R. Schneider, A. J. Smith, S. Vanacova, M. Villalvazo, B. J. Haas, M. Pertea, T. V. Feldblyum, T. R. Utterback, C. L. Shu, K. Osoegawa, P. J. D. Jong, I. Hrdy, L. Horvathova, Z. Zubacova, L. Horvathova, S. B. Malik, J. M. Logsdon, K. Henze, A. Gupta, C. C. Wang, R. L. Dunne, J. A. Upcroft, P. Upcroft, O. White, S. L. Salzberg, P. Tang, C. H. Chiu, Y. S. Lee, T. M. Embley, G. H. Coombs, J. C. Mottram, J. Tachezy, C. M. Fraser-Liggett, and P. J. Johnson. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerkasov, J., A. Cerkasovova, J. Kulda, and D. Vilhelmova. 1978. Respiration of hydrogenosomes of Tritrichomonas foetus. 1. Adp-dependent oxidation of malate and pyruvate. J. Biol. Chem. 2531207-1214. [PubMed] [Google Scholar]
- 7.Chapman, A., D. J. Linstead, and D. Lloyd. 1999. Hydrogen peroxide is a product of oxygen consumption by Trichomonas vaginalis. J. Biosci. 24339-344. [Google Scholar]
- 8.Chapman, A., D. J. Linstead, D. Lloyd, and J. Williams. 1985. C-13-NMR reveals glycerol as an unexpected major metabolite of the protozoan parasite Trichomonas vaginalis. FEBS Lett. 191287-292. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L., M. Y. Liu, J. LeGall, P. Fareleira, H. Santos, and A. V. Xavier. 1993. Purification and characterization of an NADH-rubredoxin oxidoreductase involved in the utilization of oxygen by Desulfovibrio gigas. Eur. J. Biochem. 216443-448. [DOI] [PubMed] [Google Scholar]
- 10.Chen, L., M. Y. Liu, J. LeGall, P. Fareleira, H. Santos, and A. V. Xavier. 1993. Rubredoxin oxidase, a new flavo-hemo-protein, is the site of oxygen reduction to water by the strict anaerobe Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 193100-105. [DOI] [PubMed] [Google Scholar]
- 11.Coombs, G. H., G. D. Westrop, P. Suchan, G. Puzova, R. P. Hirt, T. M. Embley, J. C. Mottram, and S. Müller. 2004. The amitochondriate eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system. J. Biol. Chem. 2795249-5256. [DOI] [PubMed] [Google Scholar]
- 12.Delgadillo, M. G., D. R. Liston, K. Niazi, and P. J. Johnson. 1997. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 944716-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Matteo, A., F. M. Scandurra, F. Testa, E. Forte, P. Sarti, M. Brunori, and A. Giuffre. 2008. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 2834061-4068. [DOI] [PubMed] [Google Scholar]
- 14.Dolezal, P., O. Smid, P. Rada, Z. Zubacova, D. Bursac, R. Sutak, J. Nebesarova, T. Lithgow, and J. Tachezy. 2005. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc. Natl. Acad. Sci. USA 10210924-10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drmota, T., P. Proost, M. VanRanst, F. Weyda, J. Kulda, and J. Tachezy. 1996. Iron-ascorbate cleavable malic enzyme from hydrogenosomes of Trichomonas vaginalis: purification and characterization. Mol. Biochem. Parasitol. 83221-234. [DOI] [PubMed] [Google Scholar]
- 16.Dyall, S. D., W. H. Yan, M. G. gadillo-Correa, A. Lunceford, J. A. Loo, C. F. Clarke, and P. J. Johnson. 2004. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature 4311103-1107. [DOI] [PubMed] [Google Scholar]
- 17.Ellis, J. E., N. Yarlett, D. Cole, M. J. Humphreys, and D. Lloyd. 1994. Antioxidant defenses in the microaerophilic protozoan Trichomonas vaginalis: comparison of metronidazole-resistant and sensitive strains. Microbiol. SGM 1402489-2494. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, D. S., and D. C. Price. 1964. Simple serum iron method using new sensitive chromogen tripyridyl-S-triazine. Clin. Chem. 1021-31. [PubMed] [Google Scholar]
- 19.Frazao, C., G. Silva, C. M. Gomes, P. Matias, R. Coelho, L. Sieker, S. Macedo, M. Y. Liu, S. Oliveira, M. Teixeira, A. V. Xavier, C. Rodrigues-Pousada, M. A. Carrondo, and J. Le Gall. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 71041-1045. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, A. M., R. A. Helmick, and P. R. Gardner. 2002. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification Escherichia coli. J. Biol. Chem. 2778172-8177. [DOI] [PubMed] [Google Scholar]
