Abstract
Previous studies showed that a Bacillus subtilis strain deficient in mismatch repair (MMR; encoded by the mutSL operon) promoted the production of stationary-phase-induced mutations. However, overexpression of the mutSL operon did not completely suppress this process, suggesting that additional DNA repair mechanisms are involved in the generation of stationary-phase-associated mutants in this bacterium. In agreement with this hypothesis, the results presented in this work revealed that starved B. subtilis cells lacking a functional error prevention GO (8-oxo-G) system (composed of YtkD, MutM, and YfhQ) had a dramatic propensity to increase the number of stationary-phase-induced revertants. These results strongly suggest that the occurrence of mutations is exacerbated by reactive oxygen species in nondividing cells of B. subtilis having an inactive GO system. Interestingly, overexpression of the MMR system significantly diminished the accumulation of mutations in cells deficient in the GO repair system during stationary phase. These results suggest that the MMR system plays a general role in correcting base mispairing induced by oxidative stress during stationary phase. Thus, the absence or depression of both the MMR and GO systems contributes to the production of stationary-phase mutants in B. subtilis. In conclusion, our results support the idea that oxidative stress is a mechanism that generates genetic diversity in starved cells of B. subtilis, promoting stationary-phase-induced mutagenesis in this soil microorganism.
Adaptive or stationary-phase mutagenesis can be defined as those mutations that permit organisms to grow and divide in response to natural or artificial selection (5) and that occur in nondividing cells during prolonged nonlethal selective pressure, e.g., starvation for an essential amino acid (32). Although this type of mutagenesis was first described in Escherichia coli (7), additional examples of adaptive mutagenesis in other prokaryotes (21, 41) and in eukaryotic organisms (14) have been published. In some cases, these mutations occurred in the absence of specific selection but in response to starvation (11). Regardless of the organisms utilized and the name used, these types of mutations and the processes that generate them are of real interest with respect to evolution and the generation of diversity across all domains of life.
Studies with the F′ lac frameshift reversion construct of E. coli (32) have demonstrated that the generation of Lac+ stationary-phase-associated revertants required functional recombination (15), as well as component(s) of the SOS system (24, 25). Further evidence suggests that the mutations generated by this lac system during stationary phase may also be the result of amplification of the plasmid-borne gene followed by SOS-induced mutagenesis and selection (18, 38). Recent studies have demonstrated that DNA double-strand-break repair, in addition to the SOS DNA damage response and the error-prone DNA polymerase, are necessary for stress-induced reversion of the E. coli F′ lac frameshift reversion construct (17, 28).
Because of its ability to form endospores, Bacillus subtilis has represented for several decades an excellent paradigm for studying prokaryotic differentiation. Interestingly, mutations that interfere with sporulation were shown in many cases to also affect other processes taking place in post-exponential-phase or transitional-growth-phase cells, such as competence, motility, secondary metabolism, and resistance to adverse environmental conditions (39). Since genetic diversity has been associated with stressed cell populations of B. subtilis (13, 19, 39), it was not a surprise to discover that mutagenesis also occurs in the post-exponential-phase of this gram-positive bacterium (41). This mutagenic process has been demonstrated in B. subtilis by means of an assay system that measures reversion in the hisC952, metB5, and leuC427 chromosomal alleles (27, 30, 41, 42). This assay system has been very valuable in demonstrating that the mechanisms underlying adaptive mutagenesis are rather different in B. subtilis and E. coli, although some similarities between the two models seem to exist (27, 30, 41, 48). While recombination-associated processes were shown to be strictly required during the generation of Lac+ stationary-phase-associated revertants in E. coli (32), adaptive mutagenesis (i.e., neither recombination nor the activation of type 1 SOS functions) in B. subtilis was not dependent upon a functional RecA protein (41). Surprisingly, the generation of B. subtilis adaptive mutants was not dependent on σB (41), the transcriptional factor which controls the general stress regulon of B. subtilis (44). Interestingly, ComA and ComK, stationary-phase active transcription factors that control competence development, influence the production of stationary-phase-induced mutants (41), and more recently, it was demonstrated that Mfd, a transcriptional repair coupling factor, is a component of this mutagenesis process (33).
It has been suggested that in B. subtilis cultures, there are cell subpopulations with suppressed DNA repair systems (41, 48) that might be responsible for some or all of the adaptive or stationary-phase-induced mutagenesis. In agreement with this hypothesis, a strain deficient in mutSL, encoding the mismatch DNA repair system of B. subtilis, showed a significant propensity to generate increased numbers of stationary-phase-induced revertants (27). Moreover, a significant decrease in the number of adaptive revertant colonies occurred in B. subtilis cells that overexpressed the mutSL operon (27). Interestingly, the single overexpression of mutS but not of mutL was sufficient to decrease the level of adaptive mutants in the reversion assay system of B. subtilis. A homolog of the MutS protein was also found to be involved in the generation of stationary-phase-induced mutants of Saccharomyces cerevisiae (14). However, in E. coli, overexpression of MutL but not of MutS decreased the number of mutations generated by the adaptive process(es) without affecting growth-dependent mutagenesis (16).
Oxidative stress is a major producer of 8-oxo-dGTP, and this oxidized precursor is frequently incorporated opposite adenine in DNA (37). However, direct oxidation of DNA also generates 8-oxo-G, which induces GC→TA and AT→CG transversions (43). The mutagenic effects of 8-oxo-G are counteracted by the base excision repair pathway utilizing the DNA glycosylases MutM and MutY (26). The former releases 8-oxo-G from 8-oxo-G:C pairs, and the latter removes adenine from 8-oxo-G:A mismatches (26). The mutagenic effects of 8-oxo-dGTP are also prevented by MutT, which hydrolyzes this triphosphate to its monophosphate version, blocking 8-oxo-G incorporation into DNA (23). The contributions of MutM, MutY, and MutT greatly reduce the mutagenic effects of 8-oxo-G in E. coli, and together these proteins make up the oxidized guanine (GO) system (12). The results of expression and genetic complementation studies of ytkD and mutT of B. subtilis suggested that these genes encode potential 8-oxo-dGTPases that can prevent the mutagenic effects of 8-oxo-dGTP (29). Recent results revealed that the loss of YtkD but not MutT increased the spontaneous mutation frequency of growing B. subtilis cells. However, cells lacking both YtkD and MutT had a higher spontaneous mutation frequency than cells lacking YtkD (8). Moreover, the loss of either YtkD or MutT sensitized growing cells to hydrogen peroxide (H2O2) and tert-butylhydroperoxide (t-BHP), and the lack of both proteins sensitized growing cells to these agents even more (8). B. subtilis cells contain a complete GO system, since it has been demonstrated that in addition to ytkD and mutT, mutM and yfhQ (which encode potential homologs of E. coli MutM and MutY, respectively) are found in the sequenced genome (35).
Our previous results showed that overexpression of the mutSL operon did not completely suppress the production of stationary-phase-induced mutations in B. subtilis (27). This result indicated that either not enough mismatch repair (MMR) proteins were generated or additional mechanisms are involved in the generation of stationary-phase mutations. To test the latter hypothesis, we investigated here whether the GO system which prevents and/or repairs oxidation-induced DNA damage is an additional component of the adaptive-mutagenesis process of B. subtilis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used are listed in Table 1. B. subtilis YB955 is a prophage-“cured” strain that contains the hisC952 (CAG→TAG at position 952; amber mutation), metB5 (GAA→TAA at position 346; ochre mutation), and leuC427 (GGA→AGA at position 427; Gly to Arg) alleles (41, 46, 47). The procedures for transformation and isolation of chromosomal and plasmid DNA were as described previously (2, 9, 34). B. subtilis strains were maintained on tryptose blood agar base medium (Difco Laboratories, Detroit, MI). Liquid cultures of B. subtilis strains were routinely grown in antibiotic medium no. 3 (A3) (Difco Laboratories, Detroit, MI) amended with different antibiotics. When required, neomycin (10 μg/ml), tetracycline (10 μg/ml), spectinomycin (100 μg/ml), chloramphenicol (5 μg/ml), rifampin (5 μg/ml), or isopropyl-β-d-thiogalactopyranoside (IPTG; 1.0 mM/ml) was added to the medium. E. coli cultures were grown in Luria-Bertani (LB) medium supplemented with ampicillin to a final concentration of 100 μg/ml.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or descriptiond | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| PERM590 | E. coli XL10-Gold carrying plasmid pPERM590 (Ampr Kmr) | 8 |
| PERM594 | E. coli XL10-Gold carrying plasmid pPERM594 (Ampr Kmr) | 8 |
| B. subtilis | ||
| PS832 | Wild-type; trp+ revertant of strain 168 | Laboratory stock |
| YB955 | hisC952 metB5 leuC427 xin-1 SpβSENS | 42 |
| PERM563 | ΔytkD::neo Neor | pPERM590→YB955b |
| PERM564 | ΔmutT::tet Tetr | pPERM594→YB955b |
| PERM597 | ΔytkD::neo ΔmutT::tet Neor Tetr | pPERM594→PERM563b |
| PERM598c | ΔyfhQ::spc Spcr | P. Setlow |
| PERM599c | ΔmutM::tet Tetr | P. Setlow |
| PERM571 | ΔmutM::tet Tetr | PS599→YB955a |
| PERM572 | ΔmutM::tet ΔyfhQ::spc Spcr Tetr | PERM599→PERM571a |
| PERM573 | ΔytkD::neo DmutM::tet ΔyfhQ::spc Neor Spcr Tetr | pPERM590→PERM572a |
| PERM731 | ΔytkD::neo DmutM::tet ΔyfhQ::spc with a Pspac-mutSL-lacI construct from pMPR002 inserted into the amyE locus; Neor Spcr Tetr Cmr | pMPR002→PERM573a |
| Plasmids | ||
| pDG148 | spac expression vector; Ampr Kmr | W. L. Nicholson |
| pDG364 | Integration vector (promotes ectopic integration into the amyE locus); Ampr Cmr | 9 |
| pPERM590 | pBEST501 containing 798-bp HindIII-SalI PCR fragment encompassing 589 bp upstream and 209 bp downstream of the ytkD translational start codon and an 866-bp BamHI-SacI PCR fragment encompassing 131 bp upstream and 735 bp downstream of the ytkD translational stop codon; Ampr | 8 |
| pPERM594 | PDG1515 containing the 986-bp BamHI-XbaI fragment encompassing 904 bp upstream and 82 bp downstream of the mutT translational start codon and 980-bp EcoRI-HindIII fragment encompassing 193 bp upstream and 787 bp downstream of the mutT translational ending codon; Ampr | 8 |
| pMPR001 | ∼4.5-kb B. subtilis mutSL open reading frame cloned into the SalI/XbaI sites of pDG148 | This study |
| pMPR002 | pDG364 with an ∼6.28-kb EcoRI/BamHI fragment from pMPR001 containing the Pspac-mutSL-lacI construct | This study |
Chromosomal DNA from the strain to the left of the arrow was used to transform the strain to the right.