- 21.Gomes, C. M., A. Giuffre, E. Forte, J. B. Vicente, L. M. Saraiva, M. Brunori, and M. Teixeira. 2002. A novel type of nitric-oxide reductase: Escherichia coli flavorubredoxin. J. Biol. Chem. 27725273-25276. [DOI] [PubMed] [Google Scholar]
- 22.Gomes, C. M., G. Silva, S. Oliveira, J. LeGall, M. Y. Liu, A. V. Xavier, C. Rodrigues-Pousada, and M. Teixeira. 1997. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J. Biol. Chem. 27222502-22508. [DOI] [PubMed] [Google Scholar]
- 23.Hampl, V., D. S. Horner, P. Dyal, J. Kulda, J. Flegr, P. G. Foster, and T. M. Embley. 2005. Inference of the phylogenetic position of oxymonads based on nine genes: support for Metamonada and Excavata. Mol. Biol. Evol. 222508-2518. [DOI] [PubMed] [Google Scholar]
- 24.Hrdy, I., R. Cammack, P. Stopka, J. Kulda, and J. Tachezy. 2005. Alternative pathway of metronidazole activation in Trichomonas vaginalis hydrogenosomes. Antimicrob. Agents Chemother. 495033-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrdy, I., R. P. Hirt, P. Dolezal, L. Bardonova, P. G. Foster, J. Tachezy, and T. M. Embley. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432618-622. [DOI] [PubMed] [Google Scholar]
- 26.Hrdy, I., and M. Müller. 1995. Primary structure and eubacterial relationships of the pyruvate, ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J. Mol. Evol. 41388-396. [PubMed] [Google Scholar]
- 27.Hrdy, I., and M. Müller. 1995. Primary structure of the hydrogenosomal malic enzyme of Trichomonas vaginalis and its relationship to homologous enzymes. J. Eukaryot. Microbiol. 42593-603. [DOI] [PubMed] [Google Scholar]
- 28.Hrdy, I., J. Tachezy, and M. Müller. 2008. Metabolism of trichomonad hydrogenosomes, p. 113-145. In J. Tachezy (ed.), Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes, vol. 9. Springer, New York, NY. [Google Scholar]
- 29.Lindmark, D. G., and M. Müller. 1973. Hydrogenosome, a cytoplasmic organelle of anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 2487724-7728. [PubMed] [Google Scholar]
- 30.Lindmark, D. G., and M. Müller. 1974. Superoxide-dismutase in anaerobic flagellates, Tritrichomonas foetus, and Monocercomonas sp. J. Biol. Chem. 2494634-4637. [PubMed] [Google Scholar]
- 31.Linstead, D. J., and S. Bradley. 1988. The purification and properties of 2 soluble reduced nicotinamide-acceptor oxidoreductases from Trichomonas vaginalis. Mol. Biochem. Parasitol. 27125-134. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd, D., and B. Kristensen. 1985. Metronidazole inhibition of hydrogen-production in vivo in drug-sensitive and resistant strains of Trichomonas vaginalis. J. Gen. Microbiol. 131849-853. [DOI] [PubMed] [Google Scholar]
- 33.Loftus, B., I. Anderson, R. Davies, U. C. M. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433865-868. [DOI] [PubMed] [Google Scholar]
- 34.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 35.Mertens, E., and M. Müller. 1990. Glucokinase and fructokinase of Trichomonas vaginalis and Tritrichomonas foetus. J. Protozool. 37384-388. [DOI] [PubMed] [Google Scholar]
- 36.Müller, M. 1993. The hydrogenosome. J. Gen. Microbiol. 1392879-2889. [DOI] [PubMed] [Google Scholar]