Plasmid DNA from the strain to the left of the arrow was used to transform the strain to the right.
The background for this strain is PS832.
Amp, ampicillin; Km, kanamycin; Neo, neomycin; Tet, tetracycline; Spc, spectinomycin; Cm, chloramphenicol.
Construction of mutant strains.
Knockouts in the genes of interest were constructed by marker exchange between the chromosome- and plasmid-borne alleles. Plasmids pPERM590 and pPERM594 (8) were used to transform B. subtilis strain YB955, generating strains PERM563 (ytkD) and PERM564 (mutT). The ytkD mutT double mutant in the YB955 background was generated by transforming strain PERM595 with plasmid pPERM564 (resulting in strain PERM597). The yfhQ mutM ytkD triple knockout was constructed as follows. Genomic DNA isolated from B. subtilis PERM599 (mutM) was used to transform strain B. subtilis YB955, generating B. subtilis PERM571 (mutM). Competent cells of this strain were transformed with genomic DNA isolated from B. subtilis PERM598 (yfhQ), generating strain PERM572 (yfhQ mutM). Finally, this strain was transformed with plasmid PERM590, giving rise to strain PERM573 (ytkD mutM yfhQ) (Table 1). The double-crossover events leading to inactivation of the appropriate genes were confirmed by PCR (data not shown).
A B. subtilis yfhQ mutM ytkD mutant overexpressing the mutSL operon was constructed as follows. The open reading frame of the mutSL operon was PCR amplified and cloned into the XbaI/SalI sites of pDG148 under the control of the IPTG-inducible Pspac promoter. The resulting plasmid (pMPPR001) was amplified in E. coli XL10-GOLD Kanr (Stratagene, Cedar Creek, TX). The mutSL operon under the Pspac promoter was released with EcoRI and BamHI and cloned into the integrative vector pDG364 to generate pMPR002. This plasmid was amplified in E. coli DH5α and was introduced by transformation into the B. subtilis strain PERM573 to generate strain B. subtilis PERM731 (Table 1).
Stationary-phase mutagenesis assays.
Essentially, cultures were grown in flasks containing A3 medium with aeration (250 rpm) at 37°C until 90 min after the cessation of exponential growth. Growth was monitored with a Pharmacia Ultrospec 2000 spectrophotometer set at 600 nm. The stationary-phase mutagenesis assays were performed as previously described (27, 41) on solid Spizizen minimal medium (SMM; 1× Spizizen salts supplemented with 0.5% glucose and either 50 μg or 200 ng of the required amino acid/ml and 50 μg each of isoleucine and glutamic acid/ml). The concentration of the amino acid used depended on the reversion that was being selected. For instance, when selecting for His+ revertants, 50 μg of methionine and leucine/ml and 200 ng of histidine/ml were added to the medium. Isoleucine and glutamic acid were added as described previously (40) in order to protect the viability of the cells. When required, the selection medium was supplemented with 1.0 mM IPTG/ml. The number of revertants was scored daily. The initial number of bacteria plated for each experiment was determined by serial dilution of the bacterial cultures and then by plating the cells on a minimal medium containing all three essential amino acids. The experiments were repeated at least three times.
The survival of the bacteria plated on the minimal selective medium was determined as follows. Three agar plugs were removed from each selection plate daily. The plugs were removed with sterile Pasteur pipettes and taken from areas of the plates where no growth of revertants was observed. The plugs were suspended in 400 μl of 1× Spizizen salts mixed, diluted, and plated on SMM containing all the essential amino acids (50 μg/ml). Again, the number of colonies was determined following 48 h of growth at 37°C.
Analysis of mutation rates.
The growth-dependent reversion rates for the His+, Met+, Leu+, and rifampin resistance phenotypes were measured by fluctuation tests with the Lea-Coulson formula, r/m − ln(m) = 1.24 (22). Three parallel cultures were used to determine the total number of CFU plated on each plate (Nt) by titration. The mutation rates were calculated as previously described with the formula m/2Nt (27, 31, 41). Strains were grown in A3 medium supplemented with the proper antibiotics for 12 h. Mutation frequencies were determined by plating aliquots on either five plates with LB medium containing rifampin to a final concentration of 5 μg/ml or five plates with SMM agar prepared as described above to select revertant colonies with His+, Met+, or Leu+ phenotypes. The revertant colonies were counted after 24 to 48 h of incubation at 37°C. These experiments were repeated at least three times (27).
RESULTS
Generation of stationary-phase mutants in a ytkD mutT-deficient B. subtilis strain.
ytkD and mutT of B. subtilis encode potential 8-oxo-dGTPases that can prevent the mutagenic effects of 8-oxo-dGTP (8). In fact, the lack of either YtkD or MutT sensitized growing cells of those strains to the oxidative-stress agents H2O2 and t-BHP and, as expected, the lack of both proteins (the double-mutant strain) showed an even more dramatic effect (Fig. 1) on the production of stationary-phase mutants. These results strongly suggested that YtkD and MutT work together to protect B. subtilis cells from the DNA damage induced by oxidative stress, as previously reported for vegetative cells (8).