- 37.Nittler, M. P., D. Hocking-Murray, C. K. Foo, and A. Sil. 2005. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol. Biol. Cell 164792-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrin, D., K. Delgaty, R. Bhatt, and G. Garber. 1998. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 11300-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pütz, S., G. Gelius-Dietrich, M. Piotrowski, and K. Henze. 2005. Rubrerythrin and peroxiredoxin: two novel putative peroxidases in the hydrogenosomes of the microaerophilic protozoon Trichomonas vaginalis. Mol. Biochem. Parasitol. 142212-223. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues, R., J. B. Vicente, R. Felix, S. Oliveira, M. Teixeira, and C. Rodrigues-Pousada. 2006. Desulfovibrio gigas flavodiiron protein affords protection against nitrosative stress in vivo. J. Bacteriol. 1882745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saraiva, L. M., J. B. Vicente, and M. Teixeira. 2004. The role of the flavodiiron proteins in microbial nitric oxide detoxification. Adv. Microb. Physiol. 4977-129. [DOI] [PubMed] [Google Scholar]
- 42.Sarti, P., P. L. Fiori, E. Forte, P. Rappelli, M. Teixeira, D. Mastronicola, G. Sanciu, A. Giuffre, and M. Brunori. 2004. Trichomonas vaginalis degrades nitric oxide and expresses a flavorubredoxin-like protein: a new pathogenic mechanism? Cell Mol. Life Sci. 61618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seedorf, H., A. Dreisbach, R. Hedderich, S. Shima, and R. K. Thauer. 2004. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F-420-dependent enzyme involved in O2 detoxification. Arch. Microbiol. 182126-137. [DOI] [PubMed] [Google Scholar]
- 44.Seedorf, H., C. H. Hagemeier, S. Shima, R. K. Thauer, E. Warkentin, and U. Ermler. 2007. Structure of coenzyme F420H2 oxidase (FprA), a di-iron flavoprotein from methanogenic Archaea catalyzing the reduction of O2 to H2O. FEBS J. 2741588-1599. [DOI] [PubMed] [Google Scholar]
- 45.Silaghi-Dumitrescu, R., E. D. Coulter, A. Das, L. G. Ljungdahl, G. N. L. Jameson, B. H. Huynh, and D. M. Kurtz. 2003. A flavodiiron protein and high molecular weight rubredoxin from Moorella thermoacetica with nitric oxide reductase activity. Biochemistry 422806-2815. [DOI] [PubMed] [Google Scholar]
- 46.Silaghi-Dumitrescu, R., K. Y. Ng, R. Viswanathan, and D. M. Kurtz. 2005. A flavo-diiron protein from Desulfovibrio vulgaris with oxidase and nitric oxide reductase activities: evidence for an in vivo nitric oxide scavenging function. Biochemistry 443572-3579. [DOI] [PubMed] [Google Scholar]
- 47.Steinbüchel, A., and M. Müller. 1986. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol. 2057-65. [DOI] [PubMed] [Google Scholar]
- 48.Steinbüchel, A., and M. Müller. 1986. Glycerol, A Metabolic end-product of Trichomonas vaginalis and Tritrichomonas foetus. Mol. Biochem. Parasitol. 2045-55. [DOI] [PubMed] [Google Scholar]
- 49.Sutak, R., P. Dolezal, H. L. Fiumera, I. Hrdy, A. Dancis, M. Delgadillo-Correa, P. J. Johnson, M. Müller, and J. Tachezy. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 10110368-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vicente, J. B., M. C. Justino, V. L. Goncalves, L. M. Saraiva, and M. Teixeira. 2008. Biochemical, spectroscopic, and thermodynamic properties of flavodiiron proteins. Methods Enzymol. 43721-45. [DOI] [PubMed] [Google Scholar]
- 51.Vicente, J. B., and M. Teixeira. 2005. Redox and spectroscopic properties of the Escherichia coli nitric oxide-detoxifying system involving flavorubredoxin and its NADH-oxidizing redox partner. J. Biol. Chem. 28034599-34608. [DOI] [PubMed] [Google Scholar]
- 52.Vidakovic, M. S., G. Fraczkiewicz, and J. P. Germanas. 1996. Expression and spectroscopic characterization of the hydrogenosomal [2Fe-2S] ferredoxin from the protozoan Trichomonas vaginalis. J. Biol. Chem. 27114734-14739. [DOI] [PubMed] [Google Scholar]
- 53.Vondruskova, E., J. van den Burg, A. Zikova, N. L. Ernst, K. Stuart, R. Benne, and J. Lukes. 2005. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 2802429-2438. [DOI] [PubMed] [Google Scholar]
- 54.Wasserfallen, A., S. Ragettli, Y. Jouanneau, and T. Leisinger. 1998. A family of flavoproteins in the domains Archaea and Bacteria. Eur. J. Biochem. 254325-332. [DOI] [PubMed] [Google Scholar]
- 55.Wolff, S. P. 1994. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233182-189. [Google Scholar]