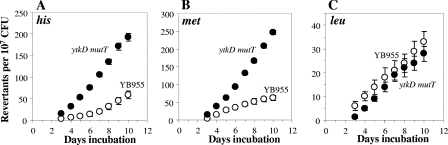
FIG. 1.
Stationary-phase-induced reversions for his (A), met (B), and leu (C) of YB955 and PERM597 (ytkD mutT) B. subtilis strains as described in Materials and Methods. Results show the average number of accumulated revertants in six selection plates; error bars show one standard error. This experiment was performed at least three times.
We used a reversion system to study stationary-phase-associated mutagenesis in B. subtilis (30, 41) to analyze the production of His+, Met+, and Leu+ revertant colonies in a mutant ytkD mutT strain. The results revealed that the loss of the YtkD and MutT proteins increased (by approximately three times) the number of His+ revertants compared to the number obtained for the isogenic-repair-proficient parent strain YB955 (Fig. 1A). Similarly, four times the number of Met+ revertant colonies were observed in the mutant strain as in YB955 (Fig. 1B). On the other hand, the ytkD mutT double mutation did not significantly increase the number of Leu+ revertant colonies compared to the number for the parental strain (Fig. 1C).
Generation of stationary-phase mutants in a ytkD mutM yfhQ-deficient B. subtilis strain.
In E. coli bacteria, some of the potential mutagenic effects of 8-oxo-dGTP are counteracted by MutT, which degrades the oxidized nucleotides to the corresponding monophosphate form, thus avoiding its incorporation into DNA (23). B. subtilis seems to contain a complete GO system since, in addition to ytkD and mutT, which encode putative 8-oxo-dGTPases (8), its genome also has mutM and yfhQ genes, encoding potential homologs of E. coli MutM and MutY, respectively (35). MutM and MutY counteract the mutagenic effects of oxidized bases on DNA. The MutM (Fpg) protein is a glycosylase which removes the 8-oxo-dGTP that has become incorporated opposite cytosine residues, whereas MutY, also a glycosylase, removes adenine when incorporated opposite 8-oxo-guanine, guanine, or cytosine (12).
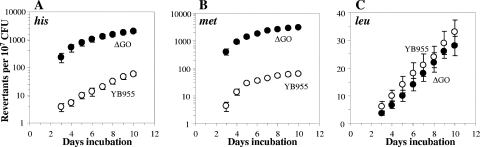
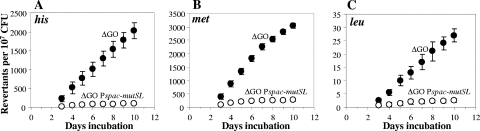
As described above, YtkD and MutT were shown to be involved in protecting cells in the stationary phase of B. subtilis from oxidative-stress-induced DNA damage. However, it was considered essential to know the effect of genetically eliminating the complete GO system from the strain YB955 on the generation of adaptive revertant colonies. To this end, a ytkD mutM yfhQ triple mutant was constructed in the genetic background B. subtilis YB955. As shown in Fig. 2, YB955 cells lacking a GO system had a dramatic propensity to increase the number of His and Met revertants during stationary phase. Specifically, with respect to the parental strain YB955, the ytkD mutM yfhQ mutant showed increases of around 22 and 50 times in the number of His and Met revertant colonies, respectively, generated during the stationary phase. Again, no significant differences were found between the ytkD mutM yfhQ mutant and its parental strain in the number of colonies with the Leu+ phenotype (Fig. 2). As previously described, both the hisC952 and metB5 alleles in strain YB955 are nonsense mutations (41, 46, 47); therefore, we investigated what types of mutations (nonsense suppressors versus true revertants) gave rise to the His+ and Met+ revertants in the GO-deficient strain. The results of this analysis revealed that 98 out of 100 His+ colonies that were picked on day five after initial plating were also Met+, whereas 94 out of 100 Met+ colonies were also His+, suggesting that the His and Met revertants generated during stationary phase in the GO-deficient strain were mainly the results of ochre suppressor mutations.
FIG. 2.
Stationary-phase-induced reversions for his (A), met (B), and leu (C) of YB955 and PERM573 (ytkD mutM yfhQ) (ΔGO) B. subtilis strains as described in Materials and Methods. Results show the average number of accumulated revertants in six selection plates; error bars show one standard error. This experiment was performed at least three times.
Our results revealed that the ytkD mutT double and ytkD mutM yfhQ triple mutants had survival rates similar to that of the parental YB955 strain during the time course of the experiments (data not shown). Thus, no net increase or decline in viable cell counts for the three strains was observed. In conclusion, the increase in the number of revertant His+ and Met+ colonies observed in both mutant strains cannot be attributed to differences in growth or survival with respect to the growth or survival of the parental strain.
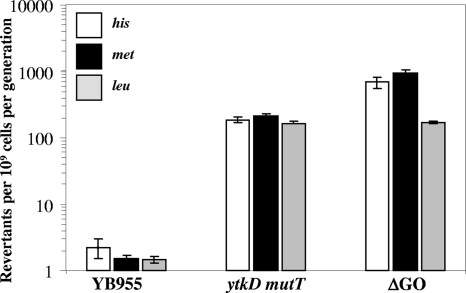
The growth-dependent mutation rates of the mutT ytkD and ytkD mutM yfhQ strains for the generation of His+, Met+, and Leu+ colonies were also determined and compared with those of the parental strain YB955. The results shown in Fig. 3 revealed that the mutation rates of the mutT ytkD double- and ytkD mutM yfhQ triple-knockout strains were in fact greater than those obtained for the strain YB955. However, it was evident that the mutation frequencies of the mutT ytkD and ytkD mutM yfhQ strains for both the his and met alleles, were two and three orders of magnitude higher during the stationary phase than in the exponential phase of growth, respectively. In contrast to the results of the assays in stationary phase, partial or full inactivation of the GO system significantly increased Leu+ reversion in cells engaged in active growth (Fig. 3).
FIG. 3.
Analysis of mutation rates. B. subtilis strains YB955, PERM597 (ytkD mutT), and PERM573 (ytkD mutM yfhQ) (ΔGO) were tested for their ability to produce His+, Met+, and Leu+ revertants during exponential growth as described in Materials and Methods. The mutation rates were calculated with the formula m/2Nt as previously described (22, 27, 41). Results show the average mutation rates from two individual fluctuation tests; error bars represent one standard error.
Growth-dependent and stationary-phase mutagenesis in a GO-deficient strain that overexpresses the mutSL operon.
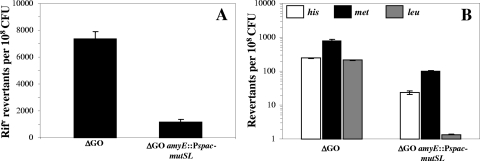
The MMR system of E. coli has been implicated in preventing growth-dependent-mutagenesis induced by oxidative stress (45). In fact, MutS and MutY have been shown to cooperatively interact to prevent the mutagenic effects caused by oxidative damage (1). Accordingly, we investigated whether overexpression of the MMR system of B. subtilis suppresses the hypermutagenic phenotype of the ytkD mutM yfhQ strain. A single copy of the mutSL operon under the control of the Pspac promoter was recombined into the amyE locus of the GO-deficient B. subtilis strain (Table 1). As previously described, IPTG was added to induce the transcription of mutSL from the spac promoter in this strain (27). The frequencies of spontaneous mutation to rifampin resistance were determined for this strain and compared to those obtained with the GO-deficient strain. Overexpression of the mutSL operon resulted roughly in a sixfold decrease in the rate of spontaneous mutation to rifampin resistance of the ytkD mutM yfhQ strain (Fig. 4A); this effect was not observed when the hypermutagenic strain only contained the pDG364 vector (data not shown). In addition, as shown in Fig. 4B, the frequencies of growth-dependent reversion to the His+, Met+, and Leu+ phenotypes decreased significantly when the mutSL operon was overexpressed in the GO-deficient strain. Taken together, our results suggest that during growth the MMR system processes not only the mismatches generated by replication errors but also those promoted by oxidative stress.
FIG. 4.
Growth-dependent reversion to rifampin resistance (A) or to His+, Met+, and Leu+ (B) of PERM573 (ytkD mutM yfhQ) (ΔGO) and PERM731 (ΔGO amyE::Pspac-mutSL). Results show the average number of accumulated revertants in six different selection plates; error bars show one standard error. This experiment was performed at least three times.
Our previous results showed that the MMR system is an important factor in the generation of stationary-phase-associated mutations, most probably due to its saturation by base mispairing occurring in starved B. subtilis cells (27). Moreover, as shown above, the amount of colonies with His+ and Met+ phenotypes increased dramatically in the strain lacking a proficient GO system (Fig. 2). These results suggest that base mismatches generated by oxidative stress during stationary phase may contribute substantially to saturation of the MMR system. In agreement with this hypothesis, our results revealed that with respect to the ytkD mutM yfhQ strain, the number of stationary-phase-associated His+ and Met+ revertants decreased by more than 1 order of magnitude in the GO-deficient strain that overexpressed mutSL (Fig. 5). Moreover, as shown in Fig. 5, overexpression of mutSL significantly decreased the number of colonies with a Leu+ phenotype in the GO-deficient strain, indicating that other types of DNA damage occur in the leu allele. The results shown in Fig. 5 cannot be attributed to a decrease in the survival rates of the strain B. subtilis PERM731 since we found no significant differences in the survival rates between this strain and the YB955 parental strain (data not shown).
FIG. 5.
Stationary-phase-induced reversions for his (A), met (B), or leu (C) of PERM573 (ytkD mutM yfhQ) (ΔGO) and PERM731 (ΔGO amyE::Pspac-mutSL) B. subtilis strains as described in Materials and Methods. Results show the average number of accumulated revertants in six different selection plates; error bars show one standard error. This experiment was performed at least three times.
DISCUSSION
Stationary-phase-induced mutagenesis of B. subtilis is a developmentally regulated process (41, 48); it has been suggested that subpopulations with suppressed DNA repair systems (30, 41) might be responsible for some or all of the adaptive or stationary-phase-induced mutagenesis in this bacterium. In agreement with this contention, our previous results suggested that MMR is an important process in the generation of mutations in nongrowing B. subtilis bacteria (27). However, overexpression of the mutSL operon did not completely suppress the production of stationary-phase-induced mutants in B. subtilis (27). These results suggested that additional DNA repair mechanisms are involved in the generation of stationary-phase mutants in B. subtilis. Previous studies have associated oxidative stress with stationary-phase mutagenesis in starved cells of E. coli (3, 4, 6) and Pseudomonas putida (36). Therefore, we investigated here whether the error prevention GO system is also part of the mechanisms which control the mutagenesis process in nondividing cells of B. subtilis.
YtkD and MutT encode putative 8-oxo-dGTPases and may function in a cooperative manner to protect B. subtilis cells from the mutagenic effects of oxidative stress (8, 29). To investigate the role played by these proteins in the stationary-phase-induced mutagenesis of B. subtilis, we genetically disrupted ytkD and mutT in the YB955 strain. The rates of spontaneous mutation to rifampin resistance in this knockout strain were around 40 times higher than those of the parental strain (Table 2). Similarly, the growth-dependent mutation rates for the production of His+, Met+, and Leu+ colonies were also higher in the ytkD mutT knockout strain than in the parental YB955 strain (Fig. 3). In the adaptive-mutagenesis reversion assay system, as shown in Fig. 1A and B, the inactivation of the ytkD and mutT genes increased the number of His+ and Met+ colonies by three to four times with respect to the number of revertants produced by the parental strain. Interestingly, the level of induction as measured by the number of His+ and Met+ colonies was very similar to that observed in YB955 cells lacking the MMR MutSL system (27). However, with respect to the YB955 strain, the absence of MutSL but not of YtkD/MutT increased the number of Leu+ colonies (27; this work).
TABLE 2.
Spontaneous mutation frequencies of YB955, ytkD mutT, and ytkD mutM yfhQ strains of B. subtilis
| Strain | Mean Rifr mutation frequency ± SD (10−9) | Relative frequency |
|---|---|---|
| YB955 | 1.7 ± 0.82 | 1 |
| ytkDmutT | 75.47 ± 0.63 | 44.39 |
| ytkDmutM yfhQ | 3,746 ± 11.6 | 2,203 |
Altogether, our results suggest that suppression of YtkD and MutT may increase the production of adaptive mutants due to the accumulation of 8-oxo-dGTP, a nucleotide analog that upon incorporating into the DNA increases the frequency of G-T tranversions (37). Previous results revealed that YtkD of B. subtilis is a protein which preferentially catalyzes the hydrolysis of 8-oxo-dGTP and 8-oxo-GTP over the respective nonoxidized substrates (29). Therefore, based on the fact that YtkD also processes the degradation of the mRNA precursor 8-oxo-GTP, the suppression of YtkD and/or absence of MutT during stationary phase could also favor transcriptional errors caused by the incorporation of 8-oxo-GTP opposite adenine in the nascent mRNA.
In agreement with our results, by using a reversion assay system to study adaptive mutagenesis in E. coli bacteria, Bridges (3) found that a mutation in mutT increased the rate of appearance of prototrophic revertants when E. coli tyrA14 (ochre) or trpA23 strains were incubated under starvation conditions. In fact, the rate of appearance of prototrophic revertants during the stationary phase in the mutT mutant of E. coli was at least 1 order of magnitude higher than the rate of production of His+ and Met+ adaptive colonies in the ytkD mutT strain of B. subtilis. However, it must be pointed out that, in contrast to results for E. coli (23), the inactivation of ytkD alone or in combination with mutT did not confer a strong mutator phenotype on B. subtilis (Table 2) (8). Therefore, either B. subtilis relies on proteins in addition to YtkD and MutT to hydrolyze oxidized nucleotides or the combined action of YtkD, MutT, and the DNA glycosylases MutM and YfhQ is sufficient to counteract the mutagenic effects induced by these potentially mutagenic precursors. We investigated this possibility in the context of the stationary-phase-induced mutagenesis of B. subtilis by constructing a triple ytkD mutM yfhQ knockout strain. Our results revealed that the absence of these genes induced a strong mutagenic phenotype in B. subtilis cells. Thus, the spontaneous mutation frequency to rifampin resistance was around 2,000 times higher in the ytkD mutM yfhQ mutant than in the parental YB955 strain (Table 2). In agreement with this result, the mutation rates of exponentially growing cells of the triple knockout for the production of His+, Met+, and Leu+ colonies were around 2 to 3 orders of magnitude higher than the mutation rates of the parental strain YB955 (Fig. 3). The ytkD mutM yfhQ triple mutant was tested by using the adaptive-mutagenesis reversion assay system with B. subtilis. The data obtained revealed a dramatic increase in the number of His+ and Met+ revertant colonies during the stationary phase with respect to the number of colonies produced by the parental strain. This effect was unprecedented since, as described above, the lack of a functional mismatch MutSL system increased, to a lesser extent than presented here, the mutation frequency of the YB955 strain during stationary phase (27). It is important to note that the suppressor analysis indicates that most mutations conferring the Met+ and His+ phenotypes are the result of ochre suppressors, even in the triple-knockout strain. Interestingly, the results of previous experiments that used a proficient strain in the GO prevention system and examined the accumulation of His+, Met+, and Leu+ reversions showed that while most of the Met+ revertants were also His+, only less than 10% of His+ revertants were Met+ (41). This suggests that tRNA genes, particularly those for lysine and glutamine, are prone to oxidative damage.
Even more interesting are the observations that deficiencies in the oxidative-damage prevention system affected the rate of the mutations conferring a Leu+ phenotype in exponential growth but not the accumulation of Leu+ mutations in stationary phase. Considering the results obtained by examining the His+ and Met+ phenotypes, it could be speculated that the GO prevention system mediates mutagenic processes that are dependent and independent of growth and that those that result in a Leu+ phenotype are exclusively produced during exponential growth. Perhaps the mutagenic changes occurring at the AGA position in the leu allele that result in a Leu+ phenotype are not generated via oxidative damage in stationary phase. The results observed with the leu allele warrant further investigation and may involve the activation of the SOS response during exponential growth.
Studies with E. coli and P. putida have shown that independent mutations in mutT and mutY but not in mutM did increase the mutation rate of starved cells (3, 4, 36). On the other hand, while a mutM mutation did not affect the mutation rate of a mutT mutant, when combined with mutY a further enhancement in the mutation rate of starved mutY cells was observed (4). However, studies using adaptive mutagenesis with cells deficient in a full GO system have not been previously published.
Several lines of evidence have implicated the MMR system in the repair of oxidative DNA damage (1, 45). The results described in this work support this idea since overexpression of the mutSL operon functionally complemented the hypermutagenic phenotype of the ytkD mutM yfhQ strain. In growing cells of B. subtilis, the spontaneous mutation rates to rifampin resistance and reversion frequencies of the his, met, and leu alleles decreased dramatically in the GO-deficient strain overexpressing the mutSL operon. Collectively, these results suggest that during growth, the MMR system of B. subtilis not only processes the DNA mismatches generated during replication but also those promoted by oxidative stress.
In the stationary-phase assay system utilized here, the high propensity of the GO-deficient B. subtilis strain to accumulate His+ and Met+ colonies was substantially decreased (by more than 1 order of magnitude) when this strain overexpressed the mutSL operon. These results suggest (i) that oxidative-stress-induced DNA mispairing may be an important source of lesions which contribute to saturation of the MMR system during stationary phase and (ii) that the type of DNA mispairs promoted by oxidative stress in starved cells may be processed in a cooperative manner by both the MMR and the GO system. Interestingly, as mentioned above with respect to YB955, the number of Leu+ colonies did not increase in the GO-deficient strain in stationary phase (Fig. 1 and 2). However, generation of these revertant colonies decreased significantly in the ytkD mutM yfhQ strain overexpressing the MMR system (Fig. 5). These results suggest that the mechanism involved in generating the Leu+ revertants during stationary phase involves other types of DNA lesions which are recognized by MMR but not oxidation-induced mutagenesis.
Based on our results with B. subtilis ytkD mutT and ytkD mutM yfhQ mutants and the role the enzymes encoded by these genes play in preventing and/or repairing oxidation-induced DNA damage, it is possible to speculate that mutations in mRNA or DNA are exacerbated by reactive oxygen species (ROS) in nondividing cells.
Overall, our results support the proposal that starved cells have increased mutation frequencies that affect the production of adaptive mutants. Furthermore, the data presented here build on previously postulated pathways of mutagenesis in B. subtilis cells experiencing starvation stress (20, 30). One assumption demonstrated here that supports these pathways is that the MMR system may be sequestered or turned off while cells experience significant oxidative damage. A major unanswered question is whether this stationary-phase process occurs in a subpopulation of the cells (30).
Acknowledgments
M.P.-R. was supported by University of Guanajuato and by Consejo Nacional de Ciencia y Tecnología (CONACYT; grants 43644 and 84482) of México. R.E.Y. and E.R. were supported by NSF and NIH grants MCB 0317076 and GM072554, respectively. This research was also supported by grant 2 P20 RR016463 from the Nevada NIH INBRE award. Luz E. Vidales and Lluvia C. Cárdenas were supported by scholarships from CONACYT.
We wish to thank Silvia J. Mellado for excellent technical assistance.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Bai, H., and A. L. Lu. 2007. Physical and functional interactions between Escherichia coli MutY glycosylase and mismatch repair protein MutS. J. Bacteriol. 189902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boylan, R. J., N. H. Mendelson, D. Brooks, and F. E. Young. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridges, B. A. 1996. Elevated mutation rate in mutT bacteria during starvation: evidence for DNA turnover? J. Bacteriol. 1782709-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridges, B. A., M. Sekiguchi, and T. Tajiri. 1996. Effect of mutY and mutM/fpg-1 mutations on starvation-associated mutation in Escherichia coli: implications for the role of 7,8-dihydro-8-oxoguanine. Mol. Gen. Genet. 251352-357. [DOI] [PubMed] [Google Scholar]
- 5.Bridges, B. A. 1998. The role of DNA damage in stationary phase (“adaptive”) mutation. Mutat. Res. 4081-9. [DOI] [PubMed] [Google Scholar]
- 6.Bridges, B. A., and S. Ereira. 1998. DNA synthesis and viability of a mutT derivative of Escherichia coli WP2 under conditions of amino acid starvation and relation to stationary-phase (adaptive) mutation. J. Bacteriol. 1802906-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, J., J. Overbaugh, and S. Miller. 1988. The origin of mutants. Nature 335142-145. [DOI] [PubMed] [Google Scholar]
- 8.Castellanos-Juárez, F. X., C. Álvarez-Álvarez, R. E. Yasbin, B. Setlow, P. Setlow, and M. Pedraza-Reyes. 2006. YtkD and MutT protect vegetative cells but not spores of Bacillus subtilis from oxidative stress. J. Bacteriol. 1882285-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Sussex, England.
- 10.Dubnau, D. 1991. Genetic competence in Bacillus subtilis. Microbiol. Rev. 55395-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, P. L., and J. Cairns. 1994. The occurrence of heritable Mu excisions in starving cells of Escherichia coli. EMBO J. 135240-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. American Society for Microbiology, Washington, DC.
- 13.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29477-508. [DOI] [PubMed] [Google Scholar]
- 14.Halas, A., H. Baranowska, and Z. Policinska. 2002. The influence of the mismatch repair system on stationary-phase mutagenesis in the yeast Saccharomyces cerevisiae. Curr. Genet. 42140-146. [DOI] [PubMed] [Google Scholar]
- 15.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264258-260. [DOI] [PubMed] [Google Scholar]
- 16.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, M. E. Winkler, and S. M. Rosenberg. 1997. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 112426-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, A. S., P. R. Rohgati, M. N. Hersh, and S. M. Rosenberg. 2005. Roles of the E. coli double strand-break repair proteins in stress-induced mutation. DNA Repair (Amsterdam) 5258-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrickson, H., E. S. Slechta, U. Bergthorsson, D. I. Andersson, and J. R. Roth. 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 992164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47441-465. [DOI] [PubMed] [Google Scholar]
- 20.Holmquist, G. P. 2002. Cell-selfish modes of evolution and mutations directed after transcriptional bypass. Mutat. Res. 510141-152. [DOI] [PubMed] [Google Scholar]
- 21.Kasak, L., R. Horak, and M. Kivisaar. 1997. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc. Natl. Acad. Sci. USA 943134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49264-285. [DOI] [PubMed] [Google Scholar]
- 23.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355273-275. [DOI] [PubMed] [Google Scholar]
- 24.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 976646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenzie, G. J., and S. M. Rosenberg. 2001. Adaptive mutations, mutator DNA polymerases and genetic change strategies of pathogens. Curr. Opin. Microbiol. 4586-594. [DOI] [PubMed] [Google Scholar]
- 26.Michaels, M., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 1746321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedraza-Reyes, M., and R. E. Yasbin. 2004. Contribution of the mismatch DNA repair system to the generation of stationary-phase-induced mutants of Bacillus subtilis. J. Bacteriol. 1866485-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponder, R. G., N. C. Fonville, and S. M. Rosenberg. 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19791-804. [DOI] [PubMed] [Google Scholar]
- 29.Ramírez, M. I., F. X. Castellanos-Juárez, R. E. Yasbin, and M. Pedraza-Reyes. 2004. The ytkD (mutTA) gene of Bacillus subtilis encodes a functional antimutator 8-oxo-(dGTP/GTP)ase and is under dual control of sigma A and sigma F RNA polymerases. J. Bacteriol. 1861050-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robleto, E. A., C. Ross, R. E. Yasbin, and M. Pedraza-Reyes. 2007. Stationary phase mutagenesis in Bacillus subtilis: a paradigm to study genetic diversity programs in cells under stress. Crit. Rev. Biochem. Mol. Biol. 42327-339. [DOI] [PubMed] [Google Scholar]
- 31.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 204-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg, S. M., C. Thulin, and R. S. Harris. 1998. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics 1481559-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross, C., C. Pybus, M. Pedraza-Reyes, H.-M. Sung, R. E. Yasbin, and E. Robleto. 2006. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 1887512-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Sasaki, M., Y. Yonemura, and Y. Kurusu. 2000. Genetic analysis of Bacillus subtilis mutator genes. J. Gen. Appl. Microbiol. 46183-187. [DOI] [PubMed] [Google Scholar]
- 36.Saumaa, S., A. Tover, M. Tark, R. Tegova, and M. Kivisaar. 2007. Oxidative DNA damage defense system in avoidance of stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 1895504-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibutani, S., M. Takeshita, and A. P. Grollman. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349431-434. [DOI] [PubMed] [Google Scholar]
- 38.Slechta, E. S., K. L. Bunny, E. Kugelberg, E. Kofoid, D. I. Andersson, and J. R. Roth. 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 10012847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonenshein, A. L., J. A. Hoch, and R. Losick (ed.). 1993. Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular genetics. American Society for Microbiology, Washington, DC.
- 40.Sung, H.-M., and R. E. Yasbin. 2000. Transient growth requirement in Bacillus subtilis following the cessation of exponential growth. Appl. Environ. Microbiol. 661220-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung, H.-M., and R. E. Yasbin. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J. Bacteriol. 1845641-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung H.-M, G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 1852153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tajiri, T., H. Maki, and M. Sekiguchi. 1995. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 336257-267. [DOI] [PubMed] [Google Scholar]
- 44.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 1813942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyrzykowski, J., and M. R. Volkert. 2003. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J. Bacteriol. 1851701-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasbin, R. E., P. I. Fields, and B. J. Andersen. 1980. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene 12155-159. [DOI] [PubMed] [Google Scholar]
- 47.Yasbin, R. E., R. Miehl-Lester, and P. E. Love. 1987. Mutagenesis in Bacillus subtilis, p. 73-84. In M. Alacevic, D. Hranueli, and Z. Tomen (ed.), Genetics of industrial microorganisms. GIM-86, Split, Yugoslavia.
- 48.Yasbin, R. E., and M. Pedraza-Reyes. 2004. Stationary phase-induced mutagenesis: is directed mutagenesis alive and well within neo-Darwinian theory. In R. Miller (ed.), Microbial evolution: gene establishment, survival, and exchange. ASM Press, Washington, DC